Abstract
Cells release RNA-carrying vesicles and membrane-free RNA/protein complexes into the extracellular milieu. Horizontal vesicle-mediated transfer of such shuttle RNA between cells allows dissemination of genetically encoded messages, which may modify the function of target cells. Other studies used array analysis to establish the presence of microRNAs and mRNA in cell-derived vesicles from many sources. Here, we used an unbiased approach by deep sequencing of small RNA released by immune cells. We found a large variety of small non-coding RNA species representing pervasive transcripts or RNA cleavage products overlapping with protein coding regions, repeat sequences or structural RNAs. Many of these RNAs were enriched relative to cellular RNA, indicating that cells destine specific RNAs for extracellular release. Among the most abundant small RNAs in shuttle RNA were sequences derived from vault RNA, Y-RNA and specific tRNAs. Many of the highly abundant small non-coding transcripts in shuttle RNA are evolutionary well-conserved and have previously been associated to gene regulatory functions. These findings allude to a wider range of biological effects that could be mediated by shuttle RNA than previously expected. Moreover, the data present leads for unraveling how cells modify the function of other cells via transfer of specific non-coding RNA species.
INTRODUCTION
Nano-sized membrane vesicles represent a recently identified class of intercellular communication vehicles operating in many organisms (1–6). Such vesicles can derive from multivesicular bodies (MVBs), which are late endosomal compartments containing multiple 50–100 nm sized intraluminal vesicles. Fusion of MVBs with the plasma membrane causes the release of their intraluminal vesicles, which are then called exosomes (7). Alternatively, vesicles can be released by cells through direct shedding from the plasma membrane (1). Cells can tightly regulate the release and molecular composition of these vesicles (8,9) and vesicle targeting depends on the type and activation status of recipient cells (10,11). Despite their early description decades ago (12,13), the wide-spread occurrence of cell-derived vesicles and their potential for tailor-made modulation of target cell behaviour has only been recognized during the last few years. It is now clear that cell-derived vesicles are not only released by almost all cultured cell types, but are also present in a wide range of body fluids (1). Since the molecular make-up and release of cell-derived vesicles is regulated by the producing cell, these vesicles are of interest for disease-related biomarkers (14). Moreover, extracellular vesicles may be used as therapeutic agents (15,16).
Besides specific sets of lipids and proteins, cells can shuttle RNA into vesicles determined for release into the extracellular space. This allows the conveyance of genetically encoded messages between cells (17). The first key publication on nucleic acids in cell-derived vesicles reported the presence of miRNA and mRNA in vesicles derived from mast cells and the functional transfer of RNA to vesicle-targeted cells (17). More recently, the luminal protein and RNA contents of cell-derived vesicles were demonstrated to be delivered into the cytoplasm of recipient cells via fusion of vesicles with these cells (18). It is currently not known whether all vesicle populations released by cells contain RNA. Although several studies indicated that the RNA composition of cell-derived vesicles is different from the parental cell (17,19–21), it is unknown how RNAs are selected for secretion into the extracellular space. Furthermore, extracellular RNA may also be associated with macromolecular complexes that are not enclosed by a vesicle. We here collectively refer to extracellular RNA as ‘shuttle RNA’. Although the majority of circulating miRNAs in human plasma and serum were found to co-fractionate with proteins such as Argonaute2 (Ago2) and nucleophosmin, which have been suggested to protect miRNAs from degradation in RNase-rich environments (22,23), it is currently unknown if vesicle-free RNAs can bind and modify the function of target cells.
Next-Generation Sequencing (NGS) techniques have led to the discovery of large numbers of unexpected non-coding RNAs (24). These transcripts were found to overlap with exons, introns and intergenic regions (24–30). Various non-coding transcripts found in close vicinity to protein coding genes, such as promoter-associated long and short RNAs, transcription start site-associated RNAs, and PROMoter uPstream Transcripts (PROMPTs) are suspected to act as regulatory elements to modulate gene activity (30). Interestingly, many small non-coding RNA species have been found that could act as regulatory RNAs similar to miRNAs. Fragments derived from small nucleolar RNA (snoRNA), vault RNA (vRNA) and transfer RNA (tRNA), for example, were shown to bind Argonaute (AGO) proteins and form RNA-induced silencing complexes (RISCs) capable of regulating expression of target mRNAs analogous to miRNA-containing RISCs (31–33).
The analysis of shuttle RNA populations has almost exclusively focused on miRNAs and mRNAs, most likely due to the availability and ease of array hybridization techniques to detect these RNAs. Which other small RNA biotypes were released by cells via vesicles or in protein complexes was unknown. We therefore aimed to comprehensively analyze small RNA species released by cells using NGS. Among the most well studied cell-derived vesicles are those produced by dendritic cells (DCs). These cells are the sentinels of the immune system and the interaction of DC with T cells is the key event in initiation of immune responses. We previously found that DC-T cell interactions induce vesicle release by DC and that DC-derived vesicles can be targeted to activated T cells (8,11). The release and targeting of RNA-containing vesicles by DC and T cells may play an important role in the communication between these cells and the ensuing immune response. Here, we extensively characterized small (<70 nt) shuttle RNA released during DC-T cell interactions and compared this RNA with small cellular RNA in order to investigate selectivity in the extracellular release of RNA species.
MATERIALS AND METHODS
Cell culture
D1 cells were maintained in Iscove’s Modified Dulbecco’s medium supplemented with 2 mM Ultraglutamine (Biowhittaker), 10% heat inactivated fetal calf serum (FCS, Sigma-Aldrich), 100 IU/ml penicillin and 100 mg/ml streptomycin (GIBCO), 50 μM β-mercaptoethanol and 30% conditioned medium from GM-CSF producing NIH 3T3 cells (R1). The p53-specific CD4+ T-cell clone, generated in a C57BL/6 p53–/– mouse (34) and provided by Prof C. Melief (Leiden University Medical Center, Leiden, The Netherlands), was cultured as described previously (8). For cognate DC/T cell co-culture conditions, DC were pre-loaded with 2.5 μM p53 peptide (amino acid 77–96) for 2 h and subsequently mixed in a 1:1 ratio with T cells, and co-cultured for 20 h in medium containing overnight ultracentrifuged (100 000g) FCS and R1 conditioned medium (to deplete bovine and R1 derived vesicles and large protein aggregates). All cultures were maintained at 37°C, 5% CO2. Cell death in DC and T cell cultures was less than 5% and cognate DC-T cell interactions did not lead to additional cell death in the indicated co-culture period. Experiments were approved by the institutional ethical animal committee at Utrecht University (Utrecht, The Netherlands).
Shuttle RNA isolation
15 × 106 p53 peptide-pulsed DCs were co-cultured with 15 × 106 T cells in 25 ml culture medium as described earlier. Supernatants were pooled to a total of 450 ml. This culture supernatant was subjected to differential centrifugation (35). In short, cells were removed by two sequential centrifugations at 200g for 10 min. Collected supernatant was subsequently centrifuged two times at 500g for 10 min, followed by 10 000g for 30 min. Vesicles and protein/RNA complexes were finally pelleted by ultracentrifugation at 100 000g for 65 min in SW28 and SW40 rotors (Beckman). When indicated, the 100 000g sedimented material was further fractionated using gradient density centrifugation. To this end, the 100 000g sedimented material was mixed with 2.5 M sucrose, overlaid with a linear sucrose gradient (2.0–0.4 M sucrose in PBS) and floated into the gradient by centrifugation in a SW40 tube (Beckman) for 16 h at 192 000g. The bottom two fractions (1.26–1.28 g/ml, as measured by refractometry) and fractions with densities of 1.12–1.18 g/ml were pooled, diluted with PBS/0.1% BSA and centrifuged again at 100 000g for 65 min. Small RNA (<200 nt) was isolated from 100 000g pellets and from co-cultured DC and T cells (15 × 106 each) using the mirVanaTM miRNA Isolation Kit (Ambion) according to the manufacturer’s procedure. The RNA integrity and size distributions were assessed using Agilent 2100 Bioanalyzer pico-RNA chips.
