Abstract
When a stop codon appears at the ribosomal A site, the class I and II release factors (RFs) terminate translation. In eukaryotes and archaea, the class I and II RFs form a heterodimeric complex, and complete the overall translation termination process in a GTP-dependent manner. However, the structural mechanism of the translation termination by the class I and II RF complex remains unresolved. In archaea, archaeal elongation factor 1 alpha (aEF1α), a carrier GTPase for tRNA, acts as a class II RF by forming a heterodimeric complex with archaeal RF1 (aRF1). We report the crystal structure of the aRF1·aEF1α complex, the first active class I and II RF complex. This structure remarkably resembles the tRNA·EF–Tu complex, suggesting that aRF1 is efficiently delivered to the ribosomal A site, by mimicking tRNA. It provides insights into the mechanism that couples GTP hydrolysis by the class II RF to stop codon recognition and peptidyl-tRNA hydrolysis by the class I RF. We discuss the different mechanisms by which aEF1α recognizes aRF1 and aPelota, another aRF1-related protein and molecular evolution of the three functions of aEF1α.
INTRODUCTION
In bacteria, eukaryotes and archaea, translation termination occurs when one of the three stop codons, UAA, UAG or UGA, appears at the ribosomal A site. This process involves the class I and II release factors (RFs) (1–5). The class I RFs enter the ribosomal A site, recognize the stop codons and hydrolyze the peptidyl-tRNA in the ribosomal P site (6). The class II RFs are translational GTPases that are required to complete the overall translation termination process (7–10).
In bacteria, the class I RFs, RF1 and RF2, recognize the UAG/UAA and UGA/UAA stop codons, respectively, by the recognition loop, including the PxT motif of RF1 or SPF motif of RF2 (11,12). After the recognition of the stop codon, they catalyze polypeptide release by their conserved GGQ motifs (13). The crystal structures of the Thermus thermophilus 70S ribosome in complexes with either RF1 or RF2 demonstrated that RF1/2 enters the ribosomal A site and hydrolyzes the peptidyl-tRNA by mimicking the shape of tRNA (14,15). The PxT/SPF motif, and the GGQ motif of RF1/2 correspond to the anticodon and the 3′-CCA end of the tRNA, respectively. After the polypeptide release by RF1/2, the class II RF (RF3) in the GDP-bound form interacts with the ribosome, and GDP is then exchanged with GTP. This induces conformational changes in RF3 and the ribosome, which result in the release of RF1/2. Finally, GTP hydrolysis causes the rapid dissociation of RF3 from the ribosome (8).
The translation termination mechanism in eukaryotes differs from that in bacteria. The eukaryotic class I RF, eRF1, adopts the overall tRNA-like configuration and catalyzes polypeptide release by the conserved GGQ motif, similarly to RF1/2 (16). However, in contrast to bacterial RF1/2, eRF1 recognizes all three stop codons (17). Various amino acid sequences within eRF1, including the conserved NIKS and YxCxxxF motifs, are implicated in stop codon recognition (18–22). In addition, eRF1 forms a heterodimeric complex with the eukaryotic class II RF, eRF3, an elongation factor 1 alpha (EF1α)-related translational GTPase (23), and the polypeptide release activity of eRF1 is enhanced by eRF3 (10). Therefore, eRF3 is considered to deliver eRF1 to ribosomes containing a stop codon in their A-sites, as EF1α delivers aminoacyl-tRNA to A-sites containing a sense codon. In the ribosome, eRF1 induces ribosome-dependent hydrolysis of GTP by eRF3 (24), which then causes eRF1 to adopt the conformation required to trigger polypeptide release (25). Recently, the crystal structures of human and Schizosaccharomyces pombe full-length eRF1 in complex with eRF3, but lacking the GTP-binding domain, were solved (26). These structures revealed that the interaction manner between eRF3 and eRF1 is partly analogous to that between EF–Tu (bacterial elongation factor) and tRNA. However, the overall structures of these complexes cannot be superposed on that of tRNA·EF–Tu (27), probably due to the absence of the GTP-binding domain. Therefore, the mechanism by which eRF3 precisely recognizes eRF1 and delivers it to ribosomes remains unclear.
The translation termination mechanism in archaea is considered to be similar to that in eukaryotes. The archaeal class I RF (aRF1), shares common sequence motifs with eRF1, including the GGQ, NIKS and YxCxxxF motifs and functions with all three stop codons on the mammalian 80S ribosome (28,29). Furthermore, the crystal structure of aRF1 is similar to that of eRF1 (29). As in the case of eRF1, the structure of aRF1 consists of three domains, A, B and C, and each domain is well superposed on the corresponding domain N, M and C of eRF1. The domain A/N harbors NIKS and YxCxxxF motifs, whereas the domain B/M harbors GGQ motif. Therefore, the domains A/N and B/M are responsible for the stop codon recognition and polypeptide release, respectively. However, a unique feature of translation termination in archaea is that the archaeal EF1α (aEF1α) acts as the class II RF. aEF1α interacts, in a GTP-dependent manner, with not only tRNA but also aRF1 and aPelota, an aRF1-related protein involved in mRNA surveillance (29). This strongly suggests that aEF1α performs three different functions: translation elongation, translation termination and mRNA surveillance. Recently, we reported the crystal structure of the aPelota·aEF1α complex (30). Based on this structure, a model of the aRF1·aEF1α complex was constructed, which is similar to the crystal structure of the tRNA·EF–Tu complex (29). Although this model is well supported by mutational analyses, the participation of the crucial amino acid residues for molecular interplay between the class I and class II eRFs, revealed in previous genetic analyses, was not consistently elucidated. Therefore, the precise mechanism by which the class I RFs are bound to the class II RFs and then delivered to the ribosome still remains to be determined, for both eukaryotes and archaea. The experimentally determined class I and II RF complex structure is thus requisite to fully understand the translation termination mechanisms in eukaryotes and archaea.
