Abstract
Alternative polyadenylation increases transcriptome diversity by generating multiple transcript isoforms from a single gene. It is thought that this process can be subject to epigenetic regulation, but few specific examples of this have been reported. We previously showed that the Mcts2/H13 locus is subject to genomic imprinting and that alternative polyadenylation of H13 transcripts occurs in an allele-specific manner, regulated by epigenetic mechanisms. Here, we demonstrate that allele-specific polyadenylation occurs at another imprinted locus with similar features. Nap1l5 is a retrogene expressed from the paternally inherited allele, is situated within an intron of a ‘host’ gene Herc3, and overlaps a CpG island that is differentially methylated between the parental alleles. In mouse brain, internal Herc3 polyadenylation sites upstream of Nap1l5 are used on the paternally derived chromosome, from which Nap1l5 is expressed, whereas a downstream site is used more frequently on the maternally derived chromosome. Ablating DNA methylation on the maternal allele at the Nap1l5 promoter increases the use of an internal Herc3 polyadenylation site and alters exon splicing. These changes demonstrate the influence of epigenetic mechanisms in regulating Herc3 alternative mRNA processing. Internal Herc3 polyadenylation correlates with expression levels of Nap1l5, suggesting a possible role for transcriptional interference. Similar mechanisms may regulate alternative polyadenylation elsewhere in the genome.
INTRODUCTION
The total number of different molecules comprising the human and mouse transcriptomes far exceeds the predicted number of genes. A single gene may give rise to multiple transcript variants that encode identical or different proteins, and the decision to express certain transcripts but not others is spatially and temporally coordinated.
Although gene expression is influenced by tissue- and developmental stage-specific transcription factors, epigenetic mechanisms also play an important role. For example, DNA methylation and a subset of histone modifications such as H3K27me3 and H3K9me3 are typically associated with transcriptional repression when they are abundant at promoters. Genes that are subject to genomic imprinting are a special case of this type of epigenetic regulation of transcription. Allele-specific DNA methylation established during gametogenesis at imprinting control regions regulates the parent-of-origin specific expression of imprinted genes (1).
In part, transcript diversity can be accounted for by differential promoter usage, and the role of epigenetic mechanisms in this process is well established. For example, at least two promoters are embedded within the gene body of Shank3, in addition to the canonical 5′ Shank3 promoter (2). DNA methylation at these internal promoters is inversely correlated with internal transcription initiation, and differs between tissues, providing a mechanism for regulating tissue-specific promoter use. Alternative splicing is another key mechanism for generating transcript diversity, with ∼95% of human multi-exonic genes estimated to produce at least two transcript isoforms by differential exon inclusion (3). Epigenetic mechanisms have recently been suggested to regulate the tissue-specific and mutually exclusive use of exons IIIb and IIIc in transcripts of the human FGFR2 gene. Alternative splicing of these exons is associated with differential enrichment for specific histone H3 modifications between tissues (4). Modulating the levels of these histone modifications results in the loss of tissue-specific splicing.
A third mechanism for generating transcript diversity is the use of alternative polyadenylation [poly(A)] sites. Little is known about the regulatory mechanisms that control this process, although the cell-specific activity of trans-acting RNA processing factors is thought to play a role (5–7). The possibility that epigenetic modifications could act in cis to regulate this process is supported by studies of an imprinted gene locus in the mouse. The locus comprises the Mcts2 retrogene that is situated in an intron of the multi-exonic ‘host’ gene H13 (8,9). Retrogenes are functional genes resulting from retrotransposition of an mRNA molecule into the genome (10). The retrogene Mcts2 is imprinted, being silenced on the maternally inherited chromosome through methylation of its promoter, which is situated 200 bp downstream of an internal H13 poly(A) site (9). When the Mcts2 promoter is unmethylated and active, H13 transcripts utilize the upstream poly(A) site, whereas the methylated and inactive allele allows transcription of H13 to proceed to a poly(A) site situated 10 kb downstream. These observations cannot be explained by cell type-specific RNA processing factors operating in trans, as both alleles would be exposed to the same complement of diffusible molecules, and hence epigenetic modifications operating in cis must be the determining factor. This could be mediated through the binding of methylation-sensitive polyadenylation factors, or through transcriptional interference from transcripts initiating at the Mcts2 promoter.
In this report, we characterize in detail another locus at which an imprinted retrogene, Nap1l5, is positioned within an intron of a host gene, Herc3, that utilizes multiple poly(A) sites. Full-length Herc3 transcripts that utilize a downstream poly(A) site encode a probable E3 ubiquitin ligase that targets proteins for proteasomal-mediated degradation (11). The gene is broadly expressed and demonstrates a subcellular localization consistent with a role in vesicular trafficking (11,12). The Nap1l5 protein is also incompletely characterized, but is likely to associate with Nap1l2 which is a chromatin-modifying protein critical for neuronal development (13,14). Consistent with this, Nap1l5 is most abundantly expressed in neural tissues (12,15).
The Nap1l5 promoter is associated with a germline differentially methylated region (gDMR), being unmethylated on the paternally derived chromosome and heavily methylated on the maternally derived chromosome. We demonstrate that polyadenylation sites of Herc3 are utilized in an allele-specific manner. A subset of transcripts arising from the paternally inherited allele, from which Nap1l5 is expressed, use internal poly(A) sites upstream of Nap1l5, whereas a downstream poly(A) site is preferentially used on the maternally inherited allele, on which Nap1l5 is silent. Ablation of Nap1l5 promoter methylation on the maternally inherited allele causes biallelic Nap1l5 expression, and increased use of an internal Herc3 poly(A) site, providing a clear example of epigenetic control of poly(A) site choice. Although in this study we do not conclusively determine the mechanism responsible for mediating this effect, we demonstrate that the abundance of Herc3 transcripts using an internal poly(A) site is not correlated to overall Herc3 transcription levels, but is closely correlated with the level of Nap1l5 transcription in vivo, supporting a role for transcriptional interference.
