Abstract
MicroRNAs (miRNAs) are major regulators of gene expression in multicellular organisms. They recognize their targets by sequence complementarity and guide them to cleavage or translational arrest. It is generally accepted that plant miRNAs have extensive complementarity to their targets and their prediction usually relies on the use of empirical parameters deduced from known miRNA–target interactions. Here, we developed a strategy to identify miRNA targets which is mainly based on the conservation of the potential regulation in different species. We applied the approach to expressed sequence tags datasets from angiosperms. Using this strategy, we predicted many new interactions and experimentally validated previously unknown miRNA targets in Arabidopsis thaliana. Newly identified targets that are broadly conserved include auxin regulators, transcription factors and transporters. Some of them might participate in the same pathways as the targets known before, suggesting that some miRNAs might control different aspects of a biological process. Furthermore, this approach can be used to identify targets present in a specific group of species, and, as a proof of principle, we analyzed Solanaceae-specific targets. The presented strategy can be used alone or in combination with other approaches to find miRNA targets in plants.
INTRODUCTION
MicroRNAs (miRNAs) are ∼21 nt small RNAs that are key regulators of gene expression in animals and plants [reviewed in (1)]. They are processed from larger precursors by ribonuclease type III enzymes that release the miRNA (2–5), which is subsequently assembled into ARGONAUTE (AGO)-containing complexes (1,6,7). MiRNAs recognize target mRNAs by base complementarity and control their translation and stability.
Animal miRNAs have only limited complementarity to their targets, and the pairing of a seed region between nt 2 and 7 at the 5′-end of the small RNA is a key feature of their interaction [reviewed in (8)]. In plants, miRNAs have an extended homology to their targets and frequently guide them to cleavage, although they can also inhibit their translation (9).
The first miRNA targets in plants were identified by allowing a maximum of three mismatches along the miRNA–target pair (10). Further strategies refined the requirements for miRNA–target interaction in plants considering larger numbers of mismatches, their position, the existence of G-U wobbles and the minimum free energy (MFE) (11–17).
The experimental validation of a plant miRNA target usually relies on the detection of the endonucleolytic cleavage guided by the small RNA. Many AGO proteins, such as Arabidopsis AGO1 (18), cleave the target RNA between positions 10 and 11 of the miRNA 5′-end (19,20). A modified rapid amplification of cDNA ends (RACE)-polymerase chain reaction (PCR) has been designed to identify mRNA fragments that are remnant of this activity in vivo (20,21). Recently, this modified RACE-PCR has been combined with next-generation sequencing techniques allowing the systematic identification of miRNA targets in plants (22,23). Bioinformatic methods have also been developed to identify potential miRNAs guiding the cleavage of these RNAs (24,25).
Many plant miRNAs are young small RNAs that have appeared recently during evolution, although their biological role is unclear (14,26–29). In contrast, ancient plant miRNAs play relevant functions in plant biology and regulate targets whose miRNA-binding sites are also conserved during evolution (30,31). This conservation, specially between Arabidopsis and rice, has been used to support the prediction of targets based on empirical approaches (10,13,17); however, it has not been fully exploited to identify new targets. Furthermore, it has been recently shown that the regulation of a target present only in species related to Arabidopsis thaliana by an ancient miRNA has biological significance (32).
In this study, we present an alternative approach to identify miRNA-regulated transcripts in plants based on conserved targeting of homologous genes present in large expressed sequence tags (EST) datasets of different species. Using this strategy, we found many potential miRNA–target interactions and experimentally validated new targets in A. thaliana. Furthermore, we were able to identify potential miRNA–target interactions that are specific to a group of related species and validated two of them. This approach represents a novel strategy to search for miRNA targets in plants.
MATERIALS AND METHODS
MiRNA consensus
The 22 conserved miRNA families in angiosperms were considered for our studies (14,30). MiR319 and miR159, which encode similar miRNAs, were considered as different families because they regulate different targets (33). We considered all members of these families, obtained from miRBASE 18.0 (http://mirbase.org/) of A. thaliana, Populus trichocarpa and Oryza sativa. Variations at positions 1, 20 and 21 are quite common among miRNA family members (32). A consensus was then defined as the most common sequence (positions 2–19) of the different members of each family. Note that the same results were obtained in the three species.
MiRNA target prediction
Plant datasets
Sequence data were extracted from libraries from the Gene Index project (http://compbio.dfci.harvard.edu/tgi/), which consist of assemblies of ESTs. We selected datasets belonging to angiosperms (see Supplementary Table S2). We also used the mRNA sequences of A. thaliana (http://arabidopsis.org) and O. sativa (http://rice.plantbiology.msu.edu/).
Target search was performed using PatMatch (34), which allows ambiguous characters, mismatches, insertions and deletions. We searched for potential targets with three mismatches to the miRNA consensus, while G:U wobbles and bulges were considered as mismatches. To perform the alignment of the miRNA–target pair, we developed an implementation of the Needleman–Wunsch dynamic programming algorithm (35) in Perl (http://www.perl.org/). Modules using BLASTX (36) against the Arabidopsis proteome and RNA hybrid (37) were integrated by developing in-house scripts.
Filters
Candidate sequences were labeled with the locus ID of the best hit in Arabidopsis using the BLASTX module (E-value cutoff of 10−5) as a tag. Genes from different species having the same tag were grouped together as they have the same homolog gene in A. thaliana. The evolutionary conservation filter referred to the minimum number of species where the same tag was present for a particular miRNA. The empirical filter was based on previous work (38) and referred to the energy of the interaction (MFE at least 72% of the perfect match) and that only one mismatch is allowed between positions 2 and 12 of the miRNA (1–11 of our modified search with the consensus sequences).
Controls
As a control, we performed the same search using shuffled miRNA sequences. For each miRNA, we generated 20 random sequences shuffling the dinucleotide composition as described previously (13). From these 20 random sequences, we chose 10 with the most similar number of total targets to the real miRNA. The signal-to-noise ratio was calculated as the relation between the number of targets for the miRNAs and the average number obtained for the shuffled sequences. As another source of control, we selected two miRNAs not conserved during evolution, miR158 and miR173.
Plant material
Arabidopsis ecotype Col-0 was used for all experiments. Plants were grown in long days (16 h light/8 h dark) at 23°C. Nicotiana tabacum (cv Petit Havana) plants were grown in long days during 8 weeks and the second leaf was used for RNA analysis.
