Abstract
MvaT from Pseudomonas aeruginosa is a member of the histone-like nucleoid structuring protein (H-NS) family of nucleoid-associated proteins widely spread among Gram-negative bacteria that functions to repress the expression of many genes. Recently, it was reported that H-NS from Escherichia coli can form rigid nucleoproteins filaments on DNA, which are important for their gene-silencing function. This raises a question whether the gene-silencing function of MvaT, which has only ∼18% sequence similarity to H-NS, is also based on the formation of nucleoprotein filaments. Here, using magnetic tweezers and atomic force microscopy imaging, we demonstrate that MvaT binds to DNA through cooperative polymerization to form a nucleoprotein filament that can further organize DNA into hairpins or higher-order compact structures. Furthermore, we studied DNA binding by MvaT mutants that fail to repress gene expression in P. aeruginosa because they are specifically defective for higher-order oligomer formation. We found that, although the mutants can organize DNA into compact structures, they fail to form rigid nucleoprotein filaments. Our findings suggest that higher-order oligomerization of MvaT is required for the formation of rigid nucleoprotein filaments that silence at least some target genes in P. aeruginosa. Further, our findings suggest that formation of nucleoprotein filaments provide a general structural basis for the gene-silencing H-NS family members.
INTRODUCTION
DNA packaging and gene regulation are critical elements in all organisms. In Escherichia coli, a nucleoid-associated protein called histone-like nucleoid structuring protein (H-NS) is involved in DNA compaction and serves as a global transcription regulator mainly by functioning as a gene silencer, especially for those genes that were acquired by horizontal transfer (1–3). H-NS is able to self-associate through its N-terminus, forming dimers and higher-order oligomers, whereas its C-terminus serves as a DNA-binding domain (4–7).
The mechanism by which H-NS performs its functions is based on its interaction with DNA. Previously, it was observed that H-NS can bridge two double-stranded DNA to form hairpin structures (8,9), or in contrast, it can polymerize along the double-stranded DNA causing DNA stiffening (10,11). Recently, it was found that the concentration of magnesium can dictate which of the two binding modes H-NS adopts; binding modes can be switched by adjusting the concentration of magnesium over a physiological range (11). It was also shown that the stiffening of the DNA backbone is caused by formation of a rigid H-NS protein filament along the DNA (11). A recent structural study has implied that H-NS has at least two oligomerization domains that can link H-NS dimers to form a helical-like filament (12). The physiological relevance of the formation of H-NS filaments is suggested by its sensitivity to salt osmolarity, temperature and pH, three factors known to modulate gene regulation by H-NS, over physiological ranges (10,11). Additional in vitro evidence was reported recently in which it was shown that SsrB, an H-NS antagonizing protein, can only compete for DNA binding with H-NS under conditions in which H-NS predominately binds DNA in the stiffening mode (13).
H-NS family members are widely spread among Gram-negative bacteria (14,15). The H-NS homology has often been defined by the capability to complement E. coli hns mutants. MvaT, a protein found in Pseudomonas strains, has been identified as an H-NS-like protein, although it only shares ∼18% sequence identity to E. coli H-NS (16–19). Despite its lack of sequence similarity to H-NS (20), MvaT is functionally similar to H-NS as a global gene regulator. In Pseudomonas aeruginosa, it controls the expression of hundreds of genes (21,22). In addition, preliminary architectural domain analysis of P16 P. mevalonii, which is 82% similar in sequence to MvaT in P. aeruginosa, revealed structural similarity to E. coli H-NS (23). Finally, like H-NS, MvaT preferentially associates with AT-rich regions of DNA (3,24).
Pseudomonas aeruginosa is a mortal pathogen in cystic fibrosis patients (25). The hundreds of genes controlled by MvaT include those involved in biofilm formation, exotoxin A production and quorum sensing (20,22,26–29). Just like H-NS, MvaT can form higher-order oligomer states mainly through oligomerization domains in its N-terminus and is predicted to contain DNA-binding domains mainly in its C-terminal region (16,30). A previous study on the MvaT-dependent cupA gene regulation suggests that the ability to form higher-order oligomers is essential for gene silencing by MvaT, as MvaT mutants that are defective for higher-order oligomer formation but are unaltered with respect to dimer formation fail to repress cupA expression (30).
MvaT, as a nucleoid-associated protein, may also play a role in physical organization of chromosomal DNA in Pseudomonas species. In agreement with this possible function, a previous atomic force microscopy (AFM) study has shown that MvaT is able to bridge the DNA and form higher-order complexes depending on protein concentration (18). However, although MvaT can bridge the DNA, it was not known whether MvaT, like H-NS, can bind the DNA in the stiffening mode and form a rigid nucleoprotein filament that is potentially important for its gene-silencing function. Furthermore, because it has been shown that higher-order oligomerization is necessary for gene silencing of cupA gene expression by MvaT (30), it raises the question of how this higher-order oligomerization affects the DNA binding properties of MvaT. As MvaT performs its functions through DNA binding, answers to this question may provide a link between MvaT nucleoprotein structure and its gene-silencing function.
In this article, we show that MvaT can form rigid nucleoprotein filaments that stiffen the DNA, and this filament can mediate further DNA organization into hairpin structure or higher-order compact structures at increased protein concentration. Importantly, we present evidence that MvaT mutants that are specifically defective for higher-order oligomer formation cannot form the rigid nucleoprotein filaments. In addition, we show that MvaT nucleoprotein filaments are insensitive to changes in salt osmolarity, pH and temperature, and are thus distinct from those formed by H-NS, which is very sensitive to these factors over physiological ranges.
MATERIALS AND METHODS
Proteins
MvaT-His6 protein and its mutants, MvaT(F36S)-His6 and MvaT(R41P)-His6, were purified as described previously (30).
AFM imaging
Freshly peeled mica was incubated with 0.01–0.1% 3-Aminopropyltriethoxysilane (APTES) in de-ionized water for 15 min. One percent glutaraldehyde was then incubated on top of the APTES layer to form glutaraldehyde mica. The DNA substrate was made by linearizing phiX174 DNA (RF 1) with Xho1 restriction enzyme (NEB). Protein (30–3000 nM) and DNA (10 ng) complex in 10 mM Tris (pH 7.4), 50 mM KCl was incubated on the glutaraldehyde mica for 15 min after being mixed.
The protein DNA complex was imaged using Molecular Imaging 5500 AFM (Molecular Imaging, Agilent Technologies) on acoustic AC mode. Silicon cantilever with a resonant frequency of ∼300 kHz and force constant of 40 N/m is used for the imaging (Photonitech, Singapore). Images were collected with a resolution of 1024 × 1024 pixels, with a scan speed of 0.5–1 lines per second. The scan size varies from 1 to 4 μm. All the images were processed with Gwyddion software (gwyddion.net).
