Abstract
We developed a highly scalable ‘shotgun’ DNA synthesis technology by utilizing microchip oligonucleotides, shotgun assembly and next-generation sequencing technology. A pool of microchip oligonucleotides targeting a penicillin biosynthetic gene cluster were assembled into numerous random fragments, and tagged with 20 bp degenerate barcode primer pairs. An optimal set of error-free fragments were identified by high-throughput DNA sequencing, selectively amplified using the barcode sequences, and successfully assembled into the target gene cluster.
INTRODUCTION
One of the fundamental quests in synthetic biology is to construct designer genes and genomes in a high-throughput manner to probe and engineer biological systems (1,2). However, typically employed DNA synthesis procedures (2) impose significant challenges for scalable construction due to the high cost of oligonucleotides, low assembly efficiency, requirement for time-consuming cloning and the high cost of sequence validation (3). In contrast to the slow progress in DNA synthesis (2), DNA sequencing technology has progressed rapidly in recent years to the level of de novo genome sequencing. One of the most important contributions of genome-level sequencing is shotgun DNA sequencing, which involves the use of computer algorithms to piece together randomly fragmented genomic DNA (4). Because random fragmentation of genomic DNA to certain sizes can be achieved easily, high-throughput sequencing of short fragments and computational analysis can be massively parallelized to reduce the costs of genome sequencing.
We hypothesized that the conceptual elegance of shotgun DNA sequencing could be adopted to synthesize long DNA fragments if an easy way to construct a pool of random DNA fragments and a method to recover computer-selected sequences in an orderly manner were combined. Furthermore, we hypothesized that shotgun sequencing could be combined with the use of microchip oligonucleotides and high-throughput sequencing technologies to achieve high-throughput DNA writing. Oligonucleotides cleaved from DNA microchips have previously been utilized to synthesize low-cost DNA (5–9). However, the low assembly efficiency of chip-derived oligonucleotides hinders target gene construction, and a laborious DNA assembly optimization process is consequently required (Supplementary Note 1) (8). To overcome the challenges associated with microchip oligonucleotides, only a small sub-pool of oligonucleotides (i.e. <20) are often amplified to ensure high assembly efficiency (7,8). Recently, a highly efficient isothermal DNA assembly method based on in vitro recombination and amplification was successfully used to synthesize the mouse mitochondrial genome (10).
Along with the use of microchip oligonucleotides, utilization of high-throughput sequencing technology (11) has great potential to decrease the cost of validation of synthetic DNA molecules. However, unlike colony-based Sanger sequencing validation, it is difficult to collect the desired DNA from high-throughput sequencing plates. In a notable report (12), chip-cleaved oligonucleotides were sequenced by 454 sequencing technology (11), and directly isolated from the sequencing plate using a bead picking pipette. These beads were subsequently used to assemble 200 bp target DNA fragments. Although this study demonstrated the utility of next-generation sequencing technology, challenges associated with the assembly of chip oligonucleotides into larger sequences along with the creation of a highly tuned bead picking instrumental set-up for an error-free oligonucleotide picking process have yet to be addressed. Furthermore, various methods have been developed to reduce the errors generated during the oligonucleotide and gene synthesis processes (3,7,13–16). However, most of these methods require laborious and time-consuming procedures for error correction.
Here, we describe a shotgun DNA synthesis technology that is a de novo high-throughput DNA writing method. This technology is based on the combination of microchip oligonucleotides and high-throughput sequencing technology. Barcodes were attached to randomly synthesized ‘shotgun’ DNA fragments, and the barcode sequences were used to recover error-free DNA fragments as validated by 454 sequencing. Although shotgun synthesis is currently limited to read lengths of 500 nt because of the current technological limits of 454 sequencing, other high-throughput sequencing technologies could also feasibly be utilized. Further, we believe that the relevance of shotgun DNA synthesis will increase as sequencing reads become longer.
MATERIALS AND METHODS
The target penicillin biosynthetic gene cluster and oligonucleotide sequence design
The N-(5-amino-5-carboxypentanoyl)-l-cysteinyl-d-valine synthase protein sequence (Supplementary Sequence 1; UniProt database entry number: P26046) from Penicillium chrysogenum was chosen as a synthetic model. A codon-optimized penicillin biosynthetic gene cluster sequence (11 376 bp: Supplementary Sequence 2) was designed using the web-based program Optimizer (17). Twenty-four nucleotides (GCAGAGTAAAGACCGTGCACTTAT) were added to the synthase sequence to ensure the total length of the sequence was in multiples of 100 bp (i.e. 11 400 bp) for convenience when designing the microchip oligonucleotides. Each Agilent chip oligonucleotide was 150 nucleotides in length and consisted of flanking sequences (2 × 25 nt) and target DNA sequences (100 nt each; Supplementary Sequences 3a and b). Oligonucleotides (114 plus and 114 minus strands) for target DNA sequences were designed in such a way that upon annealing, complementary oligonucleotides contained overlapping regions to prevent gaps during DNA assembly. These 228 oligonucleotide sequences were flanked by two sets of generic PCR primer pair sequences containing either BtsI or EarI restriction enzyme site (Primer Sets 1a and b; see Supplementary Sequence 4).