Small RNA sequencing library generation
Sequencing libraries were generated as described (36). In short, small RNA fractions were ligated to adaptors using the SOLiD Small RNA Expression Kit (Ambion). Libraries were pre-amplified using primers containing sequences that make the SREK libraries compatible with the Illumina flow cell. DNA was denatured for 30″ at 98°C followed by 18 cycles with 30″ at 98°C, 30″ at 65°C, 30″ at 72°C and final extension for 5′ at 72°C. Library fragments were separated on a native 6% gradient pre-cast PAGE gel (Novex, Invitrogen). The 100–150 bp size fractions containing inserts of 20–70 nt in length were excised, DNA was eluted from the gel, precipitated and dissolved in nuclease free water. DNA yield was quantified using an Agilent Bioanalyzer high sensitivity DNA chip. Small RNA inserts were single end sequenced for 35 cycles using standard Illumina protocols.
Small RNA sequence data analysis
Adaptor trimming, genome alignment and small RNA annotation were performed using the E-miR data analysis pipeline as described (36). Sequence reads were aligned to the Mouse Mm9 reference genome with bowtie, allowing for alignments to three loci with two mismatches, followed by a crossmapping correction as described by de Hoon et al. (37). For further annotation with coding and repeat RNAs, reads with a minimum overlap of 1 nt were clustered into regions and regions containing a total of more than five reads in shuttle and cellular RNA were annotated using custom perl scripts (38) with exon, 3′-UTR, 5′-UTR, intron and RNA biotype information from Ensembl biomart (39), all in a strand-specific manner. Intersections with repeat regions were determined using information from the repeat track in the UCSC genome browser (NCBI137/mm9) and the Galaxy interface, considering a minimum of 1 nt overlap between the sequenced and the repeat region. To determine reproducibility of the sample preparation and sequencing protocols, we have evaluated the Pearson’s correlation between the abundance of small RNAs in replicate experiments. A square root transformation on the number of counts per small RNA was applied to stabilize variance and reduce influence of high-abundant small RNAs on the Pearson’s correlation coefficient (38).
Quantitative real-time PCR
Sequencing derived expression profiles were compared with qPCR measurements on small RNA fractions from independent biological samples. cDNA was generated using the Ncode miRNA First-Strand cDNA Synthesis kit (Invitrogen). An equivalent of 100 pg RNA was used in the PCR reactions with 100 nM primers (Isogen) and 4 ul SYBR Green Sensimix (Bioline) in an 8 µl reaction. PCR amplification efficiencies were determined using 10-fold dilution series of template DNA and were between 1.8 and 2.3 for all primer sets. Transcript specific forward primers for miR-29a, miR-155 and miR-191 were based on full-length miRbase sequences (40):
3p mmu-mir-29a forward: 5′-TAGCACCATCTGAAATCGGTTA
5p mmu-mir-155 forward: 5′-GGGTTAATGCTAATTGTGATAGGG
5p mmu-mir-191 forward: 5′-CGGAATCCCAAAAGCAGCTG
All forward miRNA primers were used in combination with the NCode universal reverse primer.
Y-RNA forward: 5′-GTGTTTACAACTAATTGATCACAACC
Y-RNA reverse: NCode universal reverse primer
SRP-RNA forward: 5′-GGAGTTCTGGGCTGTAGTGC
SRP-RNA reverse: 5′-ATCAGCACGGGAGTTTTGAC
Cycling conditions were as follows: 95°C for 10 min followed by 50 cycles of 95°C for 10 s, 57°C for 30 s and 72°C for 20 s. All PCR reactions were performed on the Bio-Rad iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad). Raw threshold cycle (Ct) values were calculated using the iQ5 Optical System software v.2.0 using automatic baseline settings. Thresholds were set in the linear phase of the amplification plots.
RESULTS
Sample preparation and sequencing
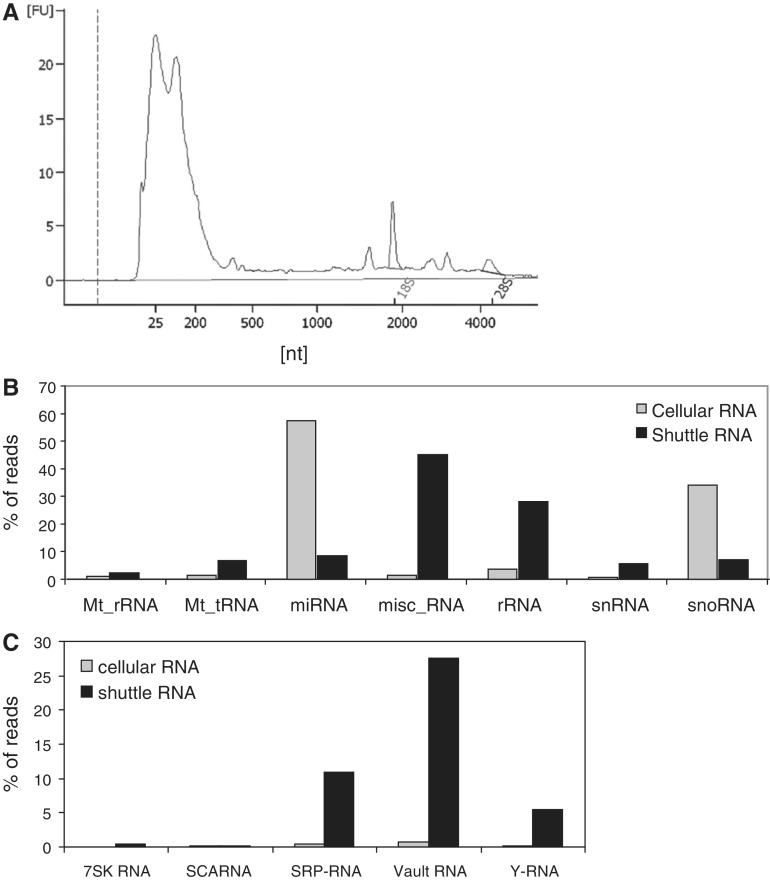
Specific antigen-bearing DCs were co-cultured with cognate T cells, leading to mutual activation of both cell types. Vesicles and large protein complexes released during these interactions were isolated from cell culture supernatant by differential centrifugation, with a final pelleting step at 100 000g (35). Total RNA was isolated from this fraction and we observed that the majority of this shuttle RNA consisted of small RNAs (<200 nt), with minor amounts of 18S and 28S rRNA (Figure 1A). Next, small (<200 nt) RNAs were isolated from the 100 000g sedimented material, and processed for analysis by NGS. To assess whether selective RNAs were released into the extracellular space, small RNA from cells that had released the shuttle RNA was analyzed in parallel. Strand specific small RNA sequencing libraries were prepared and pre-amplified as described (36). Fragments with inserts between 15 and 70 nt were gel purified, after which the first 35 bases of the library were read with a single read, with the Illumina platform. Fragments ≥70 nt were not included to prevent the large number of tRNAs (∼70 nt in size) present in the cellular RNA fraction to dominate the sequencing reaction, which would hamper quantitative comparison of the cellular and shuttle RNA fractions. Sequencing data were processed using a modified version of the E-miR pipeline (36). The sequences were mapped to the mouse genome (NCBI137/mm9), using bowtie allowing for two mismatches and three alignments per sequence read, followed by a crossmapping correction (37).