Transverse magnetic tweezers
Single λ-DNA molecules, end-labeled with biotin on both ends, were stretched in the focal plane (31). One end was tethered to streptavidin-coated cover glass edge, and the other end was bound to a 2.8 μm paramagnetic bead (Dynalbeads M-280 Streptavidin, Invitrogen, Singapore) as illustrated in Supplementary Figure S1. A home-made glass channel was made to contain these constructs in which the buffer can be changed with constant flow using a syringe pump. The end-to-end extension of the DNA was determined from the centroid of the bead to the edge of the cover glass. The details of the methods are described in the Supplementary Methods.
Binding and cooperativeness measurements
Single λ-DNA molecules are stretched by transverse magnetic tweezers as described in the previous section. We probed the persistence length of MvaT nucleoprotein filaments at various protein concentrations using the force jumping method to isolate the properties of the filaments with minimal contribution from the DNA folding. We could get the DNA occupancy from the persistence length measurement. The measured extension of the DNA is:
where 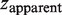 is the measured extension of the mixture of nucleoprotein filaments portion and naked DNA portion,
is the measured extension of the mixture of nucleoprotein filaments portion and naked DNA portion, 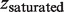 is the extension of the nucleoprotein filaments portion and
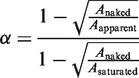
is the extension of the nucleoprotein filaments portion and 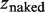 is the extension of the naked DNA portion. Re-arranging the equation to find α:
is the extension of the naked DNA portion. Re-arranging the equation to find α:
As shown in Supplementary Figure S2, a fully coated MvaT nucleoprotein filament has negligible reduction in contour length compared with the naked DNA; hence, we assumed the MvaT nucleoprotein filament has the same contour length as the naked DNA. Substituting the high force approximation of Marko–Siggia formula:
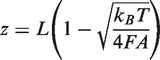 |
where 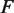 is the stretching force,
is the stretching force, 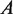 is the persistence length of the DNA,
is the persistence length of the DNA, 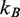 is the Boltzmann constant,
is the Boltzmann constant,  is the temperature,
is the temperature,  is the measured extension of DNA or MvaT nucleoprotein filament and
is the measured extension of DNA or MvaT nucleoprotein filament and 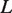 is the contour length of the DNA, which is approximately the same as the DNA coated with MvaT, we would get the occupancy α in terms of the experimentally measured persistence length:
is the contour length of the DNA, which is approximately the same as the DNA coated with MvaT, we would get the occupancy α in terms of the experimentally measured persistence length:
 |
where 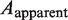 is the measured persistence length of the mixture of nucleoprotein filaments portion and naked DNA portion,
is the measured persistence length of the mixture of nucleoprotein filaments portion and naked DNA portion, 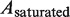 is the persistence length of the nucleoprotein filaments portion and
is the persistence length of the nucleoprotein filaments portion and 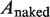 is the persistence length of the naked DNA portion.
is the persistence length of the naked DNA portion.
RESULTS
MvaT can simultaneously stiffen and fold DNA in single-molecule DNA stretching experiments
To determine whether, like H-NS, MvaT could bind DNA in the stiffening mode, we studied the mechanical response of a single λ-DNA (48 502 bp) to MvaT-binding using a transverse magnetic tweezers (31). Theoretical predictions have shown that binding of DNA-distorting proteins can change the force-extension curves of DNA, thus giving us information on the binding mechanism (32). For proteins that cause DNA bends or make DNA more flexible, the extension of DNA bound with proteins will be generally shorter than the same DNA before protein binding. On the contrary, for proteins that enhance DNA bending stiffness, the extension of DNA bound with proteins will be generally longer than the same DNA before protein binding. For proteins that change DNA contour length, the extension of DNA bound with proteins will be different from the same DNA before protein binding at higher tension range (32).
Recent experiments suggest that the DNA binding properties of E. coli H-NS family proteins, such as H-NS and StpA, depend on both monovalent cations and magnesium (11,33). To separate the possible effects of magnesium-dependent MvaT–DNA interaction, we first studied the force response of DNA to MvaT in the absence of magnesium. In our experiments, we first applied a high force (∼10 pN) to prevent DNA folding during the buffer changing process. After buffer exchange, we reduced the force successively till ∼0.1 pN, giving us the forward curve. Afterward, we increased the force successively back to high force, giving us the reverse curve. At each force, the DNA was held for 30 s and the extension was averaged from the last 15 s of the data. Overlapping forward and reverse curves means that the DNA–protein complex is at steady state over the time scale, whereas non-overlapping curves, i.e. hysteresis, hints to protein-induced non-equilibrium DNA folding. It was known previously that DNA folding by formation of hairpin or compact DNA structures often caused hysteresis in single-DNA stretching experiments (13,33).
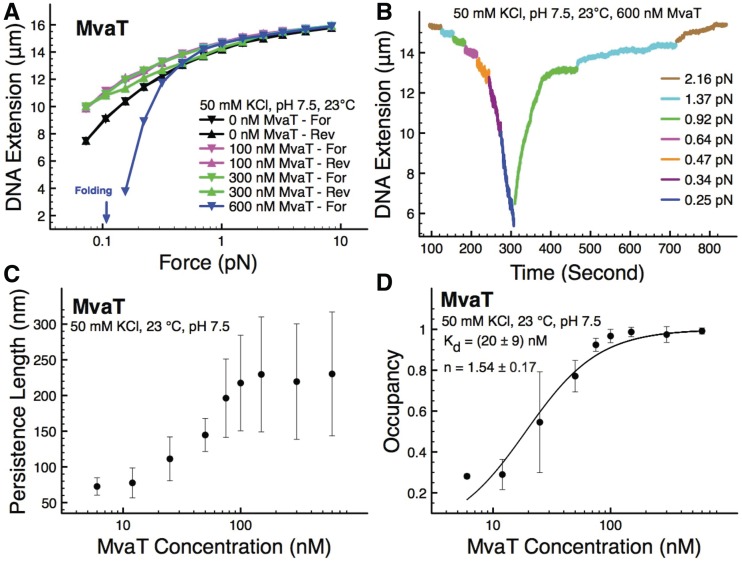
Our magnetic tweezers data show that MvaT can simultaneously stiffen and fold DNA depending on the protein concentrations. At 100 nM MvaT, stiffening dominates in the range of forces tested, indicated by higher extension of DNA compared with the naked DNA at the same force (Figure 1A). Although the hysteresis is negligible in Figure 1A for this protein concentration, varying levels of hysteresis were seen in other independent experiments under the same experimental conditions. Increasing the protein concentration to 300 nM results in a similar degree of DNA stiffening as observed at 100 nM MvaT, whereas the hysteresis was progressively larger compared with results obtained with 100 nM MvaT. Increasing the MvaT concentration further to 600 nM resulted in a degree of stiffening similar to that observed for the other concentrations of MvaT tested at forces above 0.7 pN. At forces below 0.7 pN, the pre-stiffened DNA started to fold, and the extension dropped significantly resulting in DNA that was shorter than the extended naked DNA (blue line). At 600 nM MvaT with forces ∼0.1 pN, DNA extension became too short to be recorded by our instrument (Supplementary Figure S1), so we did not record the reverse curve. Figure 1B shows the DNA folding and unfolding time course recorded on one DNA, in which apparent DNA folding started at ∼0.5 pN or less, and unfolding of partially folded DNA started at ∼1 pN or greater. Because of the non-equilibrium nature of the DNA folding and unfolding process, the values of the unfolding forces varied from one experiment to another. For folded DNA, unfolding forces often started at ∼1 pN. But to completely unfold the DNA, larger forces in the range from 2 to 10 pN were observed in different experiments (see Supplementary Figure S3B for an additional time course data on the same DNA). These force values are in general similar to those observed in DNA folding and unfolding by H-NS at ∼10 mM MgCl2 (11).
