Abstract
Bacterial spores possess an enormous resistance to ultraviolet (UV) radiation. This is largely due to a unique DNA repair enzyme, Spore Photoproduct Lyase (SP lyase) that repairs a specific UV-induced DNA lesion, the spore photoproduct (SP), through an unprecedented radical-based mechanism. Unlike DNA photolyases, SP lyase belongs to the emerging superfamily of radical S-adenosyl-l-methionine (SAM) enzymes and uses a [4Fe–4S]1+ cluster and SAM to initiate the repair reaction. We report here the first crystal structure of this enigmatic enzyme in complex with its [4Fe–4S] cluster and its SAM cofactor, in the absence and presence of a DNA lesion, the dinucleoside SP. The high resolution structures provide fundamental insights into the active site, the DNA lesion recognition and binding which involve a β-hairpin structure. We show that SAM and a conserved cysteine residue are perfectly positioned in the active site for hydrogen atom abstraction from the dihydrothymine residue of the lesion and donation to the α-thyminyl radical moiety, respectively. Based on structural and biochemical characterizations of mutant proteins, we substantiate the role of this cysteine in the enzymatic mechanism. Our structure reveals how SP lyase combines specific features of radical SAM and DNA repair enzymes to enable a complex radical-based repair reaction to take place.
INTRODUCTION
Given the myriad of DNA-damaging agents, bacteria have evolved diverse DNA repair pathways to resist and survive up to thousands of years in the extreme case of spores. This tremendous resistance is achieved by a combination of several factors including DNA packing assisted by small acid-soluble spore proteins, dehydration and the high content of dipicolinic acid (2,6-pyridinedicarboxylic acid) (1). Under these conditions, ultraviolet (UV) radiation results in the formation of one major DNA lesion, the so-called spore photoproduct (SP or 5-(R-thyminyl)-5,6-dihydrothymine) (2,3). During germination, spores have the unique ability to repair this photoproduct back to two thymidine residues (4) through a unique direct reversal mechanism using the enzyme Spore Photoproduct Lyase (SP lyase) (5,6). SP lyase is strictly specific for SP and does not repair other thymidine photoproducts like the cyclobutane pyrimidine dimer (CPD) and the 6-4 photoproduct (6-4PP) (7,8). Interestingly, unlike DNA photolyases (9), SP lyase does not use blue light energy to initiate the repair reaction and therefore constitutes a unique family of light-independent lyases involved in DNA repair (10). SP lyase belongs to the widespread radical S-adenosyl-l-methionine (SAM) superfamily of enzymes (11) involved in a wide range of biological processes (12). These enzymes share several common features, notably three strictly conserved cysteine residues generally included in the CxxxCxxC motif (11). These critical cysteines coordinate an unusual [4Fe–4S]2+/1+ cluster, while SAM serves as ligand for the fourth iron atom and acts as a co-factor or a co-substrate (12). The repair mechanism of SP lyase has been proposed to be initiated by the reduction of the [4Fe–4S]2+ cluster and subsequent transfer of an electron to SAM, inducing the reductive cleavage of the C5′-S bond and the formation of methionine and a highly reactive species, the 5′-deoxyadenosine (5′-dA) radical. This 5′-dA radical abstracts the pro-R hydrogen atom located on the C6 of the 5′-dihydrothymine residue of SP (13,14) leading to a carbon radical intermediate. β-Scission of this species results in the formation of a 5′-thymine residue and an 3′-thymineallylic radical intermediate, which was trapped by mutation of a critical and conserved cysteine residue (Cys141 in Bacillus subtilis) into alanine (15). It was suggested that this residue could be either important to seal off the enzyme active site or a hydrogen atom donor for the 3′-thymine allylic radical residue to conclude the repair reaction (14,15). The role of this cysteine and how SAM is regenerated have remained enigmatic so far. To get insight into the mechanism of this unique DNA repair enzyme, we determined the crystal structures of substrate-free and -bound SP lyase from Geobacillus thermodenitrificans at high resolution (2.2 and 2.0 Å respectively) and of several mutants (Supplementary Table S1). Together with biochemical characterization, these structures provide new insights into the repair reaction catalyzed by this radical SAM DNA lyase.
MATERIALS AND METHODS
Cloning, protein expression and purification
The gene GTNG_2348 coding for SP lyase from G. thermodenitrificans was synthesized by GeneArt and cloned into a pETM11 vector (derived from Novagene pET vectors) to express a His6-tag SP lyase fusion protein. This vector was transformed into Escherichia coli BL21 (DE3). 100 ml overnight culture of BL21 (DE3) E. coli/pETM11/His-tag-SP lyase was used to inoculate 12 l of LB medium containing 50 μg ml−1 kanamycin. The culture was incubated at 310 K and 95 rpm until the OD at 600 nm reached ∼0.8 and the protein expression was induced at 291 K by adding 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) followed by overnight incubation. Selenomethionine-labeled SP lyase was expressed using the same protocol but using a minimal medium for E. coli growth supplemented with 50 μg ml−1 l-selenomethionine (Sigma-Aldrich). Cells were harvested by centrifugation at 277 K and 5400g for 10 min and resuspended in buffer A (50 mM Tris–HCl, pH 8, 500 mM NaCl, 10 mM MgCl2 and 10% glycerol) supplemented with 5 mM 2-mercaptoethanol, 1 tablet of protease inhibitor cocktail (Complete, EDTA-free, Roche), 4 μg ml−1 of DNase I (Roche), 4 μg ml−1 of RNase (Roche) and 0.1 mg ml−1 lysozyme (Sigma-Aldrich). Cells were disrupted by sonication and centrifuged at 277 K and 186000g for 60 min. The supernatant was loaded on a 3 ml Ni-NTA (Qiagen) gel column equilibrated with buffer A containing 20 mM imidazole (Sigma-Aldrich). The brownish His-tag SP lyase fusion protein was eluted with 500 mM imidazole in buffer A. The imidazole was removed with a desalting column (Sephadex G-25, GE healthcare) equilibrated with buffer A. The protein was then loaded on a 5-ml HiTrap Heparin column (GE Healthcare) equilibrated with 50 mM Tris–HCl pH 8, 300 mM NaCl, 1% glycerol and 3 mM dithiothreitol. The fractions were analyzed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis before being pooled. The pure protein was concentrated using a 30 000-MWCO spin concentrator (Amicon, Millipore).
Site-directed mutagenesis
Site-directed mutagenesis was performed by QuickChange PCR mutagenesis on the plasmid pETM11-His-tag-SP lyase. All oligonucleotides were purchased from Eurofins MWG operon. The following primers were used to introduce the cysteine 140 mutations: for C140S, 5′-ACCCGTTTTGAAGCAGCAAGTACCAGCGATATTGTGGGT-3′ and 5′-ACCCACAATATCGCTGGTACTTGCTGCTTCAAAACGGGT-3′; for C140A, 5′-ACCCGTTTTGAAGCAGCAGCTACCAGCGATATTGTGGGT-3′ and 5′-ACCCACAATATCGCTGGTAGCTGCTGCTTCAAAACGGGT-3′. The substituted bases are shown in bold. All mutations were confirmed by DNA sequencing. All mutant proteins were expressed and purified as described for wild-type SP lyase.
Reconstitution of the iron–sulfur cluster
Reconstitution was performed under anaerobic conditions in a glove box (Belle, Weymouth, UK). SP lyase was incubated with an excess of sodium sulfide and ammonium ferrous sulfate hexahydrate (Sigma-Aldrich). The protein was desalted and concentrated with a 30 000-MWCO spin concentrator (Amicon, Millipore). The SP lyase protein concentration was determined using a Nanodrop ND-1000 spectrophotometer at 280 nm. The iron–sulfur cluster of all mutant proteins was reconstituted using the same procedure.
Crystallization and crystal harvesting
Reconstituted SP lyase was incubated with either SAM or Se–SAM and used for crystallization trials performed in anaerobic atmosphere provided by a glove box (Belle, Weymouth, UK). Crystals of the substrate-free SP lyase grew within 3 weeks at 293 K in a hanging-drop vapor diffusion setup using 70 mM octanoyl-N-hydroxyethylglucamide (Hampton Research) and a reservoir solution containing 200 mM lithium sulfate, 100 mM Tris–HCl pH 9 and 19–27% (wt/vol) PEG 8000. Single crystals were harvested into a cryoprotectant solution containing the mother liquor supplemented by 3 mM dithiothreithol, 500 mM NaCl and 15% (vol/vol) ethylene glycol before flash cooling in liquid nitrogen inside the glove box. For experimental phasing, two approaches were used: crystals of the native protein were soaked for 5 min in the cryoprotectant solution supplemented by 100 mM of 5-amino-2,4,6-triiodoisophthalic acid solution (Jena Bioscience, Jena, Germany) (16) and selenomethionine-substituted protein was crystallized and cryoprotected as described for the native protein. The substrate-bound SP lyase crystal structure was obtained by soaking crystals for 10 min in a solution containing 20 mM dinucleoside spore photoproduct (5R–SPside synthesized as described previously (17) and shown in Supplementary Figure S1), the mother liquor, 3 mM dithiothreithol, 50 mM NaCl, 15 mM pyrophosphate, 14 mM hexametaphosphate (Sigma-Aldrich) and 15% (vol/vol) ethylene glycol.
Structure determination
A highly redundant, 2.1 Å resolution dataset of ‘magic triangle’ (Jena Bioscience), soaked SP lyase crystal was collected at the PX-II beam line of the Swiss Light Source using a wavelength of 1.722 Å. Data collection statistics are given in Supplementary Table S1. Using AutoSHARP (18), seven sites for anomalous scatterers were found by SHELXD (19) corresponding to four iron atoms and three iodine atoms of the magic triangle compound (16) bound to the protein. Using these sites, excellent phases were obtained (phasing power 1.80, figure of merit (FOM) = 0.46 for acentric reflections, before solvent flattening). After solvent flattening, an electron density map of very high quality was obtained, into which the structure was built manually using Coot (20). To assist model building, an anomalous difference electron density map was calculated from data collected at the selenium peak wavelength of a selenomethionine substituted SP lyase crystal which showed the methionine positions. The model was refined to excellent geometry and R-factors (Supplementary Table S1). This initial structure was used to phase all subsequently determined complex and mutant structures by molecular replacement. All data were processed with XDS (21) and refinement of all structures was carried out using REFMAC (22). Geometric target parameters for the refinement of the SPside bound to the enzyme were obtained from (23). Other target parameters were estimated using the PRODRG server (24). All structure superpositions were performed with Coot using the SSM program and all residues (25).
Repair activity on SPside
The wild-type, reconstituted SP lyase (125 μM) was incubated with 5 mM dithiothreitol, 2 mM SAM, 250 μM SPside (5R–SPside or 5S–SPside synthesized as described previously (17) and shown in Supplementary Figure S1) in a Tris buffer (50 mM Tris–HCl pH 8 and 500 mM NaCl) and the repair reaction was started by adding 3 mM sodium dithionite. Control reactions were performed by omitting sodium dithionite. The reactions were performed at 298 K under anaerobic conditions in a glove box. Aliquots were taken at 0, 60, 120, 180, 360 and 720 min and the repair reaction was quenched by flash-freezing in liquid nitrogen. Aliquots were diluted in 0.1% aqueous trifluoroacetic acid (Sigma-Aldrich) prior to analysis by ultra-fast liquid chromatography (UFLC) (Shimadzu) using a reversed-phase C-18 column (Licrospher, 5 μm, 4 × 250 mm from VWR International, Darmstadt, Germany). The column was equilibrated with 98.9% buffer A (0.1% aqueous trifluoroacetic acid) and 1.1% buffer B (90% acetonitrile and 0.1% trifluoroacetic acid). A linear gradient from 1.1 to 11% was used at a flow rate of 1 ml min−1 and the absorbance was monitored at 260 nm. Standards of SAM, thymidine and 5′-deoxyadenosine (Sigma-Aldrich) were used for the identification of the eluted compounds, which was then confirmed by mass spectrometry as described in Supplementary Figure S8. The repair activity of C140A and C140S mutants was assayed and analyzed as described for the wild-type SP lyase.
Repair activity on SPside-containing DNA
A 12-mer oligonucleotide containing the dinucleoside SP (5′-CAGCGGT^TGCAGG-3′) was synthesized as described by Heil et al. (26). Reconstituted, wild-type SP lyase (40 μM) was incubated with 5 mM dithiothreitol, 1 mM SAM, 40 μM of 5R–SPside-containing DNA in a Tris buffer (40 mM Tris–HCl pH 8 and 410 mM NaCl) and the repair reaction was started by adding 3 mM sodium dithionite. Control reactions were performed by omitting sodium dithionite. The reactions were performed at 298 K under anaerobic conditions in a glove box. Aliquots were taken at 10 and 120 min and the reaction was quenched by flash-freezing in liquid nitrogen. The samples were desalted by ZipTipC18 (Millipore) and eluted with 50% acetonitrile. Analyses were performed by matrix-assisted laser desorption ionization – time of flight (MALDI–TOF) mass spectrometry (Shimadzu Biotech Axima performance) in reflectron/negative ion mode using a solution of 3-hydroxypicolinic acid (50 mg ml−1) resuspended in 50% acetonitrile and ammonium citrate (50 mg ml−1) as matrix. The repair activity of all mutants (C140A and C140S) was assayed and analyzed as described for the wild-type SP lyase.
RESULTS
SP lyase overall structure and active site
SP lyase was purified aerobically and the highly oxygen sensitive iron–sulfur cluster was reconstituted under anaerobic and reducing conditions, after which the protein was crystallized anaerobically. The substrate-free structure was determined using single-wavelength anomalous diffraction (SAD) phasing and the structure of SP lyase in complex with a SPside lesion was determined using Fourier synthesis. SP lyase crystals contain one monomer per asymmetric unit. All 340 amino acids were located in the electron density except the N-terminal His-tag. SP lyase shares the common fold of radical SAM enzymes and forms a partial (α/β)6 triose phosphate isomerase (TIM) barrel enabling SP lyase to accept large substrates (Figure 1). Specific features include an N-terminal α/β domain and α-helices followed by two anti-parallel β-strands which form a long β-hairpin located between strand β5 and helix α6 of the core domain (Figure 1A).
Figure 1.
Crystal structures of substrate-free and substrate-bound SP lyase. (A) SP lyase is shown with its SAM co-factor (blue, N; red, O; yellow, Se and green, C atoms) and the buried [4Fe–4S] cluster (orange, Fe and yellow, S atoms). The protein backbone of the α6/β6 partial TIM-barrel core is depicted in light purple. The N-terminal α/β domain (protein backbone rendered in salmon) is composed of a four-stranded parallel β-sheet flanked by three α-helices. At the C-terminal side, α-helices followed by the β-hairpin are shown in orange. (B) Overall structure of the lesion-bound SP lyase (protein backbone depicted in light grey) with the dinucleoside SP (blue, N; red, O and purple, C atoms) rendered in stick format. (C) Structure superposition of substrate-free and -bound SP lyase (protein backbone shown in white). Substrate-free (protein backbone colored in orange) and substrate-bound (protein backbone rendered in magenta) SP lyase differ in the conformation of the β-hairpin, the loop following helix α4′ and Tyr98. Residues Arg304 and Tyr305 are colored in magenta. (D) Active-site view of substrate-bound SP lyase. The Fo-DFc electron density contoured at 2.5 σ (blue) for the dinucleoside SP (blue, N; red, O; and white, C atoms) was calculated before the ligand was included in the model. Side-chain residues colored in purple and by atom type are involved in hydrogen-bonds and electrostatic interactions with the nucleobase residues of 5R–SPside. The interactions are depicted by black dashed lines and distances in Å are shown.
At the top of the partial TIM barrel, a single [4Fe–4S] cluster is held by the three strictly conserved cysteine residues (Cys90, Cys94 and Cys97) (27) located on a 8-residue loop connecting strand β4′ to helix α4′ surrounding the iron–sulfur cluster (Figure 1A and Supplementary Figure S2). The nature and the stoichiometry of the iron–sulfur cluster agree with results from UV–visible, electron paramagnetic resonance (EPR) and Mössbauer spectroscopy (7,28,29). Like most radical SAM enzymes, the iron–sulfur cluster is buried, surrounded by a conserved aromatic amino acid (Tyr96) and the backbone of residues composing the CxxxCxΦC motif (where Φ is an aromatic amino acid) (Figure 1A and Supplementary Figure S3). In addition, the side chains of various hydrophobic and charged residues (Leu99, Asp143, Lys174) as well as Tyr175 also contribute to the environment of the iron–sulfur cluster.
The fourth iron atom is coordinated by SAM through the amino group (N–Fe 2.3 Å) and the carboxylate (O–Fe 2.3 Å) of its methionyl moiety. The sulfur atom of SAM also interacts with this Fe atom (3.6 Å) and the adjacent sulfur atom (3.9 Å) as in other radical SAM enzymes (30). SAM is hence perfectly positioned for electron transfer from the [4Fe–4S] cluster (Supplementary Figure S2). The adenine, methionine and ribose moieties of SAM are stabilized by several interactions including salt bridges, hydrogen bonds and hydrophobic interactions. In the SP lyase sequence, no strict ‘GGE’ motif for SAM binding is found (Supplementary Figure S3) as in biotine synthase (BioB) (30). Ser142 and Asp143 substitute the glycine-rich region for the binding of the amino group of the methionyl moiety (Supplementary Figure S2). In most radical SAM enzymes, the carboxylate of SAM is coordinated by a positively charged amino acid (30), with the exception of MoaA and TYW1, which use a serine residue (31,32). In SP lyase, the orientation of this part of SAM is ensured by the side chains of Lys174 and Ser195 (Supplementary Figure S2). In addition, Ser195, the backbone atoms of Tyr96, Tyr98, Ala234 and Arg273 form hydrogen bonds with the adenine and ribose moieties of SAM. Most of these amino acids are conserved among SP lyase or replaced by similar residues (Supplementary Figure S3). The iron–sulfur cluster and the SAM co-factor are further protected from the solvent by the dinucleoside SP (5R–SPside) as shown in the lesion-bound SP lyase structure (Figure 1B).
DNA lesion binding
The 5R–SPside binds close to SAM in the lateral opening of the barrel (Figure 1B). Upon 5R–SPside binding, SP lyase undergoes distinct conformational changes (Figure 1C and Supplementary Figure S4). Differences are mainly found in and around the catalytic pocket and the β-hairpin. The Tyr98 side chain close to the substrate swivels by ∼45° to accommodate the 5R–SPside in the active site and residues Thr101 to Lys106 adopt a different conformation (Figure 1C and Supplementary Figure S4). Finally, the β-hairpin moves ∼2 Å, suggesting similar structural rearrangements upon DNA binding to seal the substrate binding pocket from the solvent. In the active site, the binding of the 5R–SPside is mostly driven by interactions with the bases (Figure 1D). The 5R–SPside interacts with the protein via the carbonyl groups and the nitrogen atom (N3) of the 5′-dihydrothymine and 3′-thymine residues. Hydrogen bonds are formed between the side chains of Arg193, Glu268, Tyr339 and the O4 atom of the 5′-dihydrothymine residue and between the side chain of Tyr339 and the nitrogen atom (N3). The planar 3′-thymine residue is held by a loop connecting the β-strands β3′ and β4′ through the backbone of Ser76, Lys77 and Ser79 (Figure 1D). This hydrogen-bonding network ensures proper orientation of the nucleobase residues in the active site. The 5′-dihydrothymine and 3′-thymine residues are at an angle of ∼45° and the deoxyribose moiety of the 5′-dihydrothymidine residue adopts a conformation which allows the 3′-hydroxyl group to be close to the 5′-OH of the deoxyribose moiety of the 3′-thymidine residue. Their distance (2.8 Å) is compatible with a phosphate group linker. Interestingly, in a crystal soaked with pyrophosphate and 5R–SPside, residual electron density was found close to the 3′-OH end of the deoxyribose moiety of the 5′-dihydrothymidine residue, which was modeled as a pyrophosphate group mimicking the phosphate bridge in the 5R–SPside lesion being coordinated by two positively charged residues, Arg273 and Lys309 (Supplementary Figure S5).
Recently, the crystal structure of a double-stranded DNA (dsDNA) containing the SPside lesion was published (26). A superposition of the 5′-dihydrothymine residues of 5R-lesion-containing DNA (26) and SP lyase-bound 5R–SPside shows few structural changes in the orientation of the 3′-thymidine residue and the deoxyribose moieties (Supplementary Figure S6). The 3′-thymidine residue of the 5R–SPside in DNA is rotated by ∼45° around the bond between the C5-methylene group and the C5 atom compared to 5R–SPside in the SP lyase complex. Interestingly, in the complex, 5R–SPside superposes perfectly to synthesized SP isostere which possesses similar properties as SP containing a phosphate linker (23) (Supplementary Figure S6). Only the deoxyribose moiety of the 5′-dihydrothymidine residue adopts a different orientation. Therefore, the lack of the phosphate bridge does not seem to affect the orientation of the nucleobase residues in the enzyme active site. In the substrate-bound enzyme, the 5R–SPside is ideally positioned and oriented to avoid any clash with the protein residues even when a lesion-containing DNA is modeled in the SP lyase structure (Figure 1B and Supplementary Figure S7).
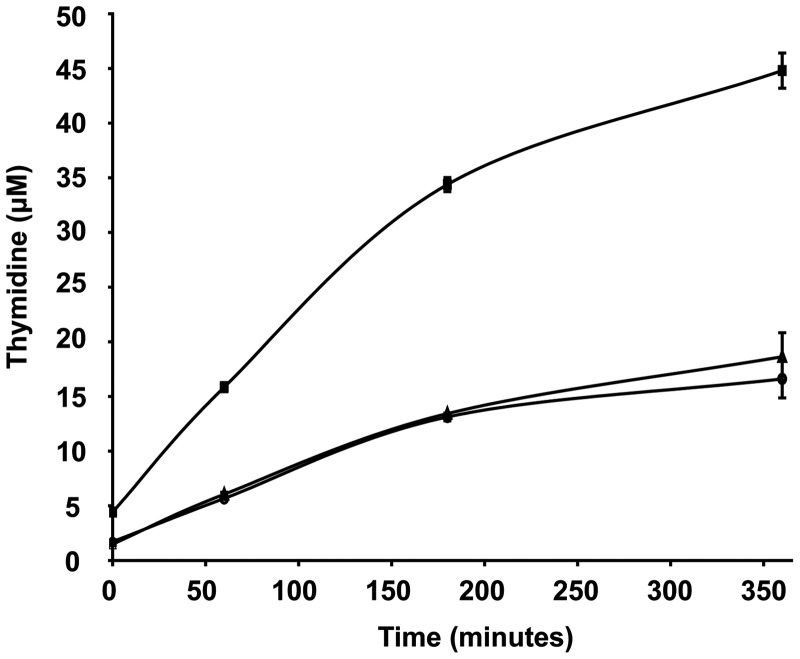
Because the C5 atom of the 5′-dihydrothymine residue can adopt two different configurations, SP synthesis can lead to two diastereomers (5R–SP and 5S–SP). However, SP lyase exhibits stereospecificity for the 5R–SP and hence does not repair the 5S–SP isomer regardless of the incorporation of this lesion into DNA or not (26,33). We tested the activity of the enzyme on both diastereomers (Supplementary Figure S8). We observed cleavage of the thymidine dimers only with the 5R–SPside with a rate of 1.3 mmol min−1 mol−1 of SP lyase (Figure 2 and Supplementary Figure S8) as reported earlier (33). The structures allow us to explain this stereoselectivity on synthesized SP, as superposition of the 5′-dihydrothymine residue of 5R–SPside in the active site with the corresponding dihydrothymine moiety in a 5S-lesion (26) shows drastic differences and either hydrogen atom abstraction or binding of the 5S–SPside is impaired (Supplementary Figure S9). Indeed, the distance between the C6 atom of the 5′-dihydrothymine residue (5S–SPside) and the C5′ atom of SAM is ∼6 Å instead of 3.9 Å in the case of 5R–SPside, far longer than in other substrate-bound structures of radical SAM enzymes (34–36). An orientation of 5S–SPside allowing correct positioning of the 5′-dihydrothymine residue for hydrogen atom abstraction is prevented by protein residues and SAM.
Figure 2.
Repair activity of wild-type and mutant SP lyase for 5R–SPside. Time-dependent formation of thymidine from 5R–SPside by reconstituted wild-type SP lyase (squares), C140A (triangles) and C140S (circles) mutants.
DNA binding mode and lesion recognition
The SP lyase structures provide new insights into the DNA binding mode. The orientation of the 5R–SPside lesion in the binding pocket (Figure 1B) indicates a base flipping mechanism as described for the 6-4 and CPD photolyases, which rotate the lesions through almost 180° into an extra-helical location (37,38). Interestingly, the β-hairpin structure protrudes from the core domain pointing into the solvent with Arg304 and Tyr305 at its tip (Figure 1C) and may assist in flipping the lesion out of the DNA double helix, either by insertion of the β-hairpin into DNA or by interactions with the DNA backbone (39). Tyrosine 305 is a highly conserved residue and substituted in some species by Phe or Trp whereas the arginine is less conserved (Supplementary Figure S3).
A large positively charged surface is displayed at the bottom of the active site (Figure 3), consistent with DNase I footprinting results showing that SP lyase protects at least nine nucleotides (8). Around the active site, numerous sulfate ions were found that interact with the protein through positively charged residues (arginine and lysine), likely mimicking DNA backbone phosphates. Based on the crystal structure of the dsDNA containing a flipped-out 6-4PP lesion (37), we generated a DNA binding model for SP lyase by overlaying the DNA backbone phosphates with the sulfate ions bound to SP lyase (Supplementary Figure S7). The 6-4PP lesion and 5R–SPside superpose as well, and SAM and the flavin cofactor of the 6-4 photolyase are in close proximity.
Figure 3.
Potential DNA binding site. (A) The electrostatic surface map is colored by the electrostatic potential distribution with positively and negatively charged regions shown in blue and red, respectively (range −10kT/e to +10kT/e). (B) View (A) rotated by 45° about the horizontal axis.
SP lyase mutant activity
The 5R–SPside-bound SP lyase structure provides a basis for assessing the proposed mechanisms (13,14). The first steps of the repair mechanism, i.e. iron–sulfur cluster reduction, the SAM cleavage and hydrogen atom abstraction from the 5′-dihydrothymine residue are well established (13,14). In the lesion-bound SP lyase structure, the substrate target carbon (C6) of the 5′-dihydrothymine residue located at a distance of 3.9 Å from the C5′ atom of SAM is perfectly positioned for hydrogen-atom abstraction (Figure 4), as in the few other radical SAM enzymes crystallized with their substrate (34–36). After β-scission of the generated radical intermediate (5-thyminyl-5,6-dihydrothymin-6-yl), the reaction is concluded by a hydrogen atom donation step, for which 5′-dA (13) or the fourth conserved cysteine (Cys141 in B. subtilis) has been suggested to be the donor in previous proposed mechanism (14,15). Interestingly, the α-methylene carbon atom of the 3′-thymidine residue is at a distance of 5.3 Å from SAM but in close proximity to the conserved Cys140 in G. thermodenitrificans (4.5 Å) (Figure 4). This latter distance will likely decrease even further after cleavage of the methylene bridge between the two nucleobase residues as the 3′-thymine moiety can then rotate enabling stacking interactions with the 5′-thymine residue as in (6-4) photolyase (37). Therefore, the SP lyase structure shows that Cys140 is ideally positioned for hydrogen-atom donation to the 3′-thymine allylic radical moiety. In Clostridium species, this cysteine is substituted by an alanine; however, another cysteine residue is found upstream and conserved throughout these species. This cysteine corresponds to serine 76 in the SP lyase structure (Supplementary Figure S3) which, interestingly, is also in close proximity to the 3′-thymidine residue of 5R–SPside (4.2 Å) (Figure 4). Therefore, although in Clostridium and Bacillus species this additional conserved cysteine has not the same position in the enzyme primary sequences, they are spatially related and may fulfill the same function (Figure 4).
Figure 4.
Structural basis of DNA lesion repair. An active site view of 5R–SPside-bound SP lyase is depicted. The iron–sulfur cluster (orange, Fe and yellow, S atoms), SAM (blue, N; red, O; yellow, Se and green, C atoms) and 5R–SPside (blue, N; red, O and white, C atoms) are depicted in stick format. The fourth conserved cysteine in SP lyase corresponds to Cys140 (purple and colored by atom type) in G. thermodenitrificans and to Cys77 in Clostridium acetobutilycum, which is substituted by a serine in G. thermodenitrificans (Ser76 depicted in pink and by atom type). The side-chain of Tyr98 is rendered in cyan and by atom type. Black dashed lines indicate the distances between protein residues, SAM and 5R–SPside.
Previous data showed that the C141A mutant in B. subtilis is impaired in its repair activity leading to the formation of various adducts (15). The authors suggest either a hydrogen atom donor function for this cysteine or a structural role. Indeed, it was proposed that in the wild-type enzyme, the active site most likely adopts a different conformation compared with the mutant and that therefore, the cysteine seals off the active site to prevent any side reactions (15).
To ascertain the role of Cys140, we determined the structure of the C140A mutant and biochemically characterized a mutant in which the cysteine residue was replaced by a serine (C140S), the closest structurally related amino acid. The crystal structures of the C140A mutant SP lyase in the absence and presence of the substrate display no difference to the wild-type enzyme (Figure 5 and Supplementary Figure S10). Furthermore, in the substrate-bound structure of the C140A mutant, 5R–SPside binding is not altered and the interactions around the substrate are conserved, demonstrating the structural integrity of this mutant (Supplementary Figure S10). The biochemical assays were performed under anaerobic, reducing conditions in the presence of SAM and monitored by high-performance liquid chromatography (HPLC) or MALDI-TOF mass spectrometry. Using 5R–SPside as substrate, we assayed the repair activity of the C140S mutant compared with the wild-type enzyme and the C140A mutant which was used here as control. All proteins produce thymidine, therefore the C5–CH2 bond linking the nucleobase residues is cleaved. However, as for the C140A mutant, the C140S mutant shows a 2.5-fold reduced repair activity compared with the wild-type (Figure 2), reflecting either a less efficient repair or, most likely, the repair of the 5′-dihydrothymine moiety and the production of a 3′-thymine sulfinic acid derivative residue as reported for the B. subtilis C141A mutant (15). We thus assayed SP lyase activity for a 5R–SPside-containing oligonucleotide (m/z = 3952.54), which facilitated product identification and analysis, as upon repair by SP lyase, two distinct DNA strands will be formed (26). Under reducing conditions, as expected, the wild-type SP lyase produces two oligonucleotides: a 5′- and a 3′-terminal-thymidine-containing DNA strands with m/z = 1830.56 and m/z = 2119.55, respectively (Figure 6 and Supplementary Figure S11). Although the C140S mutant exhibits repair activity, as previously reported for the B. subtilis C141A mutant with a dinucleotide substrate (15), additional peaks appeared for both mutants, notably one with an m/z increment of 64.0 Da (m/z = 1894.53 and 1895.06, respectively) (Figure 6 and Supplementary Figures S11–S13). This product is consistent with the formation of an SO2 adduct resulting from the reaction of dithionite with the 3′-thymine allylic radical moiety as shown previously with a dinucleotide substrate (15). Thus, like the C140A mutant, the C140S protein is also impaired in the repair of the 3′-thymine residue of 5R–SPside, even in the presence of a more physiologically relevant substrate. To exclude any structural effect, we also solved the structure of the C140S mutant and show that it superposes perfectly onto the wild-type enzyme (r.m.s.d. 0.12 Å) (Supplementary Figure S10). In particular, the side chain of the serine in the C140S mutant structure occupies exactly the same position as the thiol group of the wild-type cysteine (Figure 5), excluding a structural role for this amino acid (15). Altogether, our results demonstrate that C140 is directly involved in the repair of the 3′-thymine allylic radical residue and establish its role as a hydrogen atom donor.
Figure 5.
Superposition of active sites of substrate-free wild-type SP lyase and C140A and C140S mutants. Residue 140 is colored cyan for cysteine, pink for alanine and green for serine. Mutation of C140 induces no structural changes (see also Supplementary Figure S10).
Figure 6.
Repair activity of wild-type and mutant SP lyase. Mass spectrometry analysis of repair activity of reconstituted wild-type (A), C140A (B), C140S (C) mutant SP lyase for a DNA 12mer containing 5R–SPside after 120 min incubation. The insert shows a close-up of the mass spectra from m/z = 1890–1940. The mass was measured in negative ion mode by MALDI-TOF MS using 3-hydroxypicolinic acid (HPA) as matrix (see also Supplementary Figure S11–13).
DISCUSSION
The SP lyase structure described here is the first structure of a radical SAM DNA repair enzyme reported so far and represents a new fold among DNA lyases. SP lyase displays a (α/β)6 topology, like most radical SAM enzymes (30), with a laterally opened barrel, characteristic of radical SAM enzymes acting on macromolecules as the pyruvate–formate–lyase activating enzyme (PFL-AE) which catalyzes the formation of a glycyl radical on a protein (36) and the RNA methyltransferase RlmN involved in the methylation of 23S rRNA (40). Interestingly, the SP lyase crystal structure also shows unique structural features involved in substrate recognition that reveal structural homologies with DNA repair enzymes. Indeed, the UV-damage endonuclease structure also displays a hairpin which bears Gln and Tyr at its tip and points out from the predicted DNA binding groove, suggesting a role in DNA bending, stacking interaction and base flipping (41). Upon DNA binding to SP lyase, Tyr305 may enter the DNA helix and promote DNA unwinding activity as in RecQ1 helicase (42). The side chain of Arg304 may then facilitate nucleotide flipping by substituting the extruded nucleotides as shown in the O6-alkylguanine-DNA alkyltransferase involved in alkylation damage repair by a direct reversal mechanism (43). Therefore, the β-hairpin, Arg304 and Tyr305 are most likely the key player for DNA lesion recognition and nucleotide flipping in SP lyase. However, the DNA-bound SP lyase structure is required to firmly establish the role of these structural features in the DNA–protein interactions. Once the 5R–SPside lesion is flipped out of bound DNA into the active site, the binding of 5R–SPside is mostly driven by interactions with the bases, explaining the activity of SP lyase on structurally related substrates lacking the phosphate bridge (23,44). However, when the intra-lesion phosphate is missing or replaced by a formacetal linker (23), the repair activity decreased by 18- (33) and 2-fold (23), respectively, compared with the natural dinucleotide SP, indicating a recognition of the phosphate by the enzyme. Interestingly, two highly conserved and positively charged residues, Arg273 and Lys309, are present in the vicinity of the two deoxyribose moieties of 5R–SPside, which are most likely involved in the stabilization of the lesion in the binding pocket by electrostatic interactions with the intra-lesion phosphate. This may explain the different activities on both substrates.
In SP lyase, only four cysteine residues are strictly conserved, three of which belong to the radical SAM motif and are involved in the coordination of the [4Fe–4S] cluster, as shown here in the crystal structure. The fourth conserved cysteine located in close proximity to the lesion is found downstream or upstream of the radical SAM motif in Bacillus or Clostridium species, respectively, and its function was previously investigated (15,27). In vivo, the mutation of this cysteine (Cys141 in B. subtilis) abolishes the SP lyase repair activity (27) and in vitro, an allylic radical intermediate was trapped as a thymine sulfinic acid derivative residue (15). This result suggested the involvement of this residue in the tight control of highly reactive radical intermediates generated during the repair catalysis. Labeling studies showed no direct hydrogen atom back-transfer from 5′-dA to the 3′-thymine allylic radical moiety (14,15), but rather the involvement of an exchangeable hydrogen atom proposed to originate from the fourth conserved cysteine. Our biochemical and structural characterizations provide an explanation for these observations. SAM is located at the expected distance and orientation with respect to the bound substrate to allow direct hydrogen atom abstraction by the 5′-dA radical from the C6 atom of the 5′-dihydrothymine moiety. In this configuration, the α-methylene carbon atom of the 3′-thymidine residue is closer to Cys140, which is located in the enzyme active site, than to the C5′ atom of SAM. As with the C140A mutant, the substitution of this cysteine with a serine leads to a 2.5-fold reduced repair activity for 5R–SPside and yields to the formation of DNA adducts when using a 5R–SPside-containing oligonucleotide as substrate. We hence demonstrate that even the serine mutation cannot restore the SP lyase activity. Interestingly, the mutation to alanine or serine does not affect the enzyme structure and the serine superposes perfectly to the wild-type cysteine residue. Our results establish that Cys140 does not seal off the active site (15) but is directly involved in catalysis as a hydrogen atom donor to the 3′-thymine allylic radical moiety. This step is expected to be energetically favored as the S–H bond dissociation energy (BDE) in cysteine is 88 kcal/mol and the BDE of the methyl group of the thymine residue is 86 kcal/mol (45–47). In contrast, the high homolytic O–H BDE in serine (105 kcal/mol) (47) makes hydrogen atom abstraction by the 3′-thymine allylic radical moiety unfavorable and explains why the serine mutant cannot rescue the SP lyase activity, although its structure is identical to the wild-type enzyme.
Finally, despite the different mechanisms proposed (13,14), previous studies reached the conclusion that SAM is a true co-factor as in the well-characterized radical SAM enzyme, lysine 2,3-aminomutase (12). However, in SP lyase, this requires the back generation of SAM from the protein thiyl radical formed after the repair of SP. One possible mechanism is the direct hydrogen atom abstraction by the thiyl radical from the 5′-dA coupled to a more thermodynamically favorable reaction, the recombination of the 5′-dA radical with methionine, similar to class II ribonucleotide reductase (RNR) (48). Interestingly, in the active site of SP lyase, close to Cys140 and SAM, we noticed the presence of Tyr98, which is strictly conserved in Bacillus and Clostridium species (Figures 1C and 4). Cys140 is at a distance of 5.1 Å from Tyr98 while it is 8 Å far away from 5′-dA (Figure 4). Although we cannot so far exclude a purely structural function for this tyrosine residue, Tyr98 could be a good candidate for a proton-coupled electron transfer pathway to reduce the thiyl radical formed, similar to class I RNR (49). This step is thermodynamically favored as the O–H BDE in tyrosine is 93 kcal/mol. Furthermore, this is also consistent with previous data which showed that the 5′-dA is first accumulated and then depleted after ∼20 min while the repair reaction is still ongoing (14). This result is suggestive of the build-up of a radical intermediate concurrent with the 5′-dA which may be a tyrosyl radical. This tyrosine residue is therefore highly interesting, and we are currently investigating its functional role.
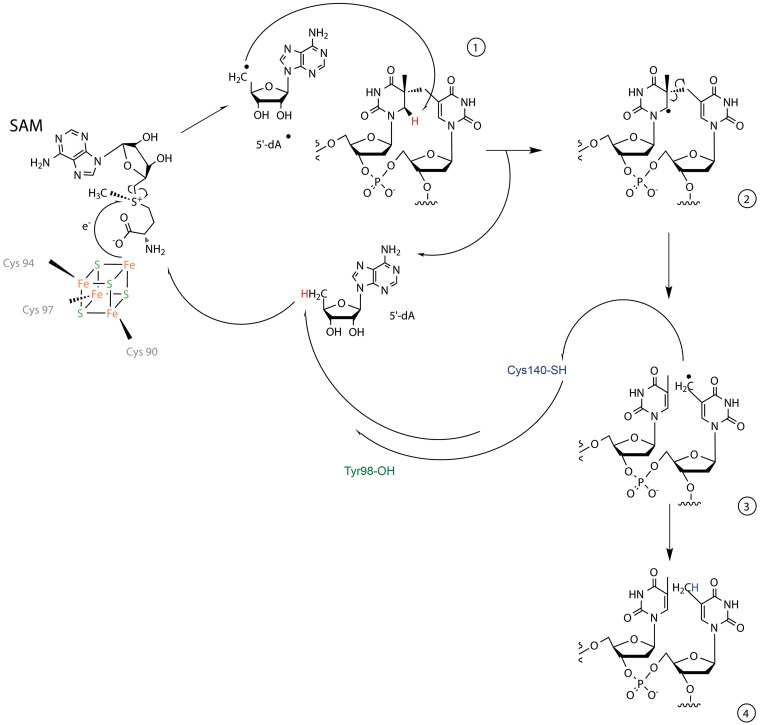
In conclusion, we report here high resolution structures of a unique DNA repair enzyme and provide new insights into its catalytic mechanism (Figure 7). Once the enzyme recognizes the DNA lesion, the β-hairpin may assist in flipping the SP out of the bound dsDNA into the binding pocket, probably helped by Arg304 and Tyr305. After lesion binding, electron transfer from the iron–sulfur cluster to SAM occurs, leading to the formation of a 5′-dA radical which abstracts the pro-R hydrogen atom from the C6 atom on the 5′-dihydrothymine moiety of 5R–SP (14). The generated radical intermediate (5-thyminyl-5,6-dihydrothymin-6-yl) splits into a repaired 5′-thymine residue and a 3′-thymine allylic radical moiety that takes a hydrogen atom from Cys140, concluding the repair reaction. The reduction of the Cys140 thiyl radical may then be mediated either by the 5′-dA or Tyr98, indicating that the regeneration of SAM is likely more complex than previously anticipated. Altogether our data demonstrate how SP lyase combines unique features to achieve a complex radical-based DNA repair mechanism. We thereby expand our understanding of spore photoresistance and of radical SAM enzyme chemistry. Given the increasing number of discovered radical SAM enzymes, the mechanistic features highlighted here will be important to investigate new chemistry in this superfamily of enzymes.
Figure 7.
Proposed mechanism for the SP lesion repair by SP lyase. Reduction of the [4Fe–4S]2+ cluster followed by electron transfer from the cluster to SAM induces cleavage of the C5′–S bond of SAM, producing 5′-dA radical. This highly reactive species abstracts the pro-R hydrogen atom from the C6 atom of the dihydrothymine residue (1) to generate the 5′-dA and a 5-thyminyl-5,6-dihydrothymin-6-yl radical intermediate (2) which then undergoes homolytic cleavage of the methylene bridge. The generated 3′-thymine allylic radical moiety (3) abstracts a hydrogen atom from the thiol group of Cys140 to produce a thiyl radical and a repaired dinucleotide (4). This thiyl radical may be reduced either by 5′-dA or a protein residue such as Tyr98. The 5′-dA radical formed may further react with methionine to regenerate SAM for the next catalytic cycle.
ACCESSION NUMBERS
The atomic coordinates and structure factor amplitudes of all structures have been deposited in the Protein Data Bank with the accession numbers: 4FHC, 4FHD, 4FHE, 4FHF, 4FHG.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–13 and Supplementary Reference [50].
FUNDING
‘European Molecular Biology Organization’ [EMBO Long-Term fellowship to A.B.]; the ‘Max Planck Society’ and the ‘Deutsche Forschungsgemeinschaft’ [SFB749]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Olivier Berteau for providing S-adenosyl-l-methionine and helpful discussions. They acknowledge Dagmar Ringe for stimulating suggestions. They also thank Tatiana Domratcheva, Anton Meinhart, Max Cryle and Anna Scherer for helpful discussions and support. They appreciate experimental assistance with mass spectrometry analysis and helpful discussions from Melanie Müller, Andreas Winkler and Robert L. Shoeman. Diffraction data were collected at the Swiss Light Source, beamline X10SA, Paul Scherrer Institute, Villigen, Switzerland. They thank the Dortmund-Heidelberg team for data collection.
REFERENCES
- 1.Desnous C, Guillaume D, Clivio P. Spore photoproduct: a key to bacterial eternal life. Chem. Rev. 2010;110:1213–1232. doi: 10.1021/cr0781972. [DOI] [PubMed] [Google Scholar]
- 2.Donnellan JE, Jr, Setlow RB. Thymine photoproducts but not thymine dimers found in ultraviolet-irradiated bacterial spores. Science. 1965;149:308–310. doi: 10.1126/science.149.3681.308. [DOI] [PubMed] [Google Scholar]
- 3.Varghese AJ. 5-Thyminyl-5,6-dihydrothymine from DNA irradiated with ultraviolet light. Biochem. Biophys. Res. Commun. 1970;38:484–490. doi: 10.1016/0006-291x(70)90739-4. [DOI] [PubMed] [Google Scholar]
- 4.Van Wang TC, Rupert CS. Evidence for the monomerization of spore photoproduct to two thymines by the light-independent “spore repair” process in Bacillus subtilis. Photochem. Photobiol. 1977;25:123–127. doi: 10.1111/j.1751-1097.1977.tb07432.x. [DOI] [PubMed] [Google Scholar]
- 5.Fajardo-Cavazos P, Salazar C, Nicholson WL. Molecular cloning and characterization of the Bacillus subtilis spore photoproduct lyase (spl) gene, which is involved in repair of UV radiation-induced DNA damage during spore germination. J. Bacteriol. 1993;175:1735–1744. doi: 10.1128/jb.175.6.1735-1744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munakata N, Rupert CS. Genetically controlled removal of “spore photoproduct” from deoxyribonucleic acid of ultraviolet-irradiated Bacillus subtilis spores. J. Bacteriol. 1972;111:192–198. doi: 10.1128/jb.111.1.192-198.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandor A, Berteau O, Douki T, Gasparutto D, Sanakis Y, Ollagnier-de-Choudens S, Atta M, Fontecave M. Dinucleotide spore photoproduct, a minimal substrate of the DNA repair spore photoproduct lyase enzyme from Bacillus subtilis. J. Biol. Chem. 2006;281:26922–26931. doi: 10.1074/jbc.M602297200. [DOI] [PubMed] [Google Scholar]
- 8.Slieman TA, Rebeil R, Nicholson WL. Spore photoproduct (SP) lyase from Bacillus subtilis specifically binds to and cleaves SP (5-thyminyl-5,6-dihydrothymine) but not cyclobutane pyrimidine dimers in UV-irradiated DNA. J. Bacteriol. 2000;182:6412–6417. doi: 10.1128/jb.182.22.6412-6417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 10.Donnellan JE, Jr, Stafford RS. The ultraviolet photochemistry and photobiology of vegetative cells and spores of Bacillus megaterium. Biophys. J. 1968;8:17–28. doi: 10.1016/S0006-3495(68)86471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 13.Cheek J, Broderick JB. Direct H atom abstraction from spore photoproduct C-6 initiates DNA repair in the reaction catalyzed by spore photoproduct lyase: evidence for a reversibly generated adenosyl radical intermediate. J. Am. Chem. Soc. 2002;124:2860–2861. doi: 10.1021/ja017784g. [DOI] [PubMed] [Google Scholar]
- 14.Yang L, Lin G, Liu D, Dria KJ, Telser J, Li L. Probing the reaction mechanism of spore photoproduct lyase (SPL) via diastereoselectively labeled dinucleotide SP TpT substrates. J. Am. Chem. Soc. 2011;133:10434–10447. doi: 10.1021/ja110196d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandor-Proust A, Berteau O, Douki T, Gasparutto D, Ollagnier-de-Choudens S, Fontecave M, Atta M. DNA repair and free radicals, new insights into the mechanism of spore photoproduct lyase revealed by single amino acid substitution. J. Biol. Chem. 2008;283:36361–36368. doi: 10.1074/jbc.M806503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck T, da Cunha CE, Sheldrick GM. How to get the magic triangle and the MAD triangle into your protein crystal. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009;65:1068–1070. doi: 10.1107/S1744309109036884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedel MG, Pieck JC, Klages J, Dauth C, Kessler H, Carell T. Synthesis and stereochemical assignment of DNA spore photoproduct analogues. Chemistry. 2006;12:6081–6094. doi: 10.1002/chem.200600169. [DOI] [PubMed] [Google Scholar]
- 18.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 19.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–806. [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Lin G, Chen CH, Pink M, Pu J, Li L. Chemical synthesis, crystal structure and enzymatic evaluation of a dinucleotide spore photoproduct analogue containing a formacetal linker. Chem. Eur. J. 2011;17:9658–9668. doi: 10.1002/chem.201101821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 26.Heil K, Kneuttinger AC, Schneider S, Lischke U, Carell T. Crystal structures and repair studies reveal the identity and the base-pairing properties of the UV-induced spore photoproduct DNA lesion. Chem. Eur. J. 2011;17:9651–9657. doi: 10.1002/chem.201100177. [DOI] [PubMed] [Google Scholar]
- 27.Fajardo-Cavazos P, Rebeil R, Nicholson WL. Essential cysteine residues in Bacillus subtilis spore photoproduct lyase identified by alanine scanning mutagenesis. Curr. Microbiol. 2005;51:331–335. doi: 10.1007/s00284-005-0052-8. [DOI] [PubMed] [Google Scholar]
- 28.Pieck JC, Hennecke U, Pierik AJ, Friedel MG, Carell T. Characterization of a new thermophilic spore photoproduct lyase from Geobacillus stearothermophilus (SplG) with defined lesion containing DNA substrates. J. Biol. Chem. 2006;281:36317–36326. doi: 10.1074/jbc.M607053200. [DOI] [PubMed] [Google Scholar]
- 29.Rebeil R, Nicholson WL. The subunit structure and catalytic mechanism of the Bacillus subtilis DNA repair enzyme spore photoproduct lyase. Proc. Natl Acad. Sci. USA. 2001;98:9038–9043. doi: 10.1073/pnas.161278998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vey JL, Drennan CL. Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 2011;111:2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanzelmann P, Schindelin H. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl Acad. Sci. USA. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, Nureki O. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J. Mol. Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 33.Chandra T, Silver SC, Zilinskas E, Shepard EM, Broderick WE, Broderick JB. Spore photoproduct lyase catalyzes specific repair of the 5R but not the 5S spore photoproduct. J. Am. Chem. Soc. 2009;131:2420–2421. doi: 10.1021/ja807375c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lepore BW, Ruzicka FJ, Frey PA, Ringe D. The x-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale. Proc. Natl Acad. Sci. USA. 2005;102:13819–13824. doi: 10.1073/pnas.0505726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vey JL, Yang J, Li M, Broderick WE, Broderick JB, Drennan CL. Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc. Natl Acad. Sci. USA. 2008;105:16137–16141. doi: 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maul MJ, Barends TR, Glas AF, Cryle MJ, Domratcheva T, Schneider S, Schlichting I, Carell T. Crystal structure and mechanism of a DNA (6-4) photolyase. Angew Chem. Int. Ed. Engl. 2008;47:10076–10080. doi: 10.1002/anie.200804268. [DOI] [PubMed] [Google Scholar]
- 38.Mees A, Klar T, Gnau P, Hennecke U, Eker AP, Carell T, Essen LO. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, et al. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst.) 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paspaleva K, Moolenaar GF, Goosen N. Damage recognition by UV damage endonuclease from Schizosaccharomyces pombe. DNA Repair (Amst.) 2009;8:600–611. doi: 10.1016/j.dnarep.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc. Natl Acad. Sci. USA. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 44.Friedel MG, Berteau O, Pieck JC, Atta M, Ollagnier-de-Choudens S, Fontecave M, Carell T. The spore photoproduct lyase repairs the 5S- and not the 5R-configured spore photoproduct DNA lesion. Chem. Commun. 2006:445–447. doi: 10.1039/b514103f. [DOI] [PubMed] [Google Scholar]
- 45.Guo JD, Luo Y, Himo F. DNA repair by spore photoproduct lyase: a density functional theory study. J. Phys. Chem. B. 2003;107:11188–11192. [Google Scholar]
- 46.Hioe J, Zipse H. Radicals in enzymatic catalysis—a thermodynamic perspective. Faraday Discuss. 2010;145:301–313. [Google Scholar]
- 47.Moore BN, Julian RR. Dissociation energies of X–H bonds in amino acids. Phys. Chem. Chem. Phys. 2012;14:3148–3154. doi: 10.1039/c2cp23443b. [DOI] [PubMed] [Google Scholar]
- 48.Lawrence CC, Stubbe J. The function of adenosylcobalamin in the mechanism of ribonucleoside triphosphate reductase from Lactobacillus leichmannii. Curr. Opin. Chem. Biol. 1998;2:650–655. doi: 10.1016/s1367-5931(98)80097-5. [DOI] [PubMed] [Google Scholar]
- 49.Stubbe J, Nocera DG, Yee CS, Chang MC. Radical initiation in the class I ribonucleotide reductase: long-range proton-coupled electron transfer? Chem. Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 50.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.