Abstract
Initiation of σ54-dependent transcription requires assistance to melt DNA at the promoter site but is impeded by numerous protein–protein and nucleo–protein interactions. To alleviate these inhibitory interactions, hexameric bacterial enhancer binding proteins (bEBP), a subset of the ATPases associated with various cellular activities (AAA+) protein family, are required to remodel the transcription complex using energy derived from ATP hydrolysis. However, neither the process of energy conversion nor the internal architecture of the closed promoter complex is well understood. Escherichia coli Phage shock protein F (PspF), a well-studied bEBP, contains a surface-exposed loop 1 (L1). L1 is key to the energy coupling process by interacting with Region I of σ54 (σ54RI) in a nucleotide dependent manner. Our analyses uncover new levels of complexity in the engagement of a multimeric bEBP with a basal transcription complex via several L1s. The mechanistic implications for these multivalent L1 interactions are elaborated in the light of available structures for the bEBP and its target complexes.
INTRODUCTION
In bacteria, the multi-subunit core RNA polymerase (RNAP or E) catalyses transcription and is directed to the promoter DNA by association with a sigma factor (σ). Bacterial σ factors fall into two classes: σ70 binds to the consensus sequences at –35 (TTGACA) and –10 (TATAAT), whereas σ54 binds to the consensus sequences at –24 (GG) and –12 (GC). In σ70-dependent transcription, RNAP forms a closed complex (RPC) on the promoter that can spontaneously isomerize to an open complex (RPO) (1). In σ54-dependent transcription, the isomerization from RPC to RPO is energetically unfavourable due to the presence of a stably engaged upstream fork junction DNA around the –12 site. Within the stable RPC, the –12 fork junction is evident (2), although the downstream DNA melting has not occurred and the +1 transcription start site is outside of the RNAP at this stage (3). The stable RPC is thought to be preceded by an unstable RPC in which the –12 fork junction has yet to form (4). The isomerization from RPC to RPO requires ATP hydrolysis by bacterial enhancer binding proteins (bEBPs), resembling in part the eukaryotic Pol II system that utilizes TFIIH and ATP for DNA melting (5). σ54-dependent transcription not only regulates various adaptive responses (6,7), but is also responsible for regulating pathogenesis determinants in disease-causing agents such as Borrelia burgdorferi (the agent of Lyme disease) and Vibrio cholera (the agent of epidemic diarrheal disease) (8,9). Therefore, an understanding of the σ54-transcription pathway is valuable for identification of new antibacterial drug targets (10).
Current information on the organization of σ54-transcription complexes have been drawn from: the low-resolution Cryo-electron microscopy (Cryo-EM) studies with purified (E)σ54–bEBP complexes (11,12), the NMR and SAXS structures of some regions of σ54 (13–16) and crystal studies of bEBPs (12,17–20). Three regions have been identified in σ54 (Figure 1B). Region I of σ54 (σ54RI) interacts with bEBPs, core RNAP and the –12 promoter region (21–23), participating in promoter melting and isomerization processes (24,25). Region II of σ54 (σ54RII) is dispensable for interactions with RNAP and DNA. Region III of σ54 (σ54RIII) contains several functionally important modules, including the RpoN box required for the recognition of the –24 promoter element (22).
Figure 1.
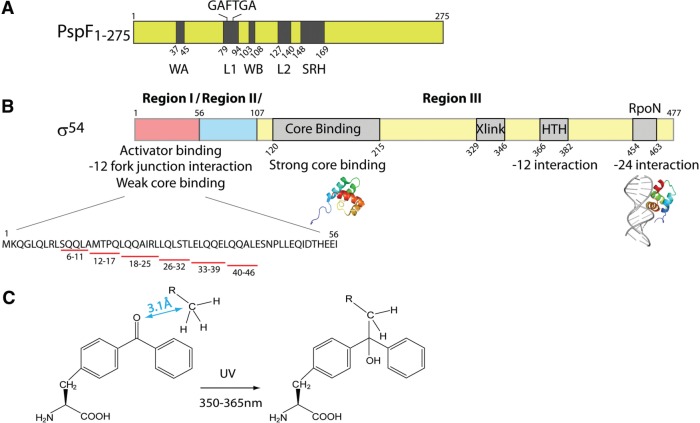
Sequence organization and functional properties of PspF1–275 and σ54. (A) Domain organization of PspF1–275. WA stands for Walker A motif, L1 for Loop 1 (containing the ‘GAFTGA’ motif), WB for Walker B motif and SRH for second region of homology. (B) Domain organization of σ54. HTH stands helix-turn-helix motif. Xlink stands for DNA cross-linking region. σ54 Regions I–III are separated by slashes. Available structures of two σ54 fragments are depicted under the corresponding sequences. Six σ54 Region I fragments (underlined in red) are generated for the following binding assays. (C) The rationale of the pBpa-based UV cross-linking assay. The ketone group in the pBpa artificial amino acid cross-links to any C–H bond within 3.1 Å under UV irradiation.
bEBPs belong to Clade 6 of the ATPases associated with various cellular activities (AAA+) protein family (26), and their functionality is dependent on an ability to self-associate, typically to form hexamers. The biochemical functions of bEBPs have been widely studied using as examples the Escherichia coli Phage shock protein F (PspF), which is involved in membrane stress responses (27), and the nitrogen control proteins NtrC and NtrC1. PspF contains an AAA+ domain (residues 1–275, PspF1–275, Figure 1A) essential for oligomerization and ATP hydrolysis and a C-terminal DNA-binding domain. Two surface-exposed loops of PspF (Figure 1A), namely Loop 1 (L1) and Loop 2 (L2), are particularly important for coupling ATP hydrolysis to the RPC remodelling event. L1 contains a highly conserved ‘GAFTGA’ motif amongst bEBPs. Residues Phe (F85) and Thr (T86) in the PspF ‘GAFTGA’ motif interact with σ54RI during RPO formation (21,28). Structural modelling indicates that L1 may have additional roles (11). These roles may be accommodated by the potential availability of up to six L1s across a PspF hexamer (11,12,29).
To seek evidence for functional specialization amongst L1s, we incorporated a photoreactive artificial amino acid, p-benzoyl-L-phenylalanine (pBpa), using an orthogonal tRNA/tRNA synthetase pair to each PspF L1 ‘GAFTGA’ position for identification of potential interacting partners (30); pBpa can cross-link to any C–H bond within 3.1 Å upon UV irradiation [Figure 1C, (31,32)]. Here we provide direct evidence that: (i) L1 contacts the DNA non-template strand immediately upstream of the –24 promoter element, and (ii) the DNA immediately upstream of the –24 element is important for the isomerization from RPC to RPO as well as formation of one transcription intermediate. Using a fragmentation approach, we were able to identify two previously unknown PspF L1-binding patches within σ54RI (residues 18–25 and 33–39). The above observations provide evidence that L1 is multifunctional, and makes at least three distinct nucleotide-dependent interactions within its target complex in driving RPO formation.
MATERIALS AND METHODS
Plasmids
Plasmid pPB1 [encoding the E. coli pspF1–275 sequence, (21)] was used as a template for the subsequent site-directed mutagenesis studies. Each ‘GAFTGA’ position was mutagenized in the context of pPB1 to an amber stop codon (TAG) to yield pET28b-pspF1–275 variant plasmids (Supplementary Table S1).
DNA probes and peptide fragments
The linear DNA probes used in this study are summarized in Supplementary Table S2. The σ54RI fragments were purchased with the highest purity level from Insight Biotechnology.
Protein expression and purification
The expression of PspF1–275 pBpa variants depends on two plasmids: (i) the pET28b-pspF1–275 variant (Supplementary Table S1) and (ii) the pDULE-pBpa [encoding the Methanococcus jannaschii tRNA/tRNA synthetase pair to specifically charge the intrinsic stop codon with pBpa, (30)]. Typically, 0.26 g of pBpa (Bachem) were dissolved under alkaline conditions and added to a L1 culture. The PspF1–275 pBpa variants were expressed and purified as previously described (28), treated with thrombin to remove the (His)x6 tag, and stored in TGED buffer 1 (20 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM DTT, 0.1 mM EDTA and 5% glycerol) at −80°C.
Klebsiella pneumoniae σ54 was purified as previously described and stored in TGED buffer 2 (20 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM DTT, 0.1 mM EDTA and 50% glycerol) at −80°C (33). K. pneumoniae heart muscle kinase (HMK) tagged full-length σ54 (HMK-σ54FL) and HMK-tagged σ54 fragments (HMK-σ54RI and HMK-σ54ΔRI) were purified and radio-labelled as previously described (34). E. coli core RNAP was purchased from Cambio.
ATPase activity assay
Typically in a 10 μl volume, 4 μM PspF1–275 was pre-incubated with the ATPase buffer (20 mM Tris-HCl pH 8.0, 50 mM NaCl, 15 mM MgCl2, 0.1 mM EDTA, 10 μM DTT) at 37°C for 5 min. ATP hydrolysis was initiated by addition of 1 mM unlabelled ATP and 0.6 μCi/μl [α-32P] ATP (3000 Ci/mmol) and incubated for various time spans at 37°C. Reactions were quenched by addition of 5 volumes of 2 M formic acid. The [α-32P] ADP was separated from the [α-32P] ATP by thin layer chromatography (Macherey–Nagel) in 0.4 M K2HPO4/0.7 M boric acid. Radioactivity was scanned by PhosphoImager (Fuji Bas-1500) and analysed by Aida software. The ATP turnover rate (kcat) of each PspF1–275 pBpa variant was expressed as a percentage of PspF1–275 wild type (WT) activity. All experiments were minimally performed in triplicate.