Figure 1.
Co-existence of DNA stiffening and folding by MvaT. (A) Apparent stiffening occurred at 100 nM MvaT, indicated by longer extension than the naked DNA at same forces. There is negligible folding over the experimental time scale because no hysteresis was observed when we compare between the forward (from higher force to lower force) and reverse curve (from lower force to higher force). Similar degree of stiffening is also observed at 300 nM MvaT, together with apparent folding indicated by the non-overlapping forward and reverse curve, i.e. hysteresis. At 600 nM MvaT concentration, the DNA is stiffened at high forces before being folded at lower forces. The reverse curve for 600 nM MvaT was not included, because at force <0.1 pN, the DNA was already folded shorter than ∼3 µm, which is outside our observation window (Supplementary Figure S1). Line is drawn connecting the data points to guide the eye. (B) Another independent experiment was done to represent the real time process of DNA folding and unfolding at 600 nM MvaT. The force required to fully unfold the partially folded DNA at this protein concentrations is ∼2 pN. (C) Quantification of MvaT filament stiffness at various protein concentrations. Extension at each force point is recorded briefly and returned back at higher force to ensure no significant folding is involved. At lower force, where folding starts to dominate, the data at that force are omitted from the persistence length measurement to ensure that we measure the persistence length of the filaments with minimal contribution from the DNA folding. The persistence length measurement is extracted from three independent data points (n = 3). (D) Measurement of MvaT cooperative binding to DNA. The data are fitted with Hill equation to measure dissociation constant and cooperativeness of MvaT binding to DNA. Three independent measurements (n = 3) give the number of dissociation constant and cooperativeness, as shown in the figure legend.
Figure 1 demonstrates that MvaT binding can cause both DNA stiffening and DNA folding. The DNA stiffening is particularly interesting, as it was also observed for H-NS and StpA binding to DNA. In those cases, it was known that the stiffening was caused by a cooperative polymerization process of H-NS and StpA, resulting in formation of rigid nucleoprotein filaments (11,33). Therefore, it will be interesting to examine whether MvaT-induced DNA stiffening is also caused by formation of an MvaT nucleoprotein filament through cooperative polymerization. The traditional electrophoretic mobility shift assay (EMSA) measurement is not suitable for the cooperativeness measurement, as the measured cooperativeness may have contribution from both protein polymerized along DNA as well as from DNA bridging and higher-order folding. Therefore, we chose to measure the cooperativeness of MvaT binding to DNA based on the apparent DNA bending persistence length measurements using single-DNA stretching experiments, as described in the next paragraph.
As shown in Figure 1A, because of the experimental timescale used (30 s at each data point), we get a mixture of signals from both DNA stiffening and folding, which will complicate our quantification of the level of DNA stiffening. Noticing that folding generally occurs at lower force and with a slower kinetics, we reasoned that it should be possible to minimize the interference from DNA folding by quickly jumping between forces. In the force-jumping measurements, we held the DNA at ∼10 pN to prevent DNA folding while allowing the rigid nucleoprotein filament to completely form. Then, we jumped the force to a lower value and recorded the DNA extension by holding the DNA for only ∼3 s to prevent significant folding. Then, we jumped back to the higher force to ensure the folding at the lower force was negligible. Repeating this process to a few other lower force values, we obtained the force-extension of the MvaT nucleoprotein filament over a much wider force range with reduced folding because of the shorter time the DNA was held at low forces (Supplementary Figure S2). From these force curves, the corresponding apparent DNA bending persistence lengths at various concentration of MvaT were determined by fitting to the Marko–Sigga formula (34,35) (Figure 1C), which reached saturation at ∼100 nM MvaT concentration with a persistence length value of ∼200 nm. The error bars were standard deviation obtained from three independent measurements. The large variation of the persistence length is unclear; however, it can be explained by interference from DNA folding because we cannot completely eliminate DNA folding by force jumping. Previously, it was reported that the nucleoprotein filament formed by H-NS has a bending persistence length of ∼130 nm at the highest protein concentration measured (10), which is slightly less stiff than that formed by MvaT.
The fraction of DNA coated with MvaT can be calculated from the apparent DNA bending persistence lengths, as described in the ‘Materials and Methods’ section. This allowed us to determine the dissociation constant Kd and cooperativeness by investigating the dependence of the DNA occupancy fraction on MvaT concentration. Figure 1D shows that the DNA occupancy fraction obtained from three independent experiments, where the error bars represent the standard deviation, as a function of MvaT concentration. The saturated persistence length used for the calculation of the occupancy was defined as the higher value of persistence length between 300 and 600 nM MvaT. Fitting these data using the Hill equation (see ‘Materials and Methods’ section), Kd and the Hill coefficient (n) were determined to be ∼20 nM and 1.54, respectively. For comparison, the value of Kd determined by EMSA is ∼148 nM (Supplementary Figure S4). Considering the different sequences and different lengths of the two DNA, the values from the two methods are consistent with each other. Importantly, the ∼1.54 Hill coefficient indicates positive cooperativeness. Hence, we conclude that MvaT-induced DNA stiffening was caused by rigid nucleoprotein filament formation through cooperative polymerization of MvaT along DNA.
In summary, Figure 1 has shown that MvaT is able to simultaneously stiffen and fold the DNA, and that DNA folding is progressively enhanced as the concentration of MvaT increases. Importantly, DNA folding can take place after DNA stiffening occurred, indicating that nucleoprotein filament formation and folding are not necessarily mutually exclusive processes. Further, MvaT was found able to form a rigid nucleoprotein filament through a cooperative polymerization process.