MATERIALS AND METHODS
Tissue sources
Mouse strains used for allele-specific assays were C57Bl6 (Bl6) and Mus musculus castaneus (cast). RNA from Dnmt3L−/+ e8.5 embryos and wild-type littermates was a kind gift of Deborah Bourc'his.
RNA isolation and 3′ rapid amplification of cDNA ends (RACE)
Total RNA was purified from a whole Day 1 mouse brain using the Qiagen RNeasy Mini kit. One microgram was used to synthesize 3′ RACE-ready cDNA using the SMARTer RACE kit (Clontech). RACE was performed using the Advantage 2 PCR kit (Clontech) and touchdown PCR, as described in the associated manual, on a MJ Research PTC-200 DNA Engine thermal cycler. 3′ RACE primers were designed to Herc3 exons 1c, 3, 4, 5, 22 and 23b. Primer sequences are presented in Supplementary Table S1. Amplicons were purified from agarose gels (Qiagen MinElute kit) and cloned into the pGEM T-Easy vector (Promega) for sequencing. Multiple clones of each amplicon were sequenced using Big Dye v3.1 (ABI) sequencing technology. Sequence data were manipulated in Sequencher.
Northern blotting
Probe A, spanning Herc3 exons 2–5, was amplified from brain cDNA using Herc3-F5 and Herc3-R5 (Supplementary Table S2) and cloned into the pGEM T-Easy vector. The 0.4 kb fragment was isolated by EcoRI digestion, gel-purified and used to generate [α-32P]dCTP-labeled probe with a High Prime DNA labeling kit (Roche) according to the associated manual. Total RNA from 2-week mouse brain was challenged with the probe as described previously (16). The relative abundance of Herc3a and Herc3b was assessed using ImageJ (http://rsbweb.nih.gov/ij/).
Allele-specific assays
RNA was purified from whole brain, heart and lung of Bl6/cast inter-subspecies hybrid Day 1 neonates as above. RNA was treated with DNase I using DNA-free (Ambion) according to the manufacturer’s instructions. cDNA was synthesized from 1 µg RNA using the SuperScript first strand synthesis kit (Invitrogen). Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed with ABgene ReddyMix and primers specific to each transcript, using multiple cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s. For Herc3a, 28 PCR cycles were used; for Herc3b transcripts using the Herc3b1 and Herc3b2 poly(A) sites and Herc3c transcripts, 35 cycles; and for Nap1l5, 30 cycles. Primers were as follows. Herc3a: Herc3-F1 in exon 20 and Herc3-R1 in exon 25. Herc3b1: Herc3-F2 in exon 20 and Herc3-R2 in exon 23b. Herc3b2: Herc3-F3 in exon 22 and Herc3-R3 in exon 23c. Herc3c: Herc3-F4 in exon 1c and Herc3-R4 in exon 25. Primer sequences are presented in Supplementary Table S2. Amplicons were sequenced over single nucleotide polymorphisms (SNPs) identified from the Sanger Mouse Genomes Project (17,18). SNPs were confirmed by amplifying from genomic DNA isolated from Bl6, cast and B × C animals; primer sequences and reaction conditions for these amplifications are available on request.
Pyrosequencing
RNA was isolated from whole brains and hearts of three B × C and three C × B inter-subspecies hybrid Day 1 neonates, and cDNA was synthesized, as discussed earlier. Pyrosequencing assays were designed for the Herc3a and Herc3c transcripts using PyroMark Assay Design 2.0 software (Qiagen). Primer sequences, as well as details on which primers were biotinylated, are provided in Supplementary Table S2. For each transcript, ‘pyro F1’ and ‘pyro R1’ were used for PCR amplification, and ‘pyro S1’ for sequencing. The SNPs quantified in the assays were the same SNPs used in the allele-specific assays described earlier. PCR was performed using ABgene ReddyMix and the same conditions as earlier, except that 30 amplification cycles were utilized for both the Herc3a and Herc3c transcripts. Reactions were prepared for analysis by using the PyroMark Q96 Vacuum Prep Workstation (Qiagen) according to the manufacturer’s instructions, and pyrosequencing was conducted on a PyroMark Q96 MD machine (Qiagen).
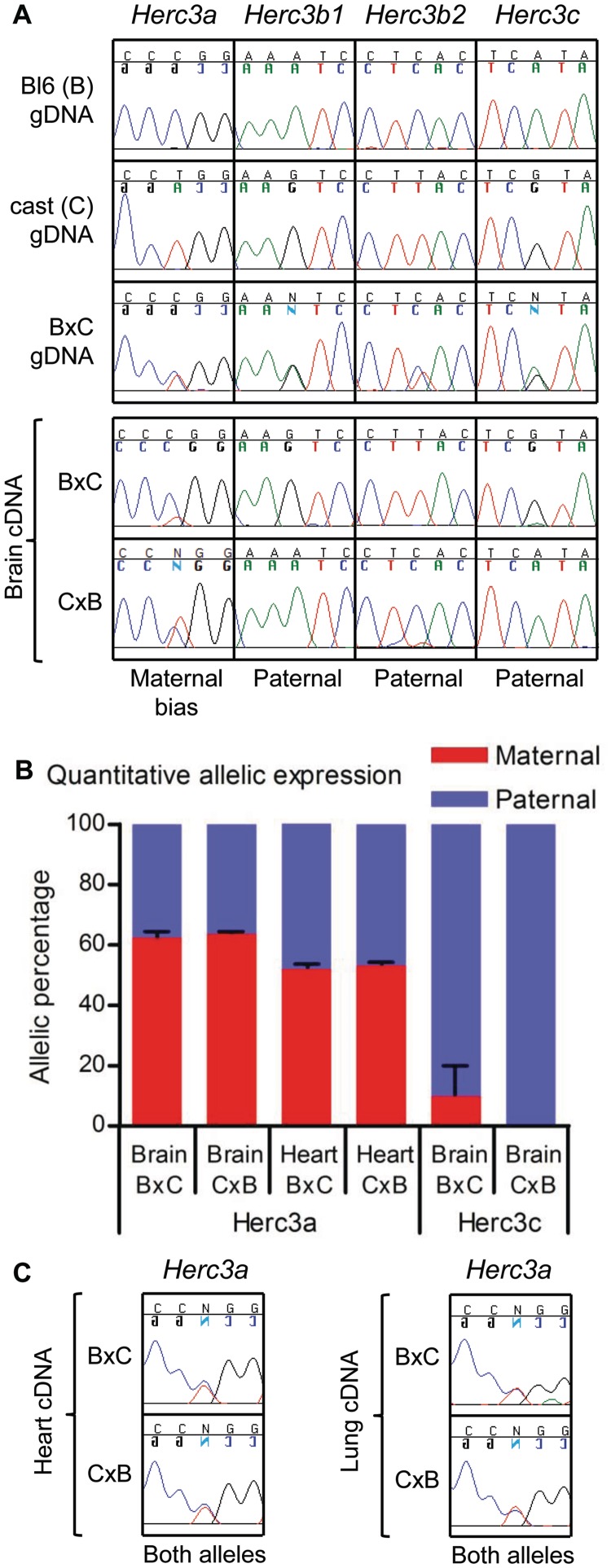
Figure 2B shows the mean of data from three biological replicates for Herc3a, and from two biological replicates for Herc3c. For one B × C and one C × B sample, allele quantification of Herc3c expression could not be determined with accuracy because the peaks were close to background levels, although both samples showed a bias consistent with the expected direction of imprinting (87.4 and 91.8% paternal allele expression in the B × C and C × B samples, respectively).
Figure 2.
Allele-specific use of poly(A) sites. (A) Primer pairs unique to each of the Herc3 transcript variants were used in RT-PCR on RNA isolated from whole brains of inter-subspecies mouse hybrids (Bl6 [B] and cast [C]). Amplicons were sequenced across SNPs to determine parent-of-origin specific expression. The SNP is the centre nucleotide of each trace. In each cross, the maternal strain is presented first. Performing the experiment on the reciprocal cross confirms true imprinted expression, rather than a preference for expression from either the Bl6 or cast alleles. The relevant genomic DNA (gDNA) sequences of the parental strains and of a B × C hybrid are shown at the top of the figure. The allele on which the poly(A) site associated with each transcript is utilized is indicated below each panel of traces (maternal bias or paternal). Some of the base calls are presented as a mirror image; this reflects the sequencing of some amplicons in the reverse orientation and does not influence the interpretation of the data. (B) Quantification of allele-specific expression of Herc3a and Herc3c using pyrosequencing over Bl6/cast SNPs (see Materials and Methods for details). In neonatal brain, Herc3a is expressed with a bias from the maternal allele (62:38 maternal:paternal ratio) but is expressed approximately equally from the two alleles in heart (52:48 maternal:paternal ratio). This is the case for samples from both B × C and C × B intercrosses. As a control, Herc3c, shown in (A) to be paternally expressed in brain, shows expression exclusively from the paternally derived allele in C × B brain and >90% paternal expression in B × C brain, confirming the sensitivity of this approach in determining allele-specific expression. Error bars indicate the standard error of the percent maternal expression values from biological replicates. (C) Sequence traces showing that transcripts utilizing the downstream poly(A) site (Herc3a transcripts) are derived equally from the two parental alleles in heart and lung.
qRT-PCR
Custom TaqMan expression assays for amplification of Herc3a, Herc3b [using the Herc3b1 poly(A) site], Herc3c and Nap1l5 were ordered from ABI. The H13a assay has been described previously (8). cDNA was synthesized from 1 µg DNase I-treated total RNA isolated from pooled e8.5 wild-type or Dnmt3L−/+ embryos, using the method described earlier. For embryo qPCR, confidence intervals in Figure 3B represent three technical replicate reactions for each transcript and template, normalized to expression of β-actin (Actb). Reactions were prepared, run and analysed as described previously (16).
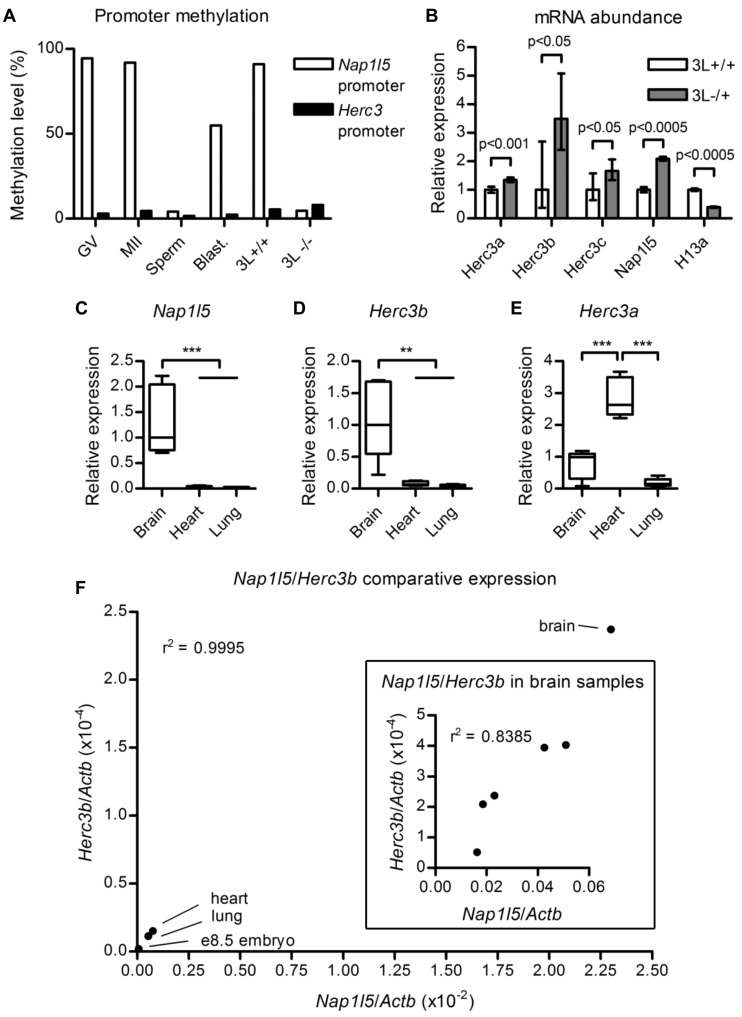
Figure 3.