Cleavage site mapping of target mRNA and expression analysis
Poly(A)+ RNA was extracted from 50 µg of total RNA of Col-0 seedlings using PolyAT trackt kit (Promega). Ligation of an RNA adaptor, reverse transcription and 5′ RACE were performed as described before (33). Two nested gene-specific reverse oligonucleotides were used for 5′ RACE. The PCR products were resolved on 2% agarose gels and detected by ethidium bromide staining. Real-Time quantitative PCR (RT–qPCR) for miR396 and miR159 targets was performed as described before (33,39). Lists of primers used for these assays are described in Supplementary Tables S7 and S8. Plants overexpressing miR396 and miR159 have been described previously (33,39).
RESULTS AND DISCUSSION
Design of an approach to identify plant miRNA targets by sequence conservation
We focused our analysis on 22 miRNAs that are conserved in angiosperms (27,30,40,41) (Table 1). In general, these miRNAs are encoded by small gene families of up to 32 members. In the fully sequenced genomes of Arabidopsis, poplar and rice, it is common to find variations in the sequence of miRNAs belonging to the same family, especially in the first, 20th and 21st positions (Supplementary Table S1) (32).
Table 1.
miRNAs and their targets in plants
| miRNA | Consensus (18 nt) | Known targetsa,b |
|---|---|---|
| miR156 | GACAGAAGAGAGTGAGCA | SPL transcription factors |
| miR159 | TTGGATTGAAGGGAGCTC | MYB transcription factors, NOZZLE (NZL) |
| miR160 | GCCTGGCTCCCTGTATGC | ARF transcription factors |
| miR162 | CGATAAACCTCTGCATCC | DCL1 |
| miR164 | GGAGAAGCAGGGCACGTG | NAC transcription factors |
| miR166 | CGGACCAGGCTTCATTCC | HDZip transcription factors |
| miR167 | GAAGCTGCCAGCATGATC | ARF transcription factors, IAA-ALANINE RESISTANT 3 (IAR3) |
| miR168 | CGCTTGGTGCAGGTCGGG | AGO1 |
| mir169 | AGCCAAGGATGACTTGCC | CCAAT-HAP2 transcription factors |
| mir171 | TTGAGCCGTGCCAATATC | GRAS transcription factors |
| miR172 | GAATCTTGATGATGCTGC | AP2 transcription factors |
| miR319 | TGGACTGAAGGGAGCTCC | TCP transcription factors |
| miR390 | AGCTCAGGAGGGATAGCG | TAS RNA |
| miR393 | CCAAAGGGATCGCATTGA | TIR1 proteins, F-BOX proteins |
| miR394 | TGGCATTCTGTCCACCTC | F-BOX proteins |
| miR395 | TGAAGTGTTTGGGGGAAC | ATP sulfurylases, sulfate transporters |
| miR396 | TCCACAGCTTTCTTGAAC | GRF transcription factors, MMG4.7, FLUORESCENT IN BLUE LIGHT (FLU) |
| miR397 | CATTGAGTGCAGCGTTGA | Laccases |
| miR398 | GTGTTCTCAGGTCACCCC | Cu/Zn SODs, CytC oxidase protein subunit, Copper chaperone (CCS) |
| miR399 | GCCAAAGGAGATTTGCCC | Ubiquitin conjugating E2 enzyme |
| miR408 | TGCACTGCCTCTTCCCTG | Blue copper proteins, Laccases, P-TYPE ATPase (PAA2), PAC1 (Proteasome component) |
| miR827 | TAGATGACCATCAGCAAA | SPX proteins |
aTarget genes were grouped according to their functions.
bNew targets experimentally validated in this study are indiacted in bold.
However, we observed that the region between positions 2 and 19 is quite conserved and we could find a consensus sequence present in the majority of the members of each miRNA family in these three species (Table 1, Supplementary Table S1). Interestingly, variable bases outside this conserved region are also prone to have mismatches to known targets (15,42), indicating that a correlation between miRNA–target pairing and miRNA sequence conservation might exist.
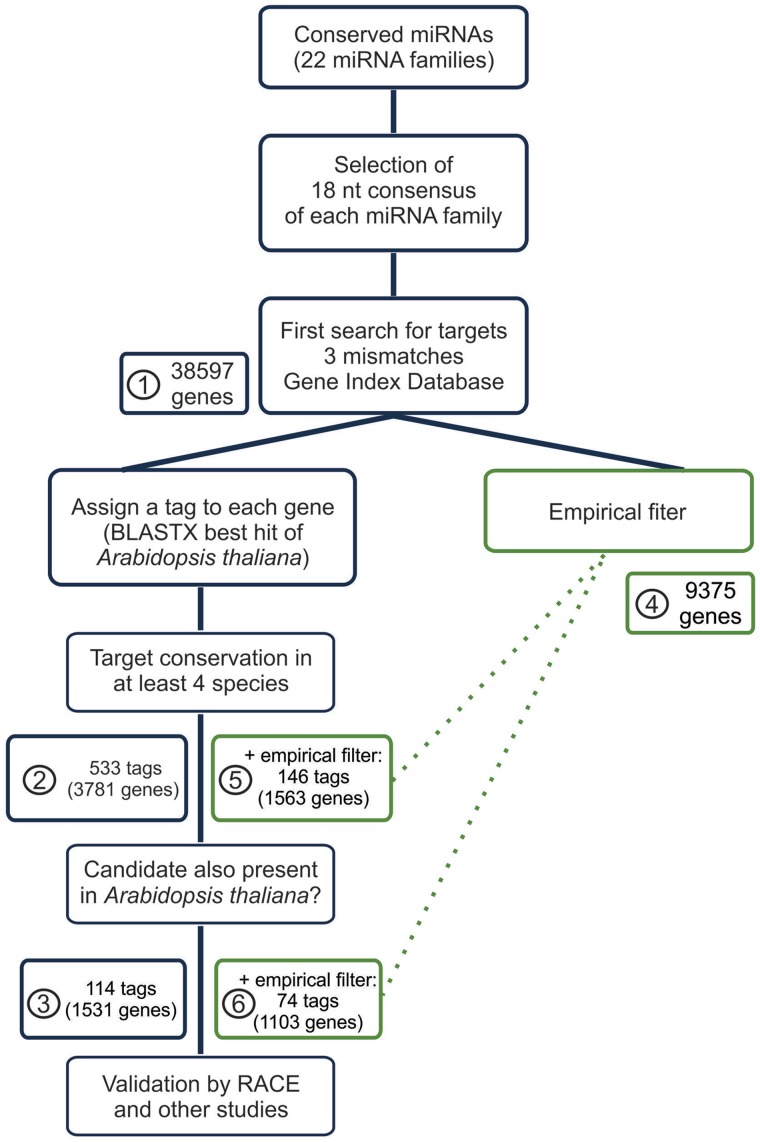
We designed a strategy to identify new miRNA–target pairs mainly based on sequence conservation (Figure 1). The 18 nt consensus sequences of each miRNA family were initially used to search for potential targets in transcript assemblies from ESTs belonging to 41 angiosperm species (Gene Index project, http://compbio.dfci.harvard.edu/tgi/), and the transcripts of the fully characterized A. thaliana (http://arabidopsis.org/) and O. sativa (http://rice.plantbiology.msu.edu/) (for a list of the species analyzed, see Supplementary Table S2). The search for target sequences of the 18 nt miRNA consensus allowing three mismatches rendered 38,597 hits distributed over the 43 species (Figure 1, bin 1). Bulges and G-U wobbles were considered as mismatches in this initial search. All up-to-date known targets of A. thaliana were identified using this approach with the exception of CSD2, a miR398 target that contains four mismatches (Supplementary Table S3).