Here, we present the complex structure of aRF1 and GTP-bound aEF1α determined at 2.3 -Å resolution. This is the first experimentally determined structure of the active class I and II RF complex, and it reveals how GTP-bound aEF1α recognizes aRF1, at an atomic resolution. Our findings also provide novel insight into the mechanism by which aRF1 recognizes stop codons and catalyzes polypeptide release in the ribosome. Furthermore, an engineered aEF1α with two mutations still binds aRF1, but no longer binds aPelota, suggesting specific recognition modes between aRF1 and aPelota and implying the molecular evolution of the three functions of aEF1α.
MATERIALS AND METHODS
Sample preparation and crystallization
aRF1 and aEF1α were expressed in Escherichia coli and purified as previously described (29). aRF1 and aEF1α were concentrated by ultrafiltration to 20 and 30 mg/ml, respectively, and then were mixed at a molar ratio of 1:1. The protein solution (8.7 mg/ml aRF1 and 9.9 mg/ml aEF1α, 5 mM GTP and 5 mM MgCl2) was used for crystallization screening of the aRF1·aEF1α·GTP complex. However, despite extensive efforts, we only obtained poorly diffracting crystals. We then tried in situ proteolysis, by incorporating a trace amount of protease in the crystallization screening (31), in which the protein was incubated with chymotrypsin (1:1000 wt/wt) and screened for crystallization. Crystals of the aRF1·aEF1α·GTP complex were grown by the hanging drop vapor diffusion method at 20°C, by mixing equal amounts of the protein solution and the reservoir solution (23% PEG4000, 200 mM (NH4)2SO4, 10 mM l-proline, 100 mM Bis–Tris, pH 5.5). The crystals were cryoprotected in 20% ethylene glycol, 27.6% PEG4000, 240 mM (NH4)2SO4, 12 mM l-proline and 120 mM Bis–Tris, pH 5.5 and were flash-cooled at 100 K.
Data collection, structure determination and refinement
Diffraction data were collected at beamline BL32XU at SPring-8 (Harima, Japan), and were processed with the program HKL2000 (HKL Research). The data collection statistics are summarized in Supplementary Table S1. Phases were obtained by molecular replacement with the program MOLREP (32), using the structure of the isolated form of aRF1 (29) and that of aEF1α in the aPelota·aEF1α complex structure (30) as search models. The crystallographic asymmetric unit contains one complex of aRF1·aEF1α. The atomic model was manually built using the program COOT (33), and iterated cycles of refinement with PHENIX (34) and manual rebuilding with COOT were performed at 2.3 Å resolution. The final model was refined to an Rwork of 20.1%, with an Rfree of 26.1%. The structure refinement statistics are summarized in Supplementary Table S2.
Yeast two-hybrid assays
For the two-hybrid assays, the genes encoding the wild-type and mutated aRF1 were constructed in the AD vectors. For the BD vectors, the wild-type and mutated aEF1α sequences were cloned into the pGBT9 vector (Clontech). The in vivo two-hybrid assay was performed with the S. cerevisiae AH 109 strain, using the same procedures and conditions as described previously (35).
Pull-down assays
The gene encoding Aeropyrum pernix aEF1α (residues 7–437) was cloned into pET26b vector and expressed in the E. coli Rosetta2(DE3) strain, as the C-terminally His-tagged protein. aEF1α was purified as previously described (30), except that the His-tag was not cleaved. The N-terminally His-tagged aPelota was expressed and purified by chromatography on a Ni–NTA Superflow column (Qiagen), as previously described (30). The His-tag was then cleaved with thrombin. Finally, the product was reapplied to the Ni–NTA Superflow column, and the flow-through fraction was collected. aRF1 was expressed and purified as previously described (29). The purified proteins were mixed at a molar ratio of aEF1α:aRF1 = aEF1α:aPelota = 1:3 in the presence of 5 mM GTP and 5 mM MgCl2, and incubated at 70°C for 1 min. MagneHis™ Ni particles (Promega) were then added to immobilize the His-tagged aEF1α. After the beads were washed, the immobilized proteins were eluted with the buffer containing 300 mM imidazole and analyzed by Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE).