Methylation and transcription analyses. (A) Percentage methylation at the internal Nap1l5 promoter and the 5′ Herc3 promoter in gametes and blastocyst (blast.). GV, germinal vesicle oocytes; MII, metaphase II oocytes. GV oocytes derived from females deficient for Dnmt3L (3L−/−) are hypomethylated at the Nap1l5 promoter relative to wild-type controls (3L+/+). The data are derived from a reduced representation bisulfite sequencing experiment performed in (24). (B) mRNA abundance determined by qRT-PCR on pooled Dnmt3L−/+ (3L−/+) e8.5 embryos relative to wild-type littermates (3L+/+), normalized to Actb. The mean values from three technical replicates are presented for each transcript. Error bars indicate the 95% confidence interval. Probability values were calculated using Student’s t test. (C) Relative abundance of Nap1l5 transcripts in brain, heart and lung. The bottom and top of the box represent the 25th and 75th percentiles, respectively; the internal line represents the median; and the whiskers represent the range of the data. Transcript abundance between the tissues was assessed using one-way analysis of variance. **P < 0.01, ***P < 0.001. (D,E) Relative abundance of Herc3b and Herc3a transcripts, as for (C). (F) Main image: Relationship between Nap1l5 expression level and use of the Herc3b1 poly(A) site in brain, heart, lung and wild-type e8.5 embryo. Inset: Association between the natural variation of Nap1l5 expression and use of the Herc3b1 poly(A) site in the brains of Day 1 littermates. Data points in the main graph indicate median values for each tissue, except for e8.5 embryo for which only one biological sample was assayed, representing cDNA from pooled embryos; this value corresponds to the mean of three pipetting replicates. Data points in the inset graph represent mean values from three pipetting replicates performed for each sample.
To compare expression of Nap1l5 and Herc3b in different tissues, cDNA was synthesized from 1 µg DNase I-treated RNA isolated from whole brains, hearts and lungs of five newborn (Day 1) B × C littermates. For each sample, reactions were performed in triplicate and normalized to expression of Actb.
Protein domain and open reading frame prediction
Herc3 transcript sequences were submitted to the EMBL SMART Tool (19,20) for domain prediction analysis. The open reading frame (ORF) of transcripts utilizing the Herc3b2 poly(A) site was predicted using NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). The ORFs of Herc3a, Herc3c and transcripts utilizing the Herc3b1 poly(A) site predicted by this program were consistent with the UCSC Known Genes annotation (21).
RESULTS
Locus organization and alternative poly(A) sites
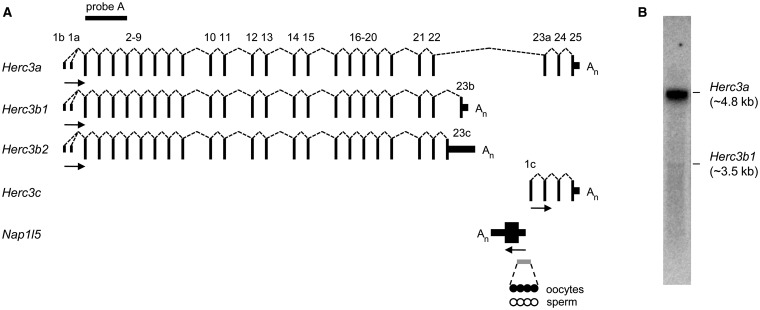
To characterize the locus fully, we focussed initially on brain where both Nap1l5 and Herc3 are abundantly expressed (12). Full-length Herc3 transcripts (Herc3a) consist of 25 exons, of which all except the first exon contribute to the ORF (Figure 1A). The UCSC Known Genes annotation (21) includes a truncated transcript isoform of Herc3 utilizing an alternative final exon (exon 23b) and internal poly(A) site. 3′ RACE (rapid amplification of cDNA ends) performed on whole brain cDNA confirms this annotation and we refer to this shorter isoform as Herc3b. Using 3′ RACE and RT-PCR, we identify an additional internal poly(A) site that is coupled to an alternative splicing event from exon 22 onto exon 23c (Figure 1A and Supplementary Figure 1A–C). We differentiate these internal poly(A) sites by calling them Herc3b1 and Herc3b2. Northern blotting with a probe complementary to exons 2–5 detects mRNA species consistent with the predicted sizes of transcripts utilizing the Herc3a and Herc3b1 poly(A) sites, but not the Herc3b2 poly(A) site, probably reflecting the low abundance of transcripts using this poly(A) site (Figure 1B). Processed Herc3a transcripts are ∼50× more abundant than Herc3b transcripts in whole brain. All Herc3 transcripts utilize canonical poly(A) signals: either AAUAAA (Herc3a and Herc3b2) or AUUAAA (Herc3b1) (Supplementary Figure 1D and data not shown).
Figure 1.
Transcription at the Herc3/Nap1l5 locus. (A) Transcripts at the locus, as determined by 3′ RACE, RT-PCR, northern blotting and expressed sequence tag data. Exons (numbered) are shown as non-coding (short vertical lines or solid boxes) or coding (tall vertical lines). Splicing events are illustrated by dashed lines. The direction of transcription is illustrated by horizontal arrows and polyadenylation sites by An. The gray bar represents the gDMR at the Nap1l5 promoter, that is methylated in oocytes (filled circles) and unmethylated in sperm (open circles). The position of probe A used in northern blotting is shown by a black bar. The diagram is not drawn to scale. (B) Northern blot of 2-week brain RNA using probe A, illustrating the relative abundance of transcripts utilizing the Herc3a and Herc3b1 poly(A) sites.
According to the UCSC Known Genes annotation, Herc3b utilizes a different initiation exon (exon 1b) to that utilized for Herc3a transcripts (exon 1a). However, by long-range PCR, we demonstrate that transcripts initiating at exon 1b can produce Herc3b transcripts or form full-length Herc3a transcripts (Figure 1A and data not shown), suggesting that the choice of poly(A) site is not dependent on the initiation exon used.
An additional Herc3 transcript, annotated as distinct by UCSC Known Genes, initiates from an alternative promoter positioned within intron 22 and splices from a unique first exon (exon 1c) onto exon 23a, producing a transcript of 2.2 kb referred to as Herc3c. The annotated transcription start site of the retrogene Nap1l5 is 306 bp upstream of the annotated transcription start site of Herc3c. However, there are expressed sequence tags that bridge this gap. Nap1l5 is transcribed in an anti-sense orientation to its host, and thus these data are consistent with a bidirectional promoter that is shared between Nap1l5 and Herc3c. Nap1l5 is monoexonic, like many products of retrotransposition, and its poly(A) site is downstream of Herc3 exon 23c (Figure 1A).
Herc3 poly(A) sites are utilized in an imprinted manner
Nap1l5 was identified as an imprinted gene in a genome-wide screen for maternal methylation (15), and we have previously shown that a promoter-associated CpG island (CGI) is differentially methylated in sperm and oocytes (9). Nap1l5 shares a number of features with the retrogene Mcts2, including a promoter-associated DMR and a position within an intron of a host gene that exhibits both upstream and downstream poly(A) sites. The host gene of Mcts2, H13, utilizes poly(A) sites in an imprinted manner. Given these similarities between the Mcts2 and Nap1l5 retrogenes, we examined each Herc3 transcript for imprinted expression. Indeed, imprinting of Herc3 has previously been suggested by RNA-seq experiments conducted on brain cDNA of inter-subspecies hybrid mice (22). In that study, exonic SNPs between the two parental strains (C57Bl6 [Bl6] and Mus musculus castaneus [cast]) indicated that most Herc3 exons are expressed from the maternally derived allele. However, exon 23b, specific to Herc3b, showed paternal allele-specific expression. To examine the imprinted status of each of the Herc3 transcripts independently, we designed primers specific to each variant. We amplified from whole neonatal brain cDNA from inter-subspecies hybrid mice and sequenced across SNPs.
In brain, full-length Herc3a transcripts are preferentially expressed from the maternally inherited allele, but there is a clear contribution from the paternally derived copy (Figure 2A). Conversely, transcripts utilizing the internal Herc3b1 and Herc3b2 poly(A) sites exhibit exclusive expression from the paternally inherited allele. Herc3c transcripts are also paternally expressed, consistent with bidirectional transcription initiation from the unmethylated allele of the Nap1l5 CGI promoter.
The biased expression of Herc3a from the maternally inherited allele is corroborated by results from pyrosequencing, a technique that permits quantification of allelic expression. In Bl6 × cast (B × C, where the maternal strain is presented first) brain samples, we find that the maternal allele contributes ∼62% of the Herc3a mRNA, and this result is reproduced in reciprocal C × B samples, confirming that this bias is due to parent-of-origin rather than a specific sequence (Figure 2B). As a control, Herc3c shows strong paternal allele-specific expression in brain (Figure 2B).
Together, these assays validate and extend the findings of (22), showing complex isoform-specific imprinting at this locus. Given that Herc3a and Herc3b transcripts initiate at a shared promoter, it is the poly(A) sites that are utilized in an allele-specific manner.
Maternal germline methylation influences Herc3 poly(A) site choice
To investigate the importance of DNA methylation at the Nap1l5 promoter in influencing Herc3 poly(A) site choice, we took advantage of mice with a mutation in the Dnmt3L gene. Female Dnmt3L−/− mice produce oocytes lacking maternal germline methylation at imprinting control regions (23). During development, embryos from Dnmt3L−/− mothers inappropriately either express or repress imprinted genes on both alleles, ultimately resulting in embryonic lethality after e9.5 (23). We first confirmed that methylation of the Nap1l5 promoter CGI is dependent on Dnmt3L using published reduced representation bisulfite sequencing data (24) (Figure 3A). Methylation levels of the Nap1l5 promoter in wild-type germinal vesicle (GV) and metaphase II (MII) oocytes exceed 90%, compared with 4% in sperm, confirming our previous observation of differential methylation in gametes (9). In wild-type blastocysts, methylation levels are ∼55%, consistent with differential methylation on the two alleles persisting after fertilization. GV oocytes depleted for Dnmt3L are <5% methylated at the Nap1l5 promoter, confirming the dependence of this gDMR on Dnmt3L activity. The CGI at the Herc3a/b promoter is <10% methylated in all cell types and stages examined.
The effects of loss of maternal germline methylation on Herc3 poly(A) site use were assessed by qRT-PCR on e8.5 Dnmt3L−/+ embryos and wild-type littermates, using primers specific to Nap1l5, Herc3a, Herc3c and transcripts using the Herc3b1 poly(A) site. Nap1l5 abundance in the Dnmt3L−/+ sample is ∼2.1× that of the wild-type control (Figure 3B), consistent with biallelic expression of this retrogene in the absence of promoter methylation on the maternally inherited chromosome. Additionally, expression of Herc3c increases by ∼1.7× in Dnmt3L−/+ embryos, providing further evidence that this transcript initiates from the differentially methylated promoter shared with Nap1l5.
Both Herc3a and Herc3b transcripts demonstrate an increase in abundance in Dnmt3L−/+ embryos. Herc3b transcripts are ∼3.5× more abundant in mutant embryos, compared with a modest increase in Herc3a abundance of ∼1.2×. In the case of H13/Mcts2, the increase in the use of upstream H13 poly(A) sites in Dnmt3L−/+ embryos is correlated with a reduction in expression of the full-length H13a form (8). This result is replicated in the present study, in which full-length H13a transcripts are reduced by ∼60% in Dnmt3L−/+ embryos (Figure 3B). The modest increase in the abundance of full-length Herc3a transcripts may be a consequence of a change in expression of a transcriptional regulator resulting from ablation of maternal germline methylation. However, given that both Herc3a and Herc3b initiate at the same promoter, the change in the Herc3a:Herc3b ratio between wild-type and Dnmt3L−/+ embryos reflects the use of alternative polyadenylation sites. Thus, allele-specific methylation at the Nap1l5 promoter influences Herc3 poly(A) site choice.
Herc3b and Nap1l5 expression levels are correlated, suggesting a role for transcriptional interference
Having established a role for DNA methylation in the regulation of alternative polyadenylation at the Herc3 locus, as well as at the H13 locus previously (8), we considered the mechanisms that could be responsible for effecting this regulation. It is interesting, and likely informative, that the use of alternative poly(A) sites at Herc3 is coupled to alternative splicing (Figure 1A and Supplementary Figure 1). Internal poly(A) sites are utilized exclusively with the incorporation of exons 23b or 23c into mature transcripts. The allele-specific inclusion of these exons may reflect differences in the activity of the transcription elongation complex on the two alleles. We reasoned that, ultimately, these differences in splicing and poly(A) site use could be caused by at least two mechanisms. The first is transcriptional interference resulting from retrogene transcription; and the second is through the binding of a methylation-sensitive polyadenylation factor at the gDMR. Transcriptional interference describes the direct negative impact of one transcriptional activity on a second transcriptional activity in cis (25). In the case of Nap1l5/Herc3, it is possible that the progression of the transcription elongation complex along the Nap1l5 gene body, or the accumulation of poised RNA polymerase II at the Nap1l5 promoter, may interfere with transcriptional elongation of Herc3.
Nap1l5 is most abundantly expressed in brain, with lower levels of expression observed in heart and lung (Figure 3C) (15,26). We hypothesized that if transcriptional interference from Nap1l5 is responsible for internal Herc3 polyadenylation, Herc3b abundance would also be greatest in brain compared with heart and lung. Using qRT-PCR, we demonstrate that Herc3b is indeed most abundant in newborn brain, where Nap1l5 transcription is greatest, and is considerably reduced in heart and lung (Figure 3D). This correlation between Nap1l5 and Herc3b cannot be attributed to an overall increase in transcription at the locus in brain, because levels of Herc3a do not follow this trend, being more abundant in heart than brain (Figure 3E). Tissue-specific differences in Nap1l5 expression between brain, heart, lung and wild-type e8.5 embryo show a positive correlation with the abundance of Herc3b transcripts (Figure 3F). Further, we show that the natural variation in Nap1l5 expression observed in brains isolated from wild-type littermates also positively correlates with Herc3b abundance (Figure 3F, inset). Although Herc3a and Herc3b transcripts initiate from the same promoter, the tissue-specific differences in Herc3b abundance are more reflective of Nap1l5 expression levels than of Herc3a.
The bias toward the expression of full-length Herc3a transcripts from the maternally derived allele, established earlier using Sanger sequencing and pyrosequencing, could be explained by the paternal allele-specific expression of Nap1l5, which might interfere with Herc3 transcriptional elongation. If this model of transcriptional interference is correct, lower levels of Nap1l5 transcription in heart and lung should correlate with a shift towards more equal Herc3a expression from both parental alleles. This prediction holds true, with biallelic Herc3a expression observed in both of these tissues using Sanger sequencing (Figure 2C). We confirmed this with a quantitative assessment in heart using pyrosequencing, which demonstrated an approximately 52:48 maternal:paternal ratio in both the B × C and C × B reciprocal intercrosses (Figure 2B).
DISCUSSION
The association between epigenetics and polyadenylation is an emerging topic with recent studies demonstrating that globally, at least in humans, poly(A) sites are depleted for nucleosomes and most post-translational histone marks, but are enriched for H3K9me3 (27,28). However, the importance of epigenetic marks in regulating alternative polyadenylation remains largely unexplored, in contrast to relatively abundant evidence for epigenetic control of alternative promoter choice and, more recently, of alternative splicing. Previously, we demonstrated a role for DNA methylation in the control of alternative polyadenylation at the H13 locus (8). Methylation at the promoter of an intronic retrogene, Mcts2, is specific to the maternal allele, causing Mcts2 to be imprinted. On the paternal allele, from which Mcts2 is expressed, H13 transcripts utilize upstream poly(A) sites. Whether this occurs due to retrogene-mediated transcriptional interference, or, alternatively, due to methylation-sensitive binding of a polyadenylation factor that is unrelated to retrogene transcription, has not been determined.
Intronic positioning within a multi-exonic gene is a common feature of several imprinted retrogenes (9,29). This is also true of retrogenes present in human but absent in mouse, such as the imprinted retrogene at the RB1 locus (30). Additionally, the imprinted human retrogene FAM50B probably lies in an intron of a non-coding RNA (BG721636) (31,32). That several imprinted retrogenes are intronic may reflect a requirement for transcription through gDMRs during maternal germline methylation establishment (24,33).
In this study, we show that the relationship between Mcts2 and H13 is not unique to that locus, but is also observed for the intronic retrogene Nap1l5 and its host Herc3 (Figure 4A). Loss of differential methylation on the parental alleles at the Nap1l5 promoter causes a change in the ratio of Herc3 transcripts using upstream and downstream poly(A) sites, demonstrating a role for epigenetic marks in regulating alternative polyadenylation. The allele-specific utilization of an internal Herc3 poly(A) site positively correlates with levels of Nap1l5 expression in different tissues, and between brain samples of wild-type littermates, but does not correlate with expression of full-length Herc3a, despite Herc3a and Herc3b initiating at the same promoter. Biallelic expression of Nap1l5 in Dnmt3L−/+ embryos also correlates with increased use of this internal poly(A) site. Further, in brain—the tissue in which Nap1l5 is most abundantly expressed—full-length Herc3a transcripts are expressed with a bias from the maternal allele on which Nap1l5 is silenced. In tissues where Nap1l5 expression is relatively low, Herc3a transcription is biallelic. These data support the hypothesis that polyadenylation can be regulated by transcriptional interference, although still cannot fully exclude the involvement of a methylation-sensitive polyadenylation factor that may demonstrate tissue-specific differences in activity. The coupling of alternative polyadenylation to the incorporation of alternative exons in Herc3 transcripts suggests that differences in the activity of transcription elongation complexes between the two alleles may form part of the mechanism. For example, the inclusion of weak alternative exons can reflect slow progress of an elongation complex (34). Our ongoing work is addressing the molecular mechanism by which methylation controls alternative polyadenylation.
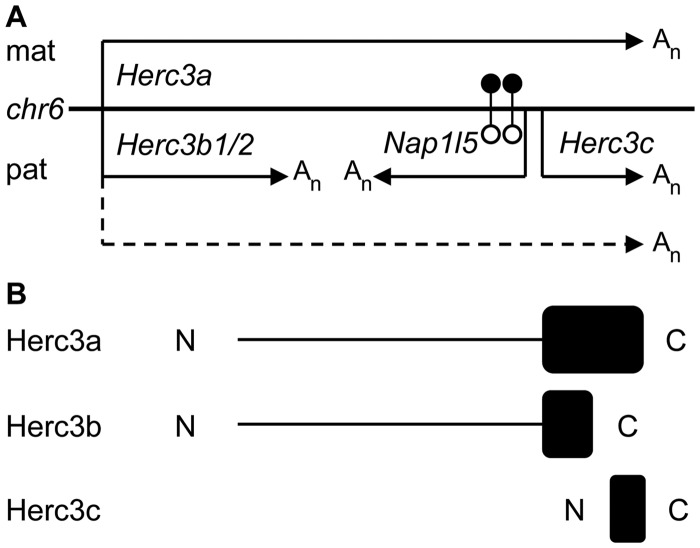
Figure 4.
Allele-specific alternative poly(A) at the Herc3/Nap1l5 locus in brain and putative proteins encoded by Herc3 transcripts. (A) In brain, Herc3a is predominantly expressed from the maternally inherited chromosome (mat) with a smaller contribution (illustrated by a dashed line) from the paternally derived copy (pat). The Nap1l5 promoter is methylated (filled lollipops) on the maternal allele and hypomethylated (open lollipops) on the paternal copy, from which Nap1l5 is expressed. The Nap1l5 promoter is likely to be a bidirectional promoter, permitting expression of Herc3c from the paternal allele. Upstream poly(A) sites are utilized by a subset of Herc3 transcripts on the paternal allele, producing Herc3b. Poly(A) sites are indicated by An. (B) Alignment of the predicted proteins encoded by Herc3a, Herc3b and Herc3c transcripts. Only Herc3a would encode a complete HECT domain (black box), important for Herc3 function as an E3 ubiquitin ligase. The N and C termini are indicated.
By northern blotting we found that, in brain, processed Herc3a transcripts represent 98% of transcripts initiating at the shared Herc3a/Herc3b promoter. Despite this, full-length Herc3a transcripts still exhibit biased expression from the maternal allele, suggesting that the internal poly(A) sites may be used by more than 2% of transcripts on the paternal allele. Many of the Herc3b transcripts may be degraded before processing, explaining their relatively low abundance as detected by northern blotting. Some of the contribution to Herc3a transcripts from the paternal allele may reflect cell type-specific imprinting, dependent on Nap1l5 expression. For example, Herc3 is abundantly expressed in the piriform cortex relative to expression of Nap1l5 (26), and this may enable biallelic expression of full-length Herc3a transcripts in these cells. However, Herc3 and H13 exhibit both maternally and paternally expressed transcripts within the same cell type. The difference between transcripts derived from the two parental alleles is the use of poly(A) sites. This is different to the imprinted gene Grb10, which gives rise to both maternal and paternal allele-specific transcripts but these are expressed in distinct cell types where they perform discrete functions (35,36). Grb10 transcripts differ in their use of promoters, but not poly(A) sites.
Human HERC3, like other HERC family members, contains a domain homologous to E6 associated protein carboxy-terminus (HECT) [reviewed in (37)]. Functional assays identify HERC3 as a probable E3 ubiquitin ligase that targets proteins for proteasomal-mediated degradation through its HECT domain, and co-localizes with proteins involved with intracellular transport (11). Murine Herc3 also contains a predicted HECT domain (Figure 4B). While Herc3b and Herc3c contain ORFs, only the Herc3a transcript would encode a full-length HECT domain necessary for ubiquitin ligase activity. Thus, the Herc3b and Herc3c transcripts may not be of direct biological importance, and this may explain the likely degradation of Herc3b transcripts discussed above. Further functional analyses are required to examine the roles of any peptides produced from these transcripts.
Recently, alternative polyadenylation of the imprinted gene Mest has been shown to influence imprinted expression of a neighboring gene (38). In this system, alternative polyadenylation produces an extended transcript of Mest, called MestXL, in the central nervous system. MestXL is expressed from the paternally inherited chromosome and transcribes into the downstream anti-sense gene Copg2. Copg2 is imprinted exclusively in the central nervous system and is expressed from the maternally inherited chromosome. Experimental truncation of MestXL causes loss of imprinting of Copg2, indicating that tissue-specific alternative polyadenylation regulates Copg2 imprinting probably by transcriptional interference. This is different to the situation with Herc3/Nap1l5 and H13/Mcts2, in which imprinted retrogene expression controls alternative polyadenylation of the host. Together, these studies illustrate the intimate, allele-specific relationships between neighboring transcripts.
We have shown, in this and a previous study, that transcript poly(A) site choice can be regulated by DNA methylation at internal CGIs. Allele-specific histone modifications are also likely to be important in this system as we have previously shown enrichment at Nap1l5 and Mcts2 of active and repressive modifications on the paternally and maternally inherited copies, respectively (39). Our work on elucidating the mechanism through which DNA methylation controls alternative polyadenylation at imprinted retrogene loci will explore the importance of other epigenetic marks.
While the relationship between Herc3 and Nap1l5, as well as H13 and Mcts2, is complicated by the imprinted nature of the retrogenes, the intimate host/retrogene relationship may not be limited to imprinted loci. Indeed, a similar situation has recently been reported at sites of mouse endogenous retrovirus (ERV) integration, where an intronic insertion can cause upstream transcriptional termination (40). In at least one case, an intronic ERV promoter exhibits variable methylation between individuals (41). When the promoter is unmethylated, the ERV is expressed and the host gene utilizes upstream poly(A) sites. However, there is no evidence for imprinting at this locus and poly(A) sites are utilized differently between individuals. Using global approaches, it will be important to evaluate the extent to which retrogenes and retrotransposons influence poly(A) site choice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figure 1.
FUNDING
Wellcome Trust [ref. no. 085448/Z/08/Z to R.J.O.]; Research Councils UK Fellowship (to R.S.); Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Deborah Bourc'his for the e8.5 Dnmt3L−/+ and wild-type embryo RNA. The authors thank the staff at the King’s College London Genomics Facility for technical support in performing pyrosequencing.
REFERENCES
- 1.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 4.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 6.Lou H, Neugebauer KM, Gagel RF, Berget SM. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol. Cell. Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:e20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowley M, Oakey RJ. Retrotransposition and genomic imprinting. Brief Funct. Genomics. 2010;9:340–346. doi: 10.1093/bfgp/elq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz C, Ventura F, Bartrons R, Rosa JL. HERC3 binding to and regulation by ubiquitin. FEBS Lett. 2001;488:74–80. doi: 10.1016/s0014-5793(00)02371-1. [DOI] [PubMed] [Google Scholar]
- 12.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attia M, Rachez C, De Pauw A, Avner P, Rogner UC. Nap1l2 promotes histone acetylation activity during neuronal differentiation. Mol. Cell. Biol. 2007;27:6093–6102. doi: 10.1128/MCB.00789-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attia M, Forster A, Rachez C, Freemont P, Avner P, Rogner UC. Interaction between nucleosome assembly protein 1-like family members. J. Mol. Biol. 2011;407:647–660. doi: 10.1016/j.jmb.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 15.Smith RJ, Dean W, Konfortova G, Kelsey G. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 2003;13:558–569. doi: 10.1101/gr.781503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz R, McCole RB, Woodfine K, Wood AJ, Chahal M, Monk D, Moore GE, Oakey RJ. Transcript- and tissue-specific imprinting of a tumour suppressor gene. Hum. Mol. Genet. 2009;18:118–127. doi: 10.1093/hmg/ddn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellaker C, Goodstadt L, Nicod J, Bhomra A, et al. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477:326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 22.Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 24.Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shearwin KE, Callen BP, Egan JB. Transcriptional interference—A crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies W, Smith RJ, Kelsey G, Wilkinson LS. Expression patterns of the novel imprinted genes Nap1l5 and Peg13 and their non-imprinted host genes in the adult mouse brain. Gene Expr. Patterns. 2004;4:741–747. doi: 10.1016/j.modgep.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Khaladkar M, Smyda M, Hannenhalli S. Epigenomic and RNA structural correlates of polyadenylation. RNA Biol. 2011;8:529–537. doi: 10.4161/rna.8.3.15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowley M, de Burca A, McCole RB, Chahal M, Saadat G, Oakey RJ, Schulz R. Short interspersed element (SINE) depletion and long interspersed element (LINE) abundance are not features universally required for imprinting. PLoS One. 2011;6:e18953. doi: 10.1371/journal.pone.0018953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanber D, Berulava T, Ammerpohl O, Mitter D, Richter J, Siebert R, Horsthemke B, Lohmann D, Buiting K. The human retinoblastoma gene is imprinted. PLoS Genet. 2009;5:e1000790. doi: 10.1371/journal.pgen.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang A, Skaar DA, Li Y, Huang D, Price TM, Murphy SK, Jirtle RL. Novel retrotransposed imprinted locus identified at human 6p25. Nucleic Acids Res. 2011;39:5388–5400. doi: 10.1093/nar/gkr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakabayashi K, Trujillo AM, Tayama C, Camprubi C, Yoshida W, Lapunzina P, Sanchez A, Soejima H, Aburatani H, Nagae G, et al. Methylation screening of reciprocal genome-wide UPDs identifies novel human-specific imprinted genes. Hum. Mol. Genet. 2011;20:3188–3197. doi: 10.1093/hmg/ddr224. [DOI] [PubMed] [Google Scholar]
- 33.Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Arnaud P, Monk D, Hitchins M, Gordon E, Dean W, Beechey CV, Peters J, Craigen W, Preece M, Stanier P, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 2003;12:1005–1019. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- 36.Garfield AS, Cowley M, Smith FM, Moorwood K, Stewart-Cox JE, Gilroy K, Baker S, Xia J, Dalley JW, Hurst LD, et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 2011;469:534–538. doi: 10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Gonzalo FR, Rosa JL. The HERC proteins: functional and evolutionary insights. Cell Mol. Life Sci. 2005;62:1826–1838. doi: 10.1007/s00018-005-5119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacIsaac JL, Bogutz AB, Morrissy AS, Lefebvre L. Tissue-specific alternative polyadenylation at the imprinted gene Mest regulates allelic usage at Copg2. Nucleic Acids Res. 2012;40:1523–1535. doi: 10.1093/nar/gkr871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monk D, Arnaud P, Frost JM, Wood AJ, Cowley M, Martin-Trujillo A, Guillaumet-Adkins A, Iglesias Platas I, Camprubi C, Bourc'his D, et al. Human imprinted retrogenes exhibit non-canonical imprint chromatin signatures and reside in non-imprinted host genes. Nucleic Acids Res. 2011;39:4577–4586. doi: 10.1093/nar/gkq1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Akagi K, Hu Y, Trivett AL, Hlynialuk CJ, Swing DA, Volfovsky N, Morgan TC, Golubeva Y, Stephens RM, et al. Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance. Genome Res. 2012;22:870–884. doi: 10.1101/gr.130740.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32: 5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.