MvaT forms nucleoprotein filaments and compact DNA structures
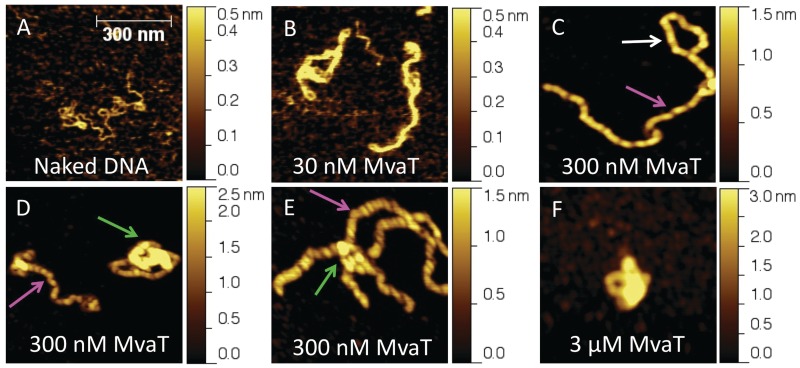
Dame et al. (18) has shown by using AFM that, in >5 mM MgCl2, MvaT is able to bridge DNA and form higher-order complexes in a protein concentration-dependent manner. To relate the DNA-binding properties of MvaT we observe in our single-molecule experiments to those observed previously, we performed AFM imaging experiments of DNA organization by MvaT at various concentrations.
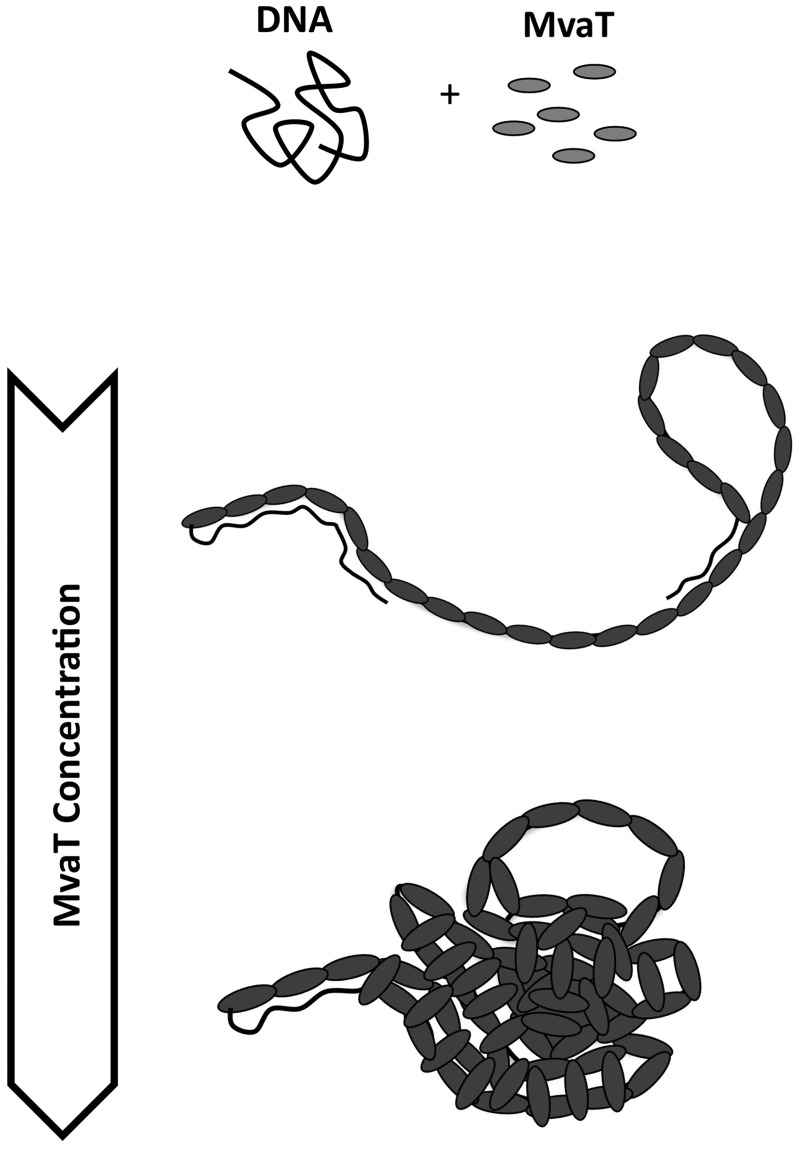
We first imaged DNA–MvaT complexes formed by incubating 0.2 ng/µl DNA in 50 mM KCl, pH 7.5, at 23°C at various MvaT concentrations. At an MvaT concentration of 30 nM, corresponding to 1 monomer of MvaT for every 10 base pairs of DNA, most of the DNA are organized into hairpin structures. These DNA hairpins either assume extended or more compact conformations. Figure 2B shows two representative DNA images, and more images can be found in Supplementary Materials (Supplementary Figures S5A). Increasing the MvaT concentration to 300 nM (corresponding to 1 monomer of MvaT to 1 bp of DNA) (Figure 2C–E and Supplementary Figures S5B) promotes formation of two distinct types of DNA organization: more extended hairpins, some of which appear helical-like (magenta arrows), and collapsed compact structures (green arrows).
Figure 2.
AFM images of linearized double-stranded phiX174 DNA complexed with MvaT at various concentrations. (A) Naked DNA image as control. (B) DNA molecules complexed with 30 nM MvaT show extended DNA hairpins. (C–E) DNA molecules complexed with 300 nM MvaT. Thick helical-like nucleoprotein filaments are generally seen at this concentration (magenta arrow). These filaments can associate with naked DNA to form large protein-coated loop (white arrow). We also observe compact DNA structures as shown by the green arrows. (F) DNA molecules complexed with 3 µM MvaT. At this higher protein concentration, most DNA molecules are condensed into such compact structures. The surface areas for all the images are 0.7 μm × 0.7 μm.
For these hairpins, loops are often found at the end of the hairpins, which are either coated by protein (Figure 2C, white arrow) or not (Supplementary Figure S5B, indicated by cyan arrows). Protein-coated loops are generally larger than the uncoated ones, suggesting that MvaT-coated DNA is likely stiffer than the naked DNA. In addition, the width of the protein-coated DNA complex in the loop is comparable with the apparent width of DNA in the hairpin, suggesting that the hairpin is not formed by association of two protein-coated DNA segments. Otherwise, one would expect double the width in the hairpin region. A plausible explanation for this observation is that the protein-coated DNA can associate with naked DNA segments to form DNA hairpins.
In addition to these regular hairpins, collapsed compact structures were also observed, which are contradictory to the above mechanism that predicts hairpin structures only. One would argue that these structures are caused by further condensation of the filaments by direct inter-filament association. This mechanism predicts progressive DNA compaction as incubation time increases, as more and more filaments will be associated with each other. However, by increasing the incubation time from 15 min to 4 h, we did not observe significant increase in the fraction of the compact DNA structures (Supplementary Figure S6A), suggesting that the more compact structures were formed by a different mechanism. Based on the observation of increasing fraction of the compact structures at higher protein concentration of 3 µM, at which the formation of DNA hairpins were no longer observed after 15 min of incubation time (Figure 2F and Supplementary Figure S5C), we speculate that free proteins in solution mediate the further DNA compaction. Details are discussed in the ‘Discussion’ section.