Native gel mobility shift assay
Reactions were performed in 10 μl volumes and supplemented with 1 μM 32P-HMK-σ54FL (or its fragments), ± 0.3 μM core RNAP, ± 50 nM radio-labelled DNA, 5 mM NaF and 4 mM nucleotides (ATP, ADP or AMP) in STA buffer [2.5 mM Tris-acetate pH 8.0, 8 mM Mg-acetate, 10 mM KCl, 1 mM DTT, 3.5% (w/v) PEG 8000] at 37°C for 5 min. Ten μM PspF1–275 and 0.4 mM AlCl3 were added for a further 15 min incubation to allow ‘trapped’ complex formation at 37°C. Complexes were either analysed on native gels or subject to UV cross-linking and then analysed.
Gel filtration assay
PspF1-275 WT or the ADP–AlFx ‘trapped’ complex was pre-incubated at 4°C with gel filtration buffer (20 mM Tri-HCl pH 8.0, 50 mM NaCl, 15 mM MgCl2) for 5 min. A Superdex 200 column (10/30, 24 ml, GE Healthcare) assembled on the AKTA system (GE Healthcare) was equilibrated with buffer. Chromatography was carried out at a flow rate of 0.5 ml/min at 4°C.
pBpa-based UV cross-linking assay
‘Trapped’ complexes were formed with either 32P-HMK-σ54 (or its fragments) or 32P-DNA. Reaction mixtures were UV irradiated at 365 nm on ice for 5 min, 15 min and 30 min then analysed on both native and SDS PAGE gels. The cross-linked protein–protein or nucleo–protein species were scanned by a Fuji PhosphoImager and analysed by Aida software.
Proteinase K-ExoIII footprinting assay
The UV cross-linked nucleo–protein species were generated with 32P-DNA and subject to Proteinase K-ExoIII footprinting assays as previously described (35). The UV-irradiated samples (20 μl) were digested with 1 μl of 20 mg/ml Proteinase K (Sigma) at 37°C for 1 h to remove the protein components. 32P-DNA containing the pBpa peptide was phenol-extracted and isopropanol-precipitated. Twenty units of exonuclease III (ExoIII, USB) were added to the DNA sample to a 10 μl final volume. The ExoIII digestion proceeded for various time spans before being quenched by 4 μl of 3X formamide stop dye (3 mg xylene cyanol, 3 mg bromophenol blue, 0.8 ml 250 mM EDTA, 10 ml deionised formamide in 10 ml). The reaction mixtures were heated at 97°C for 5 min before separated on a sequencing gel.
In vitro RPO formation assay
Open complex formation was measured in 10 μl final volumes containing: 4 μM PspF1–275, 100 nM holoenzyme (1:4 ratio of E: σ54), 20 U RNase inhibitor, 5% (v/v) glycerol, 4 mM dATP and 20 nM Sinorhizobium meliloti nifH promoter (Supplementary Table S2) in STA buffer at 37°C. Transcription was activated for various lengths of time before 0.5 mM dinucleotide primer UpG, 0.2 μCi/μl [α-32P GTP] (3000 Ci/mmol) and 0.2 mg/ml heparin were added. After extension at 37°C for 10 min, the reaction mixtures were quenched by addition of 4 μl of 3X formamide stop dye and run on a sequencing gel. The activator-bypass activities of the σ54 variants were examined in a similar experimental procedure without the addition of PspF1–275 activators and dATP.
In vitro spRNA assay
The ADP–AlFx ‘trapped’ complexes were initially formed on the late-melted –10–1/WT DNA probe. Without the addition of dATP, the ADP–AlFx complexes were allowed to synthesize a UpG-primed RNA product UpGpGpG (the spRNA) in the presence of α-32P GTP in a manner similar to the RPO formation assay as described above.
RESULTS
PspF1-275 G83pBpa can cross-link to σ54 but not to core RNAP
The photo-reactive artificial amino acid pBpa was incorporated at each L1 ‘GAFTGA’ position, generating six PspF1–275 pBpa variants (G83pBpa, A84pBpa, F85pBpa, T86pBpa, G87pBpa and A88pBpa). Characterization of each pBpa variant for bEBP functions is summarized in Table 1. Position 83 in the PspF L1 ‘GAFTGA’ motif (the position of interest, as the following experiments demonstrate) was substituted with Ala (to remove the side chain) and Phe (to mimic the pBpa cross-linker). The resulting G83A and G83F variants were also characterized (Table 1).
Table 1.
Functional characterization of PspF1–275 pBpa variants
| PspF1–275 | σ54 interaction (% of WT) |
ATPase activity (% of WT) | RPO formation (% of WT) | |
|---|---|---|---|---|
| ADP–AIFX | AMP–AIFX | |||
| G83pBpa | 50 | 0 | 10 | 0 |
| A84pBpa | 0 | 0 | 116 | 0 |
| F85pBpa | 0 | 0 | 96 | 0 |
| T86pBpa | 0 | 0 | 146 | 0 |
| G87pBpa | 0 | 0 | 138 | 0 |
| A88pBpa | 0 | 0 | 100 | 0 |
| G83A | 56 | 0 | 23 | 0 |
| G83F | 90 | 29 | 10 | 0.1 |
The PspF1–275 pBpa variant and the two G83 variants (substitution with Ala to remove the side chain and substitution with Phe to closely mimic the pBpa cross-linker) were characterized in terms of σ54 interaction (in the presence of the ADP–AlFx ‘trapping’ reagent), ATPase activity and RPO formation (on a super-coiled S. meliloti nifH promoter).
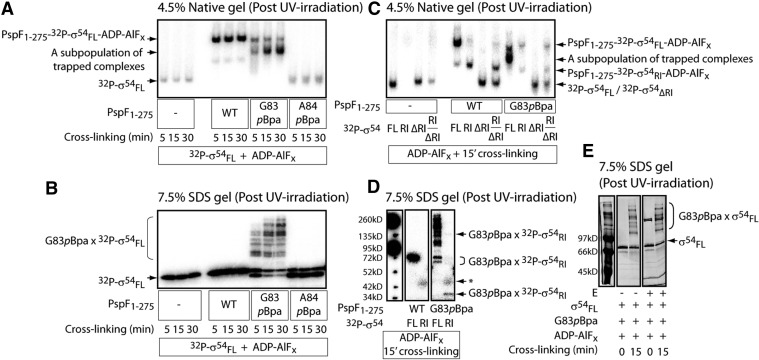
To investigate whether introducing the bulky hydrophobic pBpa cross-linker could affect the overall L1 exposure and the local σ54 interaction, we performed the well-established ‘trapping’ assay. The transient L1 ‘GAFTGA’– σ54 interaction is stabilized in the presence of an ATP transition state analogue, ADP–AlFx. The resulting ‘trapped’ complex, PspF1–275–(E)σ54–ADP–AlFx, reflects one of the intermediate states (RPIs) en route to RPO formation (36,37). G83pBpa was the only variant tested capable of maintaining the σ54 interaction in an ADP–AlFx dependent manner (Table 1 and Figure 2A). By irradiating the ‘trapped’ PspF1–275 G83pBpa-σ54–ADP–AlFx complex and analysed under denaturing conditions, multiple cross-linked PspF1–275 G83pBpa × σ54 species were observed (Figure 2B). Prolonged irradiation shifted the cross-linked species towards higher molecular forms (Figure 2B); it is likely that higher oligomeric states of G83pBpa (G83pBpa can self-associate and self-crosslink) associated with multiple σ54 regions (as demonstrated below). The cross-linked PspF1–275G83pBpa × σ54 species was only be observed in the ‘trapped’ complexes (in the presence of ADP–AlFx but not in the presence of ATP, ADP or AMP, Supplementary Figure S1), consistent with the proposal that at the point of ATP hydrolysis, the PspF L1s assume a raised conformation to contact and thereby cross-link to σ54 (20). The G83pBpa variant was unable to drive RPO formation (Table 1) possibly due to sub-optimal L1 exposure (50% of WT activity) and low ATPase hydrolysis rate (10% of WT activity). However, this derivative of PspF did support partial functionalities required for forming RPO, and was able to function for engagement of RPC and support RPO formation in a mixed oligomer with WT subunits (see below).
Figure 2.
