Abstract
MicroRNAs (miRNAs) are released from cells in association with proteins or microvesicles. We previously reported that malignant transformation changes the assortment of released miRNAs by affecting whether a particular miRNA species is released or retained by the cell. How this selectivity occurs is unclear. Here we report that selectively exported miRNAs, whose release is increased in malignant cells, are packaged in structures that are different from those that carry neutrally released miRNAs (n-miRNAs), whose release is not affected by malignancy. By separating breast cancer cell microvesicles, we find that selectively released miRNAs associate with exosomes and nucleosomes. However, n-miRNAs of breast cancer cells associate with unconventional exosomes, which are larger than conventional exosomes and enriched in CD44, a protein relevant to breast cancer metastasis. Based on their large size, we call these vesicles L-exosomes. Contrary to the distribution of miRNAs among different microvesicles of breast cancer cells, normal cells release all measured miRNAs in a single type of vesicle. Our results suggest that malignant transformation alters the pathways through which specific miRNAs are exported from cells. These changes in the particles and their miRNA cargo could be used to detect the presence of malignant cells in the body.
INTRODUCTION
MicroRNAs (miRNA) are short non-coding RNA molecules that modulate the activity of specific mRNA targets in normal development and disease, typically by compromising messenger RNA (mRNA) stability. MiRNAs are released by cells in a variety of vesicles or associated in complexes with proteins [reviewed in (1)]. Exosomes were the first extracellular vesicles shown to contain miRNA (2–12). Exosomes originate from multivesicular bodies (MVBs) of the endosomal compartment and may contain miRNA as a consequence of loading into the RNA-induced silencing complex and unloading at the MVBs (13,14).
The association of miRNA with exosomes is significant, in that exosomes can transfer cancer-specific molecules to other cells (15,16). Through this transfer of material, exosomes have been shown to contribute to tumor progression (Duelli et al., 2005), modification of the microenvironment (17) and the host (18), through induction of angiogenesis, production of niches for metastasis (19,20) and immune modulation (21–25). The packaging and export of miRNAs from cells and delivery of them to other cells may be important processes for extracellular miRNA-signaling pathways, although these mechanisms are not well understood.
Exosomes are perhaps the best studied of extracellular vesicles, although some cells, in particular tumor cells, actually release a heterogeneous population of microvesicles of uncertain subcellular origin (reviewed in (1,26)). Recent work suggests that the profile of microvesicles released from cells may be indicative of the cell type or cell state and may play an important role in cell–cell transfer and communication. For example, solid tumor cells, including breast cancer cells, release heterogeneous microvesicles that contain a variety of molecules (27,28) that can be transferred to recipient cells and influence signaling pathways (29). Although miRNA has been established as cargo of exosomes, the miRNA distribution in cells that release multiple vesicles is not known.
We recently demonstrated that miRNAs are released from breast cancer cells in a selective manner. We have identified three categories of released miRNA in mammary epithelial cells, which were classified based on the ratio between the amount of miRNA released from the cells and the amount retained in the cell (Table 1) (30). The first group is selectively released miRNAs, or ‘s-miRNA’. These miRNAs are characterized by being released excessively from breast cancer cells with relatively low concentrations of miRNA remaining in the cell. Alternatively, normal cells release nearly none of these miRNAs (30). Another group of miRNAs is in equal abundance within the cell and extracellularly, i.e. neutrally released miRNA, ‘n-miRNAs’ (30). N-miRNAs include putative biomarkers miR-16, miR-21 and other miRNAs for which the abundance in microvesicles reflects the increased abundance in the malignant cells of origin.
Table 1.
MiRNA that are differentially retained and exported in benign and malignant mammary epithelial cells (30)
| Release | Species | Benign |
Malignant |
||
|---|---|---|---|---|---|
| Cell | Microvesicle | Cell | Microvesicle | ||
| Neutral | miR-16 | +++ | ++ | +++ | ++ |
| miR-720 | ++ | ++ | +++ | +++ | |
| Selective | miR-451 | + | – | +/- | ++ |
| miR-1246 | + | – | + | ++ | |
Neutrally released miRNAs (n-miRNAs): the most consistently released miRNA of human mammary epithelial cells irrespective of malignancy is miR-16. MiR-720 is the most abundantly released miRNA of breast cancer cells tested. It is consistently released from both benign and malignant mammary epithelial cells. Selectively released miRNAs (s-miRNAs): cellular concentrations of miR-1246 and miR-451 are similar in benign and malignant cells; however, the release of these miRNAs is selective and excessive from malignant cells. + relative abundance in population by qRT–PCR and array; – not detectable in microvesicles; ± near threshold of detection.
The selectivity of release of individual miRNAs, and thereby their categorical grouping, differs depending on the cell type (6,9,12,30,31). In particular, selection is affected by malignant transformation. For example, malignant mammary epithelia release >99% of miR-451 and miR-1246 produced by the cells, while in benign epithelial cells these miRNAs are mostly retained (30). Both of these s-miRNAs are linked to cancer. MiR-451 is a tumor suppressor (32), affecting proliferation (33,34) and cell polarity (35), by deregulating several oncogenic pathways (36–39). MiR-451 also induces sensitivity to chemotherapeutic drugs (40,41), allows adaptation to metabolic stress (42,43) and can induce endocrine resistance in breast cancer cells (44). MiR-1246 induces p53-dependent apoptosis triggered by DNA damage (45). The changes in the release of cancer-related miRNAs may suggest a role for selective miRNA export in malignant transformation, and it may provide a cancer signature within the exported, circulating miRNA population.
N-miRNAs include miR-16 and miR-720 (30). MiR-16 is one of the first miRNAs identified to have tumor-suppressor activity (46,47), and circulating miR-16 is currently being evaluated as a serum marker for diagnosis or prognosis of several cancers (48,49) and pre-malignant syndromes (50). MiR-16 regulates chemosensitivity (51), apoptosis (52) and the cell cycle (53), while its biogenesis is induced by radiation (54) and is modulated by p53 (55). MiR-720 is the most abundantly released miRNA from malignant mammary epithelial cells (30). MiR-720 is up-regulated in pre-neoplastic syndromes (56) and has roles in p63 regulation and cell differentiation (57–59). The importance and roles of these vesicular miRNAs in cancer and in the extracellular environment are beginning to emerge. However, much remains to be determined regarding the nature of the vesicles that shuttle these miRNAs outside of the cell.
The observations that cells can selectively release miRNAs and also release a heterogeneous population of vesicles raise the possibility that the differential release of miRNAs is associated with different microvesicles. We thus analyzed the nature of the microvesicles that are associated with s-miRNAs. We find that s-miRNAs and n-miRNAs are released from cells in different types of particles. The identification and purification of sub-populations of extracellular microvesicles that carry distinct miRNA cargo allows for the possible assignment of the subcellular origin, extracellular function and cellular targets of these vesicles, as well as for the development of cancer diagnosis and prognosis based on the association of specific miRNA with specific extracellular vesicles.
MATERIALS AND METHODS
MDA-MB-231 and MCF7 cells were purchased from ATCC (Manassas, VA, USA) and maintained according to the provider’s recommendations. Normal skin fibroblasts were a gift from Dr. Beverly Davidson, University of Iowa, and were grown in Minimum Essential Medium Eagle With Earle’s salts (MEME) supplemented with Nonessential amino acids (NEAA), glutamine and pyruvate.
Cell culture
Exosomes and other particulates were collected from cells in culture as described (30). In brief, after 5 days of culturing in defined media, media were collected, centrifuged at 300g for 15 min and passed through a 0.45-µm filter (Pall Acrodisc, Cornwall, UK) to remove cell debris. The supernatant was centrifuged at 70 000g to collect particulates including exosomes and re-suspended with 100 µl phosphate buffered saline (PBS). Cells for miRNA analysis were cultured in defined media for 5 days prior to collection. Routinely breast cancer cell lines (1.2–1.6 × 109 cells) were maintained in Nunc Cell Factories (Nunc, Rochester, NY, USA) or 15 cm plates for each experiment.