As suggested by Figure 2C, DNA hairpins are likely formed by MvaT nucleoprotein filaments interacting with the uncoated region on the same DNA. This mechanism has two implications: (i) a cooperative polymerization process to form MvaT nucleoprotein filaments and (ii) a bridging process between the region where a nucleoprotein filament is formed and the region where DNA remains uncoated by MvaT. According to this mechanism, one would predict that, under conditions that MvaT filament formation dominates, bridging should be suppressed on shorter DNA, because on shorter DNA, polymerization has a higher chance to complete on the entire DNA before bridging occurs. To test this, we used shorter DNA (576 bp) and complexed it with 300 nM MvaT (1 monomer MvaT to 1 bp DNA). As expected from the prediction at this MvaT concentration and molar ratio, the majority of the conformations are monomeric filaments (Supplementary Figure S7A), and a smaller fraction of compact structures. We also performed the imaging experiments under conditions in which formation of compact structures dominates at high MvaT concentration and molar ratio. Increasing the MvaT concentration to 3 µM yields compact DNA structures that associate many nearby DNA species (Supplementary Figure S7B), consistent with our observation on larger phiX174 DNA that at high concentrations, MvaT promotes the formation of more compact DNA structures.
Taken together, the results shown in this section have demonstrated two distinct DNA organizations by MvaT: extended DNA hairpins and compact DNA structures, and increasing MvaT concentration and molar ratio to DNA can shift the relative abundance toward compact structures. The former is likely because of the formation of a rigid nucleoprotein filament that associates with naked DNA, and the latter is possibly because of protein-mediated inter-filament association that will be discussed in the ‘Discussion’ section. In general, these imaging results are also in agreement with the previous AFM imaging results obtained with MvaT in the presence of magnesium (18).
Environmental factors moderately modulate DNA folding by MvaT, whereas MvaT nucleoprotein filaments are generally insensitive
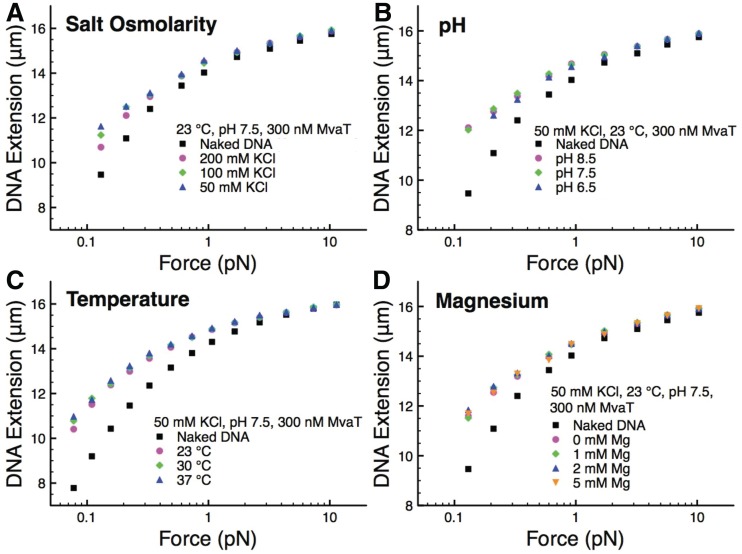
Members of the Pseudomonas genus are well known for their adaptability to a wide range of environmental conditions (36). These environmental conditions can affect the binding of many proteins to DNA. In the case of H-NS, gene expression can be modulated through environmental signals (37). One possibility of this modulation is the direct response of the nucleoprotein filament formed by H-NS to the environment. Single molecule manipulation has revealed that the formation of H-NS filaments is affected by changes in salt osmolarity, pH and temperature over physiological ranges (10,11). By comparison, H-NS-induced DNA bridging was found largely insensitive to these factors (11). Although it remains unclear whether MvaT has a regulatory response to changes in these factors in vivo, investigation of the response of the MvaT nucleoprotein filaments to experimental conditions may provide new insights to the understanding of the response of the MvaT–DNA complexes in vivo. Because MvaT has two distinct DNA organization modes (folding and stiffening), we asked how these two modes are affected by these environmental factors.
As the osmolarity of bacteria is regulated over a wide range roughly up to a few hundred mM (Christian and Waltho, 1961), it is important to see how the DNA structures organized by MvaT depend on ionic strength. We first investigated the effect of the salt osmolarity in the range of 50–200 mM KCl. Figure 3A shows the data obtained on the same DNA from higher to lower salt concentration at a fixed MvaT concentration of 300 nM. The effect of the KCl concentration appears to slightly increase the amount of hysteresis (i.e. the level of DNA folding) as the KCl concentration decreases. These results suggest the compactness of the DNA is slightly tuned by the KCl concentration, in addition to the protein concentration, over a physiologically relevant range. This observation is consistent with our AFM imaging experiments where fewer fractions of compacted DNA structures were observed at 200 mM KCl (Supplementary Figure S6E and F) compared with those observed at 50 mM KCl (Supplementary Figure S5B and C). Similar experiments were repeated to study the effects of pH in the range of 6.5–8.5 and to study the effects of temperature in the range of 23°C–37°C (Figure 3B and C). We observe an increase in hysteresis and strong DNA folding, as the pH value was decreased. The same trend goes for temperature, although with less sensitivity, as also shown in another independent experiment in Supplementary Figure S3E.
Figure 3.
Susceptibility of MvaT-DNA complexes on salt osmolarity, pH, temperature and magnesium. (A) DNA-protein complexes response to KCl concentration with the KCl concentration varied from 50 to 200 mM. Stronger folding is seen with decreasing salt osmolarity, indicated by larger hysteresis. (B) DNA-protein complexes response to pH with the pH varied from 6.5 to 8.5. Folding is stronger at lower pH (6.5), and the DNA is hardly unfolded at this condition. (C) DNA-protein complexes response to temperature, as the temperature is varied from 23°C to 37°C. Stronger folding is observed as the temperature is increased. (D) Magnesium can further enhance the DNA compaction by MvaT when MgCl2 is varied from 1 mM to 5 mM.
As previously shown by our group, magnesium has an important effect on H-NS binding to DNA. Increasing MgCl2 from lower to higher concentrations can switch the DNA conformation from extended monomeric nucleoprotein filaments to bridged DNA. This raises an interesting question regarding whether MvaT–DNA interaction is also sensitive to magnesium. As the free magnesium concentration in bacteria is in the range of a few mM (38,39), we varied the concentration of MgCl2 from 0–5 mM. Figure 3D shows that the amount of hysteresis was found to increase slightly. These observations are in agreement with AFM imaging in the presence of 2 mM MgCl2 (Supplementary Figure 6C and D).
Increased magnesium concentration, which increases the ionic strength, should increase the electrostatic screening similar to increasing the KCl concentration. The opposite trend of magnesium compared with KCl suggests that magnesium increases the level of folding likely through a different mechanism. Interestingly, it is generally consistent with the trend observed in H-NS and StpA, where higher magnesium concentration promotes more higher-order DNA conformations (DNA bridges or compact structures) (8,11,33). However, the mechanisms of magnesium-induced compaction for both H-NS and MvaT and possibly other H-NS-like protein remain unknown and may be different from each other.