Processing of sub-pools of Agilent microchip oligonucleotides
Lyophilized Agilent microchip oligonucleotides were suspended in 100 μl water. We prepared a higher concentration of the microchip oligonucleotide subpool (two sets of 228 oligonucleotides targeting the penicillin biosynthetic gene cluster) using PCR amplification with flanking primers (Primer Set 1; see Supplementary Sequence 4). The components included in each PCR reaction mixture were 8 µl water, 10 µl 2× Pfu polymerase pre-mix (Solgent, Daejeon, Korea), 0.5 µl Agilent chip oligonucleotides suspended in water, and 1 µl 10 µM forward and reverse primers [Integrated DNA Technology (IDT), Coralville, IA, USA]. Thermocycling involved a 3-min initial denaturation step at 95°C followed by 20 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final elongation step at 72°C for 10 min. To further increase the amount of oligonucleotides, the second PCR reaction was performed using 18 µl water, 25 µl 2× Pfu polymerase pre-mix, 3 µl of an aliquot from the first PCR reaction and 2 µl 10 µM forward and reverse primers. PCR conditions were the same as for the first PCR reaction with the exception of the number of reaction cycles (i.e. 12). After verification of the desired products by 4% agarose gel electrophoresis, these PCR products were used directly for restriction enzyme digestion without an extra purification step. Restriction enzyme digestion was carried out by incubating 50 µl of the amplified chip oligonucleotides with 2.5 µl EarI (or BtsI; New England Biolabs (NEB), Ipswich, MA, USA), 5 µl NEB buffer 1, 0.5 µl 100× BSA at 37°C (or 55°C for BtsI) for 3 h. The flanking sequence-cleaved oligonucleotides were purified by agarose gel (4%) electrophoresis and an AccuPrep™ gel-purification kit (Bioneer, Daejeon, Korea).
Shotgun synthesis using processed microchip oligonucleotides
We carried out the shotgun synthesis reaction using two sub-pools of processed microchip oligonucleotides by performing PCR. The PCR reaction mixture comprised 20 µl Pfu polymerase pre-mix and 20 µl of the processed microchip oligonucleotides. The conditions for PCR were as follows: 3 min of initial heating at 95°C, followed by 36 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 1 min and a final elongation step at 72°C for 10 min. We then electrophoresed the PCR products through an agarose gel (1.5%). Bands at 300–500 bp were excised and purified using an AccuPrep™ gel-purification kit.
Barcode tagging of the shotgun synthesis products
The procedure used for barcode tagging is illustrated in Figure 1. The gel-purified shotgun synthesis fragments were amplified using flanking primers to enrich DNA fragments with common flanking sequences. PCR reactions contained 10 µl water, 25 µl Pfu polymerase pre-mix, 10 µl purified shotgun synthesis fragments and 2.5 µl 10 µM forward and reverse primers (Primer Set 1; Supplementary Sequence 4). PCR conditions were as follows: an initial denaturation step at 95°C for 3 min, 18 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final elongation step at 72°C for 10 min. PCR products were electrophoresed through agarose gels (1.5%) and bands between 300 and 450 bp were excised and purified using an AccuPrep™ gel-purification kit.
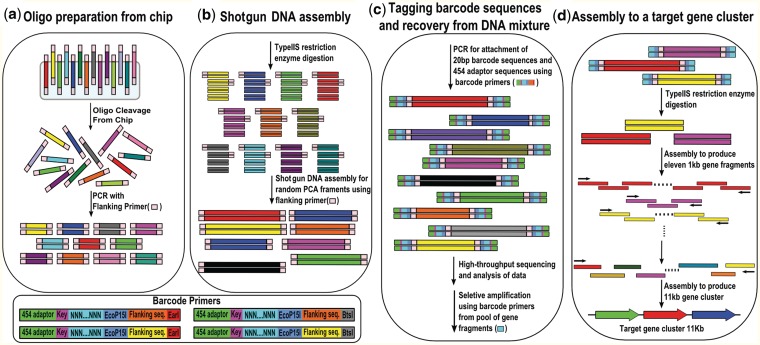
Figure 1.