Figure 1.
Scheme of the strategy to identify new miRNA targets. The number of detected target genes is indicated for each step of the analysis. After applying the conservation analysis, all genes with the same hit in the Arabidopsis proteome were considered as one target. Note that different genes with the same ID tag give only one hit, so that the total numbers of hits are reduced by this filter. Green squares refer to the target search using empirical filters: bins 5 and 6 include target genes selected by both evolutionary and empirical filters, while bins 2 and 3 have potential targets selected only by evolutionary filters.
Since most of the hits represented uncharacterized gene products, we performed a BLASTX against the A. thaliana proteome. The locus ID of the best hit in Arabidopsis was used as a tag to label the selected genes from the different species (Figure 1). Although this approach does not necessarily identify the orthologous Arabidopsis gene, it serves the purpose of classification of each potential miRNA target, as genes with the same tag are homologous to the same gene in A. thaliana. Although most of the hits could be easily assigned with a tag, a few cases including those representing non-coding RNAs were missed at this step.
The strategy allowed the selection of candidate target genes on the basis of their presence in different numbers of species. Conservation in four species, which still has a good specificity for known targets (see subsequent sections), selected 3781 genes corresponding to 533 different tags (Figure 1, bin 2).
The search could also be performed in combination with empirical rules of miRNA targeting, which take into account the energy of interaction and the position of the mismatches (see Materials and Methods). Of the initial 38,597 targets, 9375 passed this filter (Figure 1, bin 4). Combination of the empirical and evolutionary filter selected 1563 genes corresponding to 146 tags (Figure 1, bin 5).
Sequence conservation and empirical parameters can act synergistically to identify miRNA targets
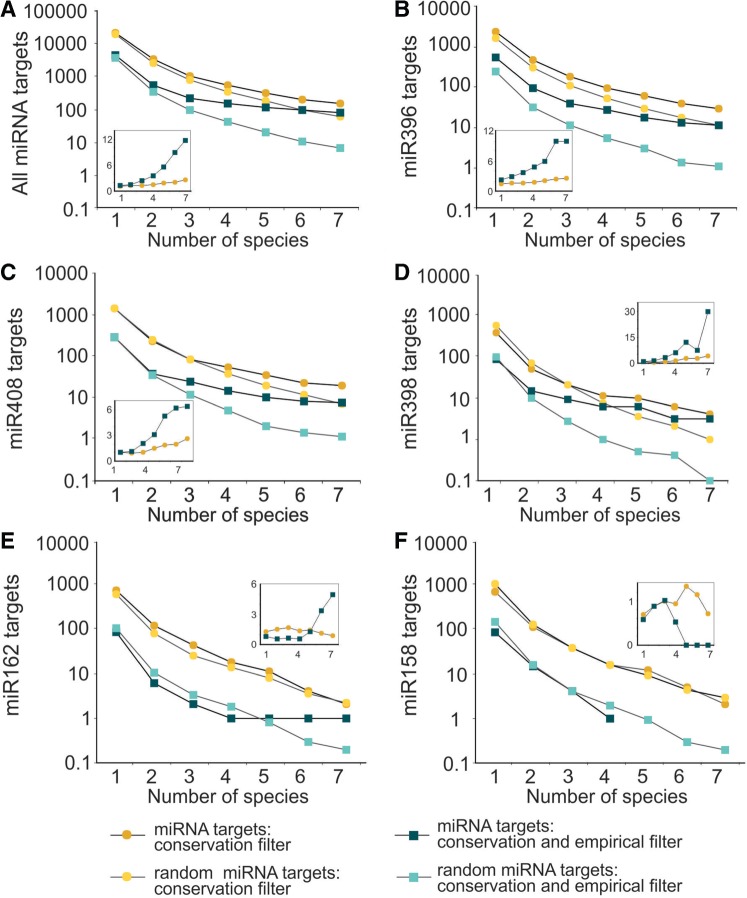
Potential miRNA targets were classified according to the minimum number of species in which they were detected (Figure 2A–E). As a control, we also generated random miRNAs shuffling the 18 nt miRNA consensus sequences (10 random sequences per miRNA). These randomized miRNAs were used to search for potential targets as was performed for the bona fide small RNAs (Figure 2A–E). The signal-to-noise ratio was calculated as the relation between the number of targets for the miRNAs and the average number obtained for the shuffled sequences (Figure 2, insets). The ratio was 1.2 for all miRNA targets without requesting any conservation and steadily increased with the number of species in which the targets are detected (Figure 2A, inset). Data for all miRNAs and their potential targets conserved in at least four species are included in Table 2.
Figure 2.
Conservation of potential miRNA targets in different species. The number of targets conserved in different species is indicated for the different miRNAs: all miRNAs (A); miR396 (B), miR408 (C), miR398 (D), miR162 (E) and miR158 (F). The ochre dots represent the targets of the miRNAs using the conservation filter; the light yellow dots show the targets for the randomized miRNAs using the conservation filter. The dark blue squares represent the targets of the miRNAs after applying empirical and evolution filters, while the light blue squares are the targets for the randomized miRNAs under the same conditions. The insets show the specificity, defined as the ratio between the number of targets for the miRNAs and their randomized sequences (ochre dots refer to the targets filtered by their presence in different number of species, while the blue square represents the targets filter by empirical parameters and number of species).
Table 2.
Detection of miRNA targets using different filters
| No filtera |
Empirical filterb |
Conservation in four speciesc |
All filtersd |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNAe | RNDf | Ratio | miRNAe | RNDf | Ratio | miRNAe | RNDf | Ratio | miRNAe | RNDf | Ratio | |||||
| mir156 | 3915 | 3994 | ±150 | 1.0 | 890 | 705 | ±45 | 1.3 | 34 | 40 | ±3.1 | 0.9 | 10 | 5 | ±1.1 | 1.9 |
| mir159 | 1663 | 1284 | ±48 | 1.3 | 472 | 255 | ±22 | 1.9 | 20 | 10 | ±1.1 | 2.0 | 6 | 2 | ±0.5 | 4.0 |
| mir160 | 793 | 696 | ±30 | 1.1 | 277 | 158 | ±29 | 1.8 | 5 | 4 | ±0.9 | 1.1 | 4 | 1 | ±0.3 | 8.0 |
| mir162 | 1191 | 930 | ±140 | 1.3 | 108 | 165 | ±24 | 0.7 | 18 | 14 | ±3.5 | 1.3 | 1 | 2 | ±0.5 | 0.6 |
| mir164 | 2486 | 1480 | ±60 | 1.7 | 678 | 333 | ±32 | 2.0 | 39 | 12 | ±1.9 | 3.1 | 12 | 2 | ±0.5 | 8.0 |
| mir166 | 879 | 816 | ±45 | 1.1 | 231 | 129 | ±14 | 1.8 | 16 | 11 | ±1.4 | 1.5 | 6 | 1 | ±0.4 | 6.7 |
| mir167 | 1777 | 1364 | ±147 | 1.3 | 478 | 215 | ±28 | 2.2 | 22 | 20 | ±3.6 | 1.1 | 4 | 2 | ±0.5 | 2.2 |
| mir168 | 962 | 798 | ±48 | 1.2 | 209 | 185 | ±14 | 1.1 | 6 | 4 | ±0.8 | 1.4 | 1 | 1 | ±0.5 | 0.9 |
| mir169 | 1540 | 1047 | ±70 | 1.5 | 464 | 181 | ±16 | 2.6 | 26 | 11 | ±2.1 | 2.3 | 10 | 1 | ±0.2 | 8.3 |
| mir171 | 884 | 723 | ±32 | 1.2 | 202 | 114 | ±13 | 1.8 | 7 | 7 | ±1.4 | 1.1 | 2 | 1 | ±0.3 | 2.9 |
| mir172 | 3007 | 1694 | ±125 | 1.8 | 540 | 288 | ±40 | 1.9 | 34 | 18 | ±1.7 | 1.9 | 5 | 2 | ±0.6 | 2.3 |
| mir319 | 1363 | 1274 | ±114 | 1.1 | 324 | 249 | ±22 | 1.3 | 18 | 15 | ±2.8 | 1.2 | 7 | 2 | ±0.5 | 3.9 |
| mir390 | 873 | 814 | ±64 | 1.1 | 335 | 173 | ±22 | 1.9 | 8 | 5 | ±1.2 | 1.7 | 3 | 1 | ±0.5 | 4.3 |
| mir393 | 986 | 845 | ±59 | 1.2 | 276 | 125 | ±11 | 2.2 | 14 | 7 | ±1.2 | 2.0 | 5 | 1 | ±0.2 | 10.0 |
| mir394 | 1569 | 1531 | ±57 | 1.0 | 188 | 237 | ±25 | 0.8 | 26 | 21 | ±2.2 | 1.2 | 3 | 3 | ±0.5 | 1.0 |
| mir395 | 1472 | 1227 | ±67 | 1.2 | 426 | 218 | ±16 | 2.0 | 11 | 9 | ±1.3 | 1.3 | 6 | 1 | ±0.3 | 4.6 |
| mir396 | 4641 | 2979 | ±247 | 1.6 | 1246 | 391 | ±39 | 3.2 | 92 | 51 | ±5.9 | 1.8 | 26 | 5 | ±1.0 | 4.8 |
| mir397 | 1426 | 1051 | ±28 | 1.4 | 368 | 237 | ±23 | 1.6 | 26 | 10 | ±0.8 | 2.7 | 10 | 2 | ±0.3 | 6.3 |
| mir398 | 935 | 834 | ±35 | 1.1 | 376 | 144 | ±18 | 2.6 | 11 | 8 | ±1.6 | 1.5 | 6 | 1 | ±0.3 | 6.0 |
| mir399 | 1192 | 1138 | ±72 | 1.0 | 275 | 208 | ±25 | 1.3 | 5 | 14 | ±1.7 | 0.4 | 1 | 2 | ±0.7 | 0.7 |
| mir408 | 2782 | 2503 | ±104 | 1.1 | 695 | 469 | ±51 | 1.5 | 51 | 35 | ±3.0 | 1.5 | 14 | 5 | ±0.8 | 3.0 |
| mir827 | 2261 | 2000 | ±120 | 1.1 | 317 | 297 | ±45 | 1.1 | 44 | 23 | ±3.9 | 1.9 | 4 | 2 | ±0.8 | 1.7 |
| Total | 38597 | 31021 | ±1860 | 1.2 | 9375 | 5473 | ±576 | 1.7 | 533 | 348 | ±47.0 | 1.5 | 146 | 42 | ±11.3 | 3.5 |
| Control | ||||||||||||||||
| mir158 | 1364 | 1463 | ±69 | 0.9 | 170 | 208 | ±16 | 0.8 | 15 | 16 | ±1.7 | 0.9 | 1 | 2 | ±0.4 | 0.5 |
| mir173 | 1386 | 1232 | ±101 | 1.1 | 243 | 216 | ±23 | 1.1 | 11 | 12 | ±2.4 | 0.9 | 1 | 1 | ±0.4 | 0.7 |
aNo filter, initial search using an 18 nt miRNA consensus sequence and three mismatches.
bEmpirical filter, an interaction energy of at least 72% of the perfect interaction and 1 mismatch in the 2–12 miRNA-target region.
cConservation of the ID tag in at least four species. Note that different genes with the same ID tag give only one hit, so that the total numbers of hits is reduced by this filter.
dAll filters, combination of the empirical and conservation filters in at least four species.
emiRNA, targets for each specific miRNA.
fRND, average targets for 10 scrambled versions of each miRNA ± standard error.
Next, we studied the selection of target candidates by empirical parameters. To do this, we applied a modified version of the filters described before and requested (i) a minimum free energy (MFE) of at least 72% of the perfect match of each 18 nt consensus and (ii) that only one mismatch was present between positions 1 and 11 of the consensus (2–12 of the miRNA). Of the initial search, 9375 genes passed this filter containing 97% of the validated Arabidopsis targets (Figure 1, bin 4).
The application of this empirical filter alone gave a signal-to-noise ratio of 1.7 when grouping all miRNAs together (Figure 2A). We observed that the simultaneous application of the empirical and conservation filters significantly increased the signal-to-noise ratio for the group of all miRNAs (Figure 2A, inset) and in individual miRNAs as well (Figure 2B-E, insets) (see also Table 2). In many cases, this ratio reached above 10 when it was requested that the targets were present in more than five species and that they pass the empirical filters (Figure 2A–D). These synergistic effects indicate that the evolutionary conservation filter and the empirical parameters might be selecting different aspects of a miRNA target interaction.
We observed that the number of target candidates and the signal-to-noise ratio varied among the different miRNAs. MiR396 had the highest number of potential targets, 92 of them being present in at least four species and 26 of them also passed the empirical filter (Table 2; Figure 2B). MiR408 and miR398 also had high numbers of potential targets and favorable signal-to-noise ratios (Figure 2C–D).
In contrast, certain miRNAs such as miR162, miR168 and miR399 had only one potential target conserved in at least four species according to our search (Table 2; Figure 2E). At least in the case of miR162 and miR168, this result might reflect their specific roles in the feedback regulation of miRNA biogenesis, as they control DCL1 and AGO1 expression levels, respectively (43,44).
As an additional control to our strategies, we searched for targets of miR158 and miR173, which are miRNAs present only in A. thaliana and closely related species (27). As expected, these miRNAs did not generate more candidate targets than their shuffled versions (Table 2; Figure 2F).
Then, we tested whether conserved miRNA–target pairs have a stronger interaction than those present in few species. To do this we calculated the MFE for each interaction detected in our assay. We observed that miRNA–target pairs present in many species tend to have stronger interaction energy than those present in only few (Figure 3A). However, the correlation was not striking and some conserved miRNA–target interactions had a low MFE (Figure 3A). These results show that a high conservation might not necessarily be equivalent to a strong interaction which might provide an explanation for the synergistic effects caused by the evolutionary and empirical filters on the signal-to-noise ratios.
Figure 3.
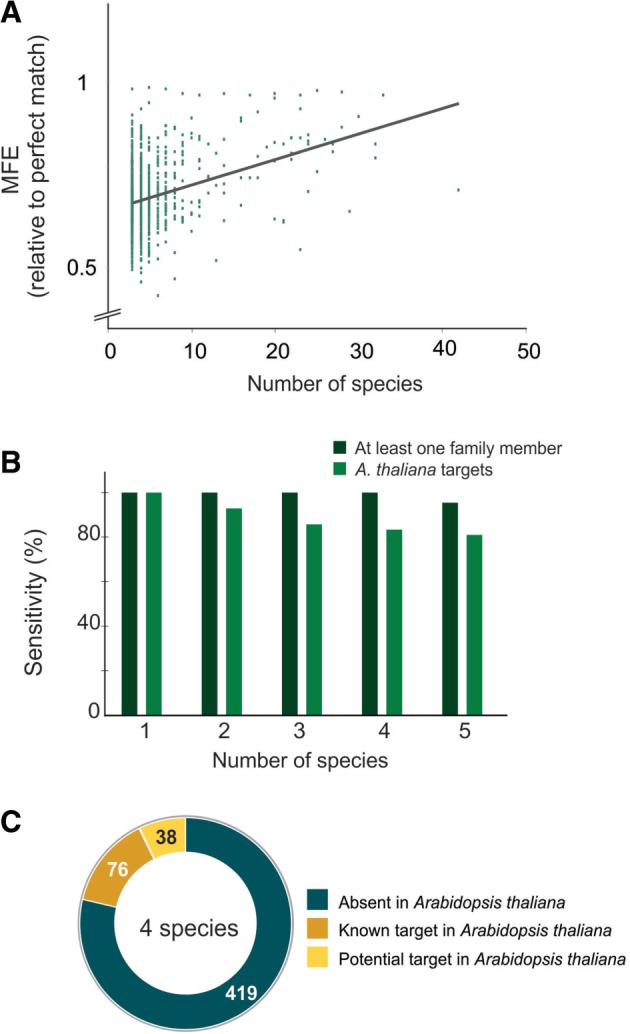
Selection of miRNA targets by sequence conservation. (A) Relationship between the MFE and the number of species where each target was detected. The MFE represents the average of all cognate target sites. A regression line is indicated. (B) Sensitivity of the approach. The sensitivity was evaluated in two ways, one analyzing the presence of validated targets in Arabidopsis thaliana (light green, described in Supplementary Table S3); and alternatively, it was assayed by the presence of at least one target of each gene family regulated by miRNAs (dark green). (C) Classification of the potential targets present in at least four species.
Identification of new miRNA targets in A. thaliana by sequence conservation
To search for new targets, we focused on the potential targets selected only by sequence conservation, as the empirical parameters have been extensively used before [e.g., (11,13,38)]. First, we analyzed the detection of targets previously validated in A. thaliana [based on (14)] using our strategy and found that 84% of them were still present in at least four species (Figure 3B). We considered these results a good outcome as not all Arabidopsis targets might be evolutionary conserved.
As plant miRNAs usually regulate genes coding for proteins of the same family, we evaluated whether at least one member of each family was detected in our approach. We found targets belonging to nearly all conserved protein-coding gene families present in at least four species (Figure 3B, Supplementary Table S4), with the exception of the miR390-regulated TAS3, which, being a non-coding RNA, is not detected by BLASTX.
To search for new miRNA-regulated genes, we focused on potential targets with miRNA-binding sites conserved in at least four species, A. thaliana being one of them (Figure 1, bin 3). MiRNA targets not present in A. thaliana might include genes that have lost their regulation during evolution or genes that gained control by a conserved miRNA more recently in other species. Conservation in four species was chosen as an evolutionary filter because it provided a good sensitivity for known targets.
We identified 114 potential targets that fulfill these criteria (Supplementary Table S4). That included 76 previously described targets or closely related genes (Figure 3C, Supplementary Table S3 and S4). Interestingly, there were 38 genes unrelated to known miRNA targets (Supplementary Table S4), and we decided to study this group in more detail. We focused first on genes present in a large number of species, as we would have a better specificity (Figure 2), and tried to validate the predicted miRNA-guided cleavage using the modified 5′ RACE PCR (20,21).
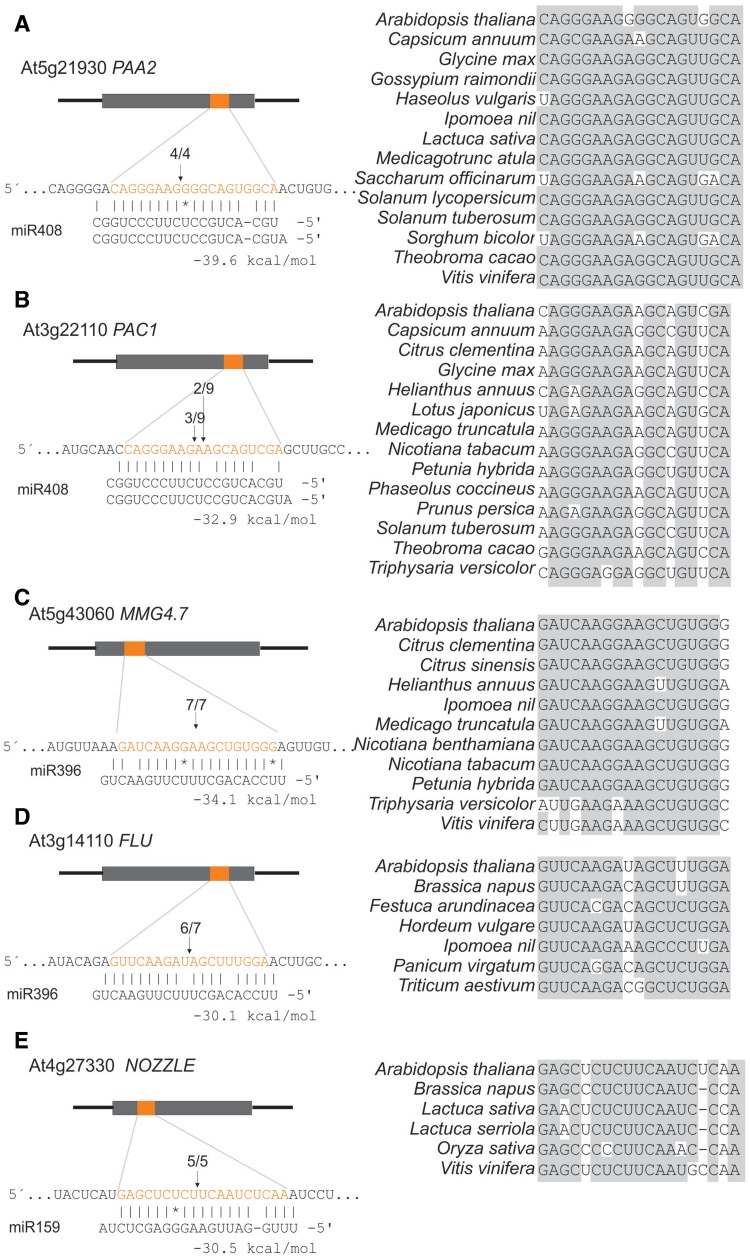
MiR408 was potentially targeting At5g21930, encoding P-TYPE ATPase OF ARABIDOPSIS 2 (PAA2), and was found in 22 different species including dicots and monocots (Supplementary Table S4). MiR408 is unusual as it has a 5′-A; however, >30% of the mature miR408 sequences correspond to a shifted variant starting with 5′-U (45) (Figure 4A). The experimental validation revealed mRNA fragments compatible with this latter cleavage site (Figure 4A). PAA2 is necessary for the transport of copper ions to PLASTOCYANIN (46), and its regulation by miR408 is related to the role of this miRNA in copper homeostasis (47).
Figure 4.
Newly validated miRNA targets in Arabidopsis thaliana. The alignments between the miRNAs and their newly identified targets are depicted on the left. The sequence conservation of the miRNA target site in selected species is shown on the right. The figure shows the interaction of miR408 with PAA2 (A); miR408 with PAC1 (B); miR396 with MMG4.7 (C); miR396 with FLU (D); miR159 with NOZZLE (E). The arrows point the position of cleavage sites as determined by 5′ RACE-PCR and the numbers indicate the cloning frequency of each fragment (21).
Another miR408 target candidate was At3g22110 that encodes PROTEASOME ALPHA SUBUNIT C1 (PAC1), present in 20 species. 5′ RACE-PCR proved that it was also a target of miR408 (Figure 4B). Interestingly, this miRNA–target interaction has three mismatches in the 5′-region that would have led to dismissal as a target if only empirical filters were applied.
The MADS box gene, SHORT VEGETATIVE PHASE (SVP) and the eukaryotic translation initiation factor SUI1 were present in 29 and 19 species, respectively, as potential targets of miR396 (Supplementary Table S4). In both cases, however, we failed to obtain a PCR product using the modified 5′ RACE (not shown). The lack of regulation by miR396 might be related to the weak MFE of these miRNA–target pairs, although we cannot rule out that miR396 is controlling their translation.
Two other potential miR396 targets were At5g43060 and At3g14110 that encode the protease MMG4.7 and FLUORESCENT IN BLUE LIGHT(FLU), respectively. These two targets had stronger interaction energies than SVP and SUI1. In these two cases, we successfully detected miR396-guided cleavage (Figure 4C and D). Determination of MMG4.7 and FLU transcript levels in 35S:miR396 plants revealed a significant decrease of FLU and a minor effect on MMG4.7 (Supplementary Figure S1).
In contrast to miR408 and miR396, which had several potential targets, miR159 hits were all but one MYB transcription factors, which regulate stamen and pollen development (48). The additional target was At4g27330, also known as NOZZLE/SPOROCYTELESS. This transcription factor, which participates in stamen and ovule development (49,50), was also validated by 5′ RACE-PCR (Figure 4E). In good agreement, 35S:miR159 caused a reduction of both MYB and NOZZLE transcript levels (Supplementary Figure S2). A miR159 target with a NOZZLE-like domain has been also recently validated in tomato (51), which together with our results point toward a general role of miR159 in the regulation of NOZZLE-like genes. Interestingly, at least the functions of NOZZLE and PAA2 can be directly related to the roles of already described targets of miR159 and miR408, respectively.
PAA2, FLU and NOZZLE transcripts with miRNA binding sites were detected in dicots and monocots, while PAC1 and MMG4.7 miRNA-binding sites were present only in dicots (Figure 4A–E). Positions in the miRNA-binding sites were highly conserved, and many of the variable positions corresponded to mismatches in the interaction with the miRNA or alternating G-C/G-U wobbles. Moreover, this method does not require that the sequence of the target site is conserved, but rather that there is a predicted interaction with the miRNA in different species. This way, the target site of NOZZLE, which changes in sequence among species (Figure 4E), could be found by this approach.
Identification of new potential targets allowing GU wobbles
The targets identified here had several mismatches and bulges with their cognate miRNAs, which might explain why they were missed by previous approaches. We also noticed that many of the new miRNA–target interactions contained positions which were alternating between G-C and G-U in different species (Supplementary Figure S3). As we considered G-U as a mismatch in our initial search, we decided to perform another search for targets of the 18-nt miRNA consensus allowing four mismatches, with at least one of them being a G-U wobble. This search would allow miRNA–target interactions with only 14 matches.
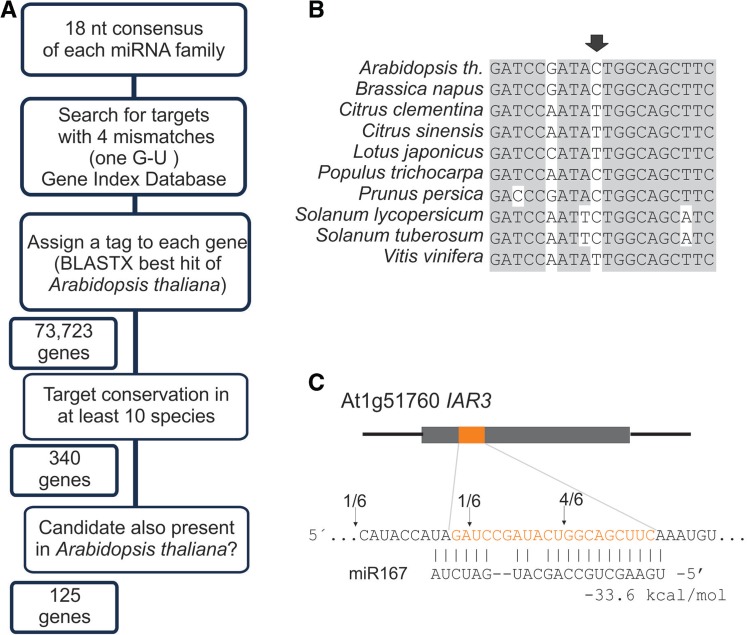
To compensate the use of these relaxed parameters in terms of mismatches, we requested that the target should appear in at least 10 different species to increase the specificity (Figure 5A). We found 125 potential targets in A. thaliana that fulfill these criteria (Figure 5A, Supplementary Table S5) and 34 of them did not appear in our previous searches. The miR398 target CSD2 that was missing in our first approach was detected with these parameters.
Figure 5.
Identification of a new target by relaxation of the interaction parameters while increasing the conservation parameter. (A) Scheme showing the strategy to identify the miRNA targets. (B) Conservation of the target site in different species. The arrow indicates a position of a G-C or G-U interaction with the miRNA depending on the species. (C) Alignment of Arabidopsis IAR3 and miR167. The position that contains a G-U wobble is indicated. The arrows show the position of cleavage sites as determined by 5′ RACE-PCR, and the numbers indicate the cloning frequency of each fragment (21).
We next screened the latter group for potential miRNA-regulated genes that were performing ancillary functions to the targets already described for each miRNA. We found that miR167 that regulates AUXIN RESPONSE FACTORS (ARFs), was potentially targeting At1g51760, IAA-ALANINE RESISTANT 3 (IAR3) (Figure 5B and C), which is involved in the control of free auxin levels (52,53).
The Arabidopsis IAR3 has three mismatches with respect to miR167, but at position 12 of this miRNA–target interaction there is a G-U wobble in several species (Figure 5B and C). Modified 5′ RACE-PCR confirmed that the gene was actually a target of miR167 (Figure 5C).
Identification of Solanaceae-specific target genes
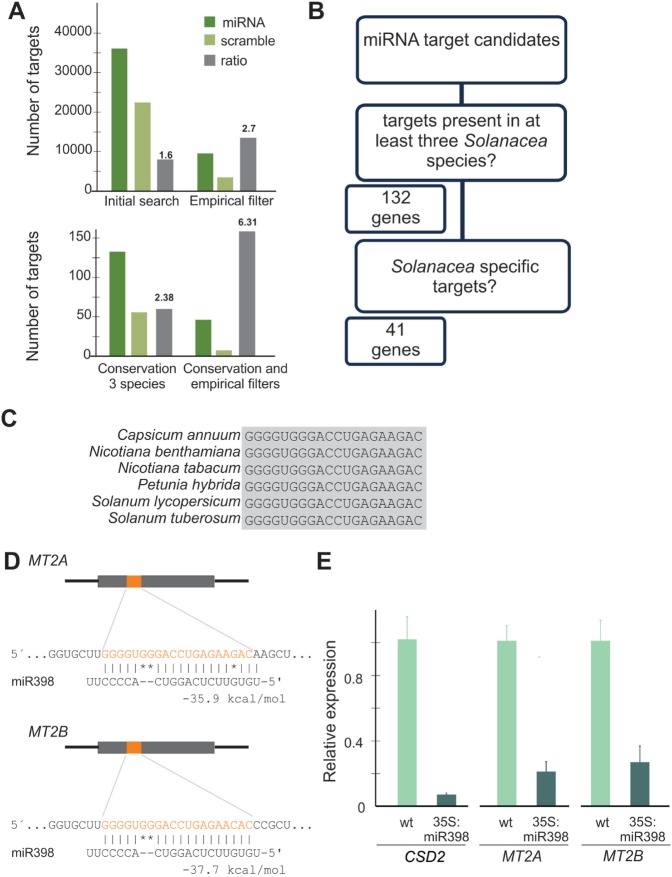
We reasoned that the strategy presented here might also be used to find targets present specifically in a group of related species. We therefore tested whether we could find potential miRNA targets specific of the Solanaceae family. We chose this particular family because six species were well represented in the analyzed libraries. The relation between the target of miRNAs and their scrambled sequences was more than two when the empirical or the conservation filters (at least three of the six Solanaceae species) were applied (Figure 6A). Interestingly, the combination of the two filters resulted in a signal-to-noise ratio above 6 (Figure 6A), confirming our previous findings that both filters enhance the detection of miRNA targets.
Figure 6.
Identification of Solanaceae-specific miRNA targets. (A) Prediction of miRNA targets in five Solanaceae species. The number of targets for all conserved miRNAs is indicated after the application of several filters. Targets obtained by the randomized sequences are also indicated. (B) Scheme showing the strategy to identify miRNA targets specific of Solanaceae species. (C) Conservation of the miR398 target site in MT2A sequences from Solanaceae species. (D) Scheme showing the miR398 binding site in tobacco MT2A and MT2B. (E) Transcript levels of CSD2, MT2A and MT2B in wild-type and transgenic tobacco plants (cv. Petit havana) overexpressing miR398. The data shown are mean ± s.e.m. of three biological replicates.