We next examined the DNA stiffening responses to environmental factors quantified by the force jumping methods mentioned earlier. As shown in Figure 4A–D, the formation of the MvaT nucleoprotein filament is insensitive to salt, osmolarity, pH, temperature and magnesium. This is different from H-NS, which respond sensitively to these factors over similar ranges. This raises a question whether the regulatory role of MvaT depends on these factors in vivo.
Figure 4.
Susceptibility of MvaT filaments to salt osmolarity (A), pH (B), temperature (C) and MgCl2 (D). Extension at each force point is recorded briefly and returned back at higher force to ensure no significant folding is involved; hence, the extension of the filaments could be estimated across a larger range of forces. We found that MvaT filaments are not responsive to the factors tested, indicated by similar amount of filament stiffness.
Taken together, our results in this section show that in the range of factors tested (KCl, pH, temperature and magnesium), the formation of the rigid MvaT nucleoprotein filament is insensitive, whereas the DNA folding is moderately modulated.
MvaT mutants that are specifically defective for higher-order oligomer formation cannot bind DNA in the stiffening mode but can still fold the DNA
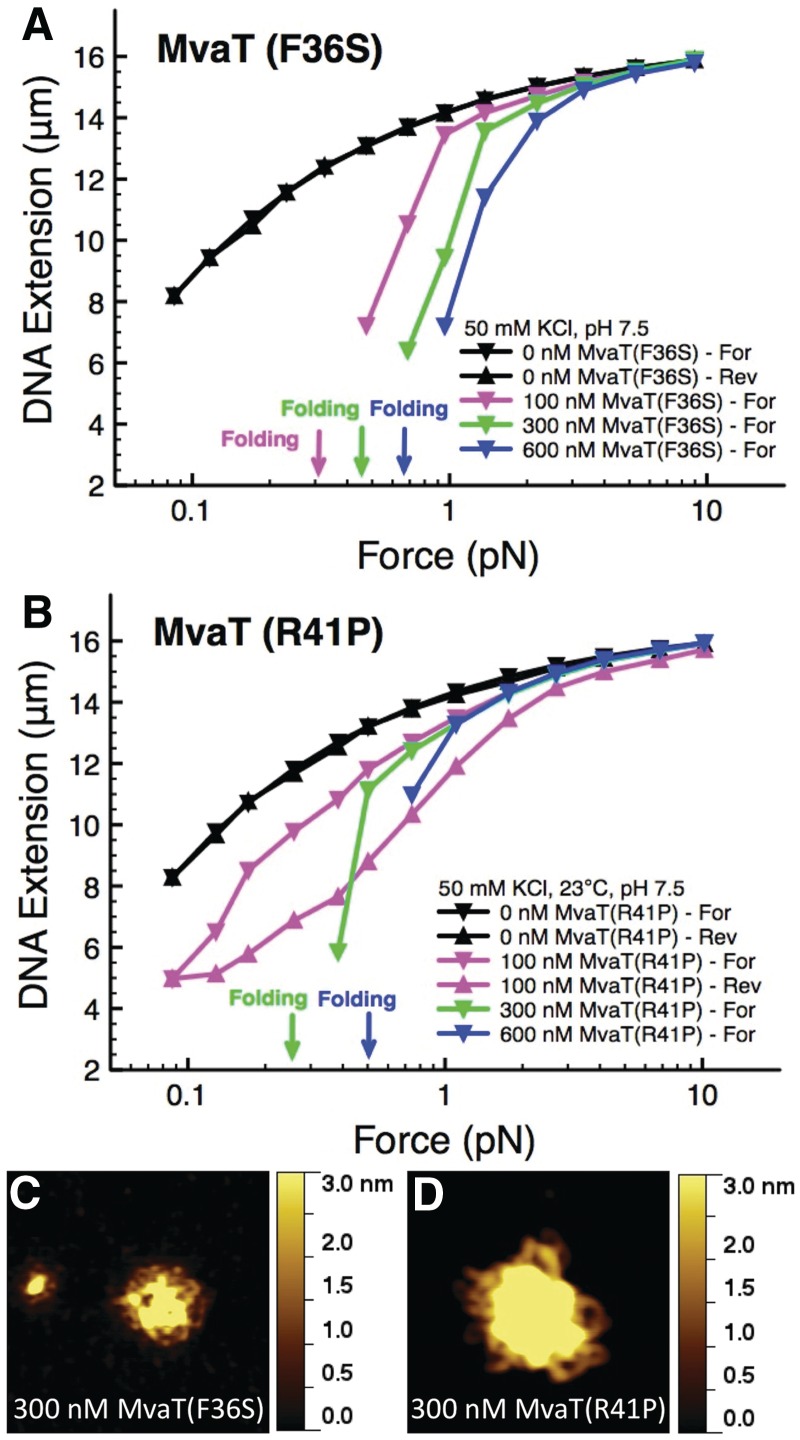
The ability of MvaT dimers to form higher-order oligomers has been shown to be essential for MvaT to function as a silencer (30). In particular, mutants of MvaT containing amino acid substitutions F36S and R41P were found to be specifically defective for higher-order oligomer formation and unable to repress expression of the cupA fimbrial genes (30). To test whether the ability of MvaT to form higher-order oligomers is important for MvaT to form a rigid nucleoprotein filament, which causes DNA stiffening in magnetic tweezers measurements, or DNA folding, or both, we measured the abilities of MvaT(F36S) and MvaT(R41P) to both stiffen and fold the DNA.
The results presented in Figure 5A and B show the impact of the mutants on DNA force response. We found that both of the mutants that are impaired for higher-order oligomer formation could not stiffen the DNA as indicated by the shorter extension of protein-bound DNA over the whole force range that we scanned. However, the mutants were still able to fold DNA, and the folding occurred at higher forces compared with the wild-type MvaT (Figure 1A and B). The absence of stiffening is not because of insufficient protein concentration because we estimate that the Kd of the MvaT mutants is similar to that of the wild-type protein (Supplementary Figure S4). From the comparison between wild-type and mutant MvaT proteins on DNA force response, we conclude that the higher-order oligomerization of MvaT is necessary for the assembly of a rigid nucleoprotein filament. In addition, because higher-order oligomerization of MvaT is necessary for its gene-silencing function, these findings suggest that the ability of MvaT to assemble a rigid nucleoprotein filament is important for silencing.
Figure 5.
Single molecule stretching experiments show that MvaT mutants are totally defective of stiffening. MvaT(F36S) (A) and MvaT(R41P) (B) can only fold the DNA at the range of concentration tested. To further support these results, we do AFM images of linearized double-stranded phiX174 DNA complexed with 300 nM MvaT(F36S) (C) and MvaT(R41P) (D). In agreement with our magnetic tweezers data, we only observed compacted DNA structures, in contrast to the extended hairpins we observed for wild-type MvaT at the same concentration (Figure 2C–E). The surface areas for the images in panel C and D are 0.7 μm × 0.7 μm.