RESULTS AND DISCUSSION
Overall structure
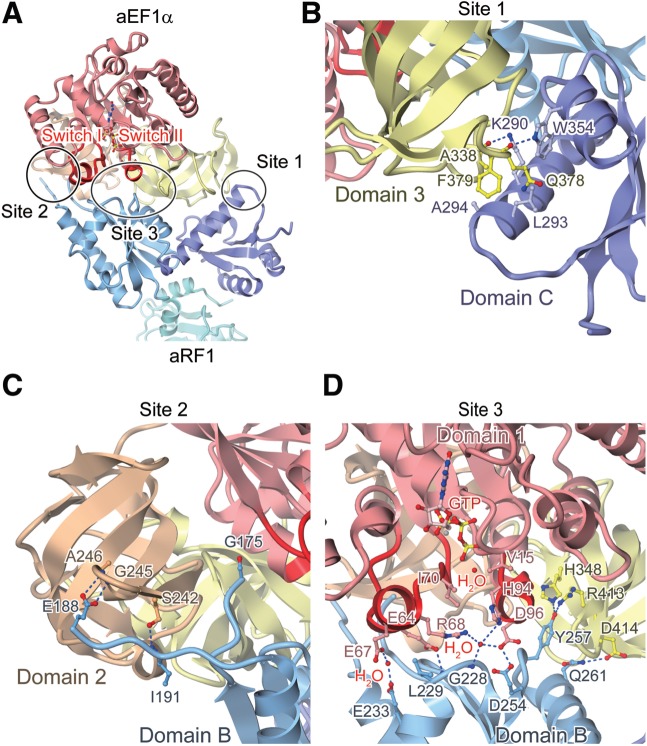
We determined the complex structure of aRF1 and GTP-bound aEF1α from A. pernix at 2.3 Å resolution, by the molecular replacement method (Figure 1; see also Supplementary Tables S1 and S2). The structural coordinates of the GTP-bound aEF1α in the aPelota·aEF1α complex structure (30) and those of the isolated form of aRF1 (29) were used as search models. The overall structure of GTP-bound aEF1α in the complex is similar to that in the complex with aPelota and consists of three domains: 1, 2 and 3 (Figure 1). Domain 1 of aEF1α, which corresponds to the GTP-binding domain of eRF3, recognizes GTP through one Mg2+ ion and several water molecules (Supplementary Figure S1). The overall structure of aRF1 also consists of three domains, A, B and C (Figure 1). The domain arrangement is similar to that in the isolated form of aRF1, with an rms deviation of 1.1 Å for all Cα atoms. In domain B, a single peptide bond is cleaved between Tyr187 and Glu188, as confirmed by the N-terminal sequence analysis, and residues Lys176–Tyr187 are structurally disordered, and thus are not included in the model. This is the result of the in situ proteolysis during crystallization (31) (see ‘Materials and Methods’ section). In addition, residues Met1–Glu3 and Lys431–Lys437 of aEF1α, and Met1–Glu8 and Thr372–Val373 of aRF1 are also structurally disordered.
Figure 1.
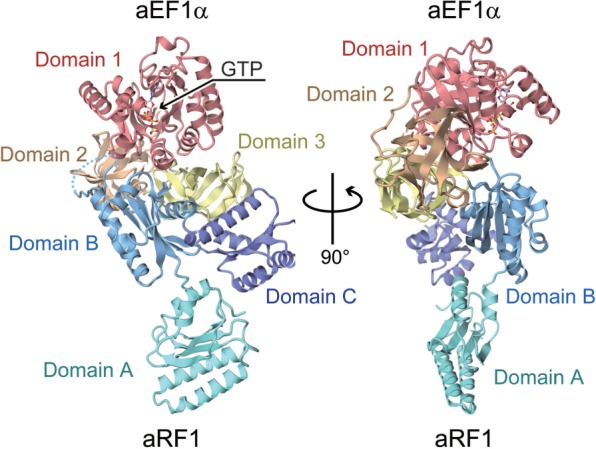
Overall structure of the aRF1·aEF1α·GTP complex, viewed from two perpendicular directions. Domains 1, 2 and 3 of aEF1α are colored red, brown and yellow, respectively; Domains A, B and C of aRF1 are colored turquoise, light blue and purple, respectively. The bound GTP is shown by a ball-and-stick model. The disordered loop of the residues Lys176–Tyr187 in aEF1α domain B is represented as the dashed line.
Interactions between aRF1 and aEF1α
In the complex structure, all three domains of aEF1α are involved in the intermolecular interactions, while the N-terminal domain A of aRF1 does not contact aEF1α. The interaction interface between aRF1 and aEF1α is divided into three sites (Sites 1, 2 and 3) (Figure 2A), and their detailed interaction manners are shown in Figure 2B–D.
Figure 2.
Interaction sites between aRF1 and aEF1α. Proteins are depicted by ribbon models, with the domains color-coded as in Figure 1. Hydrogen bonds and salt bridges are indicated by dashed blue lines. (A) The interaction interface between aRF1 and aEF1α, which is divided into three sites, sites 1, 2 and 3. The switch I and II regions of aEF1α are highlighted in red. (B) The interactions between aRF1 domain C and aEF1α domain 3 (Site 1). (C) The interactions between aRF1 domain B and aEF1α domain 2 (Site 2). (D) The interactions between aRF1 domain B and aEF1α domains 1 and 3 (Site 3). Water molecules and magnesium ions are depicted by red and grey ball-and-stick models, respectively.
In Site 1, domain C of aRF1 interacts with domain 3 of aEF1α by hydrophobic interactions and several electrostatic interactions (Figure 2B). The side chain of Phe379 of aEF1α, which is widely conserved between aEF1α and eRF3 (29), interacts with the side chains of Leu293 and Ala294 in aRF1 through hydrophobic contacts (Figure 2B). The side chain of Trp354 of aRF1 forms a hydrogen bond with the main chain oxygen of Gln378 of aEF1α, and the side chain of Lys290 of aRF1 electrostatically interacts with the main chain oxygen of Ala338 of aEF1α (Figure 2B). These observations are consistent with our previous report, showing that Ala338 and Phe379 of aEF1α domain 3, and Lys290, Leu293 and Trp354 of aRF1 domain C play important roles in complex formation (29).