Figure 1.
Shuttle RNA mainly consists of small RNA species and contains relatively low amounts of miRNA. Cell culture supernatant of DC-T cell co-cultures was subjected to several differential centrifugation steps, followed by RNA isolation from the 100 000g sedimenting material (shuttle RNA). (A) Bioanalyzer electropherogram of total shuttle RNA derived from DC-T cell co-cultures. The composition of small (<200 nt) shuttle and cellular RNA was further analyzed by deep sequencing. (B) Sequence reads uniquely aligned to the genome were annotated to known non-coding RNA transcripts as annotated in Ensembl using the E-miR pipeline. Data represent the percentages of reads in the indicated categories of annotated transcripts in shuttle RNA (grey bars) versus cellular RNA (black bars) calculated relative to the total sum of reads for Ensembl non-coding RNA transcripts. (C) Distribution of shuttle and cellular RNA transcripts over the different RNA biotypes within the Ensembl-annotated miscellaneous RNA (misc_RNA) category. Data are represented as the percentages of annotated transcripts as in (B).
To validate the robustness of our sequencing results, we replicated the presented experiment with a slightly different sample preparation method (including an extra RNA fragmentation step). We found strong concordance when ranking the different RNAs based on their abundance in the shuttle RNA. We calculated the correlation between the (ranked) small shuttle RNA sequencing data of the replicates. A high correlation was observed between the replicates (Pearson’s correlation 0.97, P-value <2.2e-16; Supplementary Figure S1).
miRNAs are underrepresented in shuttle RNA compared with cellular RNA
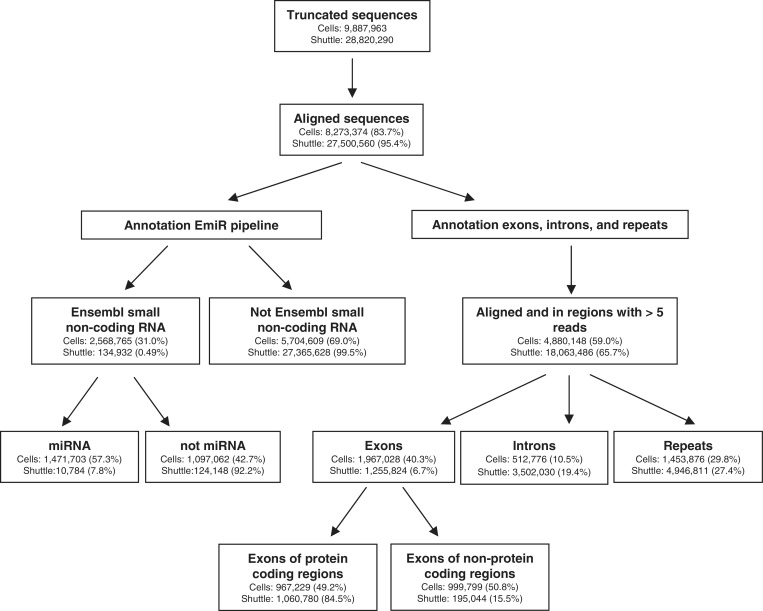
An overview of the sequencing data analysis and results can be found in Figure 2. We found that 8.3 × 10E6 cellular and 27.5 × 10E6 shuttle RNA sequences aligned to the mouse genome (Figure 2). Uniquely aligned sequence reads were annotated to known non-coding small RNA transcripts as annotated in Ensembl. Remarkably, the percentage of sequence reads mapping to this category of RNAs was much lower in shuttle RNA released from cells (0.5%) compared with cellular RNA (31%). Within this category of Ensembl-annotated non-coding small RNAs, we analyzed the distribution of RNAs over the different biotypes. We found that the majority of sequences present in the cellular small RNA population represented miRNAs (Figure 1B), which is in agreement with previous studies (36,41). In contrast, the percentage of miRNAs in shuttle RNA was very low. Instead, this fraction contained a relatively higher percentage of small ribosomal RNA (rRNA) and RNA belonging to the miscellaneous category (misc_RNA) (Figure 1B). Within this category of misc_RNA, we found that structural SRP-RNA, vRNA and Y-RNA were most abundant in shuttle RNA (Figure 1C).
Figure 2.
Overview of the sequencing data analysis and results. Indicated are the number of reads in each analysis step for small cellular versus shuttle RNA and the relative amount of reads calculated as a percentage of the reads detected in the previous steps.
The miRNA distribution in shuttle and cellular RNA is different
Since the initial discovery of miRNAs in cell-derived vesicles (17), many research groups have studied the occurrence of this type of regulatory RNAs in vesicles derived from various cell types (as listed in the ExoCarta database (42)). Although miRNAs formed only a small minority within the total pool of shuttle RNA sequences (Figure 1B), we examined whether a specific set of miRNAs was released during DC-T cell interactions. The miRNAs occurring in cells of the DC-T cell co-culture and shuttle RNA released by these cells were ranked based on the E-miR data and the 10 most prevalent miRNAs in each population were compared (Table 1). miRNAs were named according to the 5′ (5p) or 3′ (3p) arm of the hairpin where they originated from (36). Certain miRNAs, such as miR-29a and miR-31, were abundant in both cellular and shuttle RNA. However, several highly abundant cellular miRNAs, such as miR92a-1 and let-7b, occurred only in very low abundance in shuttle RNA. Conversely, the highly abundant shuttle miRNAs miR-223, miR-142 and miR-93 were less abundant in cellular RNA. These data indicate that dissemination of miRNAs into the extracellular space is not a random process, but that cells actively sort selective miRNAs for extracellular destination.
Table 1.
Enriched miRNAs in cellular and shuttle RNA
| miRNA | miRNA arm | Rank in cells | Rank in shuttle RNA |
|---|---|---|---|
| Abundant cellular miRNAs | |||
| miR-155 | 5p | 1 | 29 |
| miR-29a | 3p | 2 | 1 |
| miR-92a-1 | 3p | 3 | 860 |
| miR-31 | 5p | 4 | 5 |
| miR-15b | 5p | 5 | 46 |
| miR-744 | 5p | 6 | 47 |
| miR-let-7b | 5p | 7 | 119 |
| miR-191 | 5p | 8 | 4 |
| miR-24-2 | 3p | 9 | 10 |
| miR-24-1 | 3p | 10 | 12 |
| Abundant shuttle miRNAs | |||
| miR-29a | 3p | 2 | 1 |
| miR-21 | 5p | 15 | 2 |
| miR-223 | 3p | 41 | 3 |
| miR-191 | 5p | 8 | 4 |
| miR-31 | 5p | 4 | 5 |
| miR-142 | 5p | 47 | 6 |
| miR-93 | 5p | 48 | 7 |
| miR-103-1 | 3p | 18 | 8 |
| miR-103-2 | 3p | 14 | 9 |
| miR-24-2 | 3p | 9 | 10 |
miRNAs were ranked according to the frequency of their occurrence (number of reads). Indicated are the 10 most prevalent miRNAs in cells or shuttle RNA and the corresponding ranking in the other sample.