We next compared the abilities of wild-type MvaT and the higher-order oligomerization defective mutants to organize the DNA by AFM imaging. Figure 5C and D show the DNA structures formed at a protein concentration of 300 nM for MvaT(F36S) (Figure 5C and Supplementary Figure S5E) and MvaT(R41P) (Figure 5D and Supplementary Figure 5H), both of which only show compact DNA structures. For comparison, the wild-type MvaT under the same reaction conditions results in much more extended DNA hairpins co-existing with compact structures, as shown in Figure 1C–E and Supplementary Figure S5B. The disappearance of the extended hairpin structures, which suggests the absence of rigid filament, is consistent with our observation of the disappearance of DNA stiffening in single-DNA stretching experiments (Figure 5A and B). Similar imaging observations were also performed at a lower protein concentration of 30 nM (Supplementary Figure S5D and G) or at a higher protein concentration of 3 µM (Supplementary Figure S5F and I), at which no extended DNA conformations are observed.
DISCUSSION
The binding modes of MvaT to DNA
In this work, we have provided a comprehensive study of the binding modes of MvaT to DNA under various physiologically relevant conditions. The DNA-binding mechanism of MvaT is summarized in Figure 6. In terms of the impact of MvaT on the mechanical properties of DNA, our studies have revealed that MvaT binding can cause DNA-stiffening, suggesting the assembly of a rigid nucleoprotein filament, which is similar to what is observed with H-NS (11). To our knowledge, this is the first demonstration of the existence of rigid nucleoprotein filaments formed by a gene-silencing nucleoid-associated protein (NAP) in another species of Gram-negative bacteria, which is in a different order from the enteric bacteria, such as E. coli and Salmonella, in the bacterial kingdom. The relevance of the MvaT nucleoprotein filament to its gene-silencing function will be discussed in the next section. As H-NS and StpA are also known to form nucleoprotein filaments with some evidence of their possible role in gene silencing (11,13,33), our findings raise an important question of whether the formation of nucleoprotein filaments is a conserved and general mechanism across prokaryotes for gene silencing.
Figure 6.
A schematic figure of DNA organization by MvaT. At a concentration of a few hundred nM MvaT, the majority of the DNA are organized into rigid nucleoprotein filament that can associate with naked DNA segments to form DNA hairpins. At higher MvaT concentrations (few mM range), MvaT nucleoprotein filaments can be further compacted into more compact structures, which we speculate are because of free protein-mediated compaction through higher-order oligomerization.
We also found that a pre-stiffened DNA could be further folded at sufficiently low forces. In AFM imaging experiments, we found MvaT can organize DNA into two distinct types of conformations, i.e. extended DNA hairpin structures and more compact DNA structures. The former is consistent with rigid nucleoprotein filaments that can interact with naked DNA, whereas the latter was speculated because of protein-mediated compaction through higher-order oligomerization of the protein already bound on the DNA. One possibility is that MvaT has an additional low-affinity oligomerization domain in addition to the other two oligomerization domains that are required for filament formation. Hence, at higher protein concentration, the filaments can be further compacted by free protein through the additional oligomerization domain. This mechanism predicts increasing proportions of the compact structures at higher protein concentrations, which is consistent with our experimental observations by single-DNA stretching experiments and AFM imaging at higher MvaT concentrations (Figures 1A, 2F and Supplementary Figure S5C). In further support of this speculative mechanism, we introduced additional protein after incubating DNA and MvaT for 4 h at an MvaT concentration of 300 nM, where the conformation of the majority of DNA-containing species is rigid filament-mediated DNA hairpins as shown in Supplementary Figure S6A. After introduction of additional protein to a final concentration of 3 μM, within 15 min of incubation, all the hairpins disappeared, and only the compact structures were observed (Supplementary Figure S6B).
Implications of MvaT filament formation on gene silencing
We have found that MvaT is able to stiffen the DNA through the formation of rigid nucleoprotein filaments, hence adopting more extended conformations as seen in the AFM. In contrast, MvaT mutants that cannot form higher-order oligomers are completely defective for forming MvaT filaments on the DNA. This implies that higher-order oligomerization is necessary for MvaT to bind DNA in the stiffening mode and thus for the formation of rigid nucleoprotein filaments. Because the oligomerization-defective MvaT mutants used here have previously been shown to be incapable of repressing expression of the cupA fimbrial genes in P. aeruginosa (30), our single molecule studies suggest that at least for some genes, MvaT needs to bind DNA in the stiffening mode to function as a silencer. This view is consistent with the recent studies of H-NS and StpA, where formation of a rigid nucleoprotein filament was found relevant to its gene silencing function (11,13,33). In particular, it was reported that SsrB, a well-known anti-silencing protein that relieves H-NS mediated repression of gene expression, only competes with H-NS binding to DNA under conditions in which H-NS can form a rigid filament (13). Taken together, the results obtained with MvaT in this study and the results obtained in previous studies with H-NS, suggest that binding DNA in the stiffening mode may be a conserved feature of members of the H-NS family that is essential for these proteins to function as silencers, at least at some promoters. However, caution should be taken when the effects of the MvaT mutations on the gene-silencing capability of MvaT are generalized to H-NS family members because the MvaT mutants have only been shown incapable of repressing the expression of the cupA genes.
Rigid nucleoprotein filament formation results in DNA being covered by a continuous filament, making access to the DNA more difficult. This will also significantly reduce the dissociation rate of the protein from the DNA. With these filament properties, we suggest the following two possible mechanisms by which the nucleoprotein filament formed by H-NS-like proteins can result in gene silencing: (i) the filament formation can block DNA access preventing RNA polymerase from binding to the promoter or (ii) a stable nucleoprotein filament can block the translocation of RNA polymerase that has initiated transcription.
MvaT mutants that are specifically defective for higher-order oligomer formation cannot form rigid nucleoprotein filaments (as evident from the lack of stiffening), and therefore, they might not be able to prevent RNA polymerase access to DNA or block the translocation of RNA polymerase along the DNA. To our knowledge, this is the first demonstration by single-molecule studies that mutants, which are defective for higher-order oligomer formation, are defective for rigid nucleoprotein filament formation. These oligomerization-defective mutants of MvaT only form compact DNA structures through a mechanism that need not necessarily be the same as that used by the wild-type protein. As these mutants failed to repress cupA gene expression in vivo, this finding suggests that DNA folding per se is not a sufficient mechanism to silence cupA gene expression. Although DNA bridging is a form of DNA folding, it is important to emphasize that the DNA folding observed in our single-molecule studies need not necessarily result from DNA-bridging. It is also important to emphasize that even if the DNA folding observed in our single-molecule studies were the result of DNA bridging, our studies would not rule out the possibility that DNA bridging played an important role in repressing the expression of MvaT target genes other than cupA.