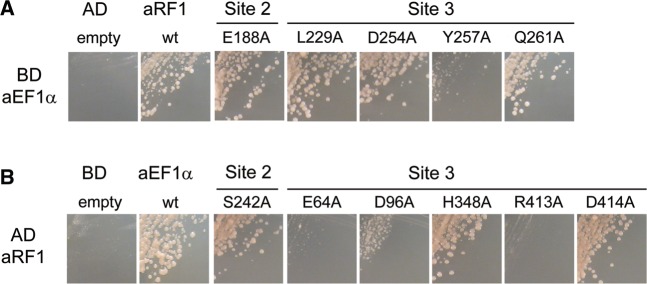
In Site 2, the loop from Gly171 to Met194 of aRF1, containing the highly conserved GGQ motif (from Gly180 to Gln182 for A. pernix aRF1), was cleaved by chymotrypsin during crystallization, and interacts with domain 2 of aEF1α (Figure 2C). The cleavage of this loop implies that it is exposed to the solvent and has a highly mobile structure. Unlike the labile ester linkage coupling an amino acid to a tRNA, the GGQ motif of aRF1 does not have to be accommodated and protected in the aEF1α pocket for the amino acid-bound A76 of the tRNA CCA end. Therefore, it is quite likely that the pocket of aEF1α is not used for the recognition of aRF1, as in the case of the recognition of aPelota (30). Consistent with this notion, the E188A mutation in aRF1, as well as the S242A mutation in aEF1α, did not affect the yeast two-hybrid interactions between aRF1 and aEF1α (Figure 3A and B), although the side chains of both residues seem to contact the counter molecules weakly (Figure 2C).
Figure 3.
Yeast two-hybrid analysis of aRF1 binding to aEF1α at Sites 2 and 3. (A) Two-hybrid analysis (3-d growth) of mutants of residues in Sites 2 and 3 in aRF1 (activation domain, AD) against aEF1α wild-type (binding domain, BD). (B) Two-hybrid analysis (3-d growth) of mutants of residues in Sites 2 and 3 in aEF1α (BD) against aRF1 wild-type (AD).
In Site 3, an extensive hydrogen-bonding network is formed between domains 1 and 3 of aEF1α, and domain B of aRF1 (Figure 2D). The side chains of Glu64, Glu67 and Asp96 of aEF1α contact Leu229, Glu233 and Asp254 of aRF1, respectively (Figure 2D). Among these residues, Glu64 and Asp96 of aEF1α play essential roles in complex formation (Figure 3B). In addition, the His94 side chain in aEF1α is fixed by a well-ordered water molecule, coordinated by the side chains of the conserved Arg68 and Asp96 of aEF1α and by the main chain nitrogen of Gly228 of aRF1 (Figure 2D). His94 of aEF1α corresponds to His84 of E. coli EF–Tu, which is considered to act as a general base to allow the catalytic water molecule to attack the γ-phosphate of GTP (36). The catalytic water molecule is present in-line with the γ-phosphate of GTP (Figure 2D). In concert with the hydrophobic gate formed by Val15 and Ile70 of aEF1α, these interactions may sequester the side chain of His94 from the catalytic site of the GTPase, thereby preventing the activation of the catalytic water molecule, and thus stabilizing the GTP form of aEF1α (Figure 2D). Furthermore, in domain 3 of aEF1α, the side chains of His348 and Arg413 interact with Tyr257 of aRF1, while the Asp414 side chain contacts Gln261 of aRF1. These residues are highly conserved between eRF3 and aEF1α, consistent with the previous consideration that the corresponding residues of S. pombe eRF3 may constitute, or exist nearby, the potential eRF1-binding site although their counterparts on eRF1/aRF1 were not identified (37). Our functional analyses revealed that while the mutations of His348 and Asp414 of aEF1α and Gln261 of aRF1 do not affect the yeast two-hybrid binding, the R413A mutation in aEF1α and the Y257A mutation in aRF1 both exhibit severe defects (Figure 3A and B).
Structural similarity of aRF1·aEF1α to tRNA ·EF–Tu and aPelota·aEF1α
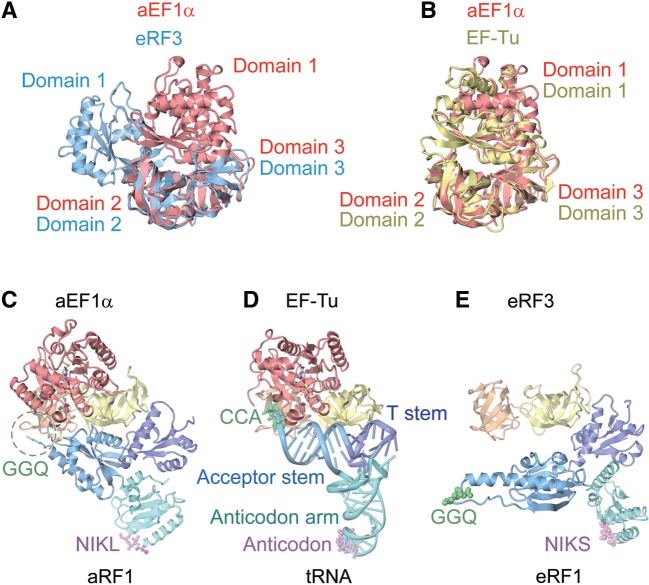
As expected from the sequence similarity between aEF1α and eRF3, the structures of the three domains of aEF1α are similar to those of the corresponding domains of the isolated eRF3 bound to the nonhydrolyzable GTP analog, GDPNP (37) (rmsd values of 1.6 Å, 1.1 Å and 1.3 Å, for domains 1, 2 and 3, respectively). However, the relative orientation of domain 1 with respect to domains 2 and 3 in aEF1α is quite different from that in eRF3 (Figure 4A). The structure of aEF1α superposes better on that of EF–Tu in the tRNA·EF–Tu·GDPNP complex (27) than on that of the isolated eRF3 (Figure 4A and B). This is probably due to the fact that, in the complex structure, the domain 1 of aEF1α interacts with aRF1 domain B through the GTP-bound switch I and switch II regions (Figure 2D). Due to this interaction, the switch I and switch II regions, which were disordered in the isolated eRF3 structure, are stabilized and clearly visible in the present structure of aEF1α in complex with aRF1 and GTP (Supplementary Figure S2A and B). This is consistent with the previous observations that eRF1 stimulates GTP binding to eRF3 (38,39) and that aEF1α interacts with aRF1 in a GTP-dependent manner (29). In addition, this interaction stabilizes the GTP form of aEF1α by sequestering the side chain of His94, a general base, from the GTPase catalytic site (Figure 2D). This may facilitate the delivery of aRF1 to the ribosomal A site by aEF1α in the GTP form.
Figure 4.
Comparison of the aRF1·aEF1α·GTP complex structure with related structures. (A) Superposition of domains 2 and 3 of aEF1α in the present complex (red) with those of the GDPNP-bound form of eRF3 (blue) (PDB ID: 1R5O). (B) Superposition of domains 2 and 3 of aEF1α in the present complex (red) with those of the GDPNP-bound form of EF–Tu (yellow) bound to tRNA (PDB ID: 1TTT). (C) The complex structure of aRF1·aEF1α·GTP (this work). Domains are color-coded as in Figure 1. (D) The complex structure of tRNA·EF–Tu·GDPNP (PDB ID: 1TTT). The domains of EF–Tu are color-coded as in aEF1α. The tRNA is colored turquoise (anticodon arm), light blue (acceptor stem) and purple (T stem). The bound GDPNP is depicted by a ball-and-stick model. (E) The complex structure of eRF1 and eRF3 lacking the GTP-binding domain (PDB ID: 3E1Y). Domains are color-coded as in aRF1·aEF1α.
As for aRF1, it is clear that aRF1 binds to the tRNA recognition site of aEF1α by mimicking the shape of tRNA, with domains A, B and C of aRF1 corresponding to the anticodon arm, acceptor stem and T stem of tRNA, respectively (Figure 4C and D). In the presence of GTP-bound domain 1 of aEF1α, the structure of aRF1 more closely resembles that of tRNA than that of eRF1 in the eRF1·eRF3 complex structure, lacking the GTP-binding domain (26) (Figure 4C–E). Furthermore, the polypeptide-hydrolyzing GGQ motif and the stop codon-recognizing NIKS motif of aRF1 (NIKL for A. pernix aRF1) correspond well to the amino acid-bound CCA end and the anticodon of tRNA, respectively (Figure 4C and D). Therefore, the overall structure of the aRF1·aEF1α complex resembles that of the tRNA·EF–Tu·GDPNP ternary complex (27). This strongly supports our proposal that aEF1α delivers both aRF1, in translation termination and tRNA, in translation elongation, to the common ribosomal A site.
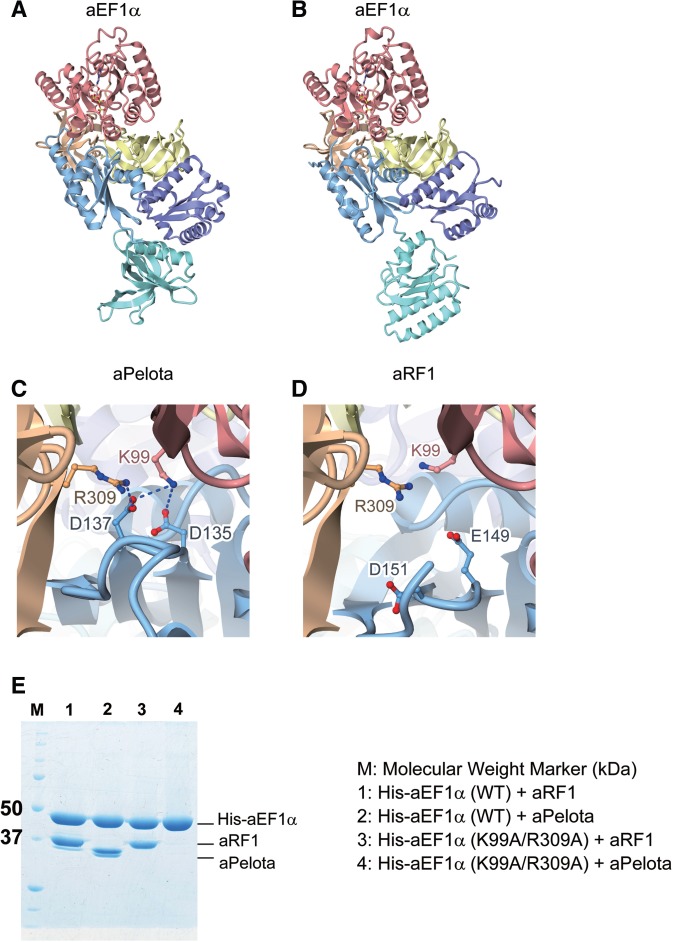
A comparison of the present structure with that of the aPelota·aEF1α·GTP ternary complex suggests that the interaction interface between aEF1α and aRF1 is similar to that between aEF1α and aPelota (30) (Figure 5A and B). Therefore, aEF1α can interact with both proteins, since the aEF1α-binding domains B and C of aRF1 and aPelota adopt similar structures. However, the detailed intermolecular interaction manners are different between these two complex structures. For instance, Lys99 and Arg309 of aEF1α form salt bridges with Asp 135 and Asp137 of aPelota, but not with the corresponding conserved residues of aRF1 (Glu149 for Asp135, and Asp151 for Asp137) (Figure 5C and D). This is supported by the results of our in vitro binding assay, which demonstrated that the K99A/R309A double mutation of aEF1α abolished its ability to bind aPelota, but not aRF1 (Figure 5E). Asp135 and Asp137 of aPelota constitute a negatively charged patch on its surface, which coincides well with the phosphate groups of the nucleotide residues at positions 1, 2, 66 and 67 in the tRNA acceptor stem (30). Lys99 of aEF1α corresponds to Lys90 of Thermus aquaticus EF–Tu, which recognizes these phosphate groups (27). Therefore, while both aPelota and aRF1 mimic the shape of tRNA and bind to the tRNA recognition site of aEF1α, aPelota further mimics the charge distribution on the surface of the tRNA molecule for the interaction, in contrast to aRF1. On the other hand, Arg413 of aEF1α recognizes aRF1 and plays essential role in complex formation (Figures 2D and 3B), but not aPelota. These imply that aEF1α can distinguish aPelota from aRF1 by the partner-specific interactions. In the molecular evolution process, these partner-specific interactions may enable omnipotent aEF1α to differentiate into three unipotent eukaryotic translational GTPases: EF1α, eRF3 and Hbs1.
Figure 5.
Comparison of the complex structure of aRF1·aEF1α·GTP with that of aPelota·aEF1α·GTP. Domains are color-coded as in Figure 1. (A) aPelota·aEF1α·GTP complex structure (PDB ID: 3AGJ). (B) aRF1·aEF1α·GTP complex structure (this work). (C) The interaction interface between aPelota and aEF1α. Lys99 and Arg309 of aEF1α, and Asp135 and Asp137 of aPelota are depicted by ball-and-stick models. (D) The interaction interface between aRF1 and aEF1α. Lys99 and Arg309 of aEF1α, and Glu149 and Asp151 of aRF1 are depicted by ball-and-stick models. (E) In vitro binding assay of wild-type (WT) and mutant (K99A/R309A) aEF1α to aRF1 and aPelota. His-tagged aEF1α (His-aEF1α) was mixed with aRF1 or aPelota, immobilized by MagneHis™ Ni particles and then eluted. Eluted fractions were analyzed by SDS–PAGE.
Furthermore, the domain A structures are totally different between aPelota and aRF1 (Figure 5A and B). aPelota is delivered by aEF1α to the vacant A site of a ribosome stalled on an aberrant mRNA, and triggers mRNA surveillance. In this process, domain A of aPelota is considered to recognize the decoding center of the stalled ribosome rather than the mRNA, in contrast to the recognition of an mRNA stop codon by aRF1 domain A. Therefore, the differences in the domain A structures of aRF1 and aPelota may explain their functional specificities.
Stop codon recognition by aRF1
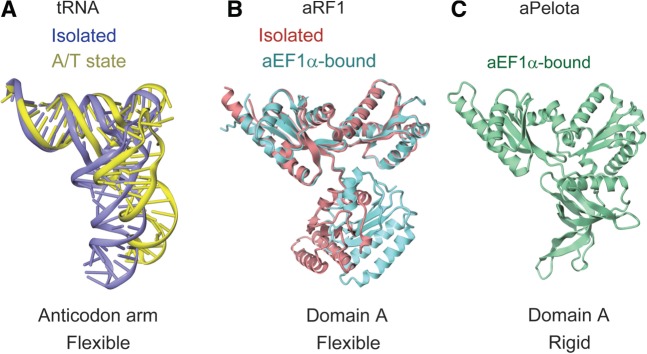
The precise mechanism of sense codon decoding by tRNA was clearly elucidated in previous studies (40,41). In the ribosome, the structure of the EF–Tu-bound tRNA is distorted, as compared to that in the isolated tRNA·EF–Tu complex. In this A/T state conformation, the tRNA anticodon arm is bent and the spatial arrangement of the D stem versus the acceptor-T stem is changed, resulting in the bending of the tRNA L-shape by ∼30° (Figure 6A). By bending its flexible anticodon arm, the tRNA correctly samples the codon–anticodon pairing, which directly couples the codon recognition to the GTPase activation of EF–Tu. Therefore, the flexibility of the tRNA anticodon arm is essential for correct decoding.
Figure 6.
Structural comparison of tRNA, aRF1 and aPelota. (A) Comparison of EF–Tu-bound tRNA in the isolated (PDB ID: 1TTT) and the ribosome-bound A/T state (PDB ID: 2XQD) forms, colored purple and yellow, respectively. (B) Comparison of aRF1 in the isolated (PDB ID: 3AGK) and aEF1α-bound (this work) forms, colored red and turquoise, respectively. (C) The structure of aPelota bound to aEF1α (PDB ID: 3AGJ).
Domain A of aRF1, which corresponds to the anticodon arm of tRNA and is involved in stop codon recognition, also seems to be flexible, since the relative orientation of domain A with respect to domains B and C is different between the isolated form of aRF1 and that in the complex with aEF1α (Figure 6B). Therefore, aRF1 is considered to mimic not only the shape but also the anticodon arm flexibility of tRNA. In this manner, aRF1 may correctly recognize stop codons by adopting a conformation similar to the A/T state tRNA, through the flexibility of domain A.
