Abstract
The Joint BioEnergy Institute Inventory of Composable Elements (JBEI-ICEs) is an open source registry platform for managing information about biological parts. It is capable of recording information about ‘legacy’ parts, such as plasmids, microbial host strains and Arabidopsis seeds, as well as DNA parts in various assembly standards. ICE is built on the idea of a web of registries and thus provides strong support for distributed interconnected use. The information deposited in an ICE installation instance is accessible both via a web browser and through the web application programming interfaces, which allows automated access to parts via third-party programs. JBEI-ICE includes several useful web browser-based graphical applications for sequence annotation, manipulation and analysis that are also open source. As with open source software, users are encouraged to install, use and customize JBEI-ICE and its components for their particular purposes. As a web application programming interface, ICE provides well-developed parts storage functionality for other synthetic biology software projects. A public instance is available at public-registry.jbei.org, where users can try out features, upload parts or simply use it for their projects. The ICE software suite is available via Google Code, a hosting site for community-driven open source projects.
INTRODUCTION
Driven by advances in DNA synthesis and assembly technologies (1–3), the ability of researchers to create and manipulate DNA has grown dramatically. As the drivers and consumers of this new capability, the synthetic biology community has experienced a surge in the number of complex engineered biological systems. Starting with basic genetic devices, such as a toggle switch (4), synthetic biologists have created more advanced and complex systems, such as cells that form patterns (5), produce biofuels (6) and fight cancer (7). Other researchers have focused on part and device characterizations (8) and interoperability (9), beginning the process of creating a body of well-characterized reusable biological components.
Information exchange and interoperability remain as some of the more persistent challenges for synthetic biology. Even today, when sequencing DNA constructs is simple and inexpensive, many prominent and well-cited synthetic biology articles typically do not include the complete DNA sequences, annotations and characteristics of all the constructs used in the publication (10). This lack of sequence information not only hampers verification of results, but also hinders the reuse of parts and inhibits better, faster and cheaper biological engineering. There are two main causes of this problem: the first is historic—traditional molecular biology journals simply could not accommodate all of the sequence information, given the limited page space. Even now with electronic publishing, complete sequence information is often missing in publications. In the past, a plasmid map with some restriction sites was sufficient. Today, however, this level of information is not adequate to engineer complex biological systems. Second, there is a noticeable lack of a coherent suite of synthetic biology software tools that work together, which means that a biological engineer has sequence information and other information scattered in different documents and software packages. Thus, when the biological constructs are about to be published, one has to laboriously reassemble all the information, which usually results in incomplete information in the published article. Tools that interoperate well would enable better coherence and organization of this information. Slowly, software applications are being developed to address various needs of synthetic biologists, many of them community driven and open source (11–14). However, the sequence information is generally exchanged in GenBank format, limiting sharing of other relevant information such as experimental data or construction details. A proposed standard for synthetic biology, Synthetic Biology Open Language (SBOL) (15), is a promising solution that will address many of these issues. However, it is still in formative stages.
Currently, the best-known information exchange mechanism for synthetic biology parts is The Registry, previously known as the MIT (Massachusetts Institute of Technology) Parts Registry (www.partsregistry.org, we will here after refer to it as the ‘MIT Registry’ to disambiguate from the Joint BioEnergy Institute Inventory of Composable Parts [JBEI-ICE]). It contains sequences and characterization data for thousands of parts, mostly created by the participants of the annual International Genetically Engineered Machine competitions. Derived from the popular MediaWiki software (the same software used by Wikipedia), each part is recorded as individual Wiki pages that may be edited by users. This system allows users to add text, graphics and charts about a part, creating a shared information repository. In addition to the Wiki system, the MIT Parts Registry has made customizations to the MediaWiki software that allows users to designate part categories, as well as annotated sequence information.
Although the MIT Parts Registry has served the community well, there are several reasons why it cannot become the standard information-sharing platform for synthetic biologists. Most significantly, it is not possible to obtain the software itself. As of this writing, the creators have not made the website engine available to others, either in source code or in binary form. Contrary to the contents of the site, which is available under an open license, the software itself is not available. Therefore, it is not possible for a laboratory or an institution to run a private instance of the MIT Parts Registry on their own servers. All data must be hosted on the MIT Parts Registry servers, which may not be suitable for everyone (e.g. for confidentiality of data reasons). Second, there are few mechanisms to access the MIT Parts Registry via automated means. There exists a very minimal application programming interface (API, http://partsregistry.org/Registry_API) that can be used to obtain xml files, but the xml files do not include the rich user-generated characterization data that would be useful for automated processing. Lastly, as neither the software nor a full API is available, it is impossible to create a plug-in or additional functionality for the MIT Parts Registry, an important requirement for automated synthetic biology design software.
We believe the synthetic biology community would benefit from an open source registry engine that can be modified, extended and used privately. As the synthetic biology community grows, multiple parts registries—public registries maintained by single Principal Investigator laboratories or institutes that share their parts, specialized registries for a particular purpose or subfield or private registries in research laboratories and companies—will be needed. In addition, a parts registry should be machine accessible via a web API to allow automated manipulation. For example, a third-party biological design program should be able to use the API to download the basic parts from a registry instance, manipulate it and upload the completed device. If this program needs to find a part with a particular feature, characteristic or sequence, it could reach out and query several registries known to it. The expansion of the idea of a registry from a single website to a distributed machine-accessible repository opens up many possibilities and creates a way to bridge the collaborative data sharing and data exchange problems faced by synthetic biologists today.
In this article, we report the creation of a new registry software, JBEI-ICE, to manage the biological constructs. It is an information repository of plasmids, strains, part libraries and Arabidopsis seeds with the following features:
ICE is licensed using the Berkeley Software Distribution (BSD) license, which allows anyone to use the software, set up their own registry or create derivative works without any onerous conditions;
ICE handles legacy parts that do not conform to a particular standard;
ICE is installation independent. Multiple ICE installations can exchange parts, and duplicate names or part numbers are handled properly;
ICE interoperates well with other software. We provide a software stack and API on which other synthetic biology software can be built, independent of the web interface;
example applications and software code are provided so that users and developers can quickly learn how to build software that works with ICE;
a public installation of ICE exists for users to try out our software, explore our toolss and use it in their research;
development tools and third-party libraries used by ICE are carefully chosen for their open source licenses and permissive redistribution terms; and
the development process is open to participation by the community.
Various commercial and noncommercial software and repositories, such as Addgene, DNASU Plasmid Repository, VectorNTI, ApE, LabLife, etc., provide some of the features present in JBEI-ICE. However, JBEI-ICE is the only open source software that brings together a variety of tools in a sharable platform. Additionally, the open source nature of JBEI-ICE prevents ‘data captivity’, where users are locked into a single vendor for their data.
MATERIALS AND METHODS
JBEI-ICE is designed along the Model-View-Controller design architecture, which separates the information, business logic and user interfaces into separate components and is written in the Java programming language. JBEI-ICE uses Hibernate object relational mapping software to communicate with a data store, Apache Wicket as the presentation framework and Lucene as the text search engine. It uses NCBI-BLAST for sequence search and comparison, Adobe Flex for rich Internet application framework and Maven for the build and dependency management system. The link to the full list of third-party software applications used in JBEI-ICE can be found in the ‘Software Availability’ section immediately below.
Software availability
The entire source code for ICE and companion Flex applications are available under the BSD license through Google Code. Source revisions, bug tracking and documentations are available through these websites:
JBEI-ICE software: http://code.google.com/p/gd-ice/;
the current manual, including instructions on getting started: https://public-registry.jbei.org/site/docbkx/html/manual/manual.html;
JavaDoc API documentation: https://public-registry.jbei.org/site/apidocs/;
license texts for third-party libraries: http://code.google.com/p/gd-ice/source/browse/trunk/ice/LIBRARY_LICENSES.txt;
VectorEditor (includes Sequence Checker): http://code.google.com/p/vectoreditor/;
stand-alone runnable version of VectorEditor: https://public-registry.jbei.org/static/vesa/VectorEditor.html;
BioFlex libraries: http://code.google.com/p/bioflex/; and
a public installation of JBEI-ICE is available at https://public-registry.jbei.org
RESULTS
We have created an open source registry platform that is feature rich, extensible and free. It can store biological parts, as well as plasmids, microbial strains and Arabidopsis seeds. The advanced search capability provides full-text search, relevance scoring and BLAST; the instant preview feature allows users to rapidly sort through large numbers of parts. The VectorEditor component allows the viewing, annotating and in silico cloning of sequences; the SequenceChecker component allows users to quickly check sequence traces against an existing record. And the GenBank and SBOL import/export filters allow users to keep using their favorite off-line tools.
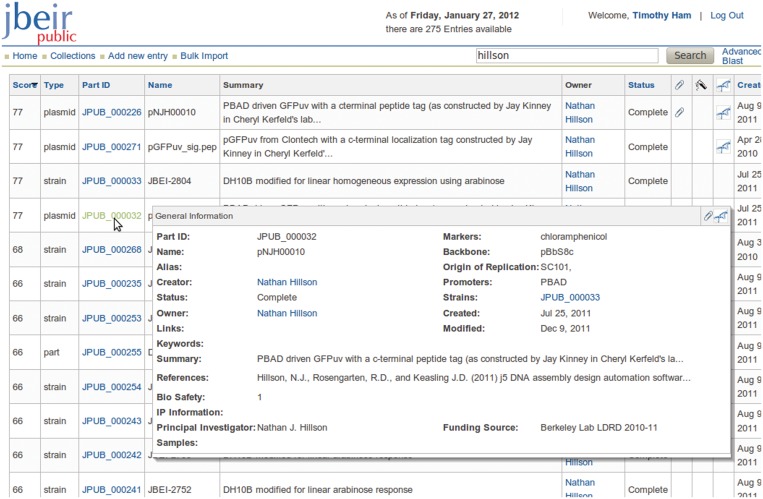
In a typical use case scenario, the user would search for a part using the search bar, entering keywords, plasmid names or other text that may be found within the description of the result. The search engine recognizes root words—that is, searching for ‘binding’ would also search for ‘bindings’, ‘bind’ and ‘bound’. The results are displayed in order of relevance, by matches in the most important fields such as part number or name, as well as frequency. In the result display, the user is able to quickly scan the short description field, which gives a better indication of the entry besides the entry name and number. Also, by hovering the mouse over the part number link, the user can see a preview of the entry, which aids in narrowing down the particular entry desired among the search results (Figure 1). The user can also search for sequence matches using the BLAST query page or filter individual fields using the advanced search page.
Figure 1.
Screen shot of a typical search result, with pop-up preview shown.
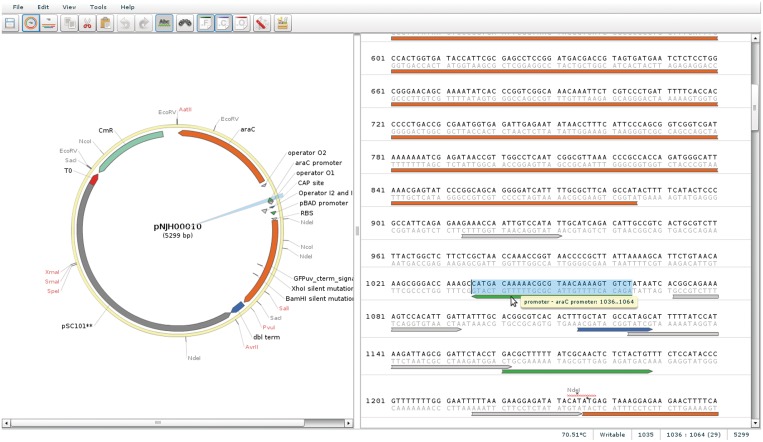
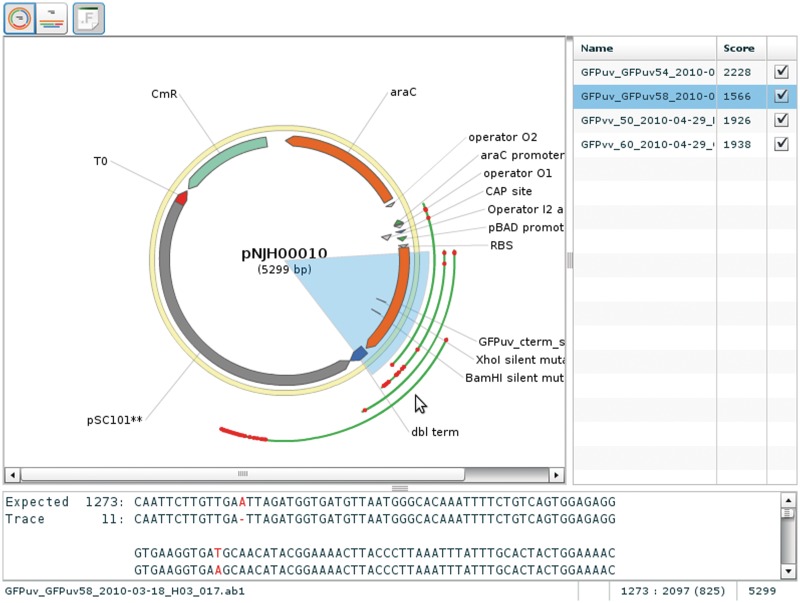
When the user clicks on the part number, he or she is shown the detailed information about the part, including an interactive graphical map of the annotated sequence, if such information had been provided. The user can select ‘Open in VectorEditor’, which will open a vector editing program with features comparable with other similar programs (Figure 2). From the detail page, the user can select ‘Seq. Analysis’, which allows him or her to upload sequence trace files (.abi files) and launch SequenceChecker (Figure 3), which displays a visual, as well as a textual, alignment of the trace files to the reference.
Figure 2.
Screen shot of VectorEditor, displaying restriction enzyme locations, open reading frames and feature annotations.
Figure 3.
Screen shot of SequenceChecker showing overlapping alignments. Green line indicates alignments of the reads, and red dots indicate misalignments to the reference sequence.
Also, from the detail page, with the proper permissions, the user is able to make changes, add attachments, change read and write permissions for other users, associate physical samples and download the sequence in GenBank, FASTA or SBOL formats.
At JBEI, the variety of tools available encourages users to enter parts into the system, as they are created at the bench, instead of waiting until the end to enter them all at once. This facilitates early data capture and preservation. Also, the extensive search capability encourages part discovery and reuse, as well as collaboration. Furthermore, the ability for users to easily transfer their information from the private instance to the public instance (detailed later) gives them a simple mechanism to publish information about their parts to the public. Some of these parts can be seen in the public site’s ‘Collections’ page.
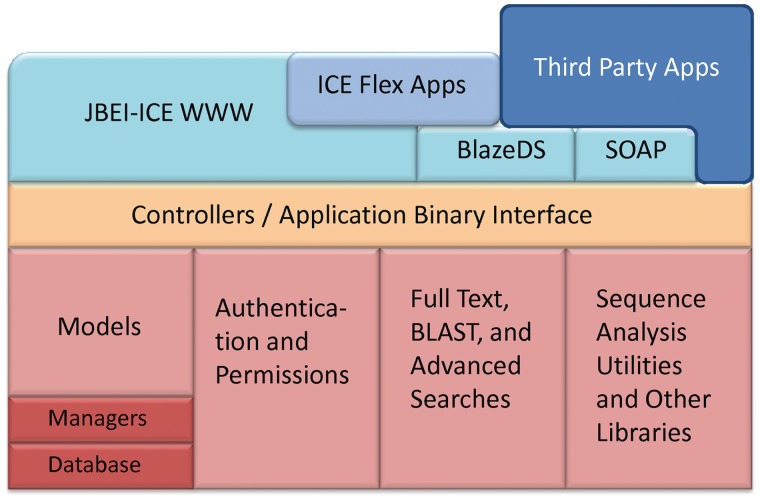
Underneath the visible functionality, the core libraries are aggregated into a service layer that hides many of the underlying architectures and technical complexities (Figure 4). With this architecture, it is possible to build third-party software that uses the service layer to interact with the repository. We also created a web interface and a suite of rich applications to take advantage of the platform, thus providing a useful resource to the synthetic biology community. As all of our software is open source, they can be used in aggregate or as components in other synthetic biology projects. JBEI-ICE has been designed from its inception to provide distributed registry software that anyone can use, customize or improve on. The BSD license used for ICE does not require modifications to be shared or place other onerous restrictions, and all third-party libraries used as part of JBEI-ICE are available under open source licenses.
Figure 4.
Schematic of the ICE software stack. Third-party applications can interface with ICE over the Internet via Soap or BlazeDS API or use ICE as a library through the Java ABI. The red shades indicate underlying systems, the orange shades indicate the ABI layer and the blue shades indicate applications using the ABI layer for functionality. Third-party applications can use any of BlazeDS, SOAP or the ABI for functionality.
JBEI-ICE provides an open, flexible and fully featured platform for synthetic biology information storage, retrieval and manipulation. It brings together different synthetic biology software by lowering the barrier of entry and providing a working solution to the part storage problem. With the current release of ICE, a developer of a new software tool does not have to write a registry-like system to store its parts—he/she can simply use ICE as a software stack or connect to an ICE instance as its data source. The ICE team is very much open to community participation in ICE and would like to encourage questions or suggestions, as well as bug reports, patches or new modules.
Design and implementation
Here, we explain in detail the principles and ideas we used to design JBEI-ICE. For the particular implementation details, we refer to the documentation and source code available via our Google Code project site mentioned in the ‘Software Availability’ section.
Overview
The core functionality of ICE is envisioned as a software library stack that can act as a black box that stores synthetic biology parts. The website is just one way to access that black box. Much care has been taken to ensure that the users of our library stack are freed from the complexities of the underlying implementation. We have also tried to ensure that the data model and the utility functions are language neutral—that is, even though our implementation is written in the Java programming language, access and manipulation of information stored in the library stack can be performed in any programming language without great effort. We carefully avoided language-specific design patterns and data types and considered the possible consumer programs written in different programming languages. By investing our time into providing a generalized storage mechanism for synthetic biology parts, it is now possible for others to simply take our library stack and build custom applications on it using their programming language of choice.
We also have developed a website interface on top of the ICE library stack, to provide simple and user friendly access to ICE, including browsing and search. Our website allows full text, BLAST and field-based searches and filters, and results are displayed with a preview mode that allows one to quickly browse through them. Administrator-created ‘collections’ provide another mechanism to organize entries for easier access. These mechanisms work well for searching through thousands of parts. However, as the number of entries grows into hundreds of thousands of parts, we plan to provide more advanced search algorithms, including the usage of content based ranking heuristics.
In addition to the web interface to access the registry information, we also provide a rich set of graphical applications for sequence manipulation, annotation and analysis. These tools help users integrate the registry into their day-to-day work flow, which increases the quality of information that is stored.
Organizations of the components
The JBEI-ICE software is organized into three distinct layers (Figure 4). The bottom layer consists of the functional components of the software, the heart of which is the data model. We have taken care to design the data model to be orthogonal and amenable for future extensions. The model instances are stored in a relational database via the Hibernate object relational mapping and persistence system. Between the model layer and the database, the ‘Manager’ layer abstracts out the database calls for handling storage and retrieval. The functionality layer also includes authentication, permissions, search algorithms and other utility functions such as BioBrick assembly, sequence comparison and GenBank file parsing, etc.
Above this layer, the ‘Controllers’ layer aggregates the functionality below it, presenting a uniform interface to the diverse underlying components. By using the controllers, ICE can behave as a black box that stores and retrieves the model objects, performs searches, executes sequence manipulation routines and so on. Users of this layer can safely ignore much of the underlying mechanisms of ICE. As such, the controllers act as the application binary interface (ABI) for JBEI-ICE.
The third major layer of ICE consists of various ways to access the ICE ABI. The website, the primary means of accessing JBEI-ICE, is built entirely on top of the ABI, thus creating a clean separation between rendering of web pages and the underlying functionality. This design allows improvements to the website without affecting the functionality of other components. ICE uses the Apache Wicket framework to render web pages and process user input.
The two other mechanisms of ICE access, the BlazeDS and Simple Object Accessible Protocol (SOAP) services, provide automated and programmable access to the ICE ABI. BlazeDS is used by Adobe Flex applications of the website (discussed later), whereas SOAP, an International Standard Organization standard that allows software written in different languages to use methods (operations) and data from a server over the Internet in a standard way, provides a generalized third-party access to the ICE ABI. In our source repository, we have provided example code in various programming languages (Python, Java, Perl, ActionScript) to facilitate the use of ICE via SOAP. If other web API access standards such as Remote Procedure Call (RPC) or Representational State Transfer (REST) are desired, we can make them available. ICE uses the Apache CXF to provide SOAP services. BlazeDS, Apache CFX and Apache Wicket are all available under open source licenses.
Using the ICE ABI interface via Java or API interfaces via SOAP or BlazeDS allows anyone interested in writing synthetic biology software to treat ICE as a software component that abstracts away all the details of the storage, retrieval, search and part manipulation operations. By using the interfaces, a developer only needs to know about the core data models and what operations are available through the controllers—other details can be safely ignored.
Graphical applications
The various rich Internet applications written in Adobe Flex comprise other major components of ICE. These graphical applications work within the browser, independent of the computer operating system. They are useful tools to help design, annotate and verify parts and also serve as illustrations on application development using the ICE APIs via the BlazeDS service.
VectorEditor is our largest and most complex application, and it is made of several components and an increasing number of features. It is a sequence display, annotation and cloning software similar in functionality to VectorNTI or ApE. Today, it is capable of real-time DNA editing, with live vector map display, sophisticated feature annotation, in silico cloning via intelligent cut and paste, easy feature manipulation, restriction site visualization, annealing temperature calculation and more. It can also display protein translation information and open reading frames. VectorEditor can import and export GenBank files for data exchange with other vector manipulation programs. Like ICE, VectorEditor is also an open source software, which means it is free to use and free to modify. It can be trivially recompiled to run as a stand-alone desktop application, and we encourage anyone interested in building an open source and free vector editing tool to add new features and capabilities to VectorEditor. We know of no other open source vector manipulation program with comparable features, especially one that can run on Windows, Mac and Linux operating systems.
SequenceChecker is a tool to align sequencing traces (.ab1 or FASTA files) onto a sequence to make easier the process of verifying DNA constructs. It overlays multiple trace files onto a plasmid vector map, visually highlighting any mismatches, making verification of a construct simple.
We also provide two minor tools, the VectorViewer, which is the VectorEditor without the DNA editing capabilities, and BioFlex. BioFlex is a set of libraries written in ActionScript that provides useful bioinformatic functionalities for other Flex-based tools, such as restriction enzyme mapper (via REBASE), ORF mapper, temperature calculator and parsers for GenBank and FASTA formats. Our Flex applications rely on it extensively.
Distributed web of registries
JBEI-ICE was designed with distributed use in mind. Any individual or group can download and install an ICE instance without centralized coordination. If a group wishes to publicize the contents of their instance, they may choose to allow others access via the website or the SOAP interface. Even if they did not coordinate their part numbering scheme with anyone else ahead of time, each entry can be distinguished by their Universally Unique Identifiers (UUIDs). Use of UUIDs overcomes the problem of name collision that comes from distributed repositories. ICE also facilitates interconnected use. If a group wishes to export some of their entries into a different publicly available ICE instance, they can export the data via xml or connect with the publicly available instance to allow access via SOAP. The private instance can then use the SOAP interface to automatically export entries into the public ICE instance.
By using a standard SOAP interface and UUIDs, it is possible to have independent ICE instances, yet allow easy sharing of information between them. If large numbers of distributed ICE instances are realized, it would not be too difficult to create a ‘web-crawler’ like service that explores known available ICE instances and indexes them, much like existing web search engines. Even if such ICE search engines are not realized, peering (establishing an automated exchange agreement) with a well-known public instance is a good way to distribute information widely.
DISCUSSION
Even as the number of standard biological parts has grown, the ways the parts are stored and managed have not advanced at the same pace. Several automation tools now exist, but they are hampered by lack of a registry that can be used programmatically to access and store part information. The current paradigm of a single parts registry for the whole world has become inadequate for the expanding field of synthetic biology, especially when the central registry cannot be extended to facilitate new subfields, new automation tools and new assembly paradigms. More than being a database of parts, a parts registry should be the mechanism by which the friction between scientists, software tools and the community is minimized. The only viable solution forward is a shift to an open distributed web of registries that can be developed by the community. As the first open source, distributed registry software that works both as a website and as a software library stack, JBEI-ICE could become the tool that bridges the gap between users, applications and institutions by providing a common platform that can bring together diverse set of resources into a shared framework. Clearly, the current iteration of JBEI-ICE is not yet ready for this monumental task. However, the open source license of JBEI-ICE enables the community to build on and expand its functionality, with or without JBEI. Of course, we wish to participate in this process, and we will provide any support as best as we can.
Furthermore, by supporting existing and emerging data exchange standards such as GenBank format and SBOL and by providing transparent conversion between different data formats, JBEI-ICE will aid the community in creating a viable information sharing medium beyond static file formats. As more information sharing is performed on the Internet by computers having APIs rather than people exchanging files, a web of registries platform like the JBEI-ICE could be an excellent resource for synthetic biologists to share their research with the larger community.
FUNDING
Office of Science, Office of Biological and Environmental Research of the U S Department of Energy [Contract No. DE-AC02-05CH11231]. Funding for open access charge: US Department of Energy.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dylan Chivian for his close reading of the manuscript and William Holtz for his extensive testing and feedback of new features and insightful discussions. We also thank Synthetic Biology Data Exchange group for their helpful discussions.
REFERENCES
- 1.Ellis T, Adie T, Baldwin GS. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr. Biol. 2011;3:109–118. doi: 10.1039/c0ib00070a. [DOI] [PubMed] [Google Scholar]
- 2.Liss M, Wagner R. Gene Synthesis - Enabling Technologies for Synthetic Biology. In: Koeppl H, Densmore D, Setti G, di Bernardo M, editors. Design and Analysis of Biomolecular Circuits. New York, NY: Springer; 2011. pp. 317–335. [Google Scholar]
- 3.Hillson NJ. DNA Assembly Method Standardization for Synthetic Biomolecular Circuits and Systems. In: Koeppl H, Densmore D, Setti G, di Bernardo M, editors. Design and Analysis of Biomolecular Circuits. New York, NY: Springer; 2011. pp. 295–314. [Google Scholar]
- 4.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 5.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 6.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 7.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 8.Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D. Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 2009;3:4. doi: 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peccoud J, Anderson JC, Chandran D, Densmore D, Galdzicki M, Lux MW, Rodriguez CA, Stan GB, Sauro HM. Essential information for synthetic DNA sequences. Nat. Biotechnol. 2011;29:22. doi: 10.1038/nbt.1753. discussion 22–23. [DOI] [PubMed] [Google Scholar]
- 11.Chandran D, Bergmann FT, Sauro HM. TinkerCell: modular CAD tool for synthetic biology. J. Biol. Eng. 2009;3:19. doi: 10.1186/1754-1611-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czar MJ, Cai Y, Peccoud J. Writing DNA with GenoCAD. Nucleic Acids Res. 2009;37:W40–W47. doi: 10.1093/nar/gkp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillson NJ, Rosengarten RD, Keasling JD. j5 DNA assembly design automation software. ACS Synth. Biol. 2012;1:14–21. doi: 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- 14.Xia B, Bhatia S, Bubenheim B, Dadgar M, Densmore D, Anderson JC. Developer's and user's guide to Clotho v2.0 A software platform for the creation of synthetic biological systems. Methods Enzymol. 2011;498:97–135. doi: 10.1016/B978-0-12-385120-8.00005-X. [DOI] [PubMed] [Google Scholar]
- 15.Galdzicki M, Rodriguez C, Chandran D, Sauro HM, Gennari JH. Standard biological parts knowledgebase. PLoS One. 2011;6:e17005. doi: 10.1371/journal.pone.0017005. [DOI] [PMC free article] [PubMed] [Google Scholar]