Implications of MvaT-induced DNA folding on chromosomal DNA compaction
In addition to establishing that MvaT forms rigid nucleoprotein filaments, our magnetic tweezers and AFM data have shown that MvaT filaments can be folded into more compact DNA structures in a manner that can be influenced by multiple factors, including protein concentration, salt concentration, temperature and pH over physiologically relevant ranges. This supports the idea that, in addition to its gene-silencing function, MvaT may be one of the NAPs in P. aeruginosa that plays a prominent role in packaging the nucleoid (18). As we have shown in our magnet tweezers experiments, rigid MvaT filaments still exist under conditions where DNA is folded into more compact structures, suggesting that the dual activities of DNA packaging and potential nucleoprotein filament-mediated gene silencing can be simultaneously achieved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7, Supplementary Methods and Supplementary Reference [40].
FUNDING
Ministry of Education of Singapore [MOE2008-T2-1-096]; Mechanobiology Institute, Singapore (internal funding to J.Y.); National Institutes of Health [AI069007 to S.L.D.]. Funding for open access charge: Mechanobiology Institute, Singapore.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Linda J. Kenney for stimulating discussions. We also thank Dr Adam Yuan and the protein expression core facility of the Mechanobiology Institute for protein purification. R.S.W. performed the experiments. J.Y. and S.L.D. conceived the research. R.S.W. and J.Y. designed the experiments and interpreted the data. S.C. and S.L.D. designed and prepared the constructs for protein expression and S.C. purified the proteins. F.W. and L.Y. contributed to experiments at the initial stage. R.S.W., S.L.D., and J.Y. wrote the article.
REFERENCES
- 1.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 2.Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali SS, Xia B, Liu J, Navarre WW. Silencing of foreign DNA in bacteria. Curr. Opin. Microbiol. 2012;15:1–7. doi: 10.1016/j.mib.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Smyth CP, Lundbäck T, Renzoni D, et al. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 2000;36:962–972. doi: 10.1046/j.1365-2958.2000.01917.x. [DOI] [PubMed] [Google Scholar]
- 5.Esposito D, Petrovic A, Harris R, et al. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 2002;324:841–850. doi: 10.1016/s0022-2836(02)01141-5. [DOI] [PubMed] [Google Scholar]
- 6.Ceschini S, Lupidi G, Coletta M, et al. Multimeric self-assembly equilibria involving the histone-like protein H-NS. A thermodynamic study. J. Biol. Chem. 2000;275:729–734. doi: 10.1074/jbc.275.2.729. [DOI] [PubMed] [Google Scholar]
- 7.Ueguchi C, Seto C, Suzuki T, Mizuno T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- 8.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucl. Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 10.Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. USA. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walthers D, Li Y, Liu Y, Anand G, et al. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertin PN, Benhabiles N, Krin E, et al. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in Gram-negative bacteria. Mol. Microbiol. 1999;31:319–329. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- 15.Dorman CJ, Hinton JC, Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- 16.Tendeng C, Bertin PN. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 2003;11:511–518. doi: 10.1016/j.tim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Tendeng C, Soutourina OA, Danchin A, Bertin PN. MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins. Microbiology. 2003;147:3047–3050. doi: 10.1099/mic.0.C0125-0. [DOI] [PubMed] [Google Scholar]
- 18.Dame RT, Luijsterburg MS, Krin E, Bertin PN, et al. DNA bridging: a property shared among H-NS-like proteins. J. Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wally H, Miller SJ, Lu C. The multifaceted proteins MvaT and MvaU, members of the H-NS Family, control arginine metabolism, pyocyanin synthesis, and prophage activation in pseudomonas aeruginosa PAO1. J. Bacteriol. 2009;191:6211–6218. doi: 10.1128/JB.00888-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diggle SP, Winzer K, Lazdunski A, Williams P, Camara M. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 2002;184:2576–2586. doi: 10.1128/JB.184.10.2576-2586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westfall LW, Carty NL, Layland N, et al. mvaT mutation modifies the expression of the Pseudomonas aeruginosa multidrug efflux operon mexEF-oprN. FEMS Microbiol. Lett. 2006;255:247–254. doi: 10.1111/j.1574-6968.2005.00075.x. [DOI] [PubMed] [Google Scholar]
- 22.Vallet I, Diggle SP, Stacey RE, et al. Biofilm formation in Pseudomonas aeruginosa: Fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 2004;186:2880–2890. doi: 10.1128/JB.186.9.2880-2890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rescalli E, Saini S, Bartocci C, et al. Novel physiological modulation of the Pu promoter of TOL plasmid - Negative regulatory role of the TurA protein of Pseudomonas putida in the response to suboptimal growth temperatures. J. Biol. Chem. 2004;279:7777–7784. doi: 10.1074/jbc.M310580200. [DOI] [PubMed] [Google Scholar]
- 24.Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2005;102:11082–11087. doi: 10.1073/pnas.0502663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westfall LW, Luna AM, San Francisco M, et al. The Pseudomonas aeruginosa global regulator MvaT specifically binds to the ptxS upstream region and enhances ptxS expression. Microbiology. 2004;186:3797–3806. doi: 10.1099/mic.0.27270-0. [DOI] [PubMed] [Google Scholar]
- 28.Schuster M, Greenberg EP. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 29.Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 2006;30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 30.Castang S, Dove SL. High-order oligomerization is required for the function of the H-NS family member MvaT in Pseudomonas aeruginosa. Mol. Microbiol. 2010;78:916–931. doi: 10.1111/j.1365-2958.2010.07378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan J, Skoko D, Marko JF. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E. 2004;70:1–5. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 32.Yan J, Marko JF. Effects of DNA-distorting proteins on DNA elastic response. Phys. Rev. E. 2003;68:1–12. doi: 10.1103/PhysRevE.68.011905. [DOI] [PubMed] [Google Scholar]
- 33.Lim CJ, Whang YR, Kenney LJ, Yan J. Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res. 2012;40:3316–3328. doi: 10.1093/nar/gkr1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;25:8759–8770. [Google Scholar]
- 35.Bustamante C, Marko JF, Siggia ED, Smith SB. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 36.Spiers AJ, Buckling A, Rainey PB. The causes of Pseudomonas diversity. Microbiology. 2000;146:2345–2350. doi: 10.1099/00221287-146-10-2345. [DOI] [PubMed] [Google Scholar]
- 37.Atlung T, Ingmer H. H-NS: A modulator of environmentally regulated gene expression. Mol. Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 38.Lusk JE, Williams RJ, Kennedy EP. Magnesium and the growth of Escherichia coli. J. Biol. Chem. 1968;243:2618–2624. [PubMed] [Google Scholar]
- 39.Martin-Orozco N, Touret N, Zaharik ML, et al. Visualization of vacuolar acidification-induced transcription of genes of pathogens inside macrophages. Mol. Biol. Cell. