Abstract
Proteins are covalently trapped on DNA to form DNA–protein crosslinks (DPCs) when cells are exposed to DNA-damaging agents. DPCs interfere with many aspects of DNA transactions. The current DPC detection methods indirectly measure crosslinked proteins (CLPs) through DNA tethered to proteins. However, a major drawback of such methods is the non-linear relationship between the amounts of DNA and CLPs, which makes quantitative data interpretation difficult. Here we developed novel methods of DPC detection based on direct CLP measurement, whereby CLPs in DNA isolated from cells are labeled with fluorescein isothiocyanate (FITC) and quantified by fluorometry or western blotting using anti-FITC antibodies. Both formats successfully monitored the induction and elimination of DPCs in cultured cells exposed to aldehydes and mouse tumors exposed to ionizing radiation (carbon-ion beams). The fluorometric and western blotting formats require 30 and 0.3 μg of DNA, respectively. Analyses of the isolated genomic DPCs revealed that both aldehydes and ionizing radiation produce two types of DPC with distinct stabilities. The stable components of aldehyde-induced DPCs have half-lives of up to days. Interestingly, that of radiation-induced DPCs has an infinite half-life, suggesting that the stable DPC component exerts a profound effect on DNA transactions over many cell cycles.
INTRODUCTION
DNA is associated with various structural and regulatory proteins in cells. Proteins are often covalently trapped on DNA, generating DNA–protein crosslinks (DPCs) when cells are exposed to DNA-damaging agents (1). The formation of DPCs was originally demonstrated for bacterial and mammalian cells that were heavily irradiated with ultraviolet light (2,3). It was subsequently shown that DPCs are produced by a number of chemical and physical agents such as aldehydes (4), metal ions (5), anticancer drugs (6,7) and ionizing radiation (8). Since crosslinked proteins (CLPs) are extremely large, steric hindrance conferred by DPCs likely interferes with many aspects of DNA transactions, including replication, transcription and repair. Recent studies with defined DPC substrates have begun to unravel the molecular mechanism by which cells respond to this type of potentially deleterious DNA damage (9). In bacteria, DPCs inhibit the replication of plasmid DNA in vivo (10–12), and replication forks stalled by DPCs are likely reactivated by RecBCD-dependent homologous recombination (HR) and the subsequent action of PriA helicase to continue DNA synthesis through DPCs (11,13). DPCs containing small (but not large) CLPs are removed from DNA by nucleotide excision repair (NER) (11,13–15). HR also deals with DPCs in mammalian cells (16,17), but there is virtually no contribution from NER due to its intrinsic low repair capacity for small CLPs (16,18–21).
The properties of DPCs vary significantly depending on the CLPs and crosslinking bonds. Recent proteomic studies have revealed numerous CLPs produced by ionizing radiation (22), formaldehyde (FA) (23), antitumor nitrogen mustards (24) and the carcinogenic metabolite of 1,3-butadiene (25). In DPCs, proteins are crosslinked to the base or sugar phosphate moiety either directly or indirectly through a linker (crosslink agent), and the half-life (stability) of crosslinking bonds ranges from hours to days (1). When studying the cytotoxic/mutagenic effects and repair mechanism of DPCs, it is crucial to detect their induction and removal in genomic DNA. The detection of DPCs requires several issues to be addressed. For instance, covalently and non-covalently bound proteins must be separated stringently: the latter are present in extreme excess over the former in the genome. Moreover, DPCs need to be detected selectively in the background of many other DNA lesions, since DNA-damaging agents concurrently induce base damage, DNA strand breaks and DNA interstrand crosslinks along with DPCs.
Several methods have been developed to detect DPCs. The alkaline elution method is based on the different elutabilities of DNA without and with CLPs from a filter under alkaline conditions (26,27). Cells are filtered onto a polyvinylchloride filter and lysed with sarkosyl. DNA retained on the filter is eluted at pH 12.1, with the adsorption of CLPs to the filter reducing the elutability of unwound single-stranded DNA, thereby changing the elution kinetics of the DNA. The nitrocellulose filter-binding method relies on the different abilities of DNA without and with CLPs to bind to a nitrocellulose filter (28,29). Cells lysed with sarkosyl are passed through the filter, which retains proteins and DNA with CLPs, but not free DNA. The amount of DNA retained on the filter via CLPs is assayed for DPCs. The sodium dodecyl sulfate (SDS)/potassium ion (K+) precipitation method is based on SDS binding tightly to proteins and forming insoluble precipitates with potassium ions (5,30). Cells are lysed with SDS, and SDS-bound proteins and DNA with CLPs, but not free DNA, are selectively precipitated by KCl. The amount of DNA precipitated due to CLPs is assayed for DPCs. In addition to these methods, it has been shown that a certain type of DPC reduces the migration of DNA in single cell gel electrophoresis assays (comet assays) (31,32). Pretreatment of lysed cells with proteinase K clearly distinguishes DNA with and without DPCs in these methods.
The aforementioned DPC detection methods have provided valuable insights into the induction and removal of genomic DPCs formed by various DNA-damaging agents. However, as well as there being many technical issues (5,30,33), the indirect detection of DPCs based on the amount of DNA employed in these methods has intrinsic drawbacks. First, the adsorption of DNA onto the filters and the precipitation of DNA by SDS/K+ depend on not only the CLPs but also on the length of DNA, which varies significantly depending on DNA-damaging agents and their doses. Second, there is no linear relationship between the amounts of DNA and CLPs. Thus, determining the amount of CLPs from that of DNA inevitably requires some assumptions (7,34,35), which have not been rigorously examined and verified by compelling evidence. Accordingly, the quantitative interpretation of data derived from the indirect measurement of DPCs is complicated and needs further support from totally independent methods.
In view of the restrictions of the currently available DPC detection methods, it would be useful to develop a direct method of detecting DPCs by virtue of the measurement of CLPs. We have recently shown that CLPs in DNA isolated from FA-treated cells can be specifically labeled with fluorescein isothiocyanate (FITC) and directly assayed for DPCs by measuring the resulting fluorescence (11,16). However, the validity and generalizability of this method have not been rigorously tested for other DPC-inducing agents. The present study extended the previous approach by developing two formats of the DPC detection method based on FITC-labeling: direct fluorometric analysis and western blotting analysis using anti-FITC antibodies. Both methods can successfully monitor the induction and elimination of DPCs in cultured cells and transplanted mouse tumors exposed to aldehydes and ionizing radiation (carbon-ion beams), respectively. The western blotting format requires one-hundredth of the amount of DNA used for the fluorometric format. Analyses of the in vitro stability of DPCs isolated from cells and tumors have revealed that aldehydes and ionizing radiation produce two types of DPC with distinct stabilities. The stable components of aldehyde-induced DPCs have in vitro half-lives of up to days. Interestingly, that of radiation-induced DPCs has an essentially infinite half-life, suggesting that the stable DPC component exerts a profound effect on DNA transactions over many cell cycles.
MATERIALS AND METHODS
Chemicals and cell culture
Acrolein (ACR) was obtained from Alexis Biochemicals, and chloroacetaldehyde (CAA) and FA were purchased from Wako. Crotonaldehyde (CRA), glutaraldehyde (GA) and trans-2-pentenal (PEN) were purchased from Tokyo Chemical Industry. MRC5-SV (an SV40-transformed human fetal lung fibroblast cell line), HeLa (a human cervical carcinoma cell line) and XP12RO (an SV40-transformed fibroblast cell line derived from a xeroderma pigmentosum (XP) complementation group A patient) were used for the experiments. Cells were cultivated in Dulbecco’s modified Eagle’s medium (Nissui) supplemented with 10% inactivated fetal bovine serum (FBS) and l-glutamine. Cells were maintained in a humidified incubator at 37°C with 5% CO2 atmosphere, and harvested with 0.05% trypsin–EDTA.
Cell survival and sister chromatid exchange assays
Colony forming assays were used for cell survival measurements. Cells were seeded into 100-mm culture dishes and incubated for 12 h. The medium was then changed to an FBS-free medium containing aldehydes at the indicated concentrations, in which cells were incubated for 3 h. Cells were washed twice with fresh medium and allowed to form colonies for ∼7 days. Colonies with more than 50 cells were counted. The physiologically relevant doses that gave a 10% survival (LD10) were determined from the survival curves. Sister chromatid exchange (SCE) induction by aldehydes in MRC5-SV cells was analyzed by the fluorescence-plus-Giemsa method (36). Well-spread metaphases (total 90) were evaluated per individual aldehyde treatments for SCEs/cell. MRC5-SV cells have a hyperdiploid karyotype with a modal number between 60 and 80.
DNA purification from aldehyde-treated cells
Cells in the midlogarithmic phase (typically four 150-mm dishes) were treated with aldehydes at LD10 concentrations as described above. Cells were recovered without or with postincubation (up to 12 h) in culture media and stored at −80°C until use. Chromosomal DNA was isolated by CsCl density gradient centrifugation as described previously (11,16) with some modifications. The cell pellet was suspended in 900 µl of a buffer containing 10 mM phosphate buffer (PB; pH 7.5), 1 mM EDTA, RNase A (20 µg, Sigma), and a protease inhibitor cocktail (Roche), and kept on ice for 10 min. The sample was mixed with sarkosyl (final concentration 1%) and kept on ice for 30 min. The lysed cells were passed through a G23-gage needle 20 times to shear DNA, and PB was added to a final volume of 9 ml. The sample was mixed with 9.3 g of CsCl (Wako) and sedimented at 500 000 × g at 20°C for 4 h with a Beckman Type 90Ti rotor. After centrifugation, the sample was fractionated (∼500 µl) through a bottom needle hole. Fractions containing DNA (determined by agarose gel electrophoresis or UV absorption) were pooled and dialyzed against PB at 4°C. The dialyzed sample was sedimented again by CsCl density gradient centrifugation as described above to ensure the complete removal of free proteins. Fractions containing DNA were combined and dialyzed against PB + 1 mM EDTA (3 h × 2), PB + 2 M NaCl (3 h × 2) and finally MilliQ water (3 h × 2) at 4°C. The sample volume was reduced to 200 µl on a centrifugation evaporator without applying heat. It is crucial for the DNA yield to completely recover viscous DNA that has adhered onto the wall of the evaporation tube. The DNA concentration was measured on a Thermo Scientific Nanodrop 2000c or Jasco V-550 UV spectrophotometer, and the DNA was stored at −20°C until use.
FITC labeling and fluorescence measurement
FITC (Dojindo) was dissolved in dimethylformamide to a final concentration of 10 mM. DNA (50 µg) in 20 mM borate buffer (pH 8.0, 100 µl) was mixed with the FITC solution (final concentration 0.1 mM) and incubated at room temperature for 1 h. DNA was precipitated by ethanol, and the resulting DNA pellet was washed twice with 70% ethanol, air dried and dissolved in MilliQ water. The concentration of DNA was measured on a UV spectrophotometer. The fluorescence of FITC-labeled DNA (30 µg) was measured on a Hitachi F-2500 fluorescence spectrophotometer.
Immunodetection of DPCs
FITC-labeled DNA (0.3 µg) was vacuum slot-blotted onto a Hybond-ECL nitrocellulose membrane (GE Healthcare), and the slot was washed with Tris-buffered saline. The membrane was blocked with 5% non-fat milk for 1 h, washed with Tris-buffered saline/Tween-20 (TBST) comprising 20 mM Tris–HCl (pH 7.6), 137 mM NaCl and 0.1% Tween-20. The membrane was incubated with a goat anti-FITC antibody (Bethyl Laboratories) at a 1/40 000 dilution in TBST for 1 h. The unbound antibodies were removed by washing in TBST (15 min × 4). The membrane was incubated with a horseradish peroxidase (HRP)-conjugated rabbit anti-goat IgG antibody (Anaspec) at a 1/20 000 dilution in TBST for 1 h. The remaining unbound antibody was washed away by TBST. The membrane was incubated with the ECL western blotting substrate (Promega) for 1 min, and the chemiluminescence was measured on a Toyobo FAS1000 luminescence analyzer. Captured images were analyzed using NIH ImageJ 1.33 software.
Irradiation of mouse tumors
Transplanted mouse tumors were irradiated with accelerated carbon ions as described previously (37,38). Briefly, squamous carcinoma cells (SCC VII) were inoculated subcutaneously into the right hind legs of syngeneic male C3H/He mice aged 8–10 weeks (Japan Animal Co.). Ten days later, tumors, ∼10 mm in diameter, were irradiated with 12C6+ (290 MeV/u) generated by the Heavy Ion Medical Accelerator in Chiba (HIMAC). A 6-cm spread-out Bragg peak with a dose-averaged linear energy transfer (LET) of 80 keV/µm was used for tumor irradiation. The dose rate was 3.6 Gy/min. For the irradiation of hypoxic tumors, tumors in mouse legs were rendered totally hypoxic by clamping at the proximal side of the tumor for 15 min before irradiation. The clamping was removed immediately after irradiation. Mice were sacrificed at the indicated times after irradiation and rapidly cooled in ice-water. Tumors were excised, washed with cold PBS and used for further analysis. All animal experiments were carried out according to the guidelines for animal experimentation of the National Institute of Radiological Sciences and Hiroshima University.
DNA purification from irradiated tumors
The excised tumor (∼ 0.13 g) was minced on ice, suspended in 1.5 ml of lysis buffer containing 5.5 M guanidine thiocyanate, 25 mM sodium citrate (pH 7.0), 0.5% sarkosyl and 100 mM 2-mercaptoethanol, and homogenized by an IKA Ultra-Turrax T25 homogenizer (10 s × 2) with cooling. The sample was centrifuged at 3000 g for 10 min, and the supernatant was recovered. The genomic DNA was purified as described for aldehyde-treated cells.
Measurement of in vitro half-lives of DPCs
The genomic DNA that was purified from cells or tumors was labeled with FITC as described above. Proteins released from DNA were separated from DNA by dialysis and the remaining CLPs were quantified by fluorescence measurements as follows. FITC-labeled DNA (typically 30–50 µg) was dissolved in 2 ml of 10 mM Tris–HCl (pH 7.5) and 1 mM EDTA (TE buffer), placed in a dialysis tube (molecular cut-off = 100 kDa) and dialyzed against TE at 37°C for up to 72 h in the dark. The dialysis solution was changed after 3 h and then every10 h thereafter. The fluorescence of the sample was measured at the indicated times. Since FITC decomposed spontaneously with a half-life of ∼183 h under the present conditions, the FITC signal of the remaining DPCs was corrected for a spontaneous decay. Free FITC-labeled bovine serum albumin and histone H2A were dialyzed out of the tube almost completely (>95%) within 6 h under these conditions. The stability of DPCs was analyzed based on a two-component model, whereby unstable and stable DPCs decay exponentially following the first-order kinetics. The compositions and half-lives of the two components were calculated by regression analysis using DeltaGraph version 5 (Red Rock Software).
Proteinase K treatment of DPCs
FITC-labeled DNA (typically 30–50 µg) was incubated with 1.5 µg of proteinase K (Wako) in PB at 37°C for 30 min. After treatment, the sample was dialyzed against cold PB using a dialysis tube with a molecular cut-off = 100 kDa for 6 h and measured for fluorescence.
Analysis of DNA double-strand breaks
The excised tumor was disaggregated by stirring for 20 min at 37°C in PBS containing 0.05% trypsin and 0.02% EDTA. DNA double-strand breaks (DSBs) in dispersed cells were analyzed by static-field gel electrophoresis as described previously (37,39). The fraction of DNA released from the plug relative to total DNA (i.e. released and retained DNA) was used as a measure of DNA DSBs.
RESULTS
Outline of the direct DPC detection methods
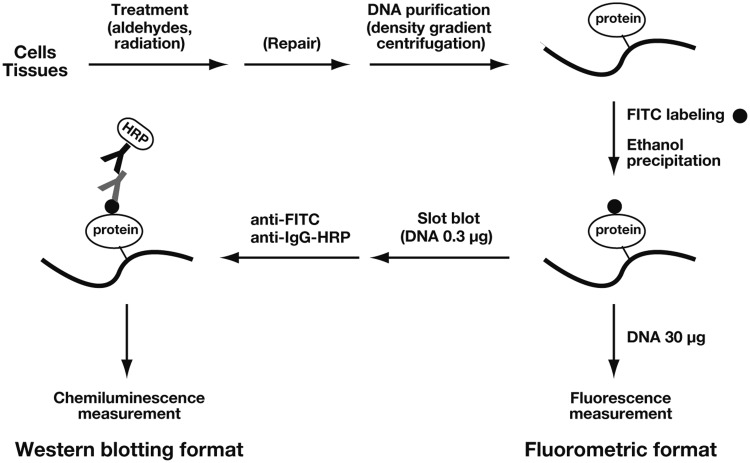
We developed two formats of the direct DPC detection method based on the FITC labeling of CLPs: fluorometric analysis and western blotting analysis using anti-FITC antibodies (Figure 1). The genomic DNA is purified from cells or tissues by CsCl density gradient centrifugation. Although we used two cycles of CsCl density gradient centrifugation to ensure the complete removal of free proteins, one cycle of DNA purification is generally sufficient for DPC detection. The DNAazol-based method may be an alternative choice for the purification of DPC-containing DNA (22,40). The CLPs in purified DNA are selectively labeled by FITC, which reacts with the primary amines of proteins but not the aromatic amines of DNA bases. Free FITC can be completely removed by the simple ethanol precipitation of DNA. In the fluorometric format, DPCs are quantified by directly measuring the fluorescence of a known amount of DNA (typically 30 µg). This method resembles the colorimetric quantitation of proteins. In the western blotting format, DNA (typically 0.3 µg) is slot-blotted onto a nitrocellulose membrane and incubated with an anti-FITC antibody followed by an HRP-conjugated secondary antibody. DPCs are quantified by measuring the chemiluminescence. As demonstrated below, the two methods yielded consistent results for the detection of DPCs induced by aldehydes and ionizing radiation.
Figure 1.
Outline of the direct methods of DPC detection. After treatment with aldehydes or ionizing radiation, genomic DNA is purified from cells or tissues using CsCl density gradient centrifugation. The CLPs in purified DNA are selectively labeled by FITC, and unbound FITC is removed by the ethanol precipitation of DNA. In the fluorometric format, DPCs are quantified by directly measuring the fluorescence of a known amount of DNA (typically 30 µg). In the western blotting format, DNA (typically 0.3 µg) is slot-blotted onto a nitrocellulose membrane and incubated with an anti-FITC antibody followed by the HRP-conjugated secondary antibody. DPCs are quantified by measuring the resultant chemiluminescence.
DPC induction can be detected at physiologically relevant aldehyde doses
We first measured the sensitivity of MRC5-SV and HeLa cells to aldehydes to determine the physiologically relevant dose ranges. Cells were treated with aldehydes for 3 h and their sensitivities were analyzed using the colony forming assay. Typical survival curves of MRC5-SV cells are shown in Supplementary Figure S1. The LD10 values (µM) were determined from the survival curves and are summarized in Table 1. The LD10 values for HeLa cells were similarly determined and are also listed in Table 1. ACR, CAA and GA exhibited strong cytotoxicity to both cell types (LD10: 5.6–16 µM), whereas CRA, FA and PEN exhibited weak cytotoxicity (LD10: 110–220 µM). Since MRC5-SV and HeLa cells share common aldehyde sensitivities, we used MRC5-SV cells for subsequent assays of DPCs.
Table 1.
LD10 values of aldehydes for MRC5-SV and HeLa cells
| Aldehydes | LD10 (µM) |
|
|---|---|---|
| MRC5-SV | HeLa | |
| ACR | 5.8 | 16 |
| CAA | 5.6 | 11 |
| CRA | 110 | 130 |
| GA | 7.1 | 11 |
| FA | 220 | 220 |
| PEN | 200 | 110 |
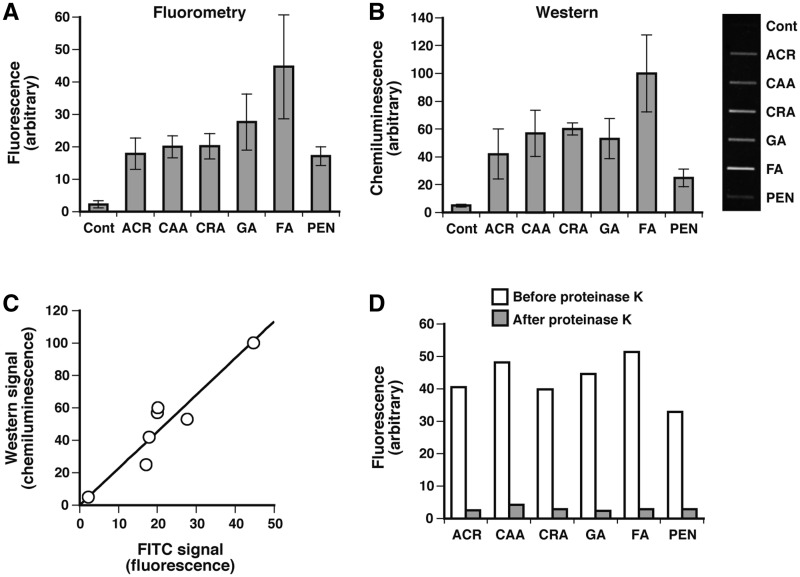
To analyze DPC induction in the genome, MRC5-SV cells were treated with aldehydes at LD10. DNA was isolated from the cells immediately after treatment and assayed for DPCs using fluorometric and western blotting formats. Both methods clearly revealed the induction of DPCs in the genome (Figure 2A and B) and the signals from the two methods exhibited a good correlation (Figure 2C). The fluorescence (Figure 2A) and chemiluminescence (Figure 2B) signals of treated cells were on average 10-fold greater than those of untreated control cells, showing that DPC-induction by aldehydes can be monitored at physiologically relevant doses. It is unknown whether the background signals for untreated control cells are due to endogenous DPCs or the non-specific background associated with FITC labeling. FA induced more DPCs than the other aldehydes, but there was a relatively small variation in the yields of DPCs among the aldehydes, indicating that the equitoxic doses of aldehydes induce the comparable amounts of genomic DPCs. To verify that the signals in the present assays were derived from DPCs, FITC-labeled DNA from aldehyde-treated cells were digested with proteinase K for 30 min and dialyzed to remove the resulting peptides. This treatment almost completely eliminated the FITC signal associated with DNA (≥94%; Figure 2D), confirming that FITC had attached to CLPs.
Figure 2.
Detection of DPC induction by aldehydes using fluorometric and western blotting formats. MRC5-SV cells were treated with aldehydes at LD10 (Table 1). DNA was isolated from the cells immediately after treatment and assayed for DPCs using (A) fluorometric and (B) western blotting formats. (C) Correlation between fluorometric and western blotting signals. Western blotting signals for aldehydes (panel B) are plotted against the corresponding FITC signals (panel A). (D) Release of the FITC signal (fluorescence) associated with DNA after treatment with proteinase K. Data points of panels A and B are means of three independent experiments with standard deviation, and those in panel D are based on a single experiment.
In vivo kinetics of the elimination of aldehyde-induced DPCs
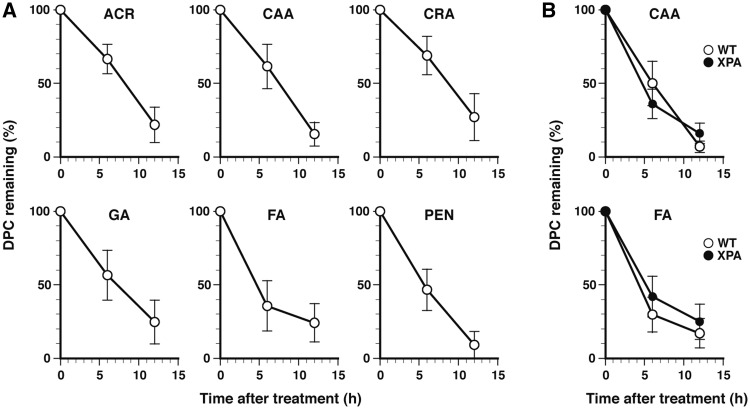
Since the induction of DPCs by aldehydes was successfully detected by the FITC-labeling methods (Figure 2), we then analyzed the elimination of DPCs in cells. MRC5-SV cells were treated with aldehydes at LD10 and incubated for up to 12 h in fresh medium. DNA was isolated from cells after 0, 6 and 12 h of postincubation, and assayed for DPCs using the fluorometric and western blotting formats. The two methods yielded consistent results. Figure 3A shows the time-dependent changes in the percentage of remaining DPCs (measured by fluorometry). With all aldehydes, the amount of DPCs decreased with postincubation time, but their changes did not follow a simple exponential decay, as was expected from the spontaneous hydrolysis (4,41–43) or excision repair of DPCs, suggesting the delayed formation of DPCs after removing aldehydes from the culture media. The apparent half-lives with 50% of the DPCs remaining were 8.2 h (ACR), 7.5 h (CAA), 8.4 h (CRA), 7.2 h (GA), 4.8 h (FA) and 5.7 h (PEN) (Figure 3A, Table 2). According to the SDS–PAGE analysis of heat-liberated CLPs, histones seem to be the major proteins that were crosslinked by FA and ACR (Supplementary Figure S2) (44). To elucidate whether the active repair of DPCs by NER was involved in the elimination of DPCs, the time-dependent changes in DPCs for selected aldehydes (CAA and FA) were followed by the western blotting format using NER-proficient MRC5-SV and NER-deficient XP12RO (XPA) cells. The two types of cell exhibited similar DPC elimination kinetics (Figure 3B) strongly suggesting that NER does not play a role in the removal of genomic DPCs. This result is consistent with the findings from both our and other laboratories of the negligible involvement of NER in the repair of DPCs in mammalian cells (16,21,41). Interestingly, mammalian cells deficient in XPF that is responsible for the 5′-incision in NER are sensitive to FA, but whether XPF plays a role in the context of NER or other repair mechanisms remains to be elucidated (16,45). Yeast cells deficient in NER factors that correspond to mammalian XPC and XPF are also sensitive to acute high-dose FA treatment (but not to chronic low-dose FA treatment). However, the NER-deficient yeast cells exhibit a similar rate of elimination of FA-induced DPCs as wild-type cells (46). Thus, it remains to be seen whether cells have repair mechanisms whereby DPCs are actively removed from the genome.
Figure 3.
In vivo kinetics of the elimination of aldehyde-induced DPCs. (A) Elimination of genomic DPCs after treatment with aldehydes. MRC5-SV cells were treated with the indicated aldehydes at LD10 (Table 1) and incubated in fresh medium. DNA was isolated from cells after 0, 6 and 12 h of postincubation, and assayed for DPCs using the fluorometric format. (B) Elimination of aldehyde-induced genomic DPCs in NER-proficient MRC5-SV (WT) and NER-deficient XP12RO (XPA) cells. Experiments were performed with selected aldehydes (CAA and FA) as described for panel A. DPCs were detected using the western blotting format. Data points of panels A and B are means of three independent experiments with standard deviation.
Table 2.
In vitro half-lives and initial compositions of unstable and stable DPCs induced by aldehydes and C-ion beams
| Agents | Half-life (h) |
Composition (%) |
Apparent half-life (h)a |
||||
|---|---|---|---|---|---|---|---|
| Unstable | Stable | Unstable | Stable | In vitro | In vivo | Ratio | |
| Aldehydes | |||||||
| ACR | 5.4 | 26.9 | 23.8 | 76.2 | 18.2 | 8.2 | 2.2 |
| CAA | 5.4 | 26.6 | 26.7 | 73.3 | 17.0 | 7.5 | 2.3 |
| CRA | 2.4 | 23.9 | 16.1 | 83.9 | 17.9 | 8.4 | 2.1 |
| GA | 5.3 | 24.1 | 12.1 | 87.9 | 20.2 | 7.2 | 2.8 |
| FA | 6.8 | 103.5 | 90.4 | 9.6 | 8.0 | 4.8 | 1.7 |
| PEN | 6.6 | 54.1 | 59.6 | 40.4 | 12.4 | 5.7 | 2.2 |
| Ionizing radiation | |||||||
| C-ion | 10.7 | 3.5 × 104 | 40.1 | 59.9 | – | – | – |
Aldehydes induce two types of DPC with distinct stabilities
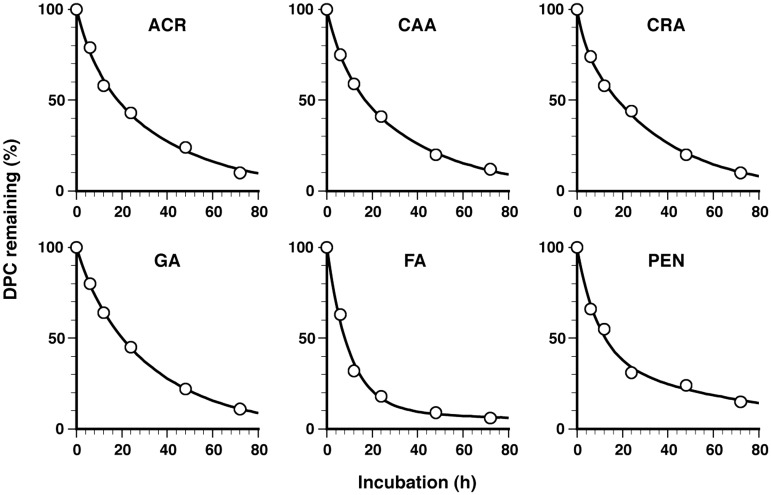
To obtain insights into the characteristics of aldehyde-induced DPCs, the in vitro stability of DPCs was analyzed. DNA isolated from cells immediately after treatment at LD10 was labeled with FITC and incubated in TE buffer (pH 7.4) at 37°C, with dialysis (molecular cut-off = 100 kDa) against the same buffer to separate the proteins released from DNA. The remaining DPCs were quantified by measuring the fluorescence of the sample inside the dialysis tube. The amount of DPCs decreased with the incubation time (Figure 4). Interestingly, with all adehydes, the observed DPC decays were fit better by an exponential model with two components than with a single component, suggesting that the aldehydes had induced two types of DPC with distinct stabilities, i.e. unstable and stable DPCs. The half-lives and initial compositions (%) of unstable and stable DPCs were calculated using a two-component model and are summarized in Table 2. The half-lives of unstable DPCs were around 6 h, and those of stable ones were around 25 h [except for FA (103.5 h) and PEN (54.1 h)]. More importantly, stable DPCs were the major initial component for ACR, CAA, CRA and GA (73.3–87.9%), whereas unstable DPCs for FA and PEN (90.4 and 59.6%). Thus, the apparent in vitro half-lives (Table 2) primarily correlate with the initial yield of dominant DPC species. The apparent in vitro half-lives were consistently about 2-fold greater than apparent in vivo half-lives (Table 2) determined from Figure 3, implying that more and/or stronger nucleophiles are involved in the hydrolysis of DPCs in actual cells than in TE buffer.
Figure 4.
In vitro kinetics of the elimination of aldehyde-induced DPCs. DNA isolated from cells immediately after aldehyde treatment (LD10) was labeled with FITC and incubated in TE buffer (pH 7.4) at 37°C, with dialysis against the same buffer (molecular cut-off = 100 kDa) to separate the proteins released from DNA. The remaining DPCs were quantified by measuring the fluorescence of the sample inside the dialysis tube. The percentage of remaining DPCs for the indicated aldehydes is plotted against the incubation time. Data points are means of two dialysis experiments. Regression curves based on a two-component exponential model are shown by the solid line. The parameters of the regression curves (half-life and initial composition) are listed in Table 2. Note that the rates of loss of DPCs were lower during ultracentrifugation and dialysis at 20 and 4°C, respectively, than the rates measured at 37°C (see ‘Discussion’ section and Supplementary Figure S3 and Supplementary Table S1).
SCE frequencies increase with increasing DPC levels
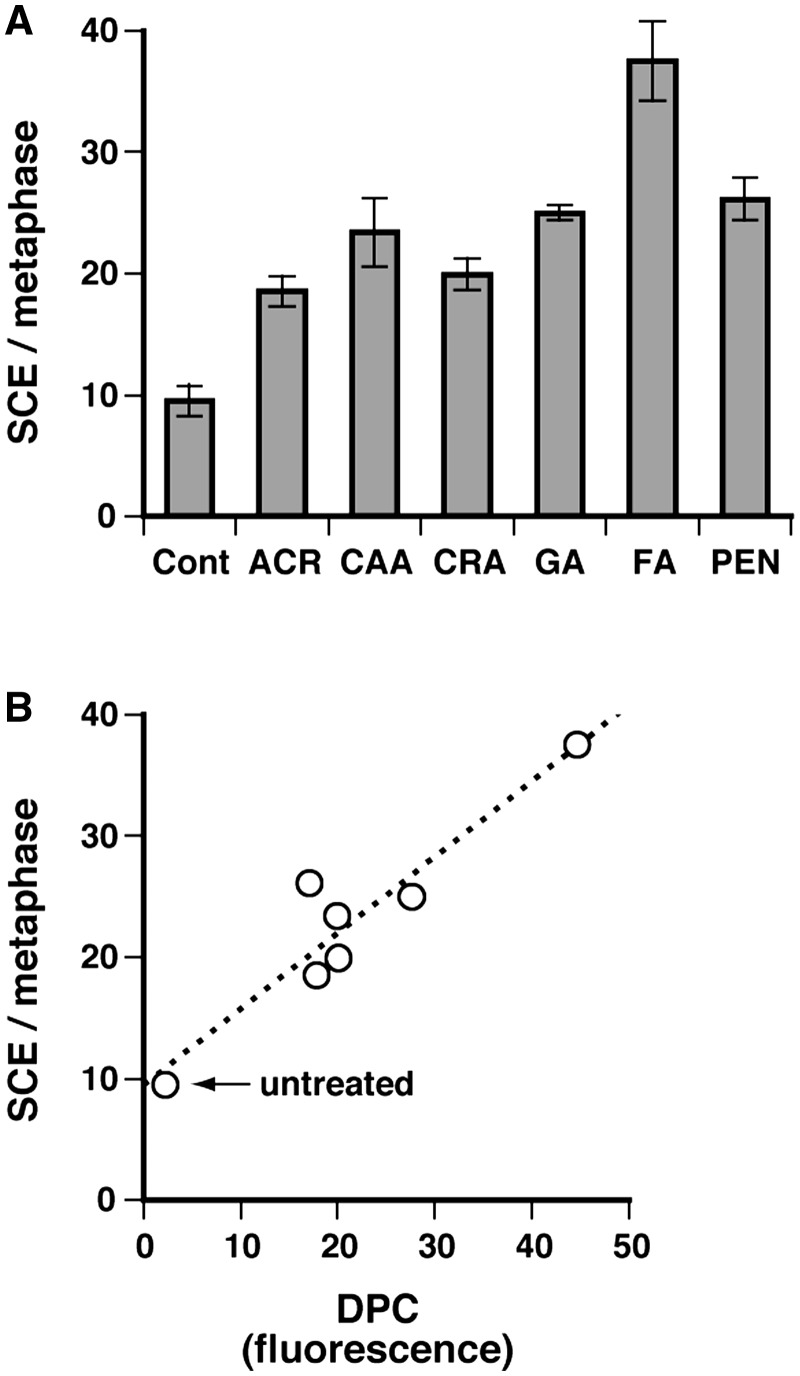
We have recently shown that DPCs are tolerated by HR, such that replication forks stalled by DPCs undergo breakage and are restored by HR (16). Since SCEs are intimately related to HR associated with fork collapse/breakage (47), we analyzed the correlation between the induction of SCEs and DPCs by aldehydes. MRC5-SV cells were treated at LD10, and SCE induction was scored (Figure 5A). The SCE frequency was increased by aldehyde treatment, and there was a positive correlation between the amount of genomic DPCs (data taken from Figure 2A) and the frequency of SCEs (Figure 5B). This observation is consistent with the finding that DPCs are processed by HR (16), when they are not reversed by spontaneous hydrolysis. Aldehydes produce DNA base-aldehyde adducts together with DPCs. However, it is unknown whether such adducts induce SCEs. In contrast, DPCs induced not only by aldehyde (FA) but also by 5-aza-2′-deoxycytidine result in replication fork breakage (16), which triggers HR and hence can lead to SCEs. Accordingly aldehyde-induced DPCs are at least in part responsible for the observed SCEs, while the role of DNA base-aldehyde adducts in SCE induction remains to be elucidated.
Figure 5.
Induction of SCEs by DPCs. (A) MRC5-SV cells were treated with the indicated aldehydes (LD10) and SCE induction was scored as described in ‘Materials and Methods’ section. The leftmost column (Cont) shows the data for untreated control cells. Data are means of three independent experiments with standard deviation. (B) Correleation between the level of genomic DPCs and the SCE frequency. The SCE frequencies for aldehydes (panel A) are plotted against the corresponding amounts of genomic DPCs shown in Figure 2A.
DPCs are preferentially formed in hypoxic tumors upon irradiation with C-ion beams
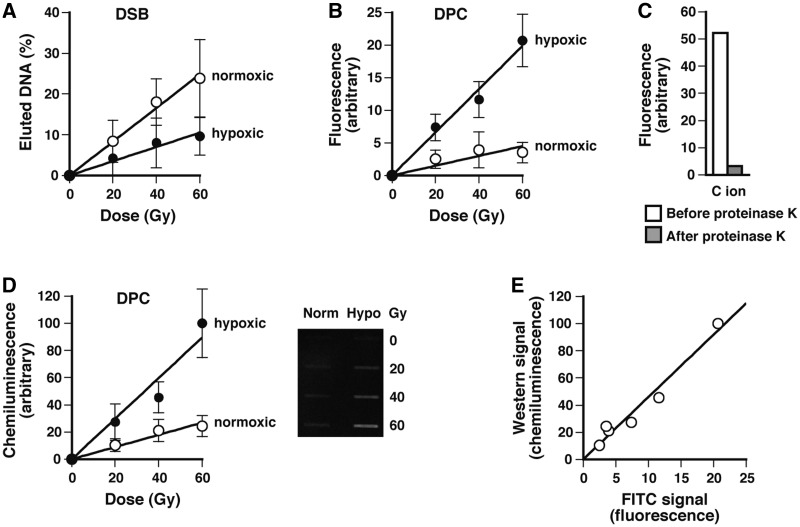
C-ion beams are successfully used in cancer therapy due to their enhanced biological effectiveness and improved beam focusing (48,49). With X-rays, DPCs are preferentially formed under hypoxic conditions (26). Keeping these in mind, we irradiated normoxic and hypoxic SCC VII mouse tumors with C-ion beams and analyzed the formation of DPCs and DNA DSBs by the FITC-labeling methods and static-field gel electrophoresis, respectively. The tumors implanted in mouse hind legs were rendered hypoxic by constricting the blood flow for 15 min before irradiation. Figure 6A shows the induction of DNA DSBs measured immediately after the irradiation of normoxic and hypoxic tumors. Consistent with the known oxygen effect on DSB formation (8), DNA DSBs were formed more efficiently in normoxic than hypoxic tumors. The oxygen enhancement ratio of DSB induction was 2.4. Figure 6B and D show the induction of DPCs measured by the fluorometric and western blotting formats, respectively. Both analyses showed that DPCs were formed more efficiently in hypoxic than normoxic tumors. According to the slopes of the dose-response plots, the induction rate of DPCs was greater in hypoxic tumors than in normoxic ones by a factor of 4.4 (fluorometry) or 3.3 (western). There was a good correlation between the results of fluorometric and western blotting analyses (Figure 6E). The fluorescence signal was almost completely lost (∼94%) after digestion of FITC-labeled purified DNA with proteinase K, substantiating that the FITC label was tethered to the CLPs (Figure 6C).
Figure 6.
Induction of DNA DSBs and DPCs in normoxic and hypoxic tumors upon irradiation with C-ion beams. Normoxic and hypoxic SCC VII mouse tumors were irradiated with C-ion beams and excised immediately after irradiation. DNA DSBs and DPCs were analyzed by static-field gel electrophoresis and the FITC-labeling methods, respectively, as described in ‘Materials and Methods’ section. (A) Dose response plots of the induction of DNA DSBs. The fraction of DNA released from the plug relative to total DNA (i.e. released and retained DNA) was used as a measure of DNA DSBs. (B) Dose response plots of the induction of DPCs measured by the fluorometric format. (C) Release of the FITC signal (fluorescence) associated with DNA after treatment with proteinase K. (D) The same as panel B except that DPCs were measured using the western blotting format. (E) Correlation between fluorometric and western blotting signals. Western blotting signals for different doses (panel D) are plotted against the corresponding FITC signals (panel B). The background signals obtained with unirradiated tumors were subtracted in panels A, B and D. Data points of panels A, B and D are means of the irradiation of three or four mice with standard deviation, and those of panel C are based on a single experiment.
C-ion beams generate extremely stable DPCs as a major component
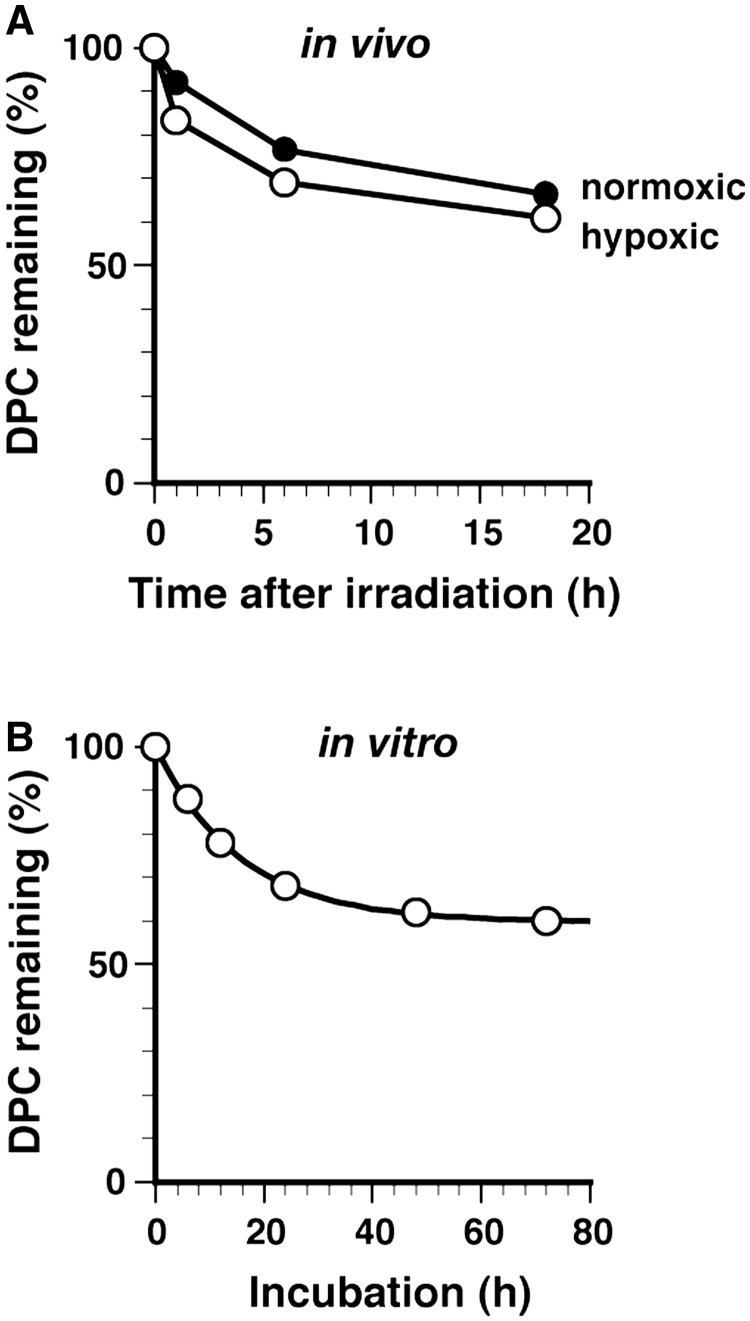
Genomic DPCs induced by aldehydes were relatively unstable and mostly lost within 12 h in vivo due to spontaneous hydrolysis (Figure 3A). We asked whether this is true for DPCs induced by C-ion beams. Normoxic and hypoxic SCC VII tumors were irradiated with 40 Gy. With hypoxic tumors, clumping was removed after irradiation to restore the blood supply. Irradiated mice were kept for 0, 1, 6 and 18 h, and tumors were excised and assayed for DPCs. The amounts of DPCs in normoxic and hypoxic tumors initially decreased to ∼70% during 6 h of postincubation, but did not change significantly with further incubation (Figure 7A). After 18 h, 60% of the DPCs remained, a level that was markedly higher than those of aldehydes (Figure 3A). Similar time-dependent kinetics of DPC elimination were obtained by western blotting assays (data not shown). Thus, C-ion beams likely generate highly stable genomic DPCs as a major component. The in vivo half-life of the unstable component for normoxic and hypoxic tumors was around 6 h (graphically determined assuming asymptotic values). The in vivo half-life of the stable component could not be determined due to the short incubation time of mice. The in vitro half-lives of unstable and stable DPCs (pH 7.4 and 37°C) were evaluated using isolated DNA and a two-component exponential model (Figure 7B) as described for aldehyde-induced DPCs (Figure 4). The unstable DPC component had a half-life of 10.7 h, which is comparable to those of aldehyde-induced DPCs, whereas the stable DPC component had an essentially infinite half-life (∼35 000 h; Table 2). The initial compositions of unstable and stable DPCs were 40.1 and 59.9%, respectively. These results suggest that C-ion beams induce essentially irreversible crosslinks between DNA and proteins as a major DPC component. Thus, the formation of irreversible crosslinks may be a characteristic of DPCs induced by C-ion beams (and perhaps other ionizing radiations).
Figure 7.
In vivo and in vitro kinetics of the elimination of DPCs induced by C-ion beams. (A) In vivo elimination of radiation-induced DPCs in tumors. Normoxic and hypoxic SCC VII mouse tumors were irradiated with C-ion beams (40 Gy) and excised after 0, 1, 6 and 18 h of postincubation. DPCs were analyzed by the fluorometric format. The percentages of remaining DPCs for normoxic and hypoxic tumors are plotted against the incubation time. Data points are means of the irradiation of two mice. Data points are simply connected without regression analysis. (B) In vitro kinetics of the elimination of radiation-induced DPCs. DNA isolated from hypoxic tumors immediately after irradiation (40 Gy) was labeled with FITC and incubated in TE buffer (pH 7.4) at 37°C, with dialysis against the same buffer (molecular cut-off = 100 kDa) to separate the proteins released from DNA. The remaining DPCs were quantified by measuring the fluorescence of the sample inside the dialysis tube. The percentage of remaining DPCs is plotted against the incubation time. Data points are means of two dialysis experiments. The regression curve based on a two-component exponential model is shown by the solid line. The parameters of the regression curve (half-life and initial composition) are listed in Table 2.
DISCUSSION
In the present study, we have developed novel methods for the direct detection of DPCs based on the FITC labeling of CLPs. The fluorometric and western blotting formats of detection brought consistent results and were successfully employed for the analysis of DPCs induced by aldehydes and ionizing radiation. Although the fluorometric format requires more DNA (typically 30 µg) than does the western blotting format (typically 0.3 µg), it provides a straightforward measurement of DPCs without any signal amplification steps that could introduce additional signal variations. Conversely, the western blotting format is highly sensitive and requires much smaller DNA samples for the assay. DPCs containing a particular protein can be detected by the western blotting method using protein-specific antibodies, e.g. DNA cytosine methyltransferases (50) and topoisomerases (51,52). However, the present FITC-labeling method allows the comprehensive detection of CLPs using a single antibody. The present methods have the advantage of enabling the linear correlation of the signal intensities of fluorometric and western blotting analyses with the amount of DPCs. Such a linear correlation cannot be obtained by the indirect measurements of DPCs using alkaline elution, filter binding, SDS/K+ precipitation, and comet methods. Conversely, indirect methods can analyze DPCs in cells without the purification of genomic DNA. Thus, the direct and indirect methods have partly complementary aspects and can be used as appropriate, depending on whether quantitative or qualitative data are sought. DPCs induced by aldehydes can be slowly reversed during the isolation of genomic DNA, and this is also true for unstable DPCs induced by C-ion beams (Table 2). To estimate the loss of DPCs during DNA isolation, we measured the stability of DPCs induced by selected aldehydes at 20°C (ultracentrifugation step) and 4°C (dialysis and most sample handling steps) (Supplementary Figure S3). The stabilities of DPCs were measured as described for those at 37°C (Figure 4). The total loss of DPCs estimated from the obtained results was 15% for FA-induced DPCs (the most unstable DPCs) and 8% for ACR-induced DPCs (fairly stable DPCs) in DNA isolation involving one ultracentrifugation (7 h at 20°C and 24 h at 4°C), and 28% for FA-induced DPCs and 15% for ACR-induced DPCs in DNA isolation involving two ultracentrifugations (14 h at 20°C and 43 h at 4°C) (Supplementary Table S1). Thus, the underestimation of the initial yield of reversible DPCs will be 8–15% and 15–28% for DNA isolations involving one and two ultracentrifugations, respectively. Although we used two cycles of ultracentrifugation to ensure the complete removal of free proteins in the present study, one cycle is generally sufficient for DPC detection. This also minimizes the underestimation of the initial yield of DPCs.
It has been shown that various aldehydes induce DPCs in cells (4). Here, we have quantified aldehyde-induced genomic DPCs upon treatment at LD10 concentrations (Figure 2). Using the data in Figure 2A and Table 1 for MRC5-SV cells, the in vivo DPC-inducing efficiencies of aldehydes can be evaluated: CAA (93), GA (68), ACR (64), CRA (5.0), FA (4.3) and PEN (1.0), where the numbers in parentheses indicate the relative amounts of DPCs induced per micromolar of aldehyde. The in vitro DPC-inducing efficiencies of these aldehydes (except for CAA) have also been determined using plasmid pUC13 and calfthymus histone (53): GA (590), ACR (37), CRA (0.72), FA (4000) and PEN (1.0), where the reported DPC-inducing efficiencies (DPCs induced per micromolar of aldehyde) were standardized relative to that of PEN for comparison with the in vivo data. Comparison of the in vivo and in vitro data points to that GA and ACR are potent DPC-inducers both in vivo and in vitro. In contrast, CRA and PEN are poor DPC-inducers in vivo and in vitro. Interestingly, FA is a very potent DPC-inducer in vitro, but is a poor DPC-inducer in vivo, indicating that FA is efficiently detoxified by metabolic enzymes and chemical reactions with cellular constituents such as glutathione (54). In the present study we systematically analyzed the in vitro and in vivo half-lives of aldehyde-induced DPCs, since they have not previously been systematically assessed in a unified way (41–43,55,56). Although rigorous analysis of in vivo DPC stability was not possible due to considerable data scatter and the delayed formation of DPCs, precise analyses of in vitro half-lives revealed the induction of unstable and stable DPCs with half-lives of 2.4–6.8 h and 23.9–103.5 h, respectively (Table 2). Both types of DPC will have an immediate impact on transcription, and stable DPCs will further affect DNA replication, which resumed 12–15 h after aldehyde treatment (data not shown). In light of the reactivity of the aldehyde group, the aldehydes used in the present study likely react with the side chains of lysine, cysteine and histidine residues to form adducts. The resulting adducts further react with the amino group of DNA bases to form various types of crosslinking bonds, which may have different stabilities. However, their relation to unstable and stable DPCs remains to be elucidated.
C-ion beams also produced unstable and stable DPCs (Figure 7). However, the nature of stable DPCs induced by C-ion beams was quite different form that of aldehyde-induced DPCs. Although the exact in vivo half-life of the stable component could not be evaluated due to its long half-life (Figure 7A), in vitro analysis revealed that these DPCs were irreversible with an essentially infinite half-life (Figure 7B, Table 2). The dose range used here results in an overkill of cells, but is clinically relevant to the radiation therapy of tumors (57,58). Although cells irradiated with 40 Gy are non-viable (i.e. unable to divide), it would require higher doses to biochemically inactivate DNA repair systems in cells. Consistent with this notion, we have previously shown that cells and tumors irradiated with 40 Gy retain a significant repair activity for DNA DSBs (37,39), suggesting that cells remain biochemically active with respect to repair. It is also noteworthy that the radiation-induced inactivation doses to reduce the activity to 37% of DNase I, ribonuclease and alkaline phosphatase in aqueous solution have been determined to be 100–1000 Gy (59–61). Accordingly it is unlikely, though not conclusive, that the in vivo observation of stable DPCs is a consequence of the inactivation of DPC repair by irradiation. The in vivo and in vitro half-lives of the C-ion-induced unstable DPC components were ∼6 and 10.7 h, respectively. The difference between the two values is likely attributable to more and/or stronger nucleophiles prensent in cells than in TE buffer, as was suggested for aldehyde-induced DPCs (Table 2). Previous studies have shown that DPCs produced by X- and γ-rays are released with biphasic kinetics. However, the in vivo half-lives of the two components were 20 min–2 h and 2–13 h (26,29,62), and much shorter than those observed in the present study. Furthermore, the release of DPCs attributed to repair in these studies at least partially results from a non-enzymatic process, as demonstrated by the spontaneous release of unstable DPCs that accounted for 40% of DPCs (Figure 7B, Table 2). The exact nature of stable (irreversible) DPCs remains to be elucidated. Some can be induced by hydroxyl radicals and likely contain a stable C–C bond between thymine–tyrosine or thymine–lysine, like those identified in the irradiation of a DNA–nucleohistone mixture (63,64) and mammalian cells (65,66). The formation of other types of stable DPC via the direct one electron oxidation of guanine has also been demonstrated in a model reaction and a reconstituted nucleosome core particle system (67,68).
Early studies showed that DPCs are formed preferentially under hypoxic conditions when cultured cells or tumors/tissues are irradiated with X- and γ-rays (26,69–71). However, the direct quantitative comparison of DPC-induction efficiencies under normoxic and hypoxic conditions is rather difficult, albeit not impossible, since DPCs were quantified by indirect methods (alkaline elution and filter binding) in these studies. The present result demonstrated that DPCs were preferentially but not exclusively formed in hypoxic tumors. The induction of DPCs in normoxic tumors was 23–30% relative to hypoxic tumors (Figures 6B and D). Implanted SCC VII tumors with a 10 mm diameter (used in this study) are reported to contain an 8.5% hypoxic fraction (72). Thus, the observed DPC-formation efficiency in normoxic tumors is slightly but significantly greater than that simply expected from the fraction of hypoxic cells, indicating the DPC formation in the normoxic fraction of cells in the tumor. It remains to be elucidated whether the detection of DPC formation in normoxic tumors is due to improvement of the detection method in this study, or is related to the radiation quality, i.e. sparsely ionizing X- and γ-rays versus densely ionizing C-ion beams. Densely ionizing radiations such as accelerated nitrogen and neon ions and α particles induce DPCs in mammalian cells (73–75), but no conclusive results have been obtained regarding the relationship between DPC induction and radiation quality such accelerated ions and LETs.
The long lasting DPCs induced by ionizing radiation or aldehydes are not repaired by NER (16), and they stall replication and transcription. The replication forks stalled by DPCs are likely reactivated by HR to continue DNA synthesis through DPCs as proposed from our previous (11,13,16) and present (Figure 5) studies. Although HR is generally an error-free process, futile cycles of HR at DPCs over many cell cycles may accidentally result in erroneous recombinations. The transcriptional arrest by persisting DPCs may attenuate the expression of proteins that are essential for cell function. Both events exert adverse effects on cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3 and Supplementary Table 1.
FUNDING
Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research (B) (to H.I.) and for Young Scientists (B) (to T.N.); Ministry of Education, Culture, Sports, Science and Technology of Japan, Grant-in-Aid for Scientific Research on Innovative Areas (to H.I.); The Support Program for Improving Graduate School Education from the Japan Society for the Promotion of Science (to M.I.S.). Funding for open access charge: Ministry of Education, Culture, Sports and Technology of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rieko Kurashige for assistance in DSB analysis and the National Institute of Radiological Sciences for the machine time of HIMAC.
REFERENCES
- 1.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Smith KC. Dose dependent decrease in extractability of DNA from bacteria following irradiation with ultraviolet light or with visible light plus dye. Biochem. Biophys. Res. Commun. 1962;8:157–163. doi: 10.1016/0006-291x(62)90255-3. [DOI] [PubMed] [Google Scholar]
- 3.Alexander P, Moroson H. Cross-linking of deoxyribonucleic acid to protein following ultra-violet irradiation different cells. Nature. 1962;194:882–883. doi: 10.1038/194882a0. [DOI] [PubMed] [Google Scholar]
- 4.Costa M, Zhitkovich A, Harris M, Paustenbach D, Gargas M. DNA-protein cross-links produced by various chemicals in cultured human lymphoma cells. J. Toxicol. Environ. Health. 1997;50:433–449. doi: 10.1080/00984109708984000. [DOI] [PubMed] [Google Scholar]
- 5.Costa M, Zhitkovich A, Gargas M, Paustenbach D, Finley B, Kuykendall J, Billings R, Carlson TJ, Wetterhahn K, Xu J, et al. Interlaboratory validation of a new assay for DNA-protein crosslinks. Mutat. Res. 1996;369:13–21. doi: 10.1016/s0165-1218(96)90043-9. [DOI] [PubMed] [Google Scholar]
- 6.Zwelling LA, Anderson T, Kohn KW. DNA-protein and DNA interstrand cross-linking by cis- and trans-platinum(II) diamminedichloride in L1210 mouse leukemia cells and relation to cytotoxicity. Cancer Res. 1979;39:365–369. [PubMed] [Google Scholar]
- 7.Woynarowski JM, Faivre S, Herzig MC, Arnett B, Chapman WG, Trevino AV, Raymond E, Chaney SG, Vaisman A, Varchenko M, et al. Oxaliplatin-induced damage of cellular DNA. Mol. Pharmacol. 2000;58:920–927. doi: 10.1124/mol.58.5.920. [DOI] [PubMed] [Google Scholar]
- 8.Frankenberg-Schwager M. Induction, repair and biological relevance of radiation-induced DNA lesions in eukaryotic cells. Radiat. Environ. Biophys. 1990;29:273–292. doi: 10.1007/BF01210408. [DOI] [PubMed] [Google Scholar]
- 9.Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, Salem AM. Repair and biochemical effects of DNA-protein crosslinks. Mutat. Res. 2011;711:113–122. doi: 10.1016/j.mrfmmm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Kuo HK, Griffith JD, Kreuzer KN. 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 2007;67:8248–8254. doi: 10.1158/0008-5472.CAN-07-1038. [DOI] [PubMed] [Google Scholar]
- 11.Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein cross-links. Mol. Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 12.Kumari A, Minko IG, Smith RL, Lloyd RS, McCullough AK. Modulation of UvrD helicase activity by covalent DNA-protein cross-links. J. Biol. Chem. 2010;285:21313–21322. doi: 10.1074/jbc.M109.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salem AM, Nakano T, Takuwa M, Matoba N, Tsuboi T, Terato H, Yamamoto K, Yamada M, Nohmi T, Ide H. Genetic analysis of repair and damage tolerance mechanisms for DNA-protein cross-links in Escherichia coli. J. Bacteriol. 2009;191:5657–5668. doi: 10.1128/JB.00417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minko IG, Zou Y, Lloyd RS. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc. Natl Acad. Sci. USA. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- 16.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, et al. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DA, Sale JE, Yamazoe M, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 18.Novakova O, Kasparkova J, Malina J, Natile G, Brabec V. DNA-protein cross-linking by trans-[PtCl2(E-iminoether)2]. A concept for activation of the trans geometry in platinum antitumor complexes. Nucleic Acids Res. 2003;31:6450–6460. doi: 10.1093/nar/gkg863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc. Natl Acad. Sci. USA. 2006;103:4056–4061. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, O'Connor TR. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 21.Zecevic A, Hagan E, Reynolds M, Poage G, Johnston T, Zhitkovich A. XPA impacts formation but not proteasome-sensitive repair of DNA-protein cross-links induced by chromate. Mutagenesis. 2010;25:381–388. doi: 10.1093/mutage/geq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- 23.Qin H, Wang Y. Exploring DNA-binding proteins with in vivo chemical cross-linking and mass spectrometry. J. Proteome Res. 2009;8:1983–1991. doi: 10.1021/pr8009319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem. Res. Toxicol. 2009;22:1151–1162. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. DNA-protein cross-linking by 1,2,3,4-diepoxybutane. J. Proteome Res. 2010;9:4356–4367. doi: 10.1021/pr1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fornace AJ, Jr, Little JB. DNA crosslinking induced by x-rays and chemical agents. Biochim. Biophys. Acta. 1977;477:343–355. doi: 10.1016/0005-2787(77)90253-2. [DOI] [PubMed] [Google Scholar]
- 27.Kohn KW, Ewig RA. DNA-protein crosslinking by trans-platinum(II)diamminedichloride in mammalian cells, a new method of analysis. Biochim. Biophys. Acta. 1979;562:32–40. doi: 10.1016/0005-2787(79)90123-0. [DOI] [PubMed] [Google Scholar]
- 28.Strniste GF, Rall SC. Induction of stable protein-deoxyribonucleic acid adducts in Chinese hamster cell chromatin by ultraviolet light. Biochemistry. 1976;15:1712–1719. doi: 10.1021/bi00653a019. [DOI] [PubMed] [Google Scholar]
- 29.Chiu SM, Sokany NM, Friedman LR, Oleinick NL. Differential processing of ultraviolet or ionizing radiation-induced DNA-protein cross-links in Chinese hamster cells. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1984;46:681–690. doi: 10.1080/09553008414551921. [DOI] [PubMed] [Google Scholar]
- 30.Zhitkovich A, Costa M. A simple, sensitive assay to detect DNA-protein crosslinks in intact cells and in vivo. Carcinogenesis. 1992;13:1485–1489. doi: 10.1093/carcin/13.8.1485. [DOI] [PubMed] [Google Scholar]
- 31.Merk O, Speit G. Detection of crosslinks with the comet assay in relationship to genotoxicity and cytotoxicity. Environ. Mol. Mutagen. 1999;33:167–172. doi: 10.1002/(sici)1098-2280(1999)33:2<167::aid-em9>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 32.Merk O, Reiser K, Speit G. Analysis of chromate-induced DNA-protein crosslinks with the comet assay. Mutat. Res. 2000;471:71–80. doi: 10.1016/s1383-5718(00)00110-8. [DOI] [PubMed] [Google Scholar]
- 33.Oleinick NL, Chiu SM, Ramakrishnan N, Xue LY. The formation, identification, and significance of DNA-protein cross-links in mammalian cells. Br. J. Cancer Suppl. 1987;8:135–140. [PMC free article] [PubMed] [Google Scholar]
- 34.Kohn KW. Principles and practice of DNA filter elution. Pharmacol. Ther. 1991;49:55–77. doi: 10.1016/0163-7258(91)90022-e. [DOI] [PubMed] [Google Scholar]
- 35.Ramakrishnan N, Chiu SM, Oteinick NL. Yield of DNA-protein cross-links in gamma-irradiated Chinese hamster cells. Cancer Res. 1987;47:2032–2035. [PubMed] [Google Scholar]
- 36.Perry P, Wolff S. New Giemsa method for the differential staining of sister chromatids. Nature. 1974;251:156–158. doi: 10.1038/251156a0. [DOI] [PubMed] [Google Scholar]
- 37.Hirayama R, Uzawa A, Matsumoto Y, Noguchi M, Kase Y, Takase N, Ito A, Koike S, Ando K, Okayasu R, et al. Induction of DNA DSB and its rejoining in clamped and non-clamped tumours after exposure to carbon ion beams in comparison to X rays. Radiat. Prot. Dosimetry. 2011;143:508–512. doi: 10.1093/rpd/ncq478. [DOI] [PubMed] [Google Scholar]
- 38.Masunaga S, Ando K, Uzawa A, Hirayama R, Furusawa Y, Koike S, Sakurai Y, Nagata K, Suzuki M, Kashino G, et al. Radiobiologic significance of response of intratumor quiescent cells in vivo to accelerated carbon ion beams compared with gamma-rays and reactor neutron beams. Int. J. Radiat. Oncol. Biol. Phys. 2008;70:221–228. doi: 10.1016/j.ijrobp.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 39.Hirayama R, Furusawa Y, Fukawa T, Ando K. Repair kinetics of DNA-DSB induced by X-rays or carbon ions under oxic and hypoxic conditions. J. Radiat. Res. 2005;46:325–332. doi: 10.1269/jrr.46.325. [DOI] [PubMed] [Google Scholar]
- 40.Barker S, Murray D, Zheng J, Li L, Weinfeld M. A method for the isolation of covalent DNA-protein crosslinks suitable for proteomics analysis. Anal. Biochem. 2005;344:204–215. doi: 10.1016/j.ab.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 41.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 42.Kuykendall JR, Bogdanffy MS. Reaction kinetics of DNA-histone crosslinking by vinyl acetate and acetaldehyde. Carcinogenesis. 1992;13:2095–2100. doi: 10.1093/carcin/13.11.2095. [DOI] [PubMed] [Google Scholar]
- 43.Voitkun V, Zhitkovich A. Analysis of DNA-protein crosslinking activity of malondialdehyde in vitro. Mutat. Res. 1999;424:97–106. doi: 10.1016/s0027-5107(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor PM, Fox BW. Isolation and characterization of proteins cross-linked to DNA by the antitumor agent methylene dimethanesulfonate and its hydrolytic product formaldehyde. J. Biol. Chem. 1989;264:6391–6397. [PubMed] [Google Scholar]
- 45.Kumari A, Lim YX, Newell AH, Olson SB, McCullough AK. Formaldehyde-induced genome instability is suppressed by an XPF-dependent pathway. DNA Repair. 2012;11:236–246. doi: 10.1016/j.dnarep.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Graaf B, Clore A, McCullough AK. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair. 2009;8:1207–1214. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson DM, III, Thompson LH. Molecular mechanisms of sister-chromatid exchange. Mutat. Res. 2007;616:11–23. doi: 10.1016/j.mrfmmm.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 48.Ando K, Kase Y. Biological characteristics of carbon-ion therapy. Int. J. Radiat. Biol. 2009;85:715–728. doi: 10.1080/09553000903072470. [DOI] [PubMed] [Google Scholar]
- 49.Allen C, Borak TB, Tsujii H, Nickoloff JA. Heavy charged particle radiobiology: using enhanced biological effectiveness and improved beam focusing to advance cancer therapy. Mutat. Res. 2011;711:150–157. doi: 10.1016/j.mrfmmm.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu K, Wang YF, Cantemir C, Muller MT. Endogenous assays of DNA methyltransferases: evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol. Cell. Biol. 2003;23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 52.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol. Cell. Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuykendall JR, Bogdanffy MS. Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat. Res. 1992;283:131–136. doi: 10.1016/0165-7992(92)90145-8. [DOI] [PubMed] [Google Scholar]
- 54.Conaway CC, Whysner J, Verna LK, Williams GM. Formaldehyde mechanistic data and risk assessment: endogenous protection from DNA adduct formation. Pharmacol. Ther. 1996;71:29–55. doi: 10.1016/0163-7258(96)00061-7. [DOI] [PubMed] [Google Scholar]
- 55.Grafstrom RC, Fornace A, Jr, Harris CC. Repair of DNA damage caused by formaldehyde in human cells. Cancer Res. 1984;44:4323–4327. [PubMed] [Google Scholar]
- 56.Cosma GN, Jamasbi R, Marchok AC. Growth inhibition and DNA damage induced by benzo[a]pyrene and formaldehyde in primary cultures of rat tracheal epithelial cells. Mutat. Res. 1988;201:161–168. doi: 10.1016/0027-5107(88)90122-4. [DOI] [PubMed] [Google Scholar]
- 57.Ando K, Koike S, Ohira C, Chen YJ, Nojima K, Ando S, Ohbuchi T, Kobayashi N, Shimizu W, Urano M. Accelerated reoxygenation of a murine fibrosarcoma after carbon-ion radiation. Int. J. Radiat. Biol. 1999;75:505–512. doi: 10.1080/095530099140438. [DOI] [PubMed] [Google Scholar]
- 58.Imadome K, Iwakawa M, Nojiri K, Tamaki T, Sakai M, Nakawatari M, Moritake T, Yanagisawa M, Nakamura E, Tsujii H, et al. Upregulation of stress-response genes with cell cycle arrest induced by carbon ion irradiation in multiple murine tumors models. Cancer Biol. Ther. 2008; 7:208–217. doi: 10.4161/cbt.7.2.5255. [DOI] [PubMed] [Google Scholar]
- 59.Akaboshi M, Kawai K, Maki H, Honda Y. Effect of dilution on thermal neutron induced inactivation of deoxyribonuclease I. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1982;42:99–104. doi: 10.1080/09553008214550951. [DOI] [PubMed] [Google Scholar]
- 60.Adams GE, Willson RL, Bisby RH, Cundall RB. On the mechanism of the radiation-induced inactivation of ribonuclease in dilute aqueous solution. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1971;20:405–415. doi: 10.1080/09553007114551311. [DOI] [PubMed] [Google Scholar]
- 61.Hasan NM, Mccall PR, Moore JS, Power DM. Radiation inactivation of bovine intestinal alkaline phosphatase. Radiat. Phys. Chem. 1994;43:233–237. [Google Scholar]
- 62.Cress AE, Bowden GT. Covalent DNA-protein crosslinking occurs after hyperthermia and radiation. Radiat Res. 1983;95:610–619. [PubMed] [Google Scholar]
- 63.Dizdaroglu M, Gajewski E. Structure and mechanism of hydroxyl radical-induced formation of a DNA-protein cross-link involving thymine and lysine in nucleohistone. Cancer Res. 1989;49:3463–3467. [PubMed] [Google Scholar]
- 64.Dizdaroglu M, Gajewski E, Reddy P, Margolis SA. Structure of a hydroxyl radical induced DNA-protein cross-link involving thymine and tyrosine in nucleohistone. Biochemistry. 1989;28:3625–3628. doi: 10.1021/bi00434a071. [DOI] [PubMed] [Google Scholar]
- 65.Olinski R, Nackerdien Z, Dizdaroglu M. DNA-protein cross-linking between thymine and tyrosine in chromatin of gamma-irradiated or H2O2-treated cultured human cells. Arch. Biochem. Biophys. 1992;297:139–143. doi: 10.1016/0003-9861(92)90651-c. [DOI] [PubMed] [Google Scholar]
- 66.Altman SA, Zastawny TH, Randers-Eichhorn L, Cacciuttolo MA, Akman SA, Dizdaroglu M, Rao G. Formation of DNA-protein cross-links in cultured mammalian cells upon treatment with iron ions. Free Radic. Biol. Med. 1995;19:897–902. doi: 10.1016/0891-5849(95)00095-f. [DOI] [PubMed] [Google Scholar]
- 67.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat JL. Characterization of lysine-guanine cross-links upon one-electron oxidation of a guanine-containing oligonucleotide in the presence of a trilysine peptide. J. Am. Chem. Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 68.Bjorklund CC, Davis WB. Stable DNA-protein cross-links are products of DNA charge transport in a nucleosome core particle. Biochemistry. 2007;46:10745–10755. doi: 10.1021/bi700475b. [DOI] [PubMed] [Google Scholar]
- 69.Meyn RE, vanAnkeren SC, Jenkins WT. The induction of DNA-protein crosslinks in hypoxic cells and their possible contribution to cell lethality. Radiat. Res. 1987;109:419–429. [PubMed] [Google Scholar]
- 70.Zhang H, Wheeler KT. Radiation-induced DNA damage in tumors and normal tissues. I. Feasibility of estimating the hypoxic fraction. Radiat. Res. 1993;136:77–88. [PubMed] [Google Scholar]
- 71.Xue LY, Friedman LR, Oleinick NL. Repair of chromatin damage in glutathione-depleted V-79 cells: comparison of oxic and hypoxic conditions. Radiat. Res. 1988;116:89–99. [PubMed] [Google Scholar]
- 72.Shibamoto Y, Yukawa Y, Tsutsui K, Takahashi M, Abe M. Variation in the hypoxic fraction among mouse tumors of different types, sizes, and sites. Jpn. J. Cancer Res. 1986;77:908–915. [PubMed] [Google Scholar]
- 73.Eguchi K, Inada T, Yaguchi M, Satoh S, Kaneko I. Induction and repair of DNA lesions in cultured human melanoma cells exposed to a nitrogen-ion beam. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1987;52:115–123. doi: 10.1080/09553008714551541. [DOI] [PubMed] [Google Scholar]
- 74.Blakely EA, Chang PY, Bjornstad KA. LET dependence of DNA–protein crosslinks. In: Hagen U, Jung H, Streffer C, editors. Radiation Research 1895–1995, Congress Proceedings. Germany: ICRR Society; 1995. pp. 136–139. [Google Scholar]
- 75.Jenner TJ, Cunniffe SM, Stevens DL, O'Neill P. Induction of DNA-protein crosslinks in Chinese hamster V79-4 cells exposed to high- and low-linear energy transfer radiation. Radiat. Res. 1998;150:593–599. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.