PspF L1 cross-links to multiple locations within σ54 Region I (σ54RI) but not to core RNAP. The HMK-tagged full-length σ54 was radio-labelled (32P-σ54FL) and used in (A–D) but not in (E). The native gel (A) and the SDS PAGE gel (B) depict the binding and cross-linking, respectively, between PspF1–275 pBpa variants and 32P-σ54FL in the presence of the ADP–AlFx ‘trapping’ reagent. A protein band (G83pBpa, Panel A) increases with UV-irradiation time, possibly corresponding to a subpopulation of the ‘trapped’ complex due to cross-linking. The A84pBpa variant was used as a negative control for cross-linking as it cannot bind to σ54. The cross-linked species between the G83pBpa variant and 32P-σ54FL were depicted as G83pBpa × 32P-σ54FL. From this point, all the following UV cross-linking reactions were performed with 15 min irradiation (the ADP–AlFx-dependent complexes are most stable within 30 min—in this case, 15 min for complex formation and 15 min for UV irradiation). To assess the interaction and cross-linking between PspF L1 and different regions of σ54, the ADP–AlFx-dependent ‘trapping’ reactions were performed with radio-labelled Region I (32P-σ54RI), Region I delete (32P-σ54ΔRI) and mixed RI/ΔRI (a 3:1 ratio). The presence of ‘trapped’ complexes was analysed on a native gel (C); the cross-linking event was analysed on an SDS PAGE gel (D). The band in the G83pBpa × 32P-σ54RI cross-linking reaction (D) indicated by an asterisk corresponds to an artefact also present in 32P-σ54RI alone. To assess whether PspF L1 cross-links to core RNAP, core RNAP was added to the ADP–AlFx-dependent ‘trapping’ reaction (E). The SDS PAGE gel was stained with Invitrogen Sypro Ruby stain and scanned by PhosphoImager. The presence of core RNAP does not seem to alter the cross-linking profile between G83pBpa and σ54. Similar results were obtained when 32P-σ54FL was used.
Based on the Cryo-EM structure, Bose et al. (11) proposed that up to three subunits in a PspF hexamer could potentially contact the RNAP holoenzyme via the PspF surface-exposed L1s. In addition, it has been shown that the AAA+ domain of the S. meliloti DctD (another well-studied hexameric bEBP) can cross-link to the core RNAP β subunit (38). To assess whether PspF L1s can directly contribute to core RNAP binding, we added core RNAP to the cross-linking reactions. As shown in Figure 2E, the cross-linking pattern between PspF1–275 G83pBpa and σ54 was not altered by the presence of core RNAP. The outcomes did not provide evidence to support a direct contact between the L1 ‘GAFTGA’ motif and core RNAP, rather the protein contacts appear to be primarily with σ54.
Taken together, formation of the ADP–AlFx ‘trapped’ complexes with the G83pBpa variant suggests it is a potentially useful reagent, as demonstrated in the following experiments, to elucidate the L1-interacting partners.
Two novel PspF L1-binding patches within σ54RI were identified
The Cryo-EM structure of the ADP–AlFx ‘trapped’ complex indicated that the PspF hexamer contacted two opposing sites in σ54 (12). If one contact site is the PspF L1 target site–σ54RI (36), the other contact site might be outside σ54RI. To explore this proposal, different radio-labelled σ54 fragments (σ54RI, σ54ΔRI and mixed σ54RI/ΔRI) were used to form the cross-linked ‘trapped’ complexes with G83pBpa. Both PspF1–275 WT and G83pBpa can form stable complexes with σ54FL and σ54RI but not with σ54ΔRI (Figure 2C). With the G83pBpa variant, a major cross-linked species of ∼39 kDa was observed (Figure 2D), corresponding to one PspF1–275 G83pBpa (33 kDa) cross-linked to one σ54RI (6 kDa). Other faint cross-linked G83pBpa × σ54RI species with higher molecular weights were also observed (Figure 2D), suggesting that more than one L1 could contact σ54RI.
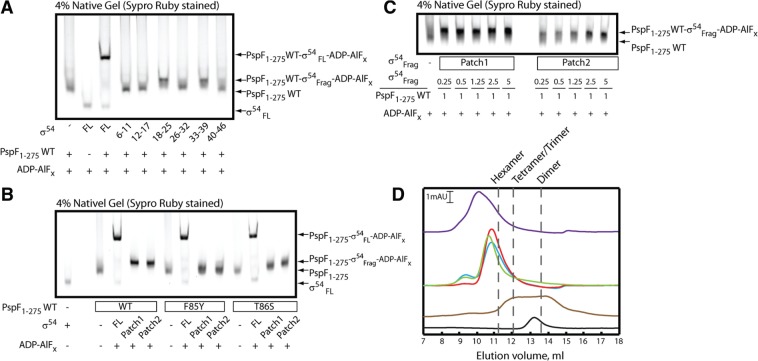
From the above observations, we set to explore the PspF L1-binding patches within σ54RI by generating six σ54RI peptide fragments (Figure 1B). As we screened the σ54RI fragments for their ability to stably bind PspF1–275 hexamers under ADP–AlFx ‘trapping’ conditions, a slower migrating complex was detected with either σ54Frag 18–25 or 33–39 but not the four other peptides tested (Figure 3A). Formation of the ‘trapped’ complexes in the presence of σ54Frag 18–25 and 33–39 was confirmed by gel filtration, providing evidence that the two peptides can associate with PspF in its ADP–AlFx-bound state (Figure 3D). Given their relatively small size, these two σ54RI fragments are unlikely to assume ‘complete’ secondary structures in solution; thus their interaction with the PspF1–275 hexamer may be largely dependent on their primary sequence and independent of their forming a well-ordered structure prior to binding to PspF. From here on, the σ54 RI residues 18–25 will be named Patch 1 and residues 33–39 Patch 2.
Figure 3.
Two σ54RI peptide fragments bind to PspF1-275 with different affinities. (A) PspF1–275 WT hexamers bind to two σ54RI fragments (residues 18–25 and 33–39) in the presence of the ADP–AlFx ‘trapping’ reagent. From here on, fragments 18–25 is depicted as Patch1 and fragments 33–39 as Patch2. (B) Mutations in the PspF ‘GAFTGA’ motif result in sensitivity to σ54RI patch binding. (C) σ54RI Patch1 fragments bind to PspF1–275 WT hexamers with a markedly higher affinity than Patch2 fragments. A titration experiment was performed with a constant PspF1–275 WT concentration while the concentration of each Patch fragment was gradually increased. (D) Gel filtration profiles of the ‘trapped’ complexes at 4°C using a Superdex 200 column. The black trace corresponds to 2 µM σ54FL. The brown trace corresponds to 20 µM PspF1-275 WT in the presence of ADP. The green trace corresponds to 20 µM PspF1–275 WT in the presence of ADP–AlFx. The red trace corresponds to 20 µM PspF1-275 WT binding to σ54RI Patch1 fragments in the presence of ADP–AlFx. The blue trace corresponds to 20 µM PspF1-275 WT binding to σ54RI Patch2 fragments in the presence of ADP–AlFx. The purple trace corresponds to 20 µM PspF1–275 WT binding to σ54FL in the presence of ADP–AlFx. The gel elution volumes corresponding to apparent hexamers, tetramers/trimers and dimers are marked by dotted lines.
To demonstrate that binding of the patch fragments to PspF1–275 is dependent on established determinants for the binding of PspF to σ54, the L1 ‘GAFTGA’ variants (F85Y and T86S) were used in the ADP–AlFx reactions (Figure 3B). The F85Y variant appears to interact with the two patch fragments (the PspF band intensity increases dramatically in the presence of these fragments). However, their binding is possibly different from that of PspF1–275 WT, as the complex band does not shift much nor into a compact band. The T86S variant can shift the complexes to the same level as WT with Patch 2 fragment but not with Patch 1 fragment. The above observations suggest that the L1 ‘GAFTGA’ motif is indeed the direct target of the σ54 RI patches. Based on the formation of stable ‘trapped’ complexes in a titration experiment (Figure 3C), the Patch 1 fragment exhibited a much higher affinity (at least 10-fold) towards PspF1–275 than did the Patch 2 fragment.
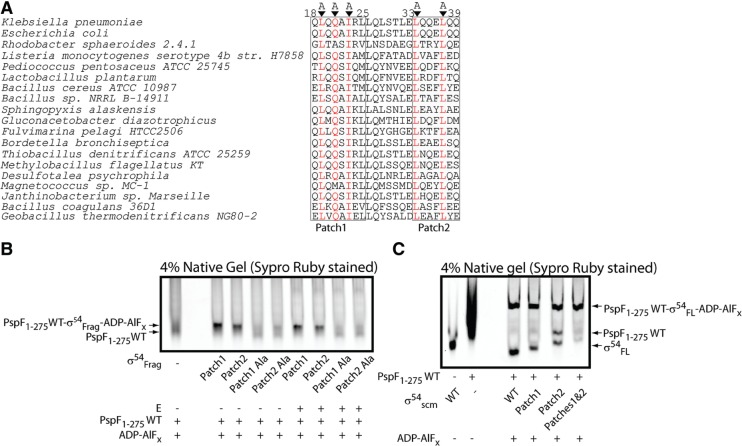
Sequence alignment of the two σ54RI patches from different organisms (Figure 4A) revealed three highly conserved residues in Patch 1 and two highly conserved residues in Patch 2. Substitutions of these residues with Ala (QAQAAARL and AQQEAQQ, as underlined) produced variant fragments unable to detectably bind the hexameric PspF1–275 (Figure 4B), suggesting that these residues are key to PspF L1 interactions with σ54.
Figure 4.
Scrambled σ54RI Patches fail to bind PspF1–275 WT in the context of fragments but can bind PspF1–275 WT in the context of full-length proteins. (A) Sequence alignment of σ54RI Patches 1 and 2 from different bacteria using NCBI BLAST. Highly conserved residues are highlighted in red and subsequently replaced with Ala (‘Scrambled’ or ‘scm’). (B) The scrambled patches failed to bind PspF1–275 WT hexamers in the context of fragments in the presence of ADP–AlFx ‘trapping’ reagents. (C) The scrambled patches were able to bind PspF1–275 WT hexamers in the context of full-length σ54 in the presence of ADP–AlFx ‘trapping’ reagents. σ54scm Patches 1 and 2 corresponds to the full-length σ54 harbouring both the scrambled patches.
Taken together, we have identified three highly conserved residues in σ54RI Patch 1 contributing to the high affinity PspF L1 binding, and two highly conserved residues in Patch 2 to a lower affinity PspF L1 binding.
L1–σ54RI sequence-specific interactions play different roles along the activation pathway
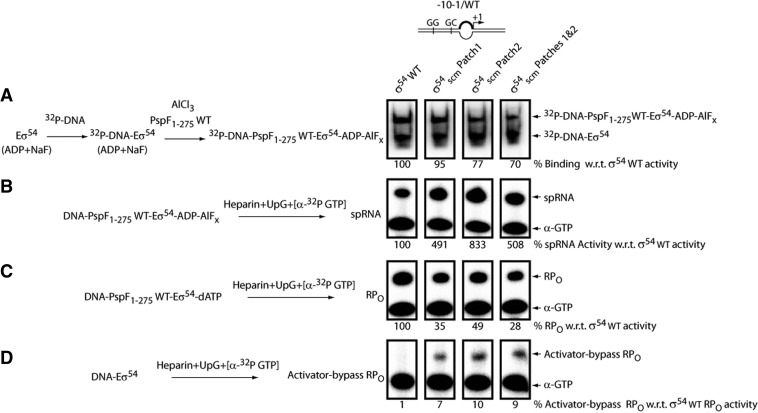
After establishing that two σ54RI patches are sequence-specific for PspF L1 interactions, we next examined their impact in the context of σ54 and RNAP holoenyzme binding interactions and different steps in the transcription activation pathway. Thus, we generated three full-length σ54 variants: σ54scm Patch 1 (harbouring the scrambled Patch 1—‘QAQAAARL’), σ54scm Patch 2 (harbouring the scrambled Patch 2—‘AQQEAQQ’) and σ54scm Patches 1 and 2 (harbouring both the scrambled patches). The initial assessment of the σ54scm patch variants revealed no large defect in forming the ‘trapped’ complexes (Figure 4C), suggesting additional sequences of the σ54RI along with promoter DNA contacts (see below) could compensate for the loss of L1 sequence-specific interactions in forming the ‘trapped’ transcription intermediate (RPI).
Burrows et al. (39) devised an assay in which the ADP–AlFx-dependent RPI could carry out dinucleotide-primed short RNA (spRNA) transcription when the ‘–10 to –1’ transcription bubble was pre-formed. Using this assay, we assessed the impact of the σ54scm patch variants on the amount of transcriptionally ‘active’ RPIs generated. As shown in Figure 5B, despite starting with a similar amount of ADP–AlFx-dependent RPIs (a slightly more pronounced reduction in RPI was observed with σ54scm Patch 2 and Patches 1 and 2, Figure 5A), all three σ54scm patch variants were able to produce significantly more spRNAs (4–8-fold) than was the σ54 WT, indicating the RPIs were more active than with σ54 WT. In contrast, the ATPase-dependent RPO formation assays replacing ADP–AlFx with dATP on the same DNA probe revealed significant defects in all three σ54scm patch variants (Figure 5C). Interestingly, all three σ54scm patch variants can generate an RPO in the absence of the PspF1–275 activator and hydrolysable nucleotides on the pre-melted DNA probe (Figure 5D), so revealing an activator-bypass phenotype (40). Considering the RPO generated from the activator-bypass activity may have contributed to the amount of RPO as observed in Figure 5C, the defect of the σ54scm patch variants in the ATPase-driven isomerization may be more pronounced.
Figure 5.
Functional importance of the PspF L1–σ54RI patch interactions along the activation pathway. A simplified reaction scheme is depicted for each assay. Full-length σ54 harbouring the scrambled patches and radio-labelled –10–1/WT DNA probes were used in the following reactions. The linear –10–1/WT DNA probe harbours a mismatch from –10 to –1 on the non-template strand to mimic the DNA conformation in RPO. (A) The ability of scrambled σ54 patch variants (σ54scm patch) to form the ADP–AlFx-dependent ‘trapped’ complexes, each expressed as a percentage of that of σ54 WT. (B) Each σ54scm patch variant was allowed to form the ADP–AlFx-dependent RPI complex (DNA–PspF1–275 WT–Eσ54–ADP–AlFx). The resulting RPI complexes were tested for their ability to support transcription in the presence of the spRNA mixture (heparin, dinucleotide primer UpG and [α-32P GTP]). The extent of transcription activity (correlates with the amount of spRNA synthesis) was expressed as a percentage of that of σ54 WT. (C) The amount of RPO generated by σ54scm patch variants in the presence of hydrolysable nucleotide dATP, each expressed as a percentage of that of σ54 WT. (D) The amount of activator-bypass RPO generated by σ54scm patch variants in the absence of PspF1–275 WT, each expressed as a percentage of that of σ54 WT in the activator-dependent assay (C).
Taken together, the above data demonstrate that the PspF L1–σ54RI sequence-specific interactions may play an inhibitory role in the activity of RPI, possibly to keep the complex in check before moving to RPO. Once the inhibitory PspF L1–σ54RI interactions were disrupted (by scrambling the Region I patches), the spontaneous transition from RPI to RPO is clearly increased as seen in the activator-bypass assays. In contrast, the PspF L1–σ54RI sequence-specific interactions are needed for ATPase-driven RPO formation, suggesting important roles for the patches in making RPI from RPC and in limiting the activity of RPI in the activator-dependent pathway. Transient interactions between RPC and PspF in the ATPase-driven reaction may therefore be more dependent upon the integrity of the σ54RI than is the stably engaged RPI created with ADP–AlFx.
‘Doped’ WT/G83pBpa heterohexamers can directly cross-link to promoter DNA
After demonstrating the G83pBpa variant can cross-link to σ54 in the ADP–AlFx ‘trapped’ complex, the DNA probe harbouring the early-melted nifH promoter (–12–11/WT, mimicking the –12 fork junction DNA in the RPC, Figure 6) was added to the reaction mixture. If this fork junction DNA conformation was successfully accommodated in the ‘trapped’ complex, the spatial proximity between G83pBpa and the corresponding promoter region could in principle be determined.
Figure 6.
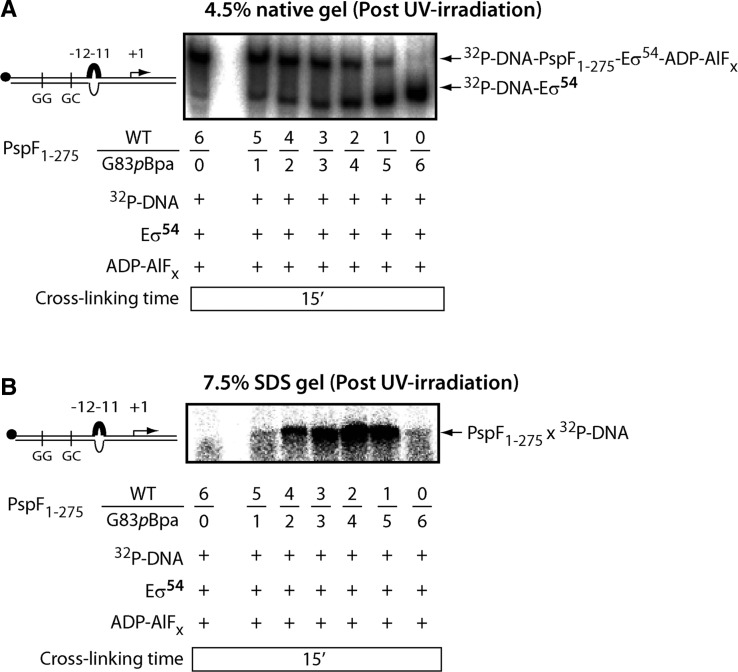
The ‘doped’ PspF1–275 WT/G83pBpa heterohexamers cross-link to promoter DNA. The ADP–AlFx-dependent ‘trapping’ reaction was performed in the presence of radio-labelled –12–11/WT DNA probe. The –12–11/WT DNA probe harbours a mismatch from –12 to –11 on the non-template strand to mimic the DNA conformation in RPC. PspF1–275 WT and G83pBpa subunits were mixed with different ratios (6/0, 5/1, 4/2, 3/3, 2/4, 1/5 and 0/6) in the ‘trapping’ reaction. Core RNAP was also added to the reaction mixture to ensure all the transcriptional components were in place. Samples were loaded on a native gel (A) and on an SDS PAGE gel (B). A single cross-linked PspF1–275 × DNA species was observed.
As shown in Figure 6, the radio-labelled –12–11/WT DNA probe was not efficiently covalently bound into the ADP–AlFx ‘trapped’ complexes with G83pBpa homohexamers (ratio of WT/G83pBpa was 0/6), resulting in a cross-linked species with abundance only slightly above the background. Since the PspF1–275 WT homohexamers (ratio of WT/G83pBpa was 6/0) were able to efficiently form the ‘trapped’ complexes in the presence of DNA (Figure 6A), we reasoned that by ‘doping’ G83pBpa with different ratios of WT subunits, accommodation of the –12–11/WT DNA may be achieved. Not only did the ‘doping’ experiment successfully restore the ‘trapped’ complex formation, but also generated a single PspF1–275 × DNA cross-linked species (Figure 6B). The above observation provides clear evidence that subunit mixing indeed occurred (see also Figure 7) and that the reconstituted WT/G83pBpa heterohexamers likely contacted the promoter DNA via position 83 or adjacent residues in the L1 ‘GAFTGA’ motif. Taken together, the above data strongly support a chemical bonding interaction between L1 ‘GAFTGA’ and the promoter DNA in the ‘doped’ WT/G83pBpa heterohexamers, as the pBpa cross-linking chemistry requires a distance of 3.1 Å (recall the H-bonding distance is 2 Å). Spatial organizations reported for RNAP-σ54 and the L1-DNA cross-linking event together suggest that L1s from different subunits of the hexamer must be involved in DNA and σ54RI contacts.
Figure 7.
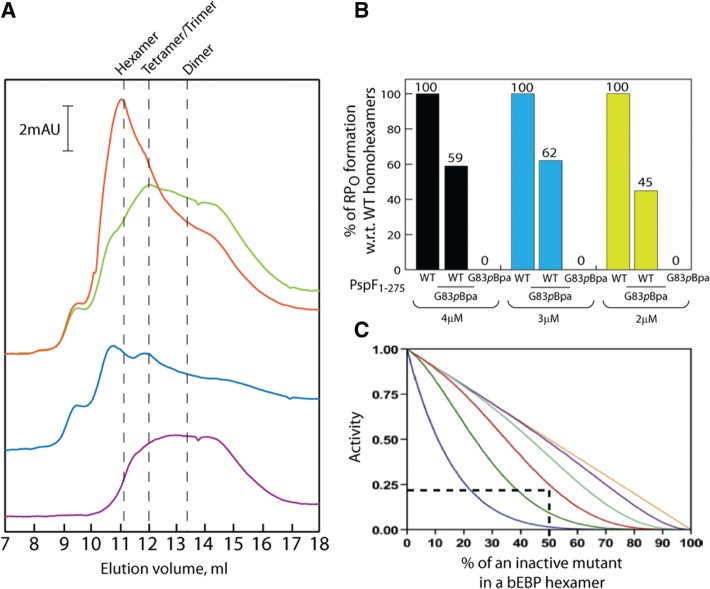
The ‘doped’ PspF1–275 WT/G83pBpa heterohexamers are transcriptionally active. (A) Gel filtration profiles of PspF1–275 WT, G83pBpa and WT/G83pBpa mixture at 4°C in the absence of nucleotides. The purple trace corresponds to 30 µM PspF1–275 WT. The blue trace corresponds to 30 µM PspF1–275 G83pBpa. The green trace corresponds to the theoretical profile of mixing 30 µM WT subunits with 30 µM G83pBpa subunits. The red trace corresponds to the experimental profile of mixing 30 µM WT subunits with 30 µM G83pBpa subunits. A prominent hexameric peak was observed in the experimental profile of the WT/G83pBpa mixture, corresponding to the presence of true heterohexamers. (B) The amount of RPO formation in the presence of WT homohexamers, WT/G83pBpa heterohexamers (an equimolar ratio of WT and G83pBpa subunits) and G83pBpa heterohexamers under three different total PspF1-275 concentrations. The –10–1/WT DNA probe was used in this assay. The average amount of RPO generated by WT/G83pBpa heterohexamer is 55% of that of WT homohexamers. (C) Theoretical activities of the ‘doped’ heterohexamers [taken from (Werbeck et al. (42)]. One inactive subunit ‘doped’ with five WT subunits (blue), two inactive subunits ‘doped’ with four WT subunits (green), three inactive subunits ‘doped’ with three WT subunits (red), four inactive subunits ‘doped’ with two WT subunits (mint), five inactive subunits ‘doped’ with one WT subunit (purple) and six inactive subunits (brown) are necessary to abolish enzyme activity. The WT/G83pBpa heterohexamers (WT/G83pBpa subunit ratio 3/3) should theoretically generate 25% the RPO of that of WT homohexamers (dotted lines).
Next we ‘doped’ the rest of the pBpa variants (recall all failed to form the ‘trapped’ complexes, Table 1 and Supplementary Figure S2) with the PspF1–275 WT subunits for DNA cross-linking complementation. Strikingly, the ‘doped’ WT/T86pBpa heterohexamers showed a comparatively strong DNA cross-linking signal with the –12–11/WT DNA probe (Supplementary Figure S2B). This outcome implies a role for the conserved residue T86 or residues adjacent to it in DNA contact in RPC. Previously, T86 has only been characterized as being a σ54-contacting residue of L1.
To assess whether the ‘doped’ WT/G83pBpa heterohexamers were biologically relevant to the bona fide WT homohexamers, we examined the activity in the context of their self-association and RPO formation by mixing an equimolar amount of PspF1–275 WT and G83pBpa subunits (Figure 7). By gel filtration of apo forms, we establish that PspF1–275 WT exists as a mixture of apparent tetramers/dimers (12 ml/13.3 ml elution volumes, Figure 7A purple trace) at a 30 µM injection concentration. The G83pBpa variant exists predominantly as apparent octamers/tetramers (10.7 ml/12 ml elution volumes, Figure 7A blue trace) at a 30 µM injection concentration. The ‘doped’ WT/G83pBpa mixture generated an apparent hexameric peak (11.2 ml elution volume, Figure 7 red trace), eluting at the same volume as the WT homohexamers [11.18 ml elution volume, (28,41)]. This apparent hexameric peak was absent in the theoretical sum of each individual subunit profile (Figure 7 green trace). We thus conclude that the ‘doped’ WT/G83pBpa heterohexamer is very similar in overall geometry to the WT homohexamer and subunit mixing indeed occurred. We also tested the ability of the WT/G83pBpa heterohexamer to form RPO under three different total concentrations (Figure 7B). The ‘doped’ WT/G83pBpa heterohexamers generated on average 55% the RPO of the WT homohexamers (Figure 7B). Based on the statistical model for mixing experiments (42), an equimolar amount of WT and an inactive variant (in this case G83pBpa) should theoretically generate ∼25% the RPO of the WT homohexamers (Figure 7C dotted lines). The above considerations imply that the reconstituted WT/G83pBpa heterohexamers are indeed active in RPO formation, and that the G83pBpa subunits contribute to this activity.
L1 cross-links to the non-template ‘–29’ region in RPC/RPI
To determine the precise DNA region cross-linked by L1, we employed a Proteinase K-ExoIII footprinting method (35). The rationale of this approach is to remove all the protein components by Proteinase K after UV irradiation; a stably cross-linked pBpa peptide will remain attached to the DNA cross-linking site and physically block the read-through of ExoIII (a 3′–5′ exonuclease).
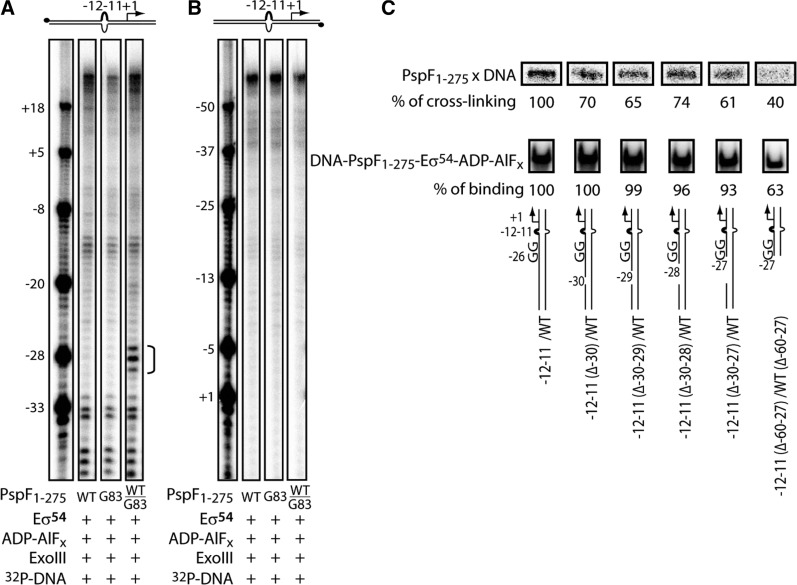
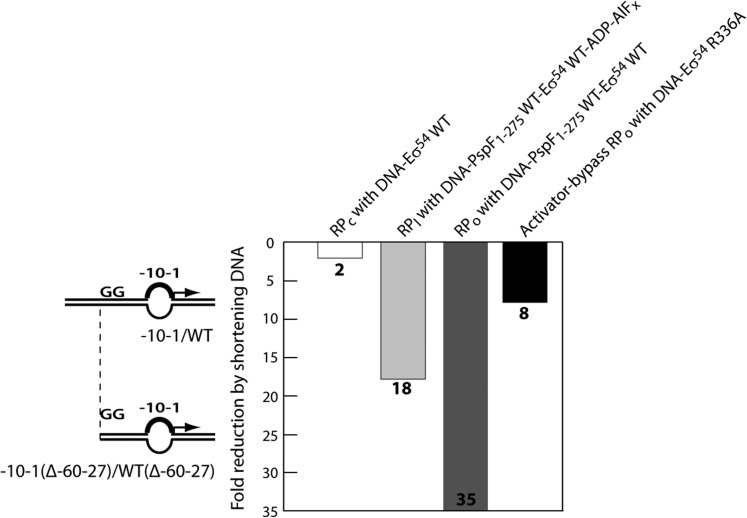
The G83pBpa homohexamer is able to weakly bind and cross-link to DNA (Figure 6B). However, owing to its inability to activate transcription (Table 1 and Figure 7B), we chose the transcriptionally active ‘doped’ WT/G83pBpa heterohexamer for footprinting. The L1 cross-linking site was mapped initially on the –12–11/WT DNA probe as this gave a very clear cross-linking signal (Supplementary Figure S2B). An ExoIII-resistant site was observed from approximately –30 to –27 on the non-template strand but not observed on the template strand (compare Figure 8A with 8B). The above data clearly demonstrate that in RPC/RPI, a L1 contacts the non-template strand of the promoter region between –30 and –27 (abbreviated as the ‘–29 region’), immediately upstream of the consensus ‘–24’ GG element (located at –26–25 in the nifH promoter). Removal of the entire upstream sequence of the consensus GG yielded a near 60% reduction in L1-DNA cross-linking, further confirming the ‘–29’ region is the major target site of L1 ‘GAFTGA’ interaction (Figure 8C).
Figure 8.
L1 cross-links to the non-template ‘–29’ region of the –12–11/WT DNA probe. (A and B) PspF1–275 WT homohexamers, WT/G83pBpa heterohexamers and G83pBpa homohexamers were allowed to form the ‘trapped’ complexes with Eσ54 in the presence of ADP–AlFx. The ‘trapped’ complexes were then subject to Proteinase K-ExoIII footprinting assays. Proteinase K removes all the protein components. The pBpa cross-linker in theory should remain attached to the promoter site and subsequently blocks the read-through of ExoIII nuclease. The cross-linking site is depicted by a half bracket from –27 to –30 (collectively called the ‘–29’ site). Both the template strand, (B) and the non-template strand (A) were radio-labelled on the –12–11/WT DNA probe (32P as black dots). (C) Nicking and truncation of the ‘–29’ region reduced the L1 cross-linking efficiency. DNA strand nicking/truncation around the non-template ‘–29’ region was made on the –12–11/WT DNA probe to assess the impact on UV cross-linking and DNA binding. Sequential nicking of the non-template ‘–29’ region reduced the UV cross-linking efficiency by ∼30–40% (top panel). The reduction in UV cross-linking was not due to unfavourable DNA binding (DNA binding was largely unaffected, middle panel), except for the –12–11(Δ–60–27)/WT(Δ–60–27) DNA probe where the entire upstream region of the consensus GG was removed. Nicking/truncation may cause the linear DNA probes to adopt slightly different confirmations to the unmodified probe (e.g. secondary sites of cross-linking), which could explain the absence of complete abolishment in UV cross-linking.
The ‘–29’ region is important for activator-dependent RPO formation
We next addressed whether the ‘–29’ region was important for isomerization from RPC to RPO and forming ‘trapped’ complexes to make RPI. The RPO formation assays were performed and the amount of RPO formed from the long DNA probe was compared with that from the short DNA. Both DNA probes harboured a mismatch from –10 to –1 on the non-template strand since this DNA conformation gave the strongest activation signal amongst the three linear probes used in this assay. As shown in Figure 9, although the RPC formation with the probe lacking DNA upstream of ‘–29’ was reduced by 2-fold (Figure 9 and Supplementary Figure S3), ‘trapped’ complexes (RPI) with activators were reduced by 18-fold (Figure 9 and Supplementary Figure S4A) and activator-dependent RPO formation was reduced by 35-fold (Figure 9 and Supplementary Figure S4B). Since truncation of the ‘–29’ region did not reduce the stability of RPO by more than 2-fold (Supplementary Figure S6), the large RPO formation defect cannot be attributed to an unstable RPO generated from the shortened DNA probe. We conclude that the cross-linking of ‘–29’ region DNA to L1 of the activator is important for forming RPI and RPO. Parallel experiments with fully duplexed probes confirmed the importance of the ‘–29’ sequence for trapping and activator-dependent formation of RPO (Supplementary Figure S5).
Figure 9.
Truncation of the ‘–29’ region affects both activator-dependent and -independent transcription activation. The –10–1/WT and –10–1(Δ–60–27)/WT(Δ–60–27) DNA probes were used in the assays to assess the impact of truncation of the ‘–29’ region on activator-dependent RPC/RPI/RPO formation and on activator-independent RPO formation. The activator-dependent RPC formation was assessed by the stability of DNA–Eσ54 WT complexes (Supplementary Figure S3). The activator-dependent RPI formation was assessed by the stability of DNA–Eσ54 WT–PspF1–275 WT–ADP–AlFx ‘trapped’ complexes (Supplementary Figure S4A). The activator-dependent RPO formation was assess by the amount of RPO generated by DNA-Eσ54 WT- PspF1-275 WT (Supplementary Figure S4B and Supplementary Figure S7 left panel). The activator-independent RPO formation was assessed by the amount of RPO generated by DNA-Eσ54 R336A (Supplementary Figure S7 right panel).
After showing that the activator-dependent RPO formation (with PspF1–275 WT-σ54 WT) involves the interaction between L1 ‘GAFTGA’ motif and the ‘–29’ region, we addressed whether the L1–DNA interaction was important for activator-independent RPO formation. Residue R336 of σ54 is located in the DNA cross-linking sequence of Region III (Figure 1B). The σ54 R336A induces an activator-bypass phenotype (40) where the RPO formation is independent of PspF hexamers and activating nucleotides on a super-coiled or on a linear –10 to –1 pre-opened nifH promoter.
In contrast to the activator-dependent RPO formation (with PspF1–275 WT-σ54 WT) which was greatly reduced when the ‘–29’ region was truncated (a 35-fold reduction on average, Figure 9 and Supplementary Figure S7), the activator-independent RPO formation with σ54 R336A remained relatively constant on both DNA probes and was only reduced by 8-fold when the ‘–29’ region was removed (Figure 9 and Supplementary Figure S7). The differences in RPO formation were not due to different affinities of Eσ54 WT and Eσ54 R336A holoenzymes towards DNA (Supplementary Figure S3). The above observations demonstrate that the activator-independent pathway for RPO formation is 4-fold less sensitive to loss of the ‘–29’ region than is the activator-dependent pathway. Clearly, the ‘–29’ region is important for σ54 WT-containing RNAP to form RPO in an activator-dependent manner.
DISCUSSION
By using a fragmentation approach, we were able to identify precisely two amino acid patches within σ54RI responsible for PspF L1 contact (residues 18–25 and 33–39). Both σ54RI patches are located within the Leu heptad/hexad repeats (residues 19–44) where the activator-bypass mutations can be found (43,44). It is not known whether these two σ54RI patches are contacted by two L1s simultaneously or in sequence when RPC passes to RPI and then to RPO. Based on the shared phenotypes of the three full-length σ54scm patch variants (Figure 5), it is possible the two σ54RI patches are contacted by two L1s from adjacent PspF subunits in a synchronized manner. This interaction with σ54RI may be responsible for initially holding the holoenzyme and the PspF hexamer together through interactions at the –12 part of the promoter DNA in RPC and then organizing the RPI to accept the melted DNA (Figure 10C). The latter reorganization may involve the LI to upstream DNA contact as described below.
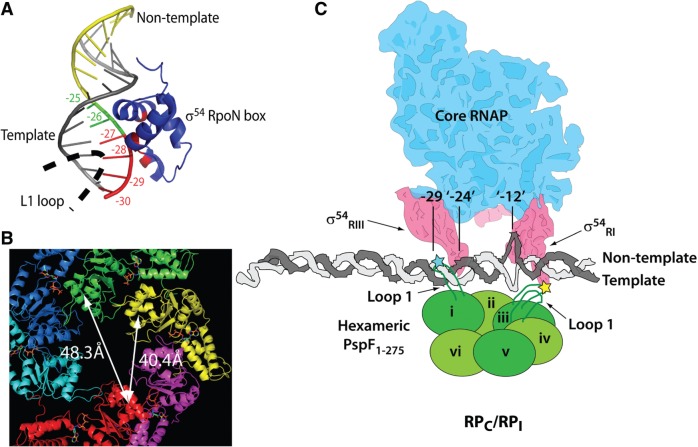
Figure 10.
The proposed organizsation of L1s in engaged RPC/RPI. (A) L1 and σ54RIII might contact the promoter ‘–29’ region from different grooves. The σ54RIII RpoN box (blue) binds to the non-template strand of the nifH promoter (PDB 2O8K). The consensus GG element at –25 and –26 is highlighted in green. The ‘–29’ region is highlighted in red. (B) The PspF1–275 WT hexameric structure with an open spiral (based on energy minimization of monomeric ATP-bound crystal structures, courtesy of M. Rappas) shows that the distances between the centres of two L1s across the hexameric plane are 40.4 Å [measured from subunit (i) to subunit (iii)] and 48.3 Å [measured from subunit (i) to subunit (iv)], consistent with the distance between the boundaries of the ‘–29’ element and the –12 fork junction (∼40–47 Å). (C) The proposed model based on the cross-linking data. The L1–DNA and L1–σ54RI cross-linking sites are depicted by the cyan and yellow stars, respectively. Alternate subunits of the PspF1–275 open spiral are highlighted by different depths of green.
Two structural features of the RPC are thought to impede spontaneous RPO formation: (i) the σ54RI which interacts with core RNAP and blocks DNA entry, functionally reminiscent to σ701.1 in RPC (45–48). (ii) The –12 DNA melting site which is modelled in an upstream position with respect to its place in RPO and is, therefore, misaligned with the active channel of the holoenzyme (11). We speculate that the contact between L1 and the upstream promoter DNA facilitates RPI and RPO formation. Although both L1 and σ54RIII contact the non-template ‘–29’ region (Figure 8 and Figure 10A), they do not appear to contact one another (Figure 2C). L1 and σ54RIII may access the ‘–29’ region from different DNA grooves to hold the promoter DNA firmly in place (Figure 10A). Doucleff et al. (14) proposed that σ54RIII may interact with the β G flap. Thus, nucleotide-dependent conformational changes directed by L1 may facilitate the re-alignment of the holoenzyme with the –12 DNA melting site via the proposed σ54RIII–β G flap interaction. In principle, reorganization of σ54RIII triggered by L1 movement could be transmitted to σ54RI and result in disruption of the previously established inhibitory interactions at the –12 fork junction DNA maintained by σ54RI. An LI-directed torsion generated on σ54RI may facilitate the propagation of DNA melting from –12 to the start site. Thus, disruption of the L1–DNA interaction around the ‘–29’ region could partially contribute to the activation defect as observed when this upstream interaction cannot be established (Figure 9). As PspF progresses through cycles of ATP hydrolysis, L1s contacting both the ‘–29’ region and σ54RI are likely to change from an extended state to a folded down state (19,20,49). These changes may move the DNA downstream to facilitate the re-alignment of the –12 fork junction with the active site of the holoenyzme (11). This is analogous to the DNA threading observed in AAA+ helicases by the ‘staircasing’ ssDNA-binding hairpins (50).
An open spiral hexameric configuration has been observed in many AAA+ proteins (51–56). Joly et al. (29) proposed that the PspF1-275 hexamer also assumed an open spiral configuration employing at least two functional L1s for σ54 binding. Thus we incorporated the PspF1–275 spiral into a proposed transcription complex organization within RPC/RPI (Figure 10C). Structural analyses indicate that the distances between the centres of the base of L1 across the PspF1–275 spiral are 40.4 Å [measured from subunit (i) to subunit (iii)] and 48.3 Å [measured from subunit (i) to subunit (iv), Figure 10B]. This fits well with a 40–47 Å span between the boundary of the ‘–29’ region (position –27, contacted by a L1) and the boundary of the ‘–12’ element [position –14, contacted by σ54RI (57)].
We believe the underlying mechanism proposed can be extended from PspF to other bEBPs. In this context, mutation of the second Gly in the ‘GAFTGA’ motif of Salmonella typhimurium NtrC results in a ‘super’ DNA binding activity (58). The authors suggested that the ‘GAFTGA’ motif may be close to DNA in such inactive bEBP dimmers, and the binding activity may be non-specific due to the additional charge introduced (Lys in place of Gly). In this study, we provide evidence to show that the L1 ‘GAFTGA’ motif is presented for a specific and direct promoter DNA engagement within active bEBP hexamers. Another bEBP that warrants discussion is the Aquifex aeolicus NtrC1 protein. NtrC1 forms closed oligomers in solution [90% heptamers and 10% hexamers (59)]. The negative-stain EM data suggest that although heptameric NtrC1 can engage σ54, a significant portion of the density for σ54 is missing in the co-complex (17). Quite how far the NtrC1 heptamer–σ54 complex might functionally deviate from the transcriptionally active complexes forming with more usual hexameric assemblies of bEBPs, such as PspF, NtrC, ZraR, DpmR, NorR and HrpR/S (18,41,60–63), is unknown. A heptameric arrangement of NtrC1 as compared to, for example, the hexameric ZraR (62) is anticipated to be distinct in terms of the details of interfacial subunit–subunit contacts, some of which are known to control the nucleotide-dependent remodelling output of PspF (64) and to precisely define sites where the ATPase can become uncoupled from remodelling by simple mutation. Although the triple L1 contacts observed in PspF is dictated by the stoichiometry and arrangement of the ring, it can still apply to NtrC1 if the exchange between heptamers and hexamers occurs frequently to allow a faithful and productive engagement of σ54 and promoter DNA.
To conclude, our work provides clear evidence that discrete L1s make interactions with three distinct and well separated elements within RPC/RPI, these are the two σ54RI patches and the ‘–29’ promoter region. The triple contacts by a single feature of a bEBP contrast directly with many AAA+ proteins (e.g. unfoldases and helicases) that contact only either protein or DNA and the classic bacterial activators (e.g. CRP-cAMP receptor protein) that contact protein and DNA via two distinct and spatially well separated domains.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–7.
FUNDING
Biotechnology and Biological Sciences Research Council project (BBSRC) [BB/J002828/1 and BB/G001278/1 to M.B.]. Funding for open access charge: BBSRC [BB/J002828/1 and BB/G001278/1].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Dr J.W. Chin and Dr D.P. Nguyen for providing the pDULE-pBpa plasmid. We thank E. Lawton and Dr T. Simpson for their stimulating comments on the manuscript. We thank all the past and present MB group members for their friendly support. N.Z., N.J. and M.B. conceived and designed the experiments included in this manuscript. N.Z. performed the experiments.
REFERENCES
- 1.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 2.Morris L, Cannon W, Claverie-Martin F, Austin S, Buck M. DNA distortion and nucleation of local DNA unwinding within sigma-54 (sigma N) holoenzyme closed promoter complexes. J. Biol. Chem. 1994;269:11563–11571. [PubMed] [Google Scholar]
- 3.Burrows PC, Wigneshweraraj SR, Buck M. Protein-DNA interactions that govern AAA+ activator-dependent bacterial transcription initiation. J. Mol. Biol. 2008;375:43–58. doi: 10.1016/j.jmb.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 4.Friedman LJ, Gelles J. Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell. 2012;148:679–689. doi: 10.1016/j.cell.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat. Struct. Mol. Biol. 2005;12:603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 6.Studholme DJ, Buck M. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol. Lett. 2000;186:1–9. doi: 10.1111/j.1574-6968.2000.tb09074.x. [DOI] [PubMed] [Google Scholar]
- 7.Wolfe AJ, Millikan DS, Campbell JM, Visick KL. Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 2004;70:2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correa NE, Lauriano CM, McGee R, Klose KE. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 2000;35:743–755. doi: 10.1046/j.1365-2958.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- 9.Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. Borrelia burgdorferi sigma54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl Acad. Sci. USA. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villain-Guillot P, Bastide L, Gualtieri M, Leonetti JP. Progress in targeting bacterial transcription. Drug Discov. Today. 2007;12:200–208. doi: 10.1016/j.drudis.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Bose D, Pape T, Burrows PC, Rappas M, Wigneshweraraj SR, Buck M, Zhang X. Organization of an activator-bound RNA polymerase holoenzyme. Mol. Cell. 2008;32:337–346. doi: 10.1016/j.molcel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rappas M, Schumacher J, Beuron F, Niwa H, Bordes P, Wigneshweraraj S, Keetch CA, Robinson CV, Buck M, Zhang X. Structural insights into the activity of enhancer-binding proteins. Science. 2005;307:1972–1975. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doucleff M, Malak LT, Pelton JG, Wemmer DE. The C-terminal RpoN domain of sigma54 forms an unpredicted helix-turn-helix motif similar to domains of sigma70. J. Biol. Chem. 2005;280:41530–41536. doi: 10.1074/jbc.M509010200. [DOI] [PubMed] [Google Scholar]
- 14.Doucleff M, Pelton JG, Lee PS, Nixon BT, Wemmer DE. Structural basis of DNA recognition by the alternative sigma-factor, sigma54. J. Mol. Biol. 2007;369:1070–1078. doi: 10.1016/j.jmb.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong E, Doucleff M, Wemmer DE. Structure of the RNA polymerase core-binding domain of sigma(54) reveals a likely conformational fracture point. J. Mol. Biol. 2009;390:70–82. doi: 10.1016/j.jmb.2009.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svergun DI, Malfois M, Koch MH, Wigneshweraraj SR, Buck M. Low resolution structure of the sigma54 transcription factor revealed by X-ray solution scattering. J. Biol. Chem. 2000;275:4210–4214. doi: 10.1074/jbc.275.6.4210. [DOI] [PubMed] [Google Scholar]
- 17.Chen B, Sysoeva TA, Chowdhury S, Guo L, De Carlo S, Hanson JA, Yang H, Nixon BT. Engagement of arginine finger to ATP triggers large conformational changes in NtrC1 AAA+ ATPase for remodeling bacterial RNA polymerase. Structure. 2010;18:1420–1430. doi: 10.1016/j.str.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SY, De La Torre A, Yan D, Kustu S, Nixon BT, Wemmer DE. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17:2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappas M, Schumacher J, Niwa H, Buck M, Zhang X. Structural basis of the nucleotide driven conformational changes in the AAA+ domain of transcription activator PspF. J. Mol. Biol. 2006;357:481–492. doi: 10.1016/j.jmb.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 21.Bordes P, Wigneshweraraj SR, Schumacher J, Zhang X, Chaney M, Buck M. The ATP hydrolyzing transcription activator phage shock protein F of Escherichia coli: identifying a surface that binds sigma 54. Proc. Natl Acad. Sci. USA. 2003;100:2278–2283. doi: 10.1073/pnas.0537525100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J. Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasse-Dwight S, Gralla JD. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc. Natl Acad. Sci. USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannon W, Gallegos MT, Buck M. DNA melting within a binary sigma(54)-promoter DNA complex. J. Biol. Chem. 2001;276:386–394. doi: 10.1074/jbc.M007779200. [DOI] [PubMed] [Google Scholar]
- 25.Gallegos MT, Buck M. Sequences in sigmaN determining holoenzyme formation and properties. J. Mol. Biol. 1999;288:539–553. doi: 10.1006/jmbi.1999.2704. [DOI] [PubMed] [Google Scholar]
- 26.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 27.Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MP, Buck M. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 2010;34:797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang N, Joly N, Burrows PC, Jovanovic M, Wigneshweraraj SR, Buck M. The role of the conserved phenylalanine in the sigma54-interacting GAFTGA motif of bacterial enhancer binding proteins. Nucleic Acids Res. 2009;37:5981–5992. doi: 10.1093/nar/gkp658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joly N, Buck M. Single chain forms of the enhancer binding protein PspF provide insights into geometric requirements for gene activation. J. Biol. Chem. 2011;286:12734–12742. doi: 10.1074/jbc.M110.203554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat. Methods. 2005;2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 31.Dorman G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 32.Lin AA, Sastri VR, Tesoro G, Reiser A. On the cross-linking mechanism of benzophenone-containing polyimides. Macromolecules. 1988;21:1165–1169. [Google Scholar]
- 33.Cannon WV, Gallegos MT, Buck M. Isomerization of a binary sigma-promoter DNA complex by transcription activators. Nat. Struct. Biol. 2000;7:594–601. doi: 10.1038/76830. [DOI] [PubMed] [Google Scholar]
- 34.Casaz P, Buck M. Probing the assembly of transcription initiation complexes through changes in sigmaN protease sensitivity. Proc. Natl Acad. Sci. USA. 1997;94:12145–12150. doi: 10.1073/pnas.94.22.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buck M, Cannon W. A simple procedure for visualising protein–nucleic acid complexes by photochemical crosslinking. Nucleic Acids Res. 1994;22:1119–1120. doi: 10.1093/nar/22.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaney M, Grande R, Wigneshweraraj SR, Cannon W, Casaz P, Gallegos MT, Schumacher J, Jones S, Elderkin S, Dago AE, et al. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev. 2001;15:2282–2294. doi: 10.1101/gad.205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leach RN, Gell C, Wigneshweraraj S, Buck M, Smith A, Stockley PG. Mapping ATP-dependent activation at a sigma54 promoter. J. Biol. Chem. 2006;281:33717–33726. doi: 10.1074/jbc.M605731200. [DOI] [PubMed] [Google Scholar]
- 38.Lee JH, Hoover TR. Protein crosslinking studies suggest that Rhizobium meliloti C4-dicarboxylic acid transport protein D, a sigma 54-dependent transcriptional activator, interacts with sigma 54 and the beta subunit of RNA polymerase. Proc. Natl Acad. Sci. USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrows PC, Joly N, Buck M. A prehydrolysis state of an AAA+ ATPase supports transcription activation of an enhancer-dependent RNA polymerase. Proc. Natl Acad. Sci. USA. 2010;107:9376–9381. doi: 10.1073/pnas.1001188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaney M, Buck M. The sigma 54 DNA-binding domain includes a determinant of enhancer responsiveness. Mol. Microbiol. 1999;33:1200–1209. doi: 10.1046/j.1365-2958.1999.01566.x. [DOI] [PubMed] [Google Scholar]
- 41.Joly N, Schumacher J, Buck M. Heterogeneous nucleotide occupancy stimulates functionality of phage shock protein F, an AAA+ transcriptional activator. J. Biol. Chem. 2006;281:34997–35007. doi: 10.1074/jbc.M606628200. [DOI] [PubMed] [Google Scholar]
- 42.Werbeck ND, Schlee S, Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J. Mol. Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 43.Bordes P, Wigneshweraraj SR, Chaney M, Dago AE, Morett E, Buck M. Communication between Esigma(54), promoter DNA and the conserved threonine residue in the GAFTGA motif of the PspF sigma-dependent activator during transcription activation. Mol. Microbiol. 2004;54:489–506. doi: 10.1111/j.1365-2958.2004.04280.x. [DOI] [PubMed] [Google Scholar]
- 44.Wigneshweraraj SR, Casaz P, Buck M. Correlating protein footprinting with mutational analysis in the bacterial transcription factor sigma54 (sigmaN) Nucleic Acids Res. 2002;30:1016–1028. doi: 10.1093/nar/30.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W. Conformational flexibility of bacterial RNA polymerase. Proc. Natl Acad. Sci. USA. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 47.Mooney RA, Landick R. Tethering sigma70 to RNA polymerase reveals high in vivo activity of sigma factors and sigma70-dependent pausing at promoter-distal locations. Genes Dev. 2003;17:2839–2851. doi: 10.1101/gad.1142203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagai H, Shimamoto N. Regions of the Escherichia coli primary sigma factor sigma70 that are involved in interaction with RNA polymerase core enzyme. Genes Cells. 1997;2:725–734. doi: 10.1046/j.1365-2443.1997.1600357.x. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Darst SA, Thirumalai D. Promoter melting triggered by bacterial RNA polymerase occurs in three steps. Proc. Natl Acad. Sci. USA. 2010;107:12523–12528. doi: 10.1073/pnas.1003533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 51.Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp–clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 52.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meinke G, Bullock PA, Bohm A. Crystal structure of the simian virus 40 large T-antigen origin-binding domain. J. Virol. 2006;80:4304–4312. doi: 10.1128/JVI.80.9.4304-4312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 56.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 57.Guo Y, Lew CM, Gralla JD. Promoter opening by sigma(54) and sigma(70) RNA polymerases: sigma factor-directed alterations in the mechanism and tightness of control. Genes Dev. 2000;14:2242–2255. doi: 10.1101/gad.794800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.North AK, Weiss DS, Suzuki H, Flashner Y, Kustu S. Repressor forms of the enhancer-binding protein NrtC: some fail in coupling ATP hydrolysis to open complex formation by sigma 54-holoenzyme. J. Mol. Biol. 1996;260:317–331. doi: 10.1006/jmbi.1996.0403. [DOI] [PubMed] [Google Scholar]
- 59.Chen B, Doucleff M, Wemmer DE, De Carlo S, Huang HH, Nogales E, Hoover TR, Kondrashkina E, Guo L, Nixon BT. ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure. 2007;15:429–440. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bush M, Ghosh T, Tucker N, Zhang X, Dixon R. Transcriptional regulation by the dedicated nitric oxide sensor, NorR: a route towards NO detoxification. Biochem. Soc. Trans. 2011;39:289–293. doi: 10.1042/BST0390289. [DOI] [PubMed] [Google Scholar]
- 61.Jovanovic M, James EH, Burrows PC, Rego FG, Buck M, Schumacher J. Regulation of the co-evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat. Commun. 2011;2:177. doi: 10.1038/ncomms1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sallai L, Tucker PA. Crystal structure of the central and C-terminal domain of the sigma(54)-activator ZraR. J. Struct. Biol. 2005;151:160–170. doi: 10.1016/j.jsb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 63.Wikstrom P, O'Neill E, Ng LC, Shingler V. The regulatory N-terminal region of the aromatic-responsive transcriptional activator DmpR constrains nucleotide-triggered multimerisation. J. Mol. Biol. 2001;314:971–984. doi: 10.1006/jmbi.2000.5212. [DOI] [PubMed] [Google Scholar]
- 64.Joly N, Buck M. Engineered interfaces of an AAA+ ATPase reveal a new nucleotide-dependent coordination mechanism. J. Biol. Chem. 2010;285:15178–15186. doi: 10.1074/jbc.M110.103150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.