We found 132 potential target genes present in at least three Solanaceae species. Of this group, 41 targets were not detected in other species (Figure 6B, Supplementary Table S6). The most common target was the metallothionein MT2A, which was present in all six Solanaceae species as a potential target of miR398, while MT2B, a homolog of this gene, was present in five species (Figure 6B–D, Supplementary Table S6).
Then, we took advantage of transgenic tobacco plants harboring a 35S:miR398 transgene (A.F.Lodeyro, N.Carrillo and J.F.Palatnik, unpublished results) and tested the expression of these genes. We found that CSD2, a broadly conserved target of miR398, decreased its expression >10-fold in 35S:miR398 transgenic plants compared with wild type (Figure 6E). Interestingly, we found that both MT2A and MT2B decreased their transcript levels >5 times in these plants (Figure 6E). These results are in agreement with the regulation of MT2A and MT2B by miR398, although they do not necessarily prove a direct interaction. Altogether, these results show that miRNA targets present in a specific group of species might be found by this strategy.
CONCLUSIONS
Here, we designed a strategy to identify miRNA-regulated genes that is mainly focused on the conservation of the potential targeting. The approach requests that the miRNA targeting should be able to occur in the context of a minimum set of interacting parameters in different species. Therefore, the sequence of the target itself does not need to be conserved. Furthermore, our approach allows adjusting the number of species requested as a filter to search with different sensitivities and signal-to-noise ratios.
Using this strategy we identified and experimentally validated new targets in A. thaliana, even though this system has already been studied in detail by several different genome-wide approaches (11,13,22,23,28,38). Three new validated targets contain bulged nucleotides. Empirical parameters have usually given a strong penalty to them, which could even be double of regular mismatches (13); however, it is possible that target sites with asymmetric bulges are more frequent than previously thought in plants.
We found that newly validated targets have functions related to those already known. MiR159 regulates MYB transcription factors (33,54,55) and NOZZLE (this work), which are involved in stamen and pollen development (48–50,55). MiR408 regulates the copper transporter PAA2 (this work) as well as copper-binding proteins (13,23,38,56). MiR167 regulates ARFs (10,57) and IAR3 (this work), and both of them participate in the control of auxin levels and activity (52,58). These results confirm the importance of miRNA regulation in plants, further indicating that a miRNA might be regulating different components of a biological pathway.
The approach offers an alternative strategy to other predictions based on empirical parameters of known miRNA–target pairs (11,13,15,38). An advantage of the strategy presented here is that conserved miRNA–target interactions might be likely involved in relevant biological processes. Furthermore, the approach could be easily modified to incorporate data from other expression libraries, and/or search for targets only present in a specific group of plant species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–8 and Supplementary Figures 1–3.
FUNDING
Argentinean National Agency of Science and Technology (PICT) and an international grant of HHMI [to J.P.] and fellowships of CONICET [to V.A.C., N.G.B., A.P.M., C.S., A.F.L., N.C. and J.F.P.]. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Edgardo Bresso, Juan Debernardi and other members of JP lab for continuous help and discussions.
REFERENCES
- 1.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 2.Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 2010;20:49–54. doi: 10.1016/j.cub.2009.10.072. [DOI] [PubMed] [Google Scholar]
- 3.Bologna NG, Mateos JL, Bresso EG, Palatnik JF. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. Embo. J. 2009;28:3646–3656. doi: 10.1038/emboj.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010;20:37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 5.Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 2010;20:42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- 6.Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22:3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axtell MJ, Westholm JO, Lai EC. Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 9.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 10.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 11.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D, et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell. 2010;22:1074–1089. doi: 10.1105/tpc.110.073999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahlgren N, Carrington JC. miRNA target prediction in plants. Methods Mol. Biol. 2010;592:51–57. doi: 10.1007/978-1-60327-005-2_4. [DOI] [PubMed] [Google Scholar]
- 16.Alves L, Jr, Niemeier S, Hauenschild A, Rehmsmeier M, Merkle T. Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Res. 2009;37:4010–4021. doi: 10.1093/nar/gkp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 21.Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 22.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Li YF, Sunkar R, Zhang W. SeqTar: an effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucleic Acids Res. 2012;40:e28. doi: 10.1093/nar/gkr1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011;23:431–442. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Z, Coruh C, Axtell MJ. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell. 2010;22:1090–1103. doi: 10.1105/tpc.110.073882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Axtell MJ. Evolution of microRNAs and their targets: are all microRNAs biologically relevant? Biochim. Biophys. Acta. 2008;1779:725–734. doi: 10.1016/j.bbagrm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF. Functional specialization of the plant miR396 regulatory network through distinct MicroRNA-target interactions. PLoS Genet. 2012;8:e1002419. doi: 10.1371/journal.pgen.1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY. PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res. 2005;33:W262–W266. doi: 10.1093/nar/gki368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137:103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. Embo. J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 44.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maunoury N, Vaucheret H. AGO1 and AGO2 act redundantly in miR408-mediated Plantacyanin regulation. PLoS One. 2011;6:e28729. doi: 10.1371/journal.pone.0028729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdel-Ghany SE, Muller-Moule P, Niyogi KK, Pilon M, Shikanai T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell. 2005;17:1233–1251. doi: 10.1105/tpc.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 2007;282:16369–16378. doi: 10.1074/jbc.M700138200. [DOI] [PubMed] [Google Scholar]
- 48.Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1999;96:11664–11669. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999;13:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buxdorf K, Hendelman A, Stav R, Lapidot M, Ori N, Arazi T. Identification and characterization of a novel miR159 target not related to MYB in tomato. Planta. 2010;232:1009–1022. doi: 10.1007/s00425-010-1231-9. [DOI] [PubMed] [Google Scholar]
- 52.Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell. 1999;11:365–376. doi: 10.1105/tpc.11.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004;135:978–988. doi: 10.1104/pp.104.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 55.Allen RS, Li J, Stahle MI, Dubroue A, Gubler F, Millar AA. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl Acad. Sci. USA. 2007;104:16371–16376. doi: 10.1073/pnas.0707653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu MF, Tian Q, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- 58.Weijers D, Jurgens G. Funneling auxin action: specificity in signal transduction. Curr. Opin. Plant Biol. 2004;7:687–693. doi: 10.1016/j.pbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.