Overall shotgun DNA synthesis scheme. (a) Preparation of microchip oligonucleotides. The oligonucleotides were designed to have flanking sequences with Type IIS restriction enzyme sites (EarI or BtsI), and were synthesized and cleaved from an Agilent DNA microarray. PCR amplification was carried out to increase the concentration of the oligonucleotides. (b) Shotgun DNA assembly. Amplified oligonucleotides were cleaved using Type IIS restriction enzymes to remove the flanking sequences. A number of oligonucleotides were simultaneously utilized for shotgun synthesis to form random fragments with sizes varying from 100 bp to 1000 bp. Because the efficiency of the restriction enzymes was less than 100%, there were still uncut flanking sequences that we could use to enrich for the shotgun synthesis products. (c) Tagging fragments with barcode sequences and recovery from the DNA mixture. To analyze the sequences using high-throughput sequencing technology, we tagged the shotgun synthesis fragments with barcode primers using PCR. These barcode primers consisted of degenerate barcode sequences and 454-adaptor sequences. The PCR products were sent for high-throughput sequencing and analyzed by a computer program to select the optimal set of error-free DNA fragments. To recover the selected error-free DNA fragments, PCR was carried out from the pool of shotgun-assembled target gene fragments using barcode primer sequences. (d) Assembly to a target gene cluster. After removing the degenerate barcode sequences from the recovered fragments by Type IIS restriction enzyme digestion, the error-free shotgun synthesis fragments were assembled into 11 ∼1 kb gene fragments. Subsequently, the 11 fragments were assembled into the full-length penicillin biosynthetic gene cluster using PCR.
The purified pool of shotgun synthesis fragments was barcoded by PCR using a barcode primer pair (Primer Set 2; Supplementary Sequence 4) that consisted of, from the 5′ to 3′ direction, a 454 DNA-sequencing adaptor sequence, a 20-mer degenerate oligonucleotide (i.e. made of poly N sites), an EcoP15I Type IIS restriction enzyme site, and the original flanking primer sequences. The original flanking sequence of the chip oligonucleotides contained either an EarI or BtsI site. The EcoP15I site was introduced into the barcode primers of the shotgun synthesis fragments as an additional Type IIS restriction enzyme site. PCR reactions contained 6 µl water, 20 µl Pfu polymerase pre-mix, 10 µl purified shotgun synthesis mixture and 2 µl of the forward and reverse barcode primers (Primer Set 2; Supplementary Sequence 4). Thermocycling conditions were as follows: 3 min of initial heating at 95°C followed by 18 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final elongation step at 72°C for 10 min. The PCR products were run on 1.5% agarose gels, and bands between 450 and 600 bp were excised and purified using an AccuPrep™ gel-purification kit. These gel-purified products (10 µl) were diluted 100-fold using water (1 ml), and the diluted products were then used for a final PCR amplification step involving 454 DNA sequencing adaptor primers. Eight replicate 50 µl PCR reactions containing 17.5 µl water, 25 µl Pfu DNA polymerase pre-mix, 2.5 µl of the 100-fold diluted gel purified products and 2.5 µl 454 DNA sequencing adaptor primers (Primer Set 3; see Supplementary Sequence 4; Macrogen, Seoul, Korea) were subjected to thermocycling using the following conditions: initial heating at 95°C for 3 min followed by 25 cycles of 95°C for 30 s, 71°C for 30 s, 72°C for 1 min and a final elongation at 72°C for 10 min. PCR products were electrophoresed through a 1.5% agarose gel and gel bands between 450 and 500 bp were excised and purified using an AccuPrep™ gel-purification kit with elution in 60 µl of water. The eight replicates were pooled (total volume ∼480 µl) prior to 454 sequencing.
Prior to 454 sequencing, we performed TOPO cloning of the barcoded shotgun synthesis fragments, and several colonies were submitted for Sanger sequencing evaluation. The detailed experimental procedures were as follows. Gel-purified and barcoded DNA fragments were cloned into the TOPO vector using the TOP Cloner™ Blunt core kit (Enzynomics, Daejeon, Korea). Chemically competent Escherichia coli cells originally derived from C2566 (NEB, Ipswich, MA, USA) were then transformed with the TOPO vectors. After overnight growth on agar plates at 37°C, several colonies were chosen for colony PCR using M13 primer pairs (M13F-pUC and M13R-pUC primers; see Primer Set 4, Supplementary Sequence 4). After confirmation of the presence of inserted DNA by colony PCR, we purified plasmids from the insert-validated colonies using an AccuPrep™ plasmid mini extraction kit (Bioneer, Daejeon, Korea). The purified plasmids were submitted for Sanger sequencing prior to Roche-454 sequencing. We analyzed the Sanger sequencing data to validate the sequences of the DNA fragments and the barcode sequences using the Lasergene program (DNAstar, Madison, WI, USA). After verification of the pool of shotgun synthesis DNA fragment sequences, we sent the pool of barcoded shotgun synthesis products for 454 FLX high-throughput sequencing. The sequencing data were analyzed using an in-house Python program, and error-free target gene fragments were selected.