Defined media
Defined media were used to replace media containing fetal bovine serum for vesicle and particle collection. Defined media were supplemented with Nutridoma-SP (Roche Applied Science, Indianapolis, IN, USA), Na-pyruvate, non-essential amino acids and L-Glutamine (all Mediatech, Manassas, VA, USA) in DMEM/high glucose (HyClone, Logan, UT, USA) in the absence of serum. MCF-7 cells were cultured as described (30).
P70/S70 preparation
Conditioned media or milk was cleared of cells and cell debris by low-speed centrifugation (300g) and filtration (0.45 µm), followed by concentration of the remaining particulates at 70 000g for 1 h. The pellet was washed in PBS by re-suspension and centrifuged again at 70 000g. We chose this centrifugation speed over conventional higher speed preparations (60) because vesicles prepared at higher speeds do not retain complete biological activity (61).
Gradient preparation and centrifugation
Sucrose density gradients ranging from 2.0 to 0.25 M sucrose in PBS were prepared. P70 samples were re-suspended in 100 µl PBS, diluted to 1 ml in 2.5 M sucrose and loaded into the bottom of the gradient. A 1 ml 2.5 M sucrose cushion was loaded below the sample. Sucrose gradients were centrifuged at 100 000g ranging from 0 h to equilibrium (48–90 h).
Negative-staining electron microscopy
All samples were adsorbed to Formvar-coated grids followed by negative staining with 2% (w/v) sodium phosphotungstate. Samples of sucrose gradient fractions were passed through MicroSpin G-25 columns (GE Healthcare, Piscataway, NJ, USA) to remove sucrose for microscopy prior to fixation. Micrographs were visualized using a transmission electron microscope (JEM-2100, Jeol, Tokyo, Japan).
Dot blots and western blots
Dot blots and western blots were prepared using antibodies as described (30). Antibodies against CD147, CD55, CD59, CD63, CD81, H2Ax and γ-H2Ax (Millipore, Billerica, MA, USA), CD44H (CD44s, R&D Systems, Temecula, CA, USA), Glut-1 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), CD98 (Pharmingen, BD Biosciences, San Diego, CA, USA) and human leucocyte antigens (HLA) (AbD Serotec, Raleigh, NC, USA) were used. In brief, P70s and other preparations were re-suspended in PBS and blotted onto Immobilon FL (Millipore) or nitrocellulose (Whatman, Kent, UK) using a Bio-Dot Filtration Apparatus (BioRad, Hercules, CA, USA). For western blots, 500 µl of each sucrose gradient fraction was diluted to 12 ml in PBS and pelleted at 100 000g for 2 h. The pellet was re-suspended in 100 µl Laemmli Buffer and 20 µl were loaded per lane onto an 18% sodium dodecyl sulphate (SDS)–polyacrylamide gel electrophoresis mini-gel and then electrophoresed at 200 V for 45 min. The gel was then transferred to Immobilon-FL (Millipore). Both fluorescently tagged secondary antibodies and horseradish peroxidase (HRP)-tagged secondary antibodies were used. For both dot blots and western blots, antibody binding was quantified using a Typhoon 9400 (GE Healthcare, Piscataway, NJ, USA) and goat-anti-mouse IgG-Alexa 488 or goat-anti-rabbit IgG-Alexa 488 (Invitrogen, Grand Island, NY, USA) and ImageQuant T software. Antigenicity was determined by quantifying antibodies bound to dot-blotted P70. Subsequently, the blots were developed for HRP activity using Luminata Classico Western HRP substrate (EMD Millipore Corporation).
Immunoprecipitation
Anti-CD59 antibodies (Millipore) were bound to Dynabeads (Invitrogen) in PBS. P70 was applied to CD59-bound Dynabeads. Supernatant containing CD59-depleated P70 was captured on magnets, washed with PBS and re-suspended in Laemmli Buffer. Captured particles were lysed from Dynabeads in Laemmli Buffer.
RNA extraction
A 300 µl aliquot of each sucrose fraction, or Dynabeads in PBS, was added to 500 µl Trizol reagent according to the manufacturer’s instructions (Invitrogen). A synthetic RNA (SYNTH, 250 fmol/µl (62)) was added as indicated as a recovery control.
MiRNA detection
End-point PCR
Ten microliters of the 20 µl total RNA preparation was used as input into a Superscript III (Invitrogen) reverse transcriptase (RT) reaction with miRNA-specific stem–loop primers as described (30).
Standardization and normalization
To ensure standardized input, exactly 300 µl of each 1 ml fraction collected from the gradients (1–12) was extracted using TRIzol as described (30,62). To assess recovery and stability of RNA, we spiked each fraction with an identical amount of SYNTH RNA (30,62) during RNA extraction. The RNA abundance in each fraction was normalized to SYNTH RNA recovery.
Quantitative PCR reactions were performed using TaqMan and miRNA-specific primers (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s instructions. Copy DNA produced in the RT reaction was amplified in MicroAmp™ optical 96-well reaction plates in triplicate 20 µl reactions on an Applied Biosystems 7900HT Thermocycler (ABI). Raw data were analyzed with SDS Relative Quantitation Software version 2.2.3 (ABI) using the Delta–Delta CT method. Absolute miRNA abundance was calculated by comparing CT values of samples to dilutions of a synthetic DNA corresponding to the complementary DNA produced by RT for each miRNA measured (63) to make a standard curve or by using an RNA oligo as a standard as described (30,62).
5′ End labeling of RNA
RNA (∼50 pmol) isolated from MDA-MB-231 P70 gradient fractions was enriched for small RNAs using PureLink followed by treatment with with Antarctic phosphatase (NEB, Ipswich, MA) according to manufacturers protocol. Dephosphorylated RNA was 5′ end labeled using T4 polynucleotide kinase (-3′ phosphatase minus; NEB) and 32P-γ-adenosine triphosphate (ATP) according to the manufacturers instructions. Radiolabeled RNA was separated on a 12% urea-polyacrylamide gel.
Size quantitation of particles
Particle sizes were determined from electron microscopy (EM) micrographs using NIS-Elements (Nikon, Melville, NY). At least 100 particles were counted for each sample.
Milk
Milk was donated from the Mother’s Milk Bank of Iowa (The University of Iowa Children’s Hospital) and the Indiana Mother’s Milk Bank, Inc. Eight hundred microliters of milk from three mothers each was analyzed to determine miRNA composition. The study was approved by the institutional review board of the Rosalind Franklin University of Medicine and Science.
In situ hybridization and immunofluorescence
In brief, MDA-MB-231 cells were grown on coverslips, fixed in paraformaldehyde and washed in imidazole buffer; miRNAs were immobilized with EDC 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide buffer (64) and hybridized with 4 pmol of biotinylated miRCURY LNA probes (Exiqon, Woburn, MA) for 16 h at 42°C and probed with avidin to detect LNA probes (65). Subsequently, cells were probed for CD63 using conventional immunofluorescence approaches (61). Cells were imaged on a Nikon Eclipse 80i confocal microscope.
Transduction
MDA-MB-231 cells were transduced with Green Fluorescent Protein (GFP), caspase-8DN, caspase-9DN or BCL2 in pBABEpuro by retroviral transduction, selected for puromycin resistance and expression of proteins confirmed as described (66).
Induction of cell death
MDA-MB-231 cells were treated with 100 µM etoposide for 24–72 h as described (66), or left untreated, and cell death was quantified using trypan blue.
RESULTS
Breast cancer cells release a variety of particles
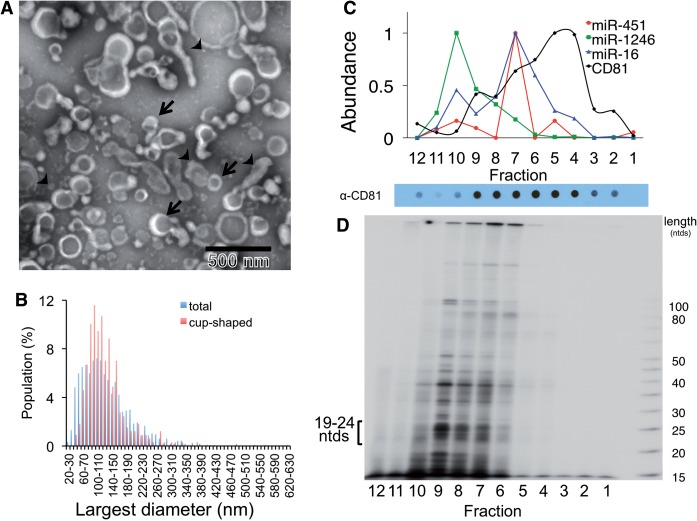
The differential release of s-miRNAs and n-miRNAs from cells could be a consequence of the mode of export. Thus, we first tested whether s-miRNAs are associated with the same extracellular particles as n-miRNAs. To do so, we performed a crude microvesicle preparation of MDA-MB-231 cells by filtration and ultracentrifugation, which yields a sample called P70 (30). We analyzed the P70 by EM and found that this preparation contained a variety of particles. Some particles had a cup shape and size (50–100 nm) typical of exosomes (Figure 1A). Other particles were either cup shaped or spherical and a wide range of sizes, measuring 20–410 nm in diameter (Figure 1B). These results indicate that malignant mammary epithelial cells release a variety of vesicles and particles into the extracellular environment. The presence of a multitude of distinct particles in the extracellular environment of breast cancer cells raises the question whether specific miRNA subspecies associate with specific microvesicles.
Figure 1.
MiRNAs exported from breast cancer cells associate with a complex population of particles. (A) Particulate complexes collected from MDA-MB-231 cells grown in defined media were cleared of debris, filtered, collected by centrifugation at 70 000g (P70) and imaged by negative-stain EM. The preparation contains cup-shaped vesicles (as indicated by arrows) and structures of different shapes (arrowheads). (B) The largest diameter (ø) of >500 structures was measured and plotted. (C) A P70 preparation of MDA-MB-231 cells was subjected to buoyant sucrose gradient centrifugation for 18 h, and 1 ml gradient fractions (12, bottom of gradient, 1, top) were assayed for miRNA and CD81 abundance. The miRNA species of each gradient fraction were quantified using TaqMan qRT–PCR, and each miRNA species is plotted as a proportion of the fraction with maximum miRNA abundance, set at 1, and CD81 was measured using a dot blot probed with anti-CD81 antibody, and binding of a secondary goat-anti-mouse IgG-Alexa 488 was measured by fluorescence quantitation as described in Materials and Methods. A dot blot developed for secondary HRP antibody binding is shown below the graph. (D) Total small RNAs were visualized after 32P end-labeling and separation by native PAGE. 19–24 ntds: RNA species with expected size for miRNAs.
MiRNAs associate with sub-populations of microvesicles
This heterogeneity of released particles from breast cancer cells (Figure 1A and B) is in contrast to the homogenous population of exosomes released by normal fibroblasts (61). Furthermore, normal fibroblasts export s-miRNAs neutrally rather than selectively (30). Therefore, we hypothesized that the diversity of vesicles could be related to the differences in the assortment of the released miRNAs in breast cancer cells. To test this idea, we determined the distribution of small RNAs among the vesicles. To do so, the P70 microvesicle preparation was separated by buoyant sucrose gradient centrifugation, which separates particles based on buoyant density (Figure 1C). RNA was isolated from all gradient fractions and the abundance of n-miRNAs and s-miRNAs in each fraction was measured by quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR) or gel electrophoresis. We found that distinct miRNAs associated with different fractions of the gradient. For example, the s-miRNA miR-1246 was present in different fractions than the other miRNAs. Furthermore, none of the measured miRNAs co-migrated with the bulk of microvesicles, as indicated by the lack of miRNA in the fractions most enriched in the cell-surface antigen CD81, a marker of many microvesicles (Figure 1C). This result supports the idea that only sub-populations of vesicles contain miRNA (67).
In order to profile the total small RNA population in each of these fractions, RNA was isolated from the P70, 5′-end labeled with radioactive ATP and imaged by autoradiography following separation on a denaturing polyacrylamide gel (Figure 1D). As is the case for miRNA, different small RNA species were enriched in different fractions; portions of the CD81-positive vesicle population were largely devoid of small RNA species (Figure 1C and D). Therefore, we conclude that different miRNAs associate with different particles and that the measured miRNAs associate with the bulk of other small RNAs exported in vesicles and particles.
Selectively and neutrally exported miRNAs associate with different released particles
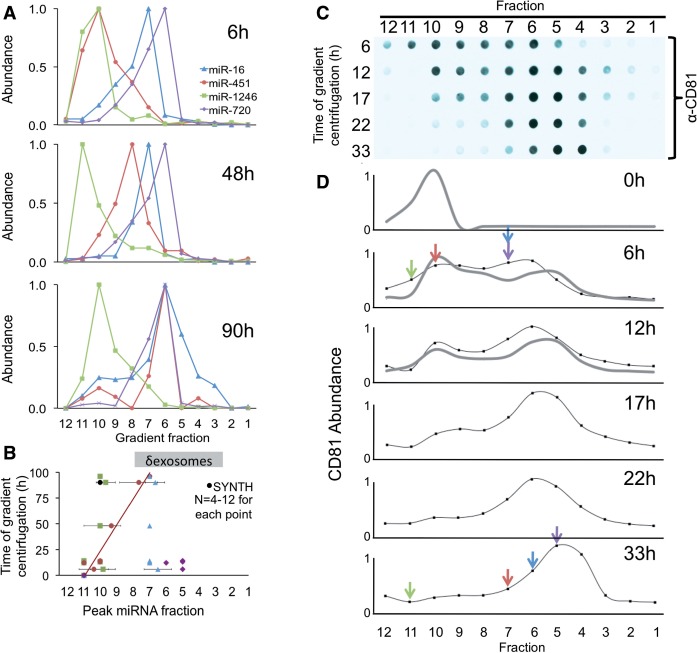
In order to systematically define the different particles released from breast cancer cells and their miRNA cargo, we developed a method to purify each particle type. This approach named ‘differential buoyant velocity centrifugation’, fractionates released vesicles by both size and density (Supplementary Figure S1, ‘Materials and Methods’ section). Using this approach, we found that each of the measured miRNAs was associated with a distinct gradient fraction, as determined by end-point RT–PCR (Supplementary Figure S2A) and qRT–PCR analysis of RNA isolated from gradient fractions (Figure 2A and Supplementary Figure S2B). A similar result was obtained with another breast cancer cell line, MCF7 (Supplementary Figure S3). These results demonstrate that each released miRNA associates with a distinct and specific sub-population of particles released by breast cancer cells.
Figure 2.
Selectively exported MiRNAs associate with different complexes than commonly released MiRNAs. (A) P70 preparations of particles exported by MDA-MB-231 cells were separated by buoyant speed centrifugation for the indicated number of hours, and the miRNA species were measured using TaqMan qRT–PCR. Each miRNA species is plotted as a proportion of the fraction with maximum miRNA abundance, set at 1. (B) Plot of peak fractions in gradients from 0 to 100 h (∂exosomes indicated the gradient density expected of exosomes [1.13–1.19 g/ml, Supplementary Figure 1]). Buoyant velocity was as follows: neutrally released MiR-16 and miR-720: 0.78 and 1.0 cm/h, respectively. Selectively exported miRNA miR-451 and miR-1246: 0.04 and 0.2 cm/h, respectively, SYNTH RNA (0.01 cm/h). n = 4–12 for each point. Error bars: standard deviation from the mean. The black symbol represents naked SYNTH RNA added to the gradient. (C) Gradient fractions (12, bottom of gradient, 1, top) as in (A) were quantified for the abundance of an extracellular vesicle surface antigen, CD81 by dot blot, and the antigen abundance was measured using fluorescent quantitation. Data are plotted in (D). Colored arrows mark peak miRNA fractions as indicated in (A). CD81 is plotted as a proportion of the fraction with maximum CD81 abundance, set at 1. Colors correspond to the same miRNA species as in (A) (miR-16 blue, miR-451 red, miR-1246 green and miR-720 purple). The gray and the black traces are from two independent experiments.
MiRNA sub-populations associate with extracellular particles of different density
To identify and characterize the particles that are associated with each of the miRNAs, we measured the buoyancy mass and buoyant velocity of the different miRNA-enriched particles. The measured density of both neutrally released miR-16/miR-720 and selectively exported miR-451 ranged from 1.13 to 1.18 g/ml (Figure 2B and Supplementary Figure S1). This density is defining for exosomes (68), yet a major difference of the s-miRNA miR-451 is that it migrated to that equilibrium density much slower than the n-miRNAs MiR-16 and miR-720 (Figure 2A and B). Furthermore, the n-miRNAs and the s-miRNAs co-migrated with the microvesicular surface marker CD81 (Figure 2C and D). Together, these results indicate that neutrally and selectively exported miRNAs are packaged in different exosome-like vesicles.
MiR-1246 had an equilibrium density of 1.25 g/ml, demonstrating that miR-1246 is not an exosomal RNA (Figure 2A and B) because exosomes, by definition, have a density 1.19 g/ml or less (68). The fact that miR-1246 exhibited minimal movement in the gradient raises the possibility that it is a non-vesicular miRNA. To test whether extracellular miR-1246 is contained within a vesicle or not, we compared its behavior on the gradient with that of a synthetic 22 nucleotide RNA (SYNTH) (62), which was added to the P70 prior to gradient centrifugation. SYNTH RNA had density equilibrium of 1.28 g/ml (Figure 2A), suggesting that the s-miRNA miR-1246 is released from cells as a complex less dense than naked RNA but denser than exosomes.
Taken together, these data indicate, that rather than being randomly distributed, miRNAs whose release is regulated by malignant transformation, associate with particles that are different than the particles associated with miRNAs whose release is not affected by malignant transformation.
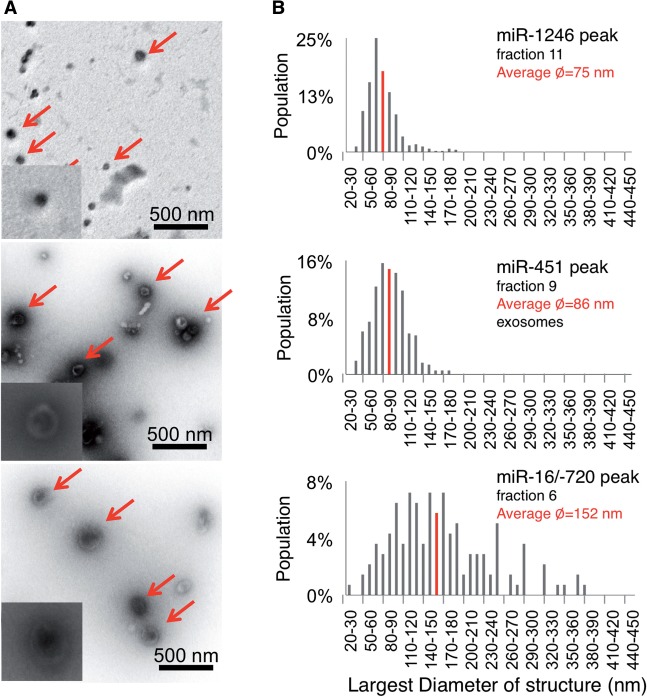
n-miRNAs and s-miRNAs associate with particles of different shape and size
To distinguish physical characteristics of the particles that are associated with each of the extracellular miRNAs, we analyzed them by EM (Figure 3A). Both s-miRNA miR-451 and n-miRNAs miR-16/miR-720 co-migrated with cup-shaped vesicles, suggestive of exosomes. However, the s-miRNA vesicles were about 86 nm in diameter, whereas n-miRNA vesicles were twice this size (Figure 3A and B). The s-miRNA miR-1246 peak fraction was enriched in spheres that were smaller than the miR-451 vesicles (Figure 3A). The association of these miRNAs with the identified particles was confirmed by the finding that these miRNAs co-partitioned with the visualized particles following filtration (Supplementary Figure S4A). The miRNAs were also resistant to ribonuclease treatment, suggesting that they are protected within vesicles or by vesicle proteins (Supplementary Figure S4B and S4C).
Figure 3.
Selectively and neutrally exported MiRNAs associate with different particles. (A) Peak miRNA fractions of a 48-h gradient were subjected to negative EM staining. Arrows indicate the typical structures detected in these preparations, and magnified in insets. (B) A plot of the largest diameters of the structure population depicted in (A). At least 200 structures were quantified for each fraction.
Overall, our data demonstrate that selectively released miR-451 associates with vesicles that are the shape and size of exosomes, while miR-1246 associates with unrelated spherical particles. In contrast, neutrally released miR-16 and miR-720 of mammary epithelial cells are released in exosome-like vesicles >100 nm.
CD44 distinguishes L-exosomes from conventional exosomes
Vesicles >100 nm are typically not considered to be exosomes (68) but may be referred to as exovesicles, ectosomes or microvesicles (69). Of these, exovesicles and ectosomes differ from exosomes in that they originate from the plasma membrane rather than the MVB and hence are devoid of the endosomal marker CD63 (69–72). To help determine the origin of the large vesicles we identified, we probed for CD63 on intact vesicles using dot blot analyses. We detected CD63 on both the large and small cup-shaped vesicles, suggesting an endosomal origin for both the s-miRNA- and n-miRNA-associated vesicles (Figure 4A–C). However, because of their distinct size, we hypothesized that s-miRNA and n-miRNA vesicles are of different subendosomal origin. To test this idea, we further analyzed the vesicle surfaces by dot blot analysis, probing for proteins preferentially transported through different endosomal pathways (73). Most antigens tested were in similar abundance in the two vesicles (Figure 4C). However, enrichment of the surface antigen CD44 on the larger exosomes distinguished them from the regular-sized exosomes. The exclusive association of miR16/miR-720 with the CD44+ vesicles (74) suggests that n-miRNAs are sorted through a different subendosomal compartment than the selectively exported miRNAs. We call these newly identified CD44-positive, large exosome-like vesicles, L-exosomes.
Figure 4.
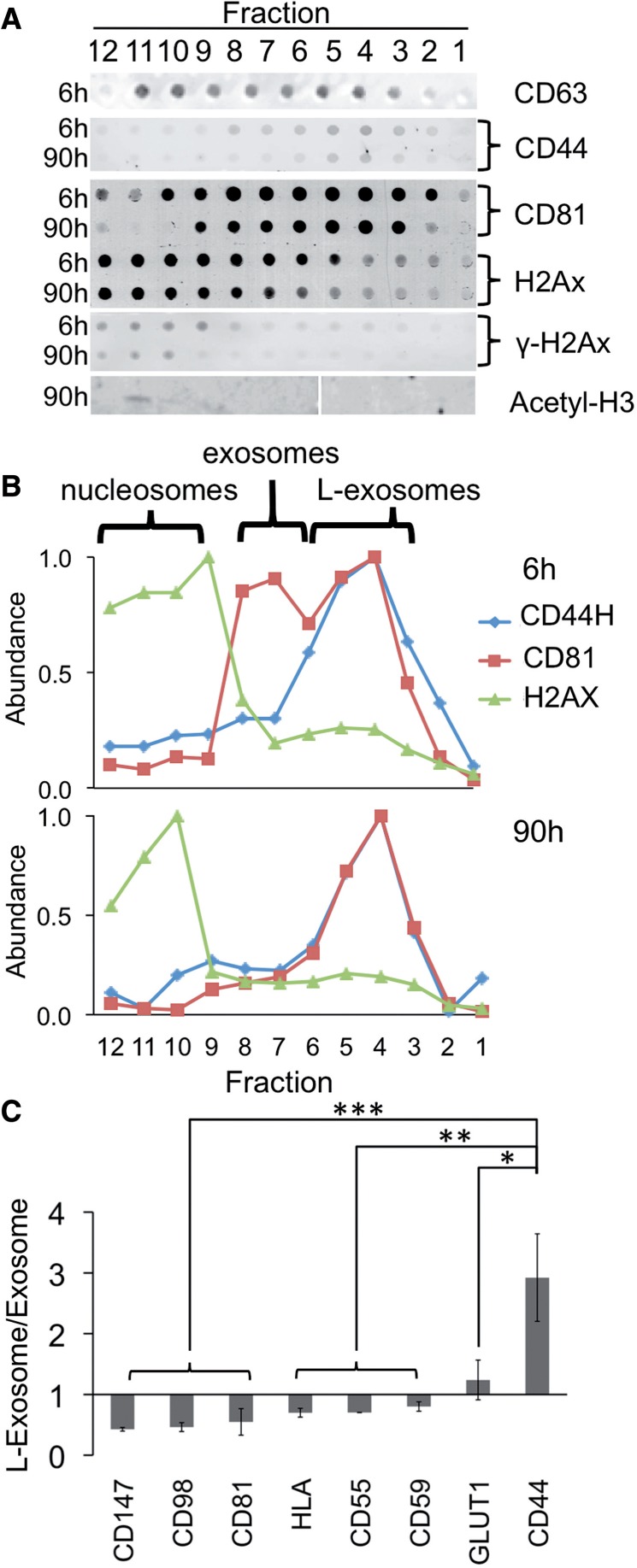
Large- and regular-sized exosomes; and nucleosomes have different proteins. (A) Fractions of 6- and 90-h gradients of P70s from MDA-MB-231 cells were probed for protein markers using quantitative dot-blot; or by western blot analysis for acetyl-H3 histone. (B) Quantitation of data in (A). (C) Comparison of abundance of exosome and L-exosome-associated antigens. *P < 0.05; **P < 0.01; ***P < 0.001, (n = 4). Bars represent standard deviation.
MiRNA species associate with distinct exosome sub-populations
Exosome sub-populations differ in their surface proteins (75), including CD59/PROTECTIN that restricts lysis by complement (76). Because of this protection, the CD59-positive (+) sub-population of exosomes is expected to be particularly useful for biomarker studies. To test whether CD59+ exosomes are enriched in a particular sub-population of miRNAs, we performed immunomagnetic separation (Figure 5A). We found that vesicles containing CD59 had 50 times more miR-16 than the total vesicle population, but the concentration of other analyzed miRNAs was not enriched (Figure 5B). This observation suggests that within the exosomal vesicle population, sub-populations exist that bear different miRNA species.
Figure 5.
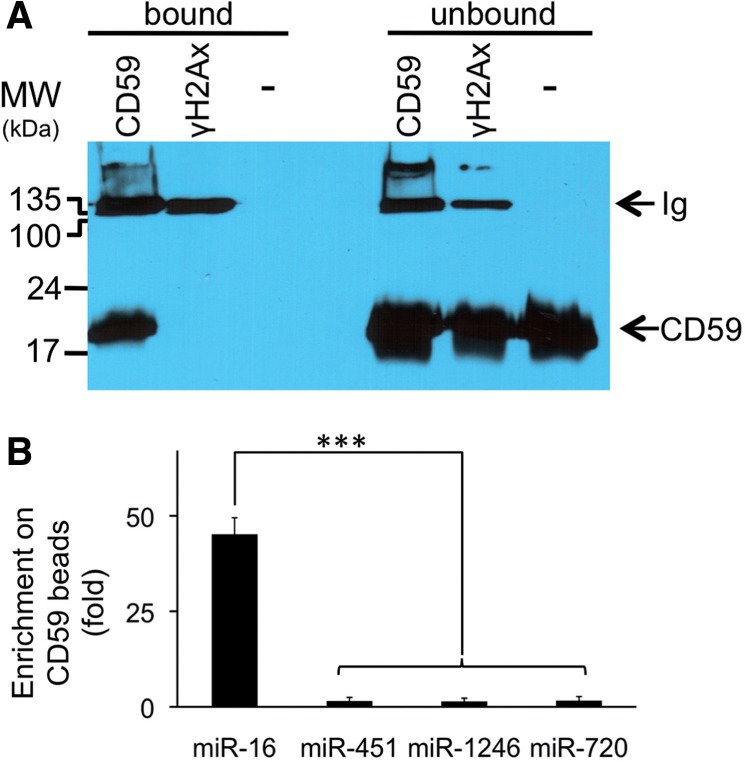
MiR-16 is enriched in CD59/PROTECTIN positive vesicles. Vesicle and particle sub-populations with CD59 or γ-H2Ax surface antigen were enriched by immunomagnetic separation. (A) Bound and remaining unbound populations of immunomagnetic beads were probed for CD59 recovery (top panel; – indicates naked beads incubated with P70. CD59 or γ-H2Ax indicates beads bound with these antibodies were incubated with P70). (B) MiRNA enrichment on beads was quantified using TaqMan qRT–PCR (bottom panel). Bars represent standard deviation (n = 4).
MiR-1246 associates with nucleosomes
We found no membrane proteins associated with the gradient fractions containing miR-1246. To define the nature of the miR-1246 particles, we probed for a number of nucleic acid–associated proteins and found that these fractions contained core histones H2A and acetylated H3 (Figure 4A and B), which are all components of nucleosomes. The size range of 30–120 nm for the majority of complexes (Figure 3B) is consistent with the miR-1246 structures being composed of single and multiple nucleosomes (77,78). Such mono- and oligonucleosomes contain RNA (79) and are released from breast cancer cells into blood plasma, where they might be useful for prognosis (80) and for monitoring chemotherapeutic outcome (81).
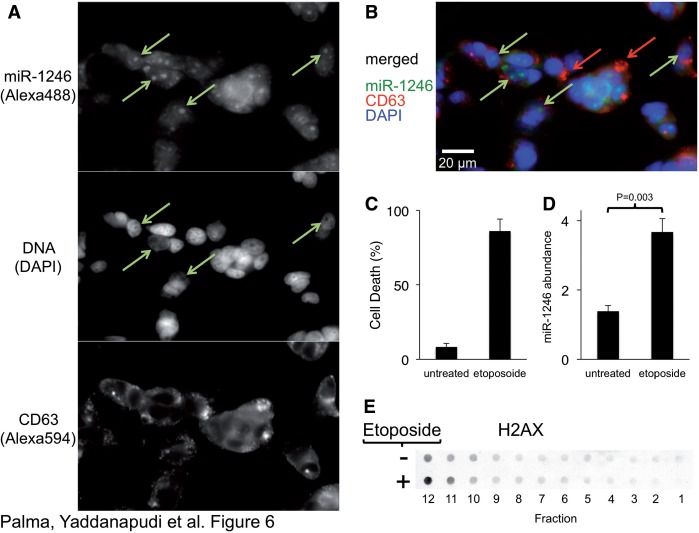
To test whether miR-1246 is associated with nucleosomes in the cell, we performed fluorescence in situ hybridization on cells using a probe to miR-1246. Consistent with the extracellular nucleosome association, we detected a sub-population of miR-1246 associated with distinct 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI)-negative nuclear bodies (nucleoli) in cells (Figure 6A and B, Supplementary Figure S5), suggesting that the association between miR-1246 and histones is physiological. We also detected γH2Ax association (Figure 4A) with the extracellular nucleosome-like particles, suggesting the presence of damaged DNA. Therefore, we tested whether DNA-damage-induced apoptosis triggers nucleosome release in the MDA-MB-231 cells. We found that apoptosis induced by DNA damage (Figure 6C) led to an increase in both the abundance of nucleosomes and miR-1246 (Figure 6D and E). Alternatively, blocking the apoptotic pathways by ectopic expression of Bcl-2 or a dominant-negative form of caspase 9 (66) did not change the release of exosomes or L-exosomes (Supplementary Figure S6). This result supports the idea that apoptosis is a source of circulating nucleosomes (80), but not exosomes (30,82) and that miR-1246 is associated with these nucleosomes.
Figure 6.
MiR-1246 associates with nucleosomes. (A) MCF7 cells were probed for miR-1246 using miRCURY LNA probes (top panel) and for CD63 using antibodies (middle panel) and dyed for DNA (with DAPI, middle panel). Arrows indicate DAPI-negative regions of the nucleus (nucleoli). (B) Overlay of the individual probes. Green arrows indicate enriched miR-1246 populations, and red arrows indicate CD63 domains. (C) MDA-MB-231 cells were treated with etoposide, and cell death was quantified using trypan blue (n = 7). (D) Exported miR-1246 abundance was measured by TaqMan qRT–PCR. The two-tailed P value was calculated by paired T-test (n = 4). (E) Probing of P70 gradient fractions with H2Ax on untreated and etoposide-treated MDA-MB-231 cells. Supplementary Figure S5 shows probing of these cells with an rRNA probe for comparison of subcellular location of these RNA species.
Normal cells release miRNAs in a single vesicle type
The finding that n-miRNAs and s-miRNAs segregate into different particles suggests that specific miRNA sub-populations are specifically diverted in breast cancer cells. To test whether this property is a hallmark of breast cancer cells, we characterized the miRNA-containing vesicles of normal fibroblast and mammary epithelial cells. We found that in normal fibroblasts, a single vesicle type was released. These vesicles resemble exosomes in their shape (Figure 7A), surface antigenicity (Figure 7A and B), density, buoyancy (Figure 7B) and size (Figure 7C), and they carry all miRNA species that we measured (Figure 7D). No nucleosomes, which are of similar size as exosomes but of different shape and density (Figure 3), were found to be released from fibroblasts. This observation suggests that the selective export of miRNAs in specialized vesicles is a feature of breast cancer cells.
Figure 7.
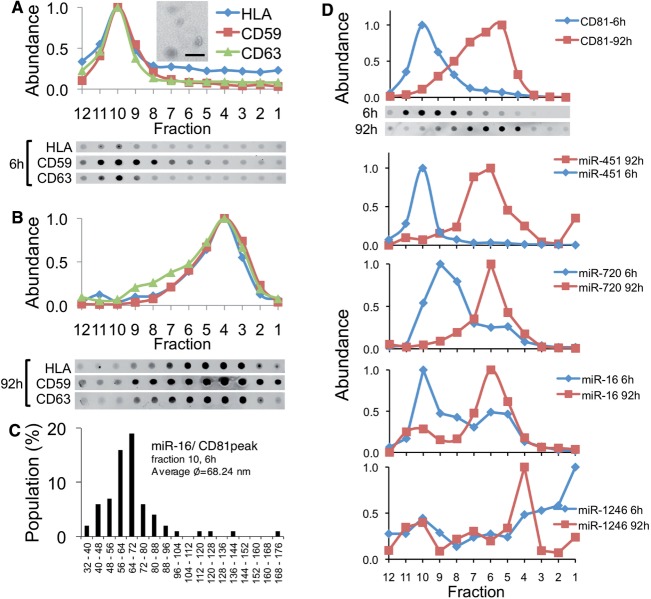
Normal human fibroblasts export MiRNAs in a single vesicle family. (A) Normal donor skin fibroblasts were grown in defined media, a P70 preparation was produced and was subjected to buoyant velocity centrifugation for 6 h or (B) 92 h and probed for CD59, CD63 and HLA by dot blot and quantified. Inset in (A): EM of peak fraction 10 of 6 h gradient. Bar = 500 nm. (C) The largest diameter of >300 vesicles was measured and summarized. (D) Distribution of measured CD81 and miRNAs within gradient, n = 2.
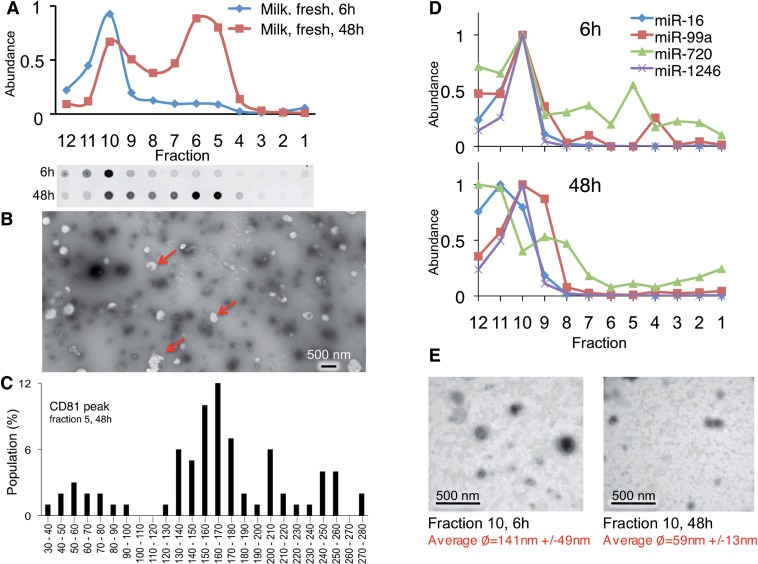
To assess miRNA export in normal mammary epithelial cells, we profiled miRNAs and vesicles in human milk, a body fluid produced mostly by mammary epithelial cells. As expected, milk contained several vesicles (2,83,84), including CD81-positive vesicles of two buoyant densities (Figure 8A) and sizes (Figure 8B and C). However, unlike breast cancer vesicles, both s-miRNAs miR-451 and miR-1246 migrated in milk vesicles together, rather than in separate vesicles as is the case during the release from malignant cells, despite the fact that both fast- and slow migrating vesicles are present in milk (Figure 8A). Furthermore, the miRNAs were associated with vesicles that barely moved in the gradient, which were enriched in 59 nm spherical particles (Figure 8D and E) and which also contain CD81 (Figure 8A). The nature of these vesicles is currently not clear. However, these results indicate that miRNAs are exported differently from malignantly transformed epithelial cells than from benign cells in the body. Overall, these data highlight the complexity of miRNA release from cells and suggest that this process is regulated in a manner that reflects the origin and transformation state of the cell.
Figure 8.
MiRNAs associate with different complexes in human milk. (A) P70 was prepared from fresh human milk and subjected to buoyant velocity centrifugation for 6 or 48 h. (B) Negative stain EM of particles in major CD81 peak void of measured miRNAs. Arrows: electron-poor globules only detectable in the peak CD81 fractions. (C) Graph of largest diameters of particles of CD81 peak fraction. (D) The abundance of indicated miRNAs were measured in each gradient fraction after 6 h or 48 h of centrifugation by TaqMan qRT–PCR. (E) Negative stain EM of particles in major miRNA containing fraction after 6 or 48 h of gradient centrifugation.
DISCUSSION
MiRNAs are released in custom-made vesicles
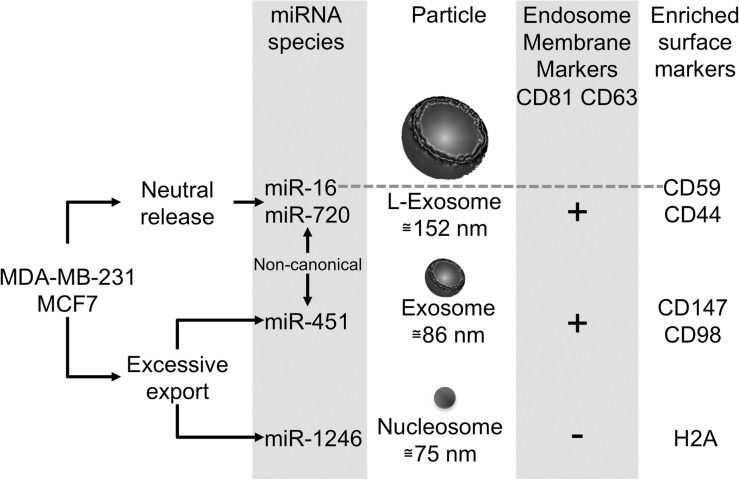
This study expands the repertoire of extracellular miRNA-associated particles to include nucleosomes and L-exosomes. A major finding of this study is that both n-miRNAs and s-miRNA species from malignant cells are exported in vesicles that are not used by normal cells (Figure 9). We discovered these miRNA-containing particles and the differential segregation of miRNA for release into these particles by developing an approach, differential buoyant velocity centrifugation.
Figure 9.
Characteristics of the major particles exported from breast cancer cells. Breast cancer cells export three families of particles containing miRNAs, L-exosomes, exosomes and dense spherical particles containing histones. Exosomes and L-exosomes contain many similar surface antigens, including CD63, an endosomal marker. However, exosomes and L-exosomes differ in the abundance of CD44, the vesicle size and the miRNA content. Sub-populations of these vesicles contain specific miRNAs. For example, miR-16, but none of the other miRNAs associate with CD59-positive vesicles. MiR-1246 associates with particles containing histones, which are void of vesicle surface markers. Interestingly, miRNAs that are selectively exported from malignant mammary epithelial cells are exported in exosomes and the spherical particles, but commonly released miRNAs miR-16 and miR-720 associate with L-exosomes. Non-canonical: miR-451 and miR-720 are processed non-canonically.
Specific miRNAs associate with some vesicles exclusively
This study demonstrates that specific miRNA species associate mutually exclusively with some vesicles. The differences in miRNA composition are more pronounced than the quantitative differences in the surface proteins of the vesicles that distinguish L-exosomes from conventional exosomes. Therefore, the miRNA cargo, rather than differences in vesicle surface antigens, may be the best markers to distinguish these vesicles. The present studies suggest that fundamental differences in miRNA sorting and trafficking in breast cancer cells exist which are linked to modifications of sub-endocytic pathways. The sorting into different vesicles appears to be sequence-specific, because s-miRNAs and n-miRNAs associate with different particles. Recently, sequence motifs in miRNAs were suggested to provide clues regarding selective release of miRNAs (85). Thus, it is possible that cell-type-specific release of miRNAs is regulated through modulation of trans-acting factors that interact with specific miRNA sequences.
MiRNAs released in nucleosomes
We found that miR-1246 is associated with nucleosomes. Nucleosomes circulating in the body are likely released as a consequence of apoptosis. The association of miR-1246 with nucleosomes fits well with our finding that miR-1246 is nearly undetectable in the extracellular environment of normal cells in culture or in mammary fluids (30), because normal cells typically undergo apoptosis at a lower frequency than cancer cells. An apoptosis-related increase of circulating nucleosomes is known to correlate with breast cancer progression (80), raising the possibility that nucleosome-associated miRNAs may correlate with breast cancer growth (81).
The vesicles subspecies contain different information regarding the cell of origin
One practical implication of this study is that it offers information, including specific surface proteins that are enriched in the different vesicles that will allow for the isolation and identification of specific vesicle types that could be useful for diagnostics, treatments and for understanding the role of circulating miRNAs in cell–cell signaling. This might be important, because defined and stable products of processes such as apoptosis and endocytosis associate with different vesicles. As all of these processes are modified in breast cancer, the presence of the vesicles suggests that a wealth of representative elements of pathways deregulated in the tumor cells of origin are present in body fluids. Thus, profiling these particles and vesicles individually may provide accurate information regarding these different functions in the cell of origin and reveal differences in functions these particles and vesicles impose upon other cells after transfer.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6 and Supplementary References [86–88].
FUNDING
US Army Medical Research and Materiel Command [W81XWH-08-1-0641], an American Cancer Society of Illinois Research [189903], Rosalind Franklin University of Medicine and Science start-up funds and a Schweppe Research Scholar Fellowship (to D.M.D.). Funding for open access charge: The American Cancer Society (1/2) The Schweppe Research Foundation (1/2). MAH was supported in part by the US National Institutes of Health grant [F31NS076237].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the women who donated human milk to the Mother’s Milk Bank of Iowa (The University of Iowa Children’s Hospital) and the Indiana Mother’s Milk Bank, Inc., and others who donated fresh milk. We thank Yuri Lazebnik for critical review of this article. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Opinions, conclusions, interpretations and recommendations are those of the author and are not necessarily endorsed by the U.S. Army. D.M.D., G.W., J.P., M.A.H., M.L.H. and S.Y. conceived and designed the experiments. B.S., D.M.D., G.W., J.P., K.W., L.P., M.A.H., M.L.H., S.J. and S.Y. performed the experiments. B.S., D.M.D., J.P., M.L.H., M.A.H., S.J. and S.Y. analyzed the data. D.M.D., J.P. and M.L.H. wrote the article.
REFERENCES
- 1.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Hata T, Murakami K, Nakatani H, Yamamoto Y, Matsuda T, Aoki N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem. Biophys. Res. Commun. 2010;396:528–533. doi: 10.1016/j.bbrc.2010.04.135. [DOI] [PubMed] [Google Scholar]
- 3.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 5.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 9.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol. Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 12.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 13.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim do H, Cho IS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell. Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 16.Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, True LD, Rubin MA, Adam RM, Beroukhim R, et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69:5601–5609. doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho JA, Park H, Lim EH, Lee KW. Exosomes from breast cancer cells can convert adipose tissue-derived. Int. J. Oncol. 2012;40:130–138. doi: 10.3892/ijo.2011.1193. [DOI] [PubMed] [Google Scholar]
- 18.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 19.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 20.Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, Tetta C, Bussolati B, Camussi G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011;71:5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- 21.Chaput N, Taieb J, Schartz NE, Andre F, Angevin E, Zitvogel L. Exosome-based immunotherapy. Cancer Immunol. Immunother. 2004;53:234–239. doi: 10.1007/s00262-003-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delcayre A, Le Pecq JB. Exosomes as novel therapeutic nanodevices. Curr. Opin. Mol. Ther. 2006;8:31–38. [PubMed] [Google Scholar]
- 23.Hao S, Moyana T, Xiang J. Review: cancer immunotherapy by exosome-based vaccines. Cancer Biother. Radiopharm. 2007;22:692–703. doi: 10.1089/cbr.2007.368-R. [DOI] [PubMed] [Google Scholar]
- 24.Hsu DH, Paz P, Villaflor G, Rivas A, Mehta-Damani A, Angevin E, Zitvogel L, Le Pecq JB. Exosomes as a tumor vaccine: enhancing potency through direct loading of antigenic peptides. J. Immunother. 2003;26:440–450. doi: 10.1097/00002371-200309000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Napoletano C, Rughetti A, Landi R, Pinto D, Bellati F, Rahimi H, Spinelli GP, Pauselli S, Sale P, Dolo V, et al. Immunogenicity of allo-vesicle carrying ERBB2 tumor antigen for dendritic cell-based anti-tumor immunotherapy. Int. J. Immunopathol. Pharmacol. 2009;22:647–658. doi: 10.1177/039463200902200310. [DOI] [PubMed] [Google Scholar]
- 26.Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 27.Palazzolo G, Albanese NN, DI Cara G, Gygax D, Vittorelli ML, Pucci-Minafra I. Proteomic analysis of exosome-like vesicles derived from breast cancer cells. Anticancer Res. 2012;32:847–860. [PubMed] [Google Scholar]
- 28.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D'Souza-Schorey C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Current Biol. 2009;19:1875–1885. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, Holowka DA, Cerione RA. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl Acad Sci. USA. 2011;108:4852–4857. doi: 10.1073/pnas.1017667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nan Y, Han L, Zhang A, Wang G, Jia Z, Yang Y, Yue X, Pu P, Zhong Y, Kang C. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 33.Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ, et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 2009;15:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- 34.Gal H, Pandi G, Kanner AA, Ram Z, Lithwick-Yanai G, Amariglio N, Rechavi G, Givol D. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem. Biophys. Res. Commun. 2008;376:86–90. doi: 10.1016/j.bbrc.2008.08.107. [DOI] [PubMed] [Google Scholar]
- 35.Tsuchiya S, Oku M, Imanaka Y, Kunimoto R, Okuno Y, Terasawa K, Sato F, Tsujimoto G, Shimizu K. MicroRNA-338-3p and microRNA-451 contribute to the formation of basolateral polarity in epithelial cells. Nucleic Acids Res. 2009;37:3821–3827. doi: 10.1093/nar/gkp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian Y, Nan Y, Han L, Zhang A, Wang G, Jia Z, Hao J, Pu P, Zhong Y, Kang C. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int. J. Oncol. 2012;40:1105–1112. doi: 10.3892/ijo.2011.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bitarte N, Bandres E, Boni V, Zarate R, Rodriguez J, Gonzalez-Huarriz M, Lopez I, Javier Sola J, Alonso MM, Fortes P, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–1671. doi: 10.1002/stem.741. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z, Luo X, Ding S, Chen J, Chen T, Chen X, Zha H, Yao L, He X, Peng H. MicroRNA-451 regulates p38 MAPK signaling by targeting of Ywhaz and suppresses the mesangial hypertrophy in early diabetic nephropathy. FEBS Lett. 2012;586:20–26. doi: 10.1016/j.febslet.2011.07.042. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Sanda T, Look AT, Novina CD, von Boehmer H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J. Exp. Med. 2011;208:663–675. doi: 10.1084/jem.20102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bian HB, Pan X, Yang JS, Wang ZX, De W. Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549) J. Exp. Clin. Cancer Res. 2011;30:20. doi: 10.1186/1756-9966-30-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem. Pharmacol. 2008;76:582–588. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, Van Brocklyn J, Ostrowski MC, Chiocca EA, Lawler SE. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell. 2010;37:620–632. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, Dore LC, Yao Y, D'Souza J, Zhang Z, et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes. Dev. 2010;24:1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3zeta and promotes breast cancer cell survival and endocrine resistance. Oncogene. 2012;31:39–47. doi: 10.1038/onc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Liao JM, Zeng SX, Lu H. p53 downregulates Down syndrome-associated DYRK1A through miR-1246. EMBO Rep. 2011;12:811–817. doi: 10.1038/embor.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 47.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl Acad. Sci. USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. Cancer. 2012;131:683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 49.Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J. Clin. Gastroenterol. 2011;45:355–360. doi: 10.1097/MCG.0b013e3181f18ac2. [DOI] [PubMed] [Google Scholar]
- 50.Zuo Z, Calin GA, de Paula M, Medeiros LJ, Fernandez MH, Shimizu M, Garcia-Manero G, Bueso-Ramos CE. Circulating microRNAs let-7a and miR-16 predict progression-free survival and overall survival in patients with myelodysplastic syndrome. Blood. 2011;118:413–415. doi: 10.1182/blood-2011-01-330704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol. Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 52.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 54.Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat. Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 56.Borze I, Scheinin I, Siitonen S, Elonen E, Juvonen E, Knuutila S. miRNA expression profiles in myelodysplastic syndromes reveal Epstein-Barr virus miR-BART13 dysregulation. Leuk. Lymphoma. 2011;52:1567–1573. doi: 10.3109/10428194.2011.568652. [DOI] [PubMed] [Google Scholar]
- 57.Sehic A, Risnes S, Khuu C, Khan QE, Osmundsen H. Effects of in vivo transfection with anti-miR-214 on gene expression in murine molar tooth germ. Physiol. Genomics. 2011;43:488–498. doi: 10.1152/physiolgenomics.00248.2010. [DOI] [PubMed] [Google Scholar]
- 58.Wenguang Z, Jianghong W, Jinquan L, Yashizawa M. A subset of skin-expressed microRNAs with possible roles in goat and sheep hair growth based on expression profiling of mammalian microRNAs. OMICS. 2007;11:385–396. doi: 10.1089/omi.2006.0031. [DOI] [PubMed] [Google Scholar]
- 59.Chikh A, Matin RN, Senatore V, Hufbauer M, Lavery D, Raimondi C, Ostano P, Mello-Grand M, Ghimenti C, Bahta A, et al. iASPP/p63 autoregulatory feedback loop is required for the homeostasis of stratified epithelia. EMBO J. 2011;30:4261–4273. doi: 10.1038/emboj.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mathias RA, Lim JW, Ji H, Simpson RJ. Isolation of extracellular membranous vesicles for proteomic analysis. Methods Mol. Biol. 2009;528:227–242. doi: 10.1007/978-1-60327-310-7_16. [DOI] [PubMed] [Google Scholar]
- 61.Duelli DM, Hearn S, Myers MP, Lazebnik Y. A primate virus generates transformed human cells by fusion. J. Cell Biol. 2005;171:493–503. doi: 10.1083/jcb.200507069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim DJ, Linnstaedt S, Palma J, Park JC, Ntrivalas E, Kwak-Kim JY, Gilman-Sachs A, Beaman K, Hastings ML, Martin JN, et al. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J. Mol. Diagn. 2012;14:71–80. doi: 10.1016/j.jmoldx.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson RC, Deo M, Turner DL. Analysis of microRNA expression by in situ hybridization with RNA oligonucleotide probes. Methods. 2007;43:153–161. doi: 10.1016/j.ymeth.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duelli DM, Lazebnik YA. Primary cells suppress oncogene-dependent apoptosis. Nat. Cell Biol. 2000;2:859–862. doi: 10.1038/35041112. [DOI] [PubMed] [Google Scholar]
- 67.Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38:215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 69.Obregon C, Rothen-Rutishauser B, Gitahi SK, Gehr P, Nicod LP. Exovesicles from human activated dendritic cells fuse with resting dendritic cells, allowing them to present alloantigens. Am. J. Pathol. 2006;169:2127–2136. doi: 10.2353/ajpath.2006.060453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miguet L, Pacaud K, Felden C, Hugel B, Martinez MC, Freyssinet JM, Herbrecht R, Potier N, van Dorsselaer A, Mauvieux L. Proteomic analysis of malignant lymphocyte membrane microparticles using double ionization coverage optimization. Proteomics. 2006;6:153–171. doi: 10.1002/pmic.200500133. [DOI] [PubMed] [Google Scholar]
- 71.Hagerstrand H, Isomaa B. Lipid and protein composition of exovesicles released from human erythrocytes following treatment with amphiphiles. Biochim. Biophys. Acta. 1994;1190:409–415. doi: 10.1016/0005-2736(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 72.Sadallah S, Eken C, Martin PJ, Schifferli JA. Microparticles (ectosomes) shed by stored human platelets downregulate macrophages and modify the development of dendritic cells. J. Immunol. 2011;186:6543–6552. doi: 10.4049/jimmunol.1002788. [DOI] [PubMed] [Google Scholar]
- 73.Eyster CA, Higginson JD, Huebner R, Porat-Shliom N, Weigert R, Wu WW, Shen RF, Donaldson JG. Discovery of new cargo proteins that enter cells through clathrin-independent endocytosis. Traffic. 2009;10:590–599. doi: 10.1111/j.1600-0854.2009.00894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eyster CA, Cole NB, Petersen S, Viswanathan K, Fruh K, Donaldson JG. MARCH ubiquitin ligases alter the itinerary of clathrin-independent cargo from recycling to degradation. Mol. Biol. Cell. 2011;22:3218–3230. doi: 10.1091/mbc.E10-11-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laulagnier K, Schieber NL, Maritzen T, Haucke V, Parton RG, Gruenberg J. Role of AP1 and Gadkin in the traffic of secretory endo-lysosomes. Mol. Biol. Cell. 2011;22:2068–2082. doi: 10.1091/mbc.E11-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clayton A, Harris CL, Court J, Mason MD, Morgan BP. Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. Eur. J. Immunol. 2003;33:522–531. doi: 10.1002/immu.200310028. [DOI] [PubMed] [Google Scholar]
- 77.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl Acad. Sci. USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sisco KL. Is RNA in serum bound to nucleoprotein complexes? Clin. Chem. 2001;47:1744–1745. [PubMed] [Google Scholar]
- 80.Roth C, Pantel K, Muller V, Rack B, Kasimir-Bauer S, Janni W, Schwarzenbach H. Apoptosis-related deregulation of proteolytic activities and high serum levels of circulating nucleosomes and DNA in blood correlate with breast cancer progression. BMC Cancer. 2011;11:4. doi: 10.1186/1471-2407-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trejo-Becerril C, Perez-Cardenas E, Trevino-Cuevas H, Taja-Chayeb L, Garcia-Lopez P, Segura-Pacheco B, Chavez-Blanco A, Lizano-Soberon M, Gonzalez-Fierro A, Mariscal I, et al. Circulating nucleosomes and response to chemotherapy: an in vitro, in vivo and clinical study on cervical cancer patients. Int. J. Cancer. 2003;104:663–668. doi: 10.1002/ijc.11003. [DOI] [PubMed] [Google Scholar]
- 82.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 83.Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, Bossios A, Sjostrand M, Gabrielsson S, Lotvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, Norman M, Neve EP, Scheynius A, Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 85.Batagov AO, Kuznetsov VA, Kurochkin IV. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics. 2011;12(Suppl. 3):S18. doi: 10.1186/1471-2164-12-S3-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012;9:e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 88.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.