Shuttle and cellular RNA contain different sets of transcripts
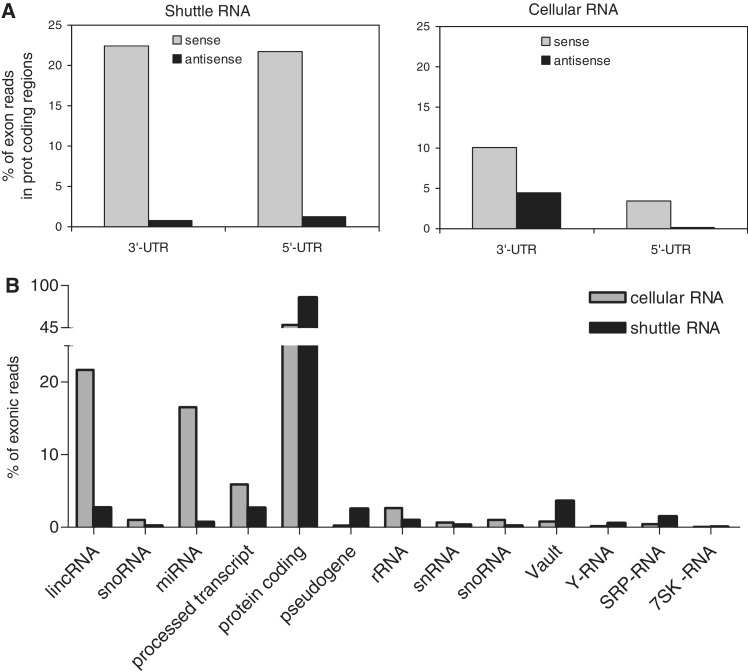
Since the shuttle RNA population contained only 0.5% Ensembl-annotated small non-coding RNA transcripts, we next aimed to define which other types of transcripts were present in this population. By comparison with genomic locations obtained from Ensembl Biomart (39), we found that 26.1% of the shuttle RNA reads (against 49.9% in cellular small RNA) mapped to introns and exons (Figure 2). Cellular small RNAs showed a higher frequency of exonic localization (including a large number of exons coding for miRNAs). However, the relative amount of exonic sequences mapping to protein coding regions was higher in shuttle RNA. In addition, shuttle RNA contained relatively more intronic sequences (Figure 2). Data have recently accumulated demonstrating that protein-coding genes can also produce a complex set of non-coding RNAs (43), including transcripts originating from introns and from the untranslated regions (UTRs) (30,43). Interestingly, we observed that the percentage of reads in protein coding loci that aligned to 3′- and 5′-UTRs was much higher in shuttle RNA compared with cellular RNA (Figure 3A). Most of these transcripts had the same directionality as the coding mRNA. Although their exact function is unknown, these UTR-derived small RNAs have been suggested to play a regulatory role in the attenuation or regulation of translation (44).
Figure 3.
Exonic hits in shuttle RNA preferentially locate to UTR regions. (A) Exonic hits were analyzed for overlap with UTR regions. Indicated is the percentage of reads in shuttle RNA (left) and cellular RNA (right) mapping to the sense or antisense strand of UTR-regions, calculated relative to the total number of reads in Ensembl annotated exons. (B) The identity of exonic hits in shuttle (black) and cellular (grey) RNA. Indicated are the percentages of reads in the different RNA biotypes calculated relative to the total number of exonic reads.
Further analysis of the exonic sequences in shuttle and cellular RNA revealed that the large majority (84%) of exonic reads in shuttle RNA annotated to protein coding transcripts (Figure 3B), preferentially locating in the UTR regions of those transcripts (Figure 3A). Also the relative abundance of the small non-coding vRNA, Y-RNA and SRP-RNA was higher in shuttle RNA compared with cellular RNA. In contrast, lincRNA and miRNA are less abundant in shuttle RNA compared with cellular RNA (Figure 3B). Taken together, these results suggest that shuttle RNA is enriched in non-coding RNAs other than miRNAs and lincRNAs and that many of these are still to be annotated.
The distribution of RNA repeat sequences is different in shuttle and cellular RNA
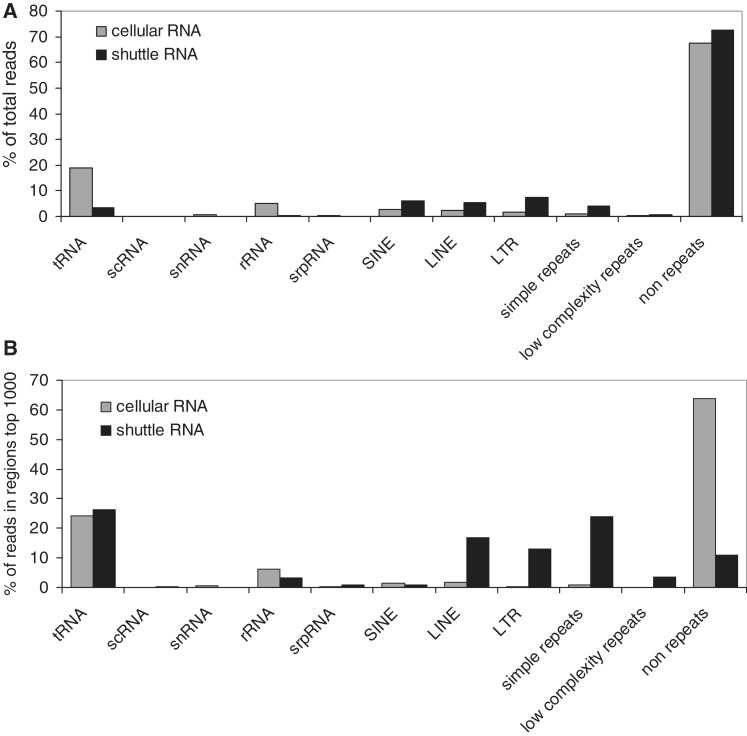
Since the majority of small shuttle RNA transcripts did not correspond to Ensembl-annotated small non-coding or protein-coding RNAs, we next investigated whether the as yet undefined transcripts corresponded to repeat-derived RNAs. The relative abundance of repeat RNAs in shuttle and cellular RNA was approximately the same (+/− 25%, Figure 4A). However, the distribution over the type of repeat sequences in these two RNA populations was different. Shuttle RNA contained relatively large numbers of Long Interspersed Elements (LINEs), Long Terminal Repeats (LTRs) and simple repeat sequences, whereas the majority of repeat sequences in the cellular RNA consisted of tRNAs (Figure 4A). To further classify the most abundant repeat-associated RNA species, we next considered only the top 1000 of most prominently present shuttle and cellular RNA. The differences in the distribution of the most abundant shuttle and cellular RNA were much more pronounced. Almost 90% of the 1000 most abundant sequences in shuttle RNA (versus 33% in cells) consisted of repeat sequences (Figure 4B). The most prevalent types of repeats in shuttle RNA were tRNAs, LTRs, LINEs and simple repeats. LTRs, LINEs and simple repeats were relatively much more abundant in shuttle RNA compared with cellular RNA (Figure 4B).
Figure 4.
Different distribution of RNA repeat sequences in shuttle and cellular RNA. Abundance and classification of sequencing reads that match different types of RNA repeats. Indicated are the percentages of all reads (A) or reads in the top 1000 of most abundant regions (B) in the indicated repeat types, as detected in shuttle (black) and cellular (grey) RNA.
Specific tRNA fragments are abundantly and selectively present in shuttle RNA
To investigate in greater detail the most abundant sequences in shuttle RNA, all unique regions were ranked by abundance (number of reads found per region) and the RNA biotypes of the top 75 highest ranked hits were analyzed. For comparison, a similar top 75 ranking was made for the sequences found in cellular RNA (Table 2). We observed that specific snoRNA and miRNAs were among the most abundant sequences in the cellular small RNA fraction, but not in shuttle RNA. However, as expected from previous data (Figure 4B), the shuttle RNA was more abundant in different types of repeat sequences. The high abundance of tRNA hits in both the shuttle and cellular RNA fractions was remarkable, since we restricted our sequencing analysis to <70 nt RNAs. Consequently, the sequence reads were not likely to represent mature full-length tRNAs. We next explored whether these reads could represent tRNA fragments (tRFs), which have recently gained interest due to their suspected regulatory nature (45). tRNA fragmentation is a specific process, with the composition, abundance and cleavage site varying per organism and cell type. Fragments of 18–22 nt, 30–35 nt and ∼50 nt locating at the 3′- or 5′- end of mature tRNAs have previously been described (45–47). In cellular RNA, we observed fragments located at the 5′ or 3′end of the mature tRNA (Table 3). However, the most abundant tRNA hits in shuttle RNA were all located at the 5′end of mature tRNAs. Differences were also observed in the type of tRNAs occurring in the top 75 most abundant hits of cellular and shuttle RNA; whereas fragments of tRNA-Lys-AAA were abundant in both cellular and shuttle RNA, fragments of tRNA-Asp-GAY were only abundant in cellular RNA. We observed that the majority of abundant tRNA hits in shuttle RNA covered larger regions of about 40–50 nt (Table 3 and Figure 5), caused by the presence of reads representing tRNA fragments (rather than full-length tRNAs) missing the first 5–15 nt of the mature tRNA. Remarkably, at the same genomic locations, the coverage in cellular RNA was restricted to ∼30–35 nt fragments (Figure 5), indicating the presence of two different fragments of the same tRNA, of which one is uniquely present in shuttle RNA.
Table 2.
Most abundant biotypes in cellular and shuttle RNA
| RNA biotype | No. of reads | No. of regions |
|---|---|---|
| Cellular RNA | ||
| snoRNA | 615 728 | 20 |
| miRNA | 599 709 | 20 |
| tRNA repeat | 586 583 | 27 |
| Unclassified | 379 199 | 4 |
| rRNA | 107 855 | 2 |
| Protein coding | 103 877 | 1 |
| vRNA | 15 776 | 1 |
| Shuttle RNA | ||
| tRNA repeat | 670 645 | 47 |
| Simple repeat | 484 945 | 6 |
| LINE | 325 549 | 5 |
| rRNA | 120 797 | 5 |
| vRNA | 45 546 | 1 |
| Protein coding | 26 492 | 4 |
| SRP-RNA | 18 553 | 2 |
| Y-RNA | 5929 | 1 |
Regions were ranked by abundance (number of reads per region) and the top 75 highest ranked hits were categorized based on RNA biotype. Indicated are the collective number of reads mapping to the different biotypes and the number of different regions over which these reads were distributed.
Table 3.
Most abundant tRNA fragments in cellular and shuttle RNA
| tRNA fragment | Location | No. of reads | No. of regions | Region length (nt) |
|---|---|---|---|---|
| Abundant cellular tRNA hits | ||||
| tRNA-Asp-GAY | 3′-end | 389 792 | 14 | 35 |
| tRNA-Lys-AAG | 5′-end | 106 551 | 6 | 30–35 |
| tRNA-Lys-AAA | 5′-end | 36 995 | 3 | 30 |
| tRNA-Lys-AAG | 3′-end | 18 079 | 1 | 35 nt |
| Abundant shuttle tRNA hits | ||||
| tRNA-Lys-AAA | 5′-end | 246 384 | 6 | 35 |
| tRNA-Lys-AAG | 5′-end | 134 033 | 9 | 50 |
| tRNA-Gly-GGA | 5′-end | 85 930 | 7 | 50 nt |
| tRNA-Gly-GGY | 5′-end | 81 737 | 9 | 40 |
| tRNA-Val-GTG | 5′-end | 44 160 | 5 | 50 |
| tRNA-Glu-GAG | 5′-end | 27 900 | 5 | 40 |
| tRNA-Val-GTA | 5′-end | 13 198 | 2 | 40–50 |
Hits in tRNAs in the top 75 highest ranked hits were annotated with anticodon, location of the region within the tRNA coding sequence (3′- or 5′-end), collective number of reads (accumulative for different copies of the same type of tRNA), the number of regions over which these reads were distributed, and the length of the regions.
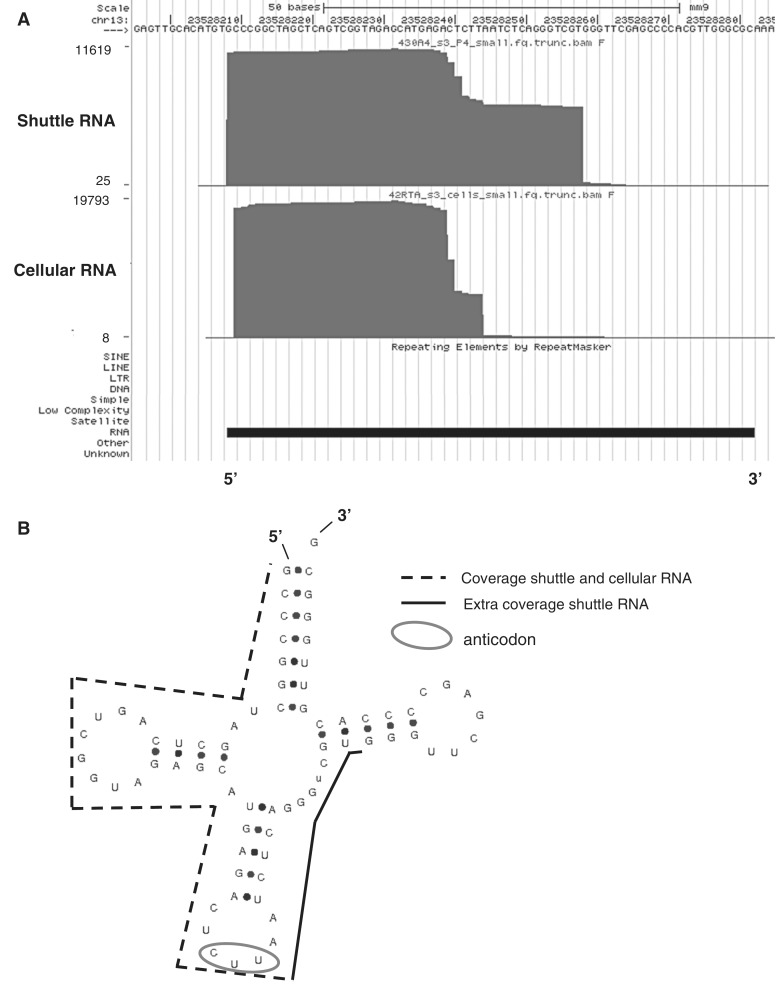
Figure 5.
Different coverage of tRNA genes in shuttle and cellular RNA. (A) Screen shots were taken from the UCSC genome browser (http://genome.ucsc.edu/) and show examples of tRNAs. Custom tracks were added to show sequence coverage in shuttle and cellular RNA. An example is shown of a tRNA gene (tRNA-Lys-AAG) on which differential coverage was detected in shuttle RNA (top) and cellular RNA (bottom). The y-axis represents the coverage at each genomic position. (B) tRNAscan-SE-predicted (http://lowelab.ucsc.edu/tRNAscan-SE) secondary structure of the mouse tRNA gene (tRNA-Lys-AAG) presented in (A). Indicated are the regions with coverage in shuttle and cellular RNA (dotted line), the additional coverage in shuttle RNA (black line), and the anticodon (circle).
A specifically cleaved part of vRNA is abundantly present in shuttle RNA
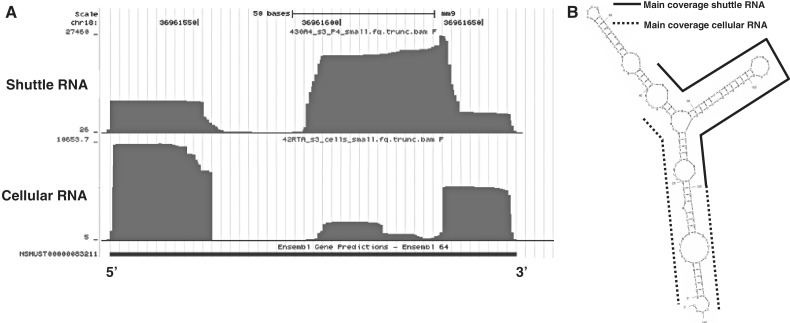
Apart from the repeat and tRNA fragments, three other RNA biotypes were abundantly present in shuttle RNA (Table 2): (i) vRNA; (ii) SRP-RNA (7SL-RNA); and (iii) Y-RNA. All three RNAs are non-coding polymerase III RNA transcripts. Due to our <70 nt size restriction, we did not expect to find full-length transcripts of these three RNA biotypes. vRNAs are a family of RNAs found associated with the vault ribonucleoprotein complex located in the cytoplasm (48). The function of these vault particles are largely unknown, but they are thought to play a role in transportation of molecules, such as mRNA, from the nucleus to the cytoplasm and in drug metabolism (e.g. in tumor cells). Mouse vRNA is 141 nt long, but it was recently discovered that small vault RNAs (svRNAs) can be generated from vault non-coding RNAs through a DICER-dependent and DROSHA-independent mechanism (32). These svRNAs can downregulate expression of specific genes, similar to miRNAs. Interestingly, we found that the predominant coverage on the vRNA in exosomes was in only one of the internal stem loop structures (Figure 6A, B). In contrast, the coverage in cellular RNA was predominantly at the 3′ and 5′ ends of vRNA, resembling the localization of the described human svRNAs (32). These data indicate that a specifically cleaved part of the vRNA is preferentially shuttled into the extracellular space.
Figure 6.
Different coverage of vRNA in shuttle and cellular RNA. (A) Screenshot from the UCSC genome browser showing the sequence coverage on the vRNA gene in shuttle RNA (top) and cellular RNA (bottom). The y-axis represents the coverage at each genomic position. (B) MFOLD-predicted (http://mfold.rna.albany.edu/) secondary structure of mouse vRNA at 21°C. Indicated are the regions with predominant coverage in shuttle RNA (black line) and cellular RNA (dotted lines).
SRP- and Y-RNA are highly enriched in cell-derived vesicles
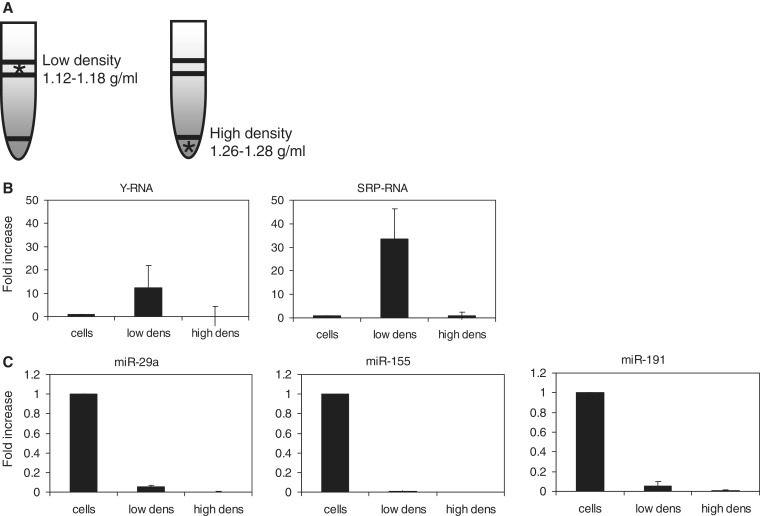
SRP-RNA (bound to the signal recognition particle) and Y-RNA (bound to the Ro ribonucleoprotein complex (49)) both function in intracellular transport of proteins and RNAs (50). We found a 28 nt Y-RNA fragment and 26–50 nt SRP-RNA covering regions in shuttle RNA. The abundance of SRP- and Y-RNA in material released by cells into the extracellular space is remarkable, since these RNAs belong to a small group of host RNAs that is also selectively incorporated together with viral genomes into the capsids of several different viruses (51,52). To establish a link between viruses and cell-derived vesicles regarding incorporation of these RNAs, we investigated whether full-length SRP- and Y-RNA released in our DC-T cell co-cultures were indeed associated to membrane vesicles. Hereto, the 100 000g material that was sedimented from the culture supernatant was further fractionated by gradient density ultracentrifugation. This allowed separation of RNA associated to large protein complexes, which are retained in the bottom of the gradient, and RNA enclosed in membrane vesicles, which float to low-buoyant densities (1.12–1.18 g/ml (35); Figure 7A). Subsequently, the presence of SRP- and Y-RNA in the different density fractions was analyzed by RT-qPCR in independent experiments. For comparison, the presence of these RNAs was analyzed in the cellular <200 nt RNA fraction obtained from the DC-T cell co-culture from which the shuttle RNA was derived. Invariant endogenous controls (reference genes) for qPCR analysis of vesicle-enclosed RNA are unknown. As an alternative, all samples were normalized to the total input quantity of RNA. Various miRNAs, which were relatively abundant in cellular and/or shuttle RNA (Table 1), were analyzed in parallel as a control. We observed that full-length SRP- and Y-RNA indeed associated to low-density cell-derived vesicles and not to high-density complexes. Moreover, these RNAs were highly enriched in vesicles compared with cells (Figure 7B). In contrast, all tested miRNAs were relatively more abundant in cells compared with cell-derived vesicles, confirming the sequencing data (Figure 7C). Conclusively, SRP- and Y-RNA are highly abundant in cell-derived vesicles, similar to the enrichment of these host RNAs in virions. These findings may point to an evolutionary conserved mechanism by which cellular RNA and viral genomes are selected and/or stabilized in membrane vesicles with extracellular destination or by which transferred RNA can function in cells targeted by these vesicles.
Figure 7.
Culture supernatant of DC-T cell co-cultures was subjected to consecutive differential centrifugation steps. The 100 000g pelleted material was loaded at the bottom of a sucrose gradient, after which low-density vesicles were floated to equilibrium density by ultracentrifugation. Low-density fractions (1.12–1.18 g/ml) and high-density (1.26–1.28 g/ml) bottom fractions of the gradient were collected (A). Small RNA (<200 nt) was isolated from the 100 000g pelleted material present in these fractions. (B, C) RT-qPCR analysis of Y- and SRP-RNA (B) and different miRNAs (C) in small cellular RNA (cells), 100 000g pelleted vesicles floating to low-density fractions (low dens) and 100 000g pelleted non-floating high-density material (high dens). All samples were normalized to input quantity of RNA. Relative expression levels were calculated based on the Ct values. Indicated is the fold increase ± s.d. of PCR product in floating or non-floating material relative to cells (set to 1).
DISCUSSION
The complexity and diversity of the pools of extracellular RNA in cell-derived vesicles and vesicle-free complexes is daunting. Many research groups have focused on analyzing miRNAs in material released from a broad range of cell types. However, the size distribution of shuttle RNA, as shown here and by others (53,54), extends well beyond the 20–23 nt size of miRNAs and is mostly in the range of ∼20 to ∼200 nt. This indicates that also other small RNA species are released by cells into the extracellular space. By deep sequencing of the <70 nt fraction of this shuttle RNA, we found a large variety of small non-coding RNA species representing pervasive transcripts or cleavage products overlapping with protein coding regions, repeat sequences, or structural RNAs. We extended this analysis with detailed evaluation of the sequence coverage of several of the most abundant RNA biotypes in shuttle RNA. This led to the discovery that cellular and shuttle RNA can contain different small RNAs derived from the same non-coding RNA, as was the case for tRNA and vRNA. Although no conclusions can be drawn regarding the absolute concentration of different RNA species, the data allow comparison of the relative amounts of RNA types in the pools of cellular and shuttle RNA. The unequal distribution of the detected RNA species over cellular and shuttle RNA, combined with increasing evidence for their role in gene regulation strongly suggest that cells specifically release these RNAs to modify the function of target cells.
miRNA
From both the sequencing and RT-qPCR data it became clear that miRNAs were underrepresented in shuttle RNA compared with cellular RNA (Figures 1B and 7C). However, the sequencing data revealed that the miRNA composition in shuttle RNA is not a mere reflection of the cellular miRNA (Table 1), indicating that a specific set of miRNAs is selected for extracellular release. Evidence exists that miRNAs associated to cell-derived vesicles are functional upon transfer to target cells (17,18). Although the relative amounts of released miRNAs detected here may appear low, significant effects on target cell functioning could be imposed due to efficient and specific cellular targeting of vesicles and the capacity of miRNAs to cause large-scale and long-term modulation of gene expression. Interestingly, 5 out of the 10 most abundant shuttle miRNAs have validated target genes that play important roles in immune regulation. miR-93, for example, targets Stat3, which is involved in regulation of T cell responses (55), and miR-223 targets Mef2c, which is necessary for the transcriptional activation of interleukins during peripheral T cell activation (56). The sequencing and qPCR studies described here have been performed on total shuttle RNA obtained from DC-T cell co-culture supernatant. Future studies will reveal which miRNAs (and other small non-coding RNAs) are released by DC or by T cells and which target cells can be modified by this shuttle RNA.
RNA repeats
We found that repeat sequences were highly enriched in shuttle RNA compared with cells and that especially the LINE, LTR and simple repeats were more abundant in shuttle RNA. Due to the size restriction of RNAs selected for sequencing, the detected transcripts do not represent full-length LINE or LTR. Small RNAs derived from these repeats (repeat-associated small interference RNA) have previously been shown to originate from various scattered regions within repeats such as SINE, LINE and LTR (57,58). Interestingly, retrotransposon RNA was recently found enriched in microvesicles derived from tumor cells (59). It is currently unknown whether this retrotransposon RNA can be transferred to other cells and whether it can insert into the target cell genome. However, the dissemination and active targeting of retrotransposable elements or fragments thereof to other cells may be an effective strategy of cells to modify genes and regulate gene expression in other cells.
A large number of sequences detected in shuttle RNA mapped to tRNA loci. Full-length tRNAs were, however, excluded from analysis by size selection of RNAs smaller than 70 nt. Although we cannot exclude low-level contamination with full-length tRNA, most tRNA hits will represent tRNA fragments (tRFs). Many recent publications have demonstrated that tRFs are not merely degradation products but are specific cleavage products with versatile functions, such as the inhibition of translation and guidance of other RNAs (60). We here demonstrated that the tRF compositions in cellular and shuttle RNA were different, indicating selectivity in released tRFs. Earlier evidence that tRFs can be released from cells and transported to distant cells comes from the field of plant physiology, where specific tRFs were identified in the phloem sap of several different plants (61). The 5′ end located tRNA fragments observed in shuttle RNA could represent tRNA halves, which are produced by a single cleavage event in the anticodon loop that cuts the mature tRNA into one 5′ and one 3′ halve. It has been shown that these halves are often not present at equal quantities, differ in functions, such as the induction of stress granule assembly (62), and can be recruited to distinct cytoplasmic structures (63). Our observation that different cleavage products of the same RNAs distributed unequally over cellular and shuttle RNA further strengthens the concept that cells select specific RNAs for extracellular release. We also observed that the sequence coverage on particular tRNA genes was different in shuttle RNA compared with cellular RNA. This could indicate that specific tRNA cleavage processes are involved in the generation of tRFs for dissemination to the extracellular space.
Infrastructural RNAs
We observed a remarkable enrichment of sequences matching SRP-RNA and the less studied infrastructural RNAs vault- and Y-RNA in shuttle RNA. Similar to tRNAs, all of these structural RNAs are transcribed by Polymerase III and function in association with cytoplasmic proteins. The full-length forms of these RNAs are longer than 70 nt, and only fragments were detected by sequencing. Small vRNA fragments (svRNA) covering the 5′ and 3′ end of the vRNA have been detected in human cells (32), and we detected similar fragments in cellular RNA derived from mouse DC-T cell co-cultures. Interestingly, in shuttle RNA we detected coverage on a different stem-loop structure. Future studies should reveal whether this specific fragment of vRNA can also be processed by Dicer and whether it can function in regulation of gene expression. Indications based on qPCR analysis suggest that not only fragments, but also full-length forms of SRP- and Y-RNA were present in shuttle RNA, more specifically, in low-density vesicles released by the cells. The relative high amounts of full-length SRP- and Y-RNA (Figure 6B) in cell-derived vesicles may explain the higher enrichment of SRP- and Y-RNA observed by qPCR in comparison with fragment sequencing. Y- and SRP-RNAs belong to a small group of 5–6 host RNAs that are also selectively incorporated together with viral genomes into capsids of several different viruses (51,52). It is suggested that these host RNAs are encapsidated to enhance virus assembly, virion stability and/or viral infectivity. Many enveloped viruses exploit the existing routes of membrane traffic to leave the host cell. A relationship between endosomal and/or plasma membrane routing of viruses and cell-derived vesicles can therefore be envisioned (64). We hypothesize that these RNAs could play a role in the specific sorting of regulatory RNAs into cell-derived vesicles. Alternatively, these RNAs could stabilize the RNA content of cell-derived vesicles or guide regulatory RNAs for efficient functioning upon release into the cytoplasm of target cells.
The abovementioned RNA biotypes make up only around 50% of the total transcripts present in shuttle RNA. We found that many of the other transcripts located to intergenic regions. One example is a highly abundant transcript matching a region located 3 kB upstream of the Tia1 gene, coding for T cell intracellular antigen 1, which is involved in apoptosis and mRNA sorting into stress granules. This non-annotated transcript locates to a highly conserved region and might be classified as PROMPT (28). The function of PROMPs is largely unknown, but could involve positively or negatively influencing the expression of downstream located genes. Future studies on the large number of (non-)annotated intergenic elements enriched in shuttle RNA may uncover additional gene regulatory sequences that are transferred between cells. Other issues that need to be addressed in the future include the deep sequencing of the larger, 70–200 nt fraction of small shuttle RNA and determining the cellular origin (DC or T cell) of the observed RNA species. The RNA in DC- and T cell-derived vesicles could for example be separately analyzed after absorption of vesicles released in DC-T cell co-cultures onto beads coated with cell-type specific antibodies.
Since the relative amount of small cellular RNA sequences that map to 18S and 28S rRNA is low, we expect that experimentally induced RNA degradation in our studies is limited. Although some of the abundant RNA species detected in shuttle RNA are cleavage products derived from mature non-coding RNAs, it is not likely that shuttle RNA represents non-selective disposal of RNAs that have been degraded inside cells. Non-selective disposal of degraded RNAs would lead to a much more similar distribution of the degradation products over cells and vesicles than indicated by our deep sequencing results. In fact, we even observed a highly unequal distribution of different degradation products of the same RNAs over cellular and shuttle RNA. Furthermore, evidence accumulates that (partly) degraded RNA fragments, such as tRFs, can also act as regulatory RNAs influencing gene expression. Uncontrolled release of these fragments would therefore impose a risk on disturbing cellular homeostasis.
By classification and quantification of the deep sequencing data, we gained important information on the type of RNAs selected by immune cells for extracellular release. This study revealed that shuttle RNA contains a wealth of different non-coding small RNAs. Many of the highly abundant small non-coding transcripts present in shuttle RNA are evolutionary well conserved and have previously been associated to gene regulatory functions. These findings allude to a wider range of biological effects that could be mediated by shuttle RNA than previously anticipated. Gene regulatory functions of shuttle RNA should therefore also be sought beyond the action of specific miRNAs and mRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Netherlands Genomics Initiative (NGI) Horizon Breakthrough Project [93519013]. Funding for open access charge: The Netherlands Organization for Scientific Research (NWO).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank H. de Boer-Brouwer and Y. Ariyurek for technical assistance and M.A. Nolte for critical reading of the manuscript. M.H.M. Wauben and P.A.C. 't Hoen contributed equally as senior authors.
REFERENCES
- 1.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 2.Silverman JM, Reiner NE. Exosomes and other microvesicles in infection biology: organelles with unanticipated phenotypes. Cell Microbiol. 2011;13:1–9. doi: 10.1111/j.1462-5822.2010.01537.x. [DOI] [PubMed] [Google Scholar]
- 3.Sadallah S, Eken C, Schifferli JA. Ectosomes as immunomodulators. Semin. Immunopathol. 2010;33:487–495. doi: 10.1007/s00281-010-0232-x. [DOI] [PubMed] [Google Scholar]
- 4.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 6.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 7.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 8.Buschow SI, Nolte-'t Hoen EN, van Niel G, Pols MS, ten Broeke T, Lauwen M, Ossendorp F, Melief CJ, Raposo G, Wubbolts R, et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic. 2009;10:1528–1542. doi: 10.1111/j.1600-0854.2009.00963.x. [DOI] [PubMed] [Google Scholar]
- 9.Nolte-'t Hoen EN, van der Vlist EJ, Aalberts M, Mertens HC, Bosch BJ, Bartelink W, Mastrobattista E, van Gaal EV, Stoorvogel W, Arkesteijn GJ, et al. Quantitative and qualitative flow cytometric analysis of nanosized cell-derived membrane vesicles. Nanomedicine. 2011;8:712–720. doi: 10.1016/j.nano.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segura E, Guerin C, Hogg N, Amigorena S, Thery C. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J. Immunol. 2007;179:1489–1496. doi: 10.4049/jimmunol.179.3.1489. [DOI] [PubMed] [Google Scholar]
- 11.Nolte-'t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113:1977–1981. doi: 10.1182/blood-2008-08-174094. [DOI] [PubMed] [Google Scholar]
- 12.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J. Biol. Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 14.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escudier B, Dorval T, Chaput N, Andre F, Caby MP, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of the first phase I clinical trial. J. Transl. Med. 2005;3:10. doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaput N, Taieb J, Schartz N, Flament C, Novault S, Andre F, Zitvogel L. The potential of exosomes in immunotherapy of cancer. Blood Cells Mol. Dis. 2005;35:111–115. doi: 10.1016/j.bcmd.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 18.Montecalvo A, Larregina AT, Shufesky WJ, Beer Stolz D, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2011;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 20.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 25.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 29.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 31.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 32.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat. Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 33.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauwen MM, Zwaveling S, de Quartel L, Ferreira Mota SC, Grashorn JA, Melief CJ, van der Burg SH, Offringa R. Self-tolerance does not restrict the CD4+ T-helper response against the p53 tumor antigen. Cancer Res. 2008;68:893–900. doi: 10.1158/0008-5472.CAN-07-3166. [DOI] [PubMed] [Google Scholar]
- 35.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buermans HP, Ariyurek Y, van Ommen G, den Dunnen JT, t Hoen PA. New methods for next generation sequencing based microRNA expression profiling. BMC Genomics. 2010;11:716. doi: 10.1186/1471-2164-11-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Hoon MJ, Taft RJ, Hashimoto T, Kanamori-Katayama M, Kawaji H, Kawano M, Kishima M, Lassmann T, Faulkner GJ, Mattick JS, et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20:257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hestand MS, Klingenhoff A, Scherf M, Ariyurek Y, Ramos Y, van Workum W, Suzuki M, Werner T, van Ommen GJ, den Dunnen JT, et al. Tissue-specific transcript annotation and expression profiling with complementary next-generation sequencing technologies. Nucleic Acids Res. 2010;38:e165. doi: 10.1093/nar/gkq602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2011;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Curr. Opin. Cell Biol. 2008;20:274–280. doi: 10.1016/j.ceb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5'- and 3'-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kedersha NL, Rome LH. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J. Cell Biol. 1986;103:699–709. doi: 10.1083/jcb.103.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol. Biol. Cell. 2009;20:1555–1564. doi: 10.1091/mbc.E08-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Garcia EL, Onafuwa-Nuga A, Sim S, King SR, Wolin SL, Telesnitsky A. Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J. Virol. 2009;83:12526–12534. doi: 10.1128/JVI.01219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Onafuwa-Nuga AA, King SR, Telesnitsky A. Nonrandom packaging of host RNAs in moloney murine leukemia virus. J. Virol. 2005;79:13528–13537. doi: 10.1128/JVI.79.21.13528-13537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE. 2010;5:e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng F, Wang HW, Cuenca A, Huang M, Ghansah T, Brayer J, Kerr WG, Takeda K, Akira S, Schoenberger SP, et al. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19:425–436. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- 56.Pan F, Ye Z, Cheng L, Liu JO. Myocyte enhancer factor 2 mediates calcium-dependent transcription of the interleukin-2 gene in T lymphocytes: a calcium signaling module that is distinct from but collaborates with the nuclear factor of activated T cells (NFAT) J. Biol. Chem. 2004;279:14477–14480. doi: 10.1074/jbc.C300487200. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 59.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hurto RL. Unexpected functions of tRNA and tRNA processing enzymes. Adv Exp. Med. Biol. 2011;722:137–155. doi: 10.1007/978-1-4614-0332-6_9. [DOI] [PubMed] [Google Scholar]
- 61.Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol. Biochem. Parasitol. 2010;171:64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.