In-house Python script to analyze the 454 high-throughput sequencing data
The primary task of the Python program was to select error-free shotgun synthesis fragments for assembly. The computer program first scanned for the restriction enzyme sites from the 454 sequencing reads. The flanking sequences were removed in silico, and internal sequences were aligned to the reference penicillin biosynthetic gene cluster sequence. In detail, DNA fragments with EcoP15I and either, EarI or BtsI sites at both ends of the read were selected. Flanking sequences containing the enzyme site were eliminated from the DNA fragments, and the internal sequences [only for reads with a high quality score (Phred-like consensus quality ≥30)] were aligned to the target penicillin biosynthetic gene cluster sequence. When these internal sequences matched perfectly with the reference sequence, the aligned sequences were graphically listed along with their target gene cluster sequence (Figure 3b). Subsequently, the program determined the optimal set of internal sequences that overlapped by more than 15 bp with other fragments. These selected shotgun synthesis fragments were mapped onto the target gene cluster sequence (Figure 3c). The Python scripts used for the analysis are available upon request to D.B.
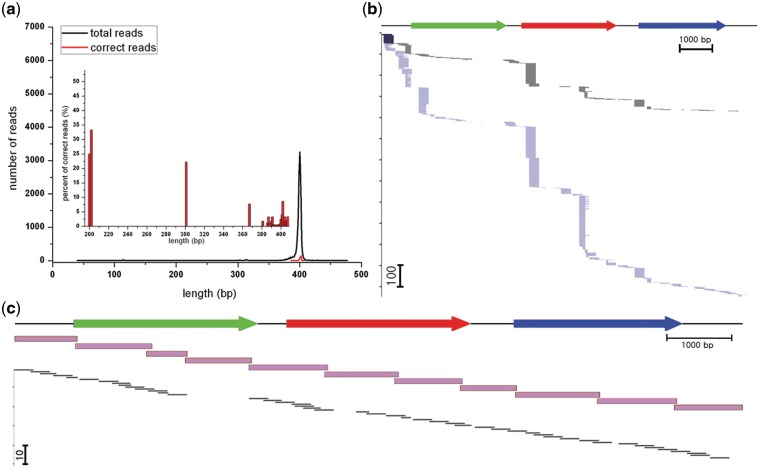
Figure 3.
Computational analysis of the 454 sequencing data from shotgun synthesis. (a) The number of 454 sequencing reads versus the length of the shotgun synthesis random fragments. The black and red lines show the number of total 454 sequencing reads and the error-free fragment reads, respectively. The most abundant and correct reads had a length of ∼400 bp (including barcoding regions; they were typically ∼300 bp without barcode flanking regions). The inset in a shows that the percentage of error-free random fragments tended to decrease as the length of the fragments increased. (b) Computational analysis of two independent experiments (black and blue), and graphically aligned error-free gene fragments after the removal of the flanking barcode sequences. The green, red and blue arrows on top of the figure represent clusters of genes (adipate-activating, cysteine-activating and valine-activating domains, respectively). The y-axis indicates the number of error-free gene fragments corresponding to various parts of the target gene. (c) Hierarchical gene cluster synthesis. Selected shotgun synthesis fragments (∼300 bp; in black) were assembled into ∼1000 bp gene fragments (in pink), which were subsequently assembled to the target ∼11.4 kbp penicillin biosynthetic gene cluster.
Assembly of the shotgun synthesis fragments to the target penicillin biosynthetic gene cluster
Amplification of the desired shotgun synthesis products using barcode primer pairs
As described above, we used an in-house Python program to select overlapping sets of shotgun synthesis fragments. This analysis resulted in the selection of error-free shotgun synthesis fragments covering 88% of the 11 376 bp target sequence. These overlapping error-free DNA fragments were individually PCR-amplified from a pool of shotgun synthesis DNA mixtures using selected barcode primer pairs (Supplementary Table S1). PCR reactions comprised 8 µl water, 10 µl Phusion polymerase pre-mix (NEB, Ipswich, MA, USA), 1 µl barcode primer pairs and 1 µl of the shotgun synthesis DNA mixture. Thermocycling conditions were as follows: 3 min of heating at 95°C followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 1 min and a final elongation step at 72°C for 10 min. For the 12% of target gene cluster regions that exhibited low coverage (with only error-containing DNA fragments), we first amplified these error-containing fragments with barcode primers (Supplementary Table S1). The fragments were then re-amplified using error-correcting primers to fill the 12% low coverage regions of the target gene cluster. We carried out the two-step PCR reactions using the same PCR protocol as was used to synthesize the error-free shotgun synthesis fragments for two rounds of 30 cycles. PCR reaction mixes comprised 8 µl water, 10 µl Phusion polymerase pre-mix, 1 µl barcode primer pairs (Supplementary Table S1) and 1 µl shotgun synthesis DNA mixture. For the second round of PCR amplification, we used 1 µl nested PCR primer pairs and 1 µl of the PCR products from the first amplification step (Supplementary Table S1). The desired PCR amplification product sequences are listed in Supplementary Table S2. The error-free PCR amplification products were electrophoresed through an agarose gel (Supplementary Figure S1g), and selected bands were excised and purified using an AccuPrep™ gel-purification kit.
PCR assembly for 1 kb DNA synthesis using flanking sequence removed shotgun synthesis products
We assembled the flanking sequence removed shotgun synthesis products to make 11 gene cluster fragments (645–1325 bp). For the construction of 11 ∼1 kb DNA fragments, gel purified error-free fragments were pooled together based on the use of restriction enzyme types (Supplementary Table S1). For each pool, restriction enzyme digestion was carried out by incubating 2 µl EarI (or EcoP15I), 5 µl NEB buffer 1, 0.5 µl 100× BSA and 10 µl water with 30 µl of purified (and pooled) shotgun synthesis fragments at 37°C for 3 h. For EcoP15I (NEB, Ipswich, MA, USA) restriction enzyme digestion, we used NEB buffer 3 and added 10× ATP. We then electrophoresed the restriction digest products through 1.5% agarose gels and excised the DNA bands at ∼300 bp (Figure 1h and Supplementary Figure S1h). The expected DNA fragment sequences after restriction enzyme digestion are listed in Supplementary Table S3. Eleven gene cluster fragments (645–1325 bp; Supplementary Table S4) were constructed by PCR using 3 µl water, 10 µl Phusion polymerase pre-mix, 5 µl of 3–7 flanking sequence-cleaved DNA fragments (or error-corrected fragments) and 1 µl PCR primer sequences (Figure 1i, Supplementary Figure S1i and primer Set 5 in Supplementary Sequence 4). PCR products were run through 1% agarose gels and selected bands (Figure 1i and Supplementary Figure S1i) were excised and purified using a gel purification kit. These ∼1 kb DNA fragments were cloned into the TOPO vector using the TOP Cloner™ Blunt core kit (Enzynomics, Daejeon, Korea) and submitted for Sanger sequencing. A few colonies were chosen for colony PCR using M13 primer pairs (M13F-pUC and M13R-pUC primers). We used the Lasergene program (DNAstar, Madison, WI, USA) to analyze the DNA sequence data.
PCR assembly of an 11.4 kb gene cluster using flanking sequence removed shotgun synthesis products
We used a PCR method to assemble 11 error-free ∼1 kb fragments into the full-length penicillin biosynthetic gene cluster. The PCR reaction mixture consisted of 11 ∼1 kb fragments (each 1 µl) and 15 µl of Phusion polymerase pre-mix (NEB, Ipswich, MA, USA) in the absence of primers. The reaction mixture was heated at 95°C for 3 min, followed by 10 cycles of 95°C for 30 s, 70°C for 30 s, 72°C for 3 min 30 s, followed by a final elongation step at 72°C for 5 min. We then added 1 µl primer pairs containing restriction enzyme sites (BglII, NotI) (Primer Set 6 at the Supplementary Sequence 4) to the PCR reaction mixture and performed 25 more PCR cycles. The PCR products were electrophoresed through an agarose gel and bands of the desired size (Figure 2j) were excised and purified using a gel purification kit. We cloned the products into a plasmid (pBK3) using BglII and NotI restriction enzymes (Enzynomics, Korea) based on protocols provided by the vendor, and transformed C2566 E. coli competent cells (NEB, Ipswich, MA, USA) with the vector. After overnight growth at 37°C, we screened a few colonies for pBK3 vector containing the desired DNA using colony PCR. Several colonies were grown in Luria-Bertani media for plasmid extraction using an AccuPrep™ plasmid extraction kit (Bioneer, Daejeon, Korea). One plasmid was extracted and submitted for Sanger sequencing with multiple sequencing primers (Primer Set 7; Supplementary Sequence 4). Sequencing data were analyzed using the Lasergene program (DNAstar, Madison, WI, USA).
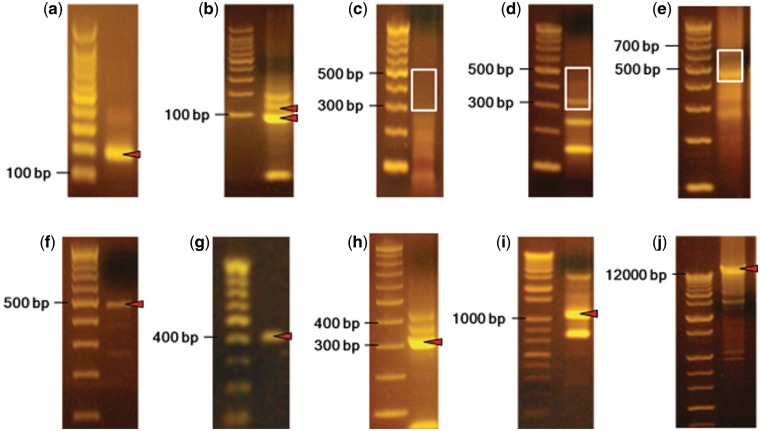
Figure 2.
Gel data from the shotgun DNA synthesis experiment. (a) PCR products of the microchip oligonucleotides using flanking primers. (b) Removal of the flanking sequences from the microchip oligonucleotides prior to shotgun assembly. DNA bands at 100 and 125 bp (red triangles) were excised and gel-purified together. (c) The shotgun synthesis reaction using the processed pool of 228 microchip oligonucleotides. Bands ranging in size from 300 bp to 500 bp (white box) were isolated from the smear and gel-purified. (d) The gel-purified products from c were re-amplified using microchip flanking primers. The bands in the white box were excised and gel-purified. (e) The shotgun synthesis fragments (from d) were PCR-tagged using barcode primers. Bands between 450 and 600 bp (white box) were excised and purified. (f) Re-amplification of barcoded shotgun synthesis products using 454-adaptor primers. DNA from the 500 bp region (red triangle) was excised and purified for 454 sequencing. (g) One of the 61 desired DNA PCR recovery reactions using degenerate barcode sequences (the remaining gel data is shown in Supplementary Figure S1g). (h) Removal of the flanking sequences from a pool of 3–7 shotgun synthesis products using a Type IIS restriction enzyme (the remaining gel data is shown in Supplementary Figure S1h). (i) One of the PCR products (∼1 kb DNA fragments) using flanking sequence removed shotgun synthesis products (the remaining gel data is shown in Supplementary Figure S1i). (j) PCR assembly of 11 ∼1 kb DNA fragments to construct the target 11.4 kb gene cluster.
RESULTS AND DISCUSSION
We set out to develop a shotgun DNA synthesis technology as illustrated in Figure 1. While we focused on employing shotgun synthesis, we also wanted to overcome the challenges of high-throughput DNA construction using both microchip oligonucleotides and high-throughput DNA sequencing. The shotgun method is based on the hypothesis that a pool of oligonucleotides can be synthesized in one pot to produce randomly assembled products, and that each one of these heterogeneous products can be identified by high-throughput sequencing. Computational analysis is subsequently utilized to select the optimal set of synthetic DNA fragments for target assembly.
We designed two sets of 228 microchip oligonucleotides to construct a codon-optimized penicillin biosynthetic gene cluster [N-(5-amino-5-carboxypentanoyl)-l-cysteinyl-d-valine synthase, 11 376 bp] (17,18) (Supplementary Sequences 1 and 2). For each set, two different Type IIS restriction enzymes (i.e. EarI and BtsI) were used in common flanking sequences to ensure that at least one set of microchip oligonucleotides remained intact after restriction enzyme digestion. We carried out selective amplification of sub-pool oligonucleotides cleaved from a 55 K Agilent DNA microchip using two sets of flanking primer pairs (Supplementary Sequence 4: primer set 1), and removed common sequences using the Type IIS restriction enzymes as previously illustrated (Figure 2a and b) (8,9). We then carried out shotgun DNA synthesis by utilizing microchip oligonucleotides with cleaved flanking sequences. As expected, the shotgun synthesis of hundreds of oligonucleotides resulted in highly heterogeneous DNA fragments ranging in size from 100 bp to 1000 bp (Figure 2c). We isolated DNA corresponding to the 300–500 bp region using agarose gel electrophoresis. These sizes of DNA fragments were chosen because the limit of 454 high-throughput sequencing was 500 bp at the time of this study.
We then focused on developing a method to identify random DNA fragments using high-throughput sequencing technology, as well as a method to obtain sequence-validated error-free fragments from the pool of DNA fragments (Figure 1c). The gel-purified shotgun synthesis DNA fragments were tagged with barcodes through amplification with barcode PCR primers that consisted of three functional parts: the original primer sequences used for the amplification of the chip oligonucleotides, 20 bp degenerate-barcode sequences and 454 adaptor sequences. Because the efficiency of restriction enzyme digestion was <100%, the shotgun synthesis DNA fragments were amplified using the remaining uncut original flanking primer sequences. We found that using the same microchip flanking primer sequences for PCR amplification of the shotgun-synthesized DNA fragments was the key for successful preparation of barcode-tagged random DNA molecules (Figures 1c and 2d–f, and Supplementary Note 2).
We sequenced a mixture of shotgun synthesis products (∼400 bp including barcode flanking sequences) from two independent experiments. It turned out that for this length of products, ∼3% of the fragments were error-free (Figure 3a and Supplementary Figure S2). Calculated error value using an equation for error rate evaluation was 8.4% (Supplementary Note 3). The rest of the fragments (i.e. 97%) containing errors were analyzed as well (Supplementary Figures 3 and 4). Out of 56 632 reads of error-containing shotgun products, 1750 reads failed to align to the reference sequence of the target gene cluster. This may be because these reads were incorrectly overlapped during the shotgun assembly process. Of the reads, 12 495 reads did not meet the minimum quality score criterion (Phred-like consensus quality ≥30) and the rest of the reads among the error-containing shotgun products (40 653 reads/56 632 reads, i.e. 72%) had errors in the internal sequences. A detailed analysis of the error-containing reads is provided in Supplementary Figures 3 and 4. We then wrote a computer program to determine the optimal set of overlapping error-free DNA fragments that could be used to advance the assembly process (Figure 3 and Supplementary Methods). Eighty-eight percent of the 11 376 bp target sequences exhibited enough synthetic coverage to allow the extraction of error-free shotgun synthesis DNA fragments (∼300 bp each). In contrast, 12% of the target DNA regions showed low coverage from the assembly steps, but still contained error-containing DNA fragments. To determine whether the missing 12% of sequences were difficult regions to synthesize due to biased GC content, we analyzed the missing 12% of sequences. The GC content of the missing sequences ranged from 49.6% to 60.0%, which is a typical GC content for bacterial DNA (Supplementary Table S5). We evaluated the error-containing fragments and determined which sequences could be re-amplified using error-correcting primers to fill in the 12% missing regions of the target gene cluster.
A total of 61 pairs of barcode primer sequences were selected, of which 14 primer pairs were used for both the selection and correction of DNA fragments (Supplementary Table S1). Based on the gel data (∼400 bp; Supplementary Figure S1g), 77% (47 out of 61) of selective amplification reactions using barcode primer sequences resulted in the desired sequences (Supplementary Table S2). Alternative PCR primer sequences were utilized successfully to obtain the remaining target fragments. We processed these amplified sequences to remove the barcode sequences using a Type IIS restriction enzyme prior to the second round of DNA assembly (Supplementary Table S3).
We pooled 3–7 fragments with cleaved flanking sequences (∼300 bp each) and constructed 11 ∼1 kb fragments by PCR (Figure 2i, Supplementary Figure S1i and Supplementary Table S4). We cloned these 1 kb DNA fragments and submitted them for Sanger sequencing for validation (Supplementary Table S4). In summary, 16 out of 21 colonies (i.e. one to three colonies per construct) were error-free with an average synthetic error rate of 0.022% (i.e. 5 errors per 22 903 bp; Supplementary Table S6). Note that we used a Phred-like quality score >30, which corresponded to a base call accuracy of >99.9% (i.e. one incorrect base call per 1000 bases) when we evaluated the 300 bp shotgun fragments using 454 sequencing. The intrinsic sequencing error rate might affect the error rate of the 1 kb synthetic fragments, because the 300 bp fragments used for ∼1 kb construction were selected based on sequencing data with a base call accuracy of >99.9%. We used 11 error-free DNA fragments for the final PCR assembly to construct the biosynthetic gene cluster of penicillin (Figure 2j). Subsequent cloning and sequencing showed that we successfully obtained the desired penicillin gene cluster (i.e. 0 errors per 11 376 bp).
In summary, we developed a shotgun DNA synthesis technology for high-throughput assembly of large DNA molecules that involved the use of a novel, random assembly process to synthesize DNA intermediates. The shotgun synthesis products were barcoded and profiled by high-throughput sequencing. The sequencing information was analyzed by a computer program to ensure the accuracy of the DNA molecules prior to further assembly into target DNA sequences. We believe that our high-throughput-based sequencing method provides a way to reduce the costs of synthetic DNA associated with sequence validation. Furthermore, no cloning is required early in the process of shotgun synthesis, and our use of random barcodes to identify error-free products is unique.
We ran a cost analysis of the shotgun DNA synthesis of an 11.4 kb gene cluster based on several factors including the cost of microchip oligonucleotides, 454 high-throughput sequencing, Sanger sequencing, primers and PCR reagents, among other considerations (Supplementary Note 4). Based on the cost analysis, we believe that as 454 sequencing becomes cheaper and gene syntheses can be multiplexed, sequencing costs will decrease. Our shotgun synthesis method can be performed using other sequencing platforms [i.e. Illumina or Pacific Biosciences (PacBio)] to further reduce synthesis costs. In particular, the use of Illumina platforms in shotgun synthesis may dramatically lower the sequencing costs if the Illumina read length becomes longer (Supplementary Table S7). We also note that for each target cluster, new barcode primers will have to be ordered for extraction of DNA segments, which is expensive and will make the process slower. However, selection primers can be reused if thousands (e.g. 3000) of forward barcode primers and thousands (e.g. 3000) of reverse barcode primers are used to tag the shotgun synthesis fragments. The possible combinations of these barcode primers are sufficient (i.e. 3000 × 3000 = 9 × 106) to ensure unique selection of fragments from a pool of DNA for 454 high-throughput sequencing.
Due to the low error rate associated with the use of Agilent microchip oligonucleotides, cloning of DNA fragments using a high-fidelity DNA microchip-based assembly method (7) should not be necessary for more than a couple of colonies per fragment to find error-free fragments. Although the use of Agilent microchip oligos makes high-throughput sequencing-based shotgun DNA synthesis less attractive, combining the shotgun method with ultra-deep sequencing would make low-fidelity DNA microchips relevant for further DNA assembly steps. For example, Illumina’s MiSeq platform can produce 4 GB of data based on 150 paired-end reads (i.e. 300 bp), and the read length is expected to reach 500 bp (250 bp paired-end) in 2012. The long read length and massive throughput of the MiSeq platform will facilitate shotgun synthesis using not only oligos from Agilent OLS-based microarrays, but also from more error-prone microchips provided by other microchip vendors. The PacBio sequencing platform could also potentially be utilized to obtain longer synthetic DNA fragments. However, the error rate of this sequencing platform is still 87%. Thus, four circular consensus reads from PacBio are needed to accurately determine sequencing errors (19) and to select synthetic DNA fragments with an error rate better than 1 in 1000 bp. Recently developed gene synthesis platforms (454, MiSeq and PacBio platforms) that could be used in shotgun DNA synthesis are compared in Supplementary Table S8.
Furthermore, it is worthwhile to note that shotgun synthesis can provide a solution for low DNA assembly efficiency. Highly heterogeneous by-products from less efficient DNA assembly processes can be isolated and utilized for further DNA assembly procedures. We found that shotgun DNA synthesis using high-throughput sequencing resulted in one error per 4581 bp, which was comparable to the error rate with additional error correction procedures (Supplementary Table S9) (20). Thus, we expect that shotgun synthesis based on currently available error correction/screening methods would facilitate the multiplexing of DNA assembly at a given sequencing throughput. Shotgun sequencing has made a significant contribution to de novo genome sequencing. Considering the fact that current high-throughput DNA sequencing technology is approaching read lengths of 1000 bp (454 sequencings FLX+ system) (11) and 10 kb (Pac Bio) (21), and that microchip DNA density is approaching a few million spots, we envision that shotgun synthesis will greatly benefit genome writing and synthetic biology projects that require a large number of highly variable synthetic DNA molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–9, Supplementary Figures 1–4, Supplementary Methods, Supplementary Sequences 1–4 and Supplementary Notes 1–4.
FUNDING
This work was supported by the Intelligent Synthetic Biology Center of the Global Frontier Project funded by the Ministry of Education, Science and Technology, Korea [2011-0031956]. Hwangbeom Kim was supported by a National Junior Research Fellowship, National Research Foundation of Korea [2012-002388]. Funding for open access charge: Ministry of Education, Science and Technology [2011-0031956].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jahyang Jung for her critical reading of this manuscript.
REFERENCES
- 1.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 2.Carr PA, Church GM. Genome engineering. Nat. Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 3.Bang D, Church GM. Gene synthesis by circular assembly amplification. Nat. Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 4.Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. Nucleotide sequence of bacteriophage lambda DNA. J. Mol. Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 5.Quan J, Saaem I, Tang N, Ma S, Negre N, Gong H, White KP, Tian J. Parallel on-chip gene synthesis and application to optimization of protein expression. Nat. Biotechnol. 2011;29:449–452. doi: 10.1038/nbt.1847. [DOI] [PubMed] [Google Scholar]
- 6.Borovkov AY, Loskutov AV, Robida MD, Day KM, Cano JA, Le Olson T, Patel H, Brown K, Hunter PD, Sykes KF. High-quality gene assembly directly from unpurified mixtures of microarray-synthesized oligonucleotides. Nucleic Acids Res. 2010;38:e180. doi: 10.1093/nar/gkq677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosuri S, Eroshenko N, Leproust EM, Super M, Way J, Li JB, Church GM. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat. Biotechnol. 2010;28:1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H, Jeong J, Bang D. Hierarchical gene synthesis using DNA microchip oligonucleotides. J. Biotechnol. 2011;151:319–324. doi: 10.1016/j.jbiotec.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 10.Gibson DG, Smith HO, Hutchison CA, III, Venter JC, Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods. 7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- 11.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matzas M, Stahler PF, Kefer N, Siebelt N, Boisguerin V, Leonard JT, Keller A, Stahler CF, Haberle P, Gharizadeh B, et al. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing. Nat. Biotechnol. 2010;28:1291–1294. doi: 10.1038/nbt.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. 2004;32:e162. doi: 10.1093/nar/gnh160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Han H, Shin D, Bang D. A fluorescence selection method for accurate large-gene synthesis. Chembiochem. 11:2448–2452. doi: 10.1002/cbic.201000368. [DOI] [PubMed] [Google Scholar]
- 15.Saaem I, Ma S, Quan J, Tian J. Error correction of microchip synthesized genes using Surveyor nuclease. Nucleic Acids Res. 40:e23. doi: 10.1093/nar/gkr887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linshiz G, Yehezkel TB, Kaplan S, Gronau I, Ravid S, Adar R, Shapiro E. Recursive construction of perfect DNA molecules from imperfect oligonucleotides. Mol. Syst. Biol. 2008;4:191. doi: 10.1038/msb.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diez B, Gutierrez S, Barredo JL, van Solingen P, van der Voort LH, Martin JF. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J. Biol. Chem. 1990;265:16358–16365. [PubMed] [Google Scholar]
- 19.Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW. A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 38:e159. doi: 10.1093/nar/gkq543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma S, Saaem I, Tian J. Error correction in gene synthesis technology. Trends Biotechnol. 30:147–154. doi: 10.1016/j.tibtech.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.