In contrast to aRF1, domain A of aPelota adopts a rigid conformation by forming several hydrogen bonds with its domain C (Figure 6C) (30). Due to the rigid conformation of domain A, aPelota strictly mimics the A/T state tRNA prior to the interaction with the ribosome. The A/T state tRNA mimicry is essential for mRNA surveillance by aPelota because it may enable aPelota to enter the ribosomal A site and directly recognize the decoding center, regardless of the codon sequence at the A site. In order to enter the vacant A site of a ribosome stalled by an aberrant mRNA, which may occur irrespective of the mRNA codon in the A site, and to facilitate mRNA surveillance, aPelota should be able to recognize the decoding center of the ribosome in an A site codon-independent manner. Therefore, aPelota accomplishes its function by mimicking A/T state tRNA. In contrast, translation termination by aRF1, as well as translation elongation by an aminoacyl-tRNA, should strictly occur in a codon-dependent manner in the ribosomal A site. In this sense, aRF1 is functionally more similar to tRNA than aPelota. Therefore, although both aRF1 and aPelota bind to aEF1α and are delivered to the ribosomal A site by mimicking the shape of tRNA, at the functional level, the stop codon-specific aRF1 mimics an isolated tRNA by its flexible domain A, whereas the codon-nonspecific aPelota strictly mimics A/T tRNA by its rigid domain A.
Peptidyl-tRNA hydrolysis by aRF1
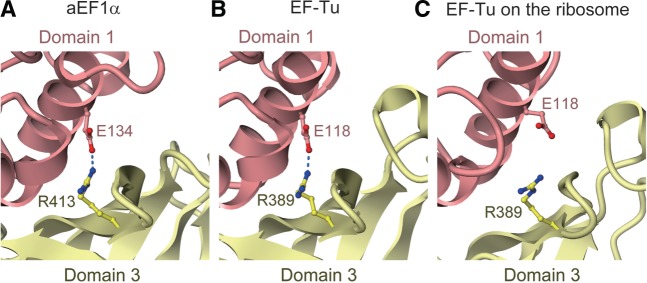
By analogy with the translation elongation cycle, the recognition of a stop codon by aRF1 may primarily cause conformational changes of aEF1α in the switch I and switch II regions, thus triggering the GTPase activity of aEF1α via catalytic water molecule activation by His94. Intriguingly, Arg413, which resides in domain 3 of aEF1α and contacts Tyr257 of aRF1 (Figure 2D), forms an additional contact with Glu134 in domain 1 of aEF1α (Figure 7A). This inter-domain contact of aEF1α is also observed in the tRNA·EF–Tu·GDPNP ternary complex (27), while they are sequestered from each other in the tRNA·EF–Tu·GDPCP complex on the ribosome, due to the conformational change between domains 1 and 3 of EF–Tu (41) (Figure 7B and C). The side chain contact between Arg413 of aEF1α and Tyr257 of aRF1 is necessary for the binding between aEF1α and aRF1 (Figure 3A and B), although no interaction between the corresponding residues was observed in the crystal structures of human and S. pombe full-length eRF1 in complexes with eRF3 lacking the GTP-binding domain (26). Tyr257 of aRF1 is not conserved in aPelota, and no similar contact between aPelota and aEF1α was observed in the aPelota·aEF1α·GTP complex, which exhibited much lower GTP hydrolysis activity on the Pyrococcus horikoshii ribosome (29). These observations suggested that Arg413 of aEF1α, which mediates the interaction between aRF1 and aEF1α domain 1, couples the stop codon recognition by aRF1 with the GTP hydrolysis by aEF1α on the ribosome. Subsequently, following GTP hydrolysis on the ribosome, aEF1α in the GDP form probably loses its affinity for aRF1 and may dissociate from the ribosome. The question then arises as to how aRF1 hydrolyzes the peptidyl-tRNA in the ribosomal P site by its GGQ motif. The crystal structure of the T. thermophilus ribosome in complex with RF1 revealed that in the bacterial system, the main chain amide group of the Gln residue in the GGQ motif plays a crucial role in the catalysis, by either coordinating a water molecule to attack the P site tRNA or stabilizing the transition state (14). Since aRF1s also contain the GGQ motif conserved among the archaeal species, this motif may play essentially the same role as that in the bacterial system. Therefore, in the ribosome, aRF1 may adopt a conformation that places its GGQ motif in close proximity to the amino acid-bound CCA end of the tRNA.
Figure 7.
Inter-domain interaction between domains 1 and 2 of aEF1α and EF–Tu through conserved arginine and glutamate residues (Glu134 and Arg413 for aEF1α, and Glu118 and Arg389 for EF–Tu). Domains are color-coded as in Figure 1. (A) aEF1α in the aRF1·aEF1α·GTP complex structure (this work). (B) EF–Tu in the tRNA·EF–Tu·GDPNP complex structure (PDB ID: 1TTT). (C) EF–Tu in the tRNA·EF–Tu·GDPCP complex structure on the ribosome (PDB ID: 2XQD).
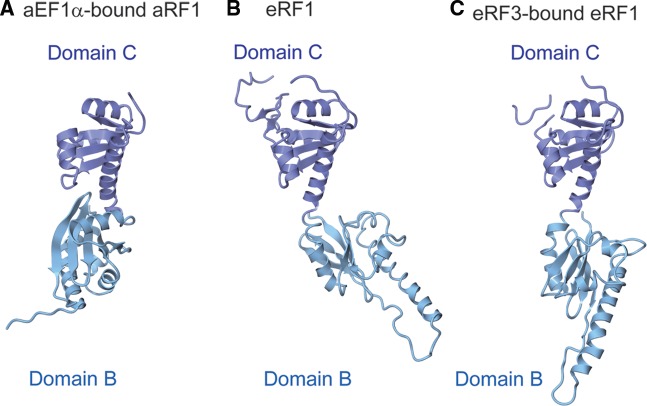
One of the possible conformational changes enabling GGQ motif relocation may occur in the linker helices connecting domains B and C. The arrangement of domains B and C of aRF1 in the present structure is considerably different from that in eRF1 (Figure 8A–C). The linker helix connecting them is bent in the structure of aRF1, whereas it is straightened in eRF1. However, the amino acid sequences of these linker helices are well conserved between aRF1 and eRF1 (29). Therefore, it is likely that domains B and C of aRF1 can also be arranged similarly to those of eRF1. This may reflect the intrinsic flexibility of this region. In the ribosome, the linker helices may provide the flexibility required to properly position the GGQ motif for peptidyl-tRNA hydrolysis. Further clarification of the mechanism of anticodon recognition and peptidyl-tRNA hydrolysis coupled with GTP hydrolysis will require the structure determination of the aRF1·aEF1α complex bound to the ribosome.
Figure 8.
Comparison of the arrangements of domains B and C in (A) aRF1 bound to aEF1α (this work), (B) eRF1 (PDB ID: 1DT9) and (C) eRF1 bound to eRF3 lacking the GTP-binding domain (PDB ID: 3E1Y). Domains are color-coded as in Figure 1.
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3VMF).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2 and Supplementary Tables 1 and 2.
FUNDING
The Japan Society for the Promotion of Science (JSPS) through its ‘Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST program)’ (to O.N.); a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to O.N.); a Grant-in-Aid for Scientific Research on Innovative Areas from MEXT (to R.I. and O.N.) and grants from The Uehara Memorial Foundation (to O.N.). Funding for open access charge: The Japan Society for the Promotion of Science (JSPS) through its ‘Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST program)’.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the beam-line staff at BL32XU of SPring-8 for assistance in data collection, and to the RIKEN BioResource Center (Ibaraki, Japan) for providing the A. pernix genomic DNA. The authors also thank Dr Naoshi Dohmae for the N-terminal sequence analysis of aRF1.
REFERENCES
- 1.Capecchi MR. Polypeptide chain termination in vitro: isolation of a release factor. Proc. Natl Acad. Sci. USA. 1967;58:1144–1151. doi: 10.1073/pnas.58.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grentzmann G, Brechemierbaey D, Heurgue V, Mora L, Buckingham RH. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc. Natl Acad. Sci. USA. 1994;91:5848–5852. doi: 10.1073/pnas.91.13.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikuni O, Ito K, Moffat J, Matsumura K, Mccaughan K, Nobukuni T, Tate W, Nakamura Y. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc. Natl Acad. Sci. USA. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Teravanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 Genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhouravleva G, Frolova L, Legoff X, Leguellec R, Ingevechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura Y, Ito K. Making sense of mimic in translation termination. Trends Biochem. Sci. 2003;28:99–105. doi: 10.1016/S0968-0004(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 7.Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 8.Gao HX, Zhou ZH, Rawat U, Huang C, Bouakaz L, Wang CH, Cheng ZH, Liu YY, Zavialov A, Gursky R, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 9.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 11.Ito K, Uno M, Nakamura Y. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 12.Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurberg M, Asahara H, Korostelev A, Zhu JY, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 15.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song HW, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 17.Frolova L, Legoff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 18.Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seit-Nebi A, Frolova L, Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolosov P, Frolova L, Seit-Nebi A, Dubovaya V, Kononenko A, Oparina N, Justesen J, Efimov A, Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lekomtsev S, Kolosov P, Bidou L, Frolova L, Rousset JP, Kisselev L. Different modes of stop codon restriction by the stylonychia and paramecium eRF1 translation termination factors. Proc. Natl Acad. Sci. USA. 2007;104:10824–10829. doi: 10.1073/pnas.0703887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL. Termination of translation in eukaryotes is mediated by the quaternary eRF1·eRF3·GTP·Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res. 2006;34:3947–3954. doi: 10.1093/nar/gkl549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frolova L, LeGoff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 25.Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol. Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong CG, Lim M, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BFC, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 28.Dontsova M, Frolova L, Vassilieva J, Piendl W, Kisselev L, Garber M. Translation termination factor aRF1 from the archaeon Methanococcus jann aschii is active with eukaryotic ribosomes. FEBS Lett. 2000;472:213–216. doi: 10.1016/s0014-5793(00)01466-6. [DOI] [PubMed] [Google Scholar]
- 29.Saito K, Kobayashi K, Wada M, Kikuno I, Takusagawa A, Mochizuki M, Uchiumi T, Ishitani R, Nureki O, Ito K. Omnipotent role of archaeal elongation factor 1 alpha (EF1α) in translational elongation and termination, and quality control of protein synthesis. Proc. Natl Acad. Sci. USA. 2010;107:19242–19247. doi: 10.1073/pnas.1009599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi K, Kikuno I, Kuroha K, Saito K, Ito K, Ishitani R, Inada T, Nureki O. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1α complex. Proc. Natl Acad. Sci. USA. 2010;107:17575–17579. doi: 10.1073/pnas.1009598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong A, Xu X, Edwards AM, Chang C, Chruszcz M, Cuff M, Cymborowski M, Di Leo R, Egorova O, Evdokimova E, et al. In situ proteolysis for protein crystallization and structure determination. Nat. Methods. 2007;4:1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Ito K, Ebihara K, Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarano G, Krab IM, Bocchini V, Parmeggiani A. Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: its substitution by glutamine and alanine. FEBS Lett. 1995;365:214–218. doi: 10.1016/0014-5793(95)00469-p. [DOI] [PubMed] [Google Scholar]
- 37.Kong CG, Ito K, Walsh MA, Wada M, Liu YY, Kumar S, Barford D, Nakamura Y, Song HW. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 38.Hauryliuk V, Zavialov A, Kisselev L, Ehrenberg M. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 2006;88:747–757. doi: 10.1016/j.biochi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Pisareva VP, Pisarev AV, Hellen CUT, Rodnina MV, Pestova TV. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J. Biol. Chem. 2006;281:40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- 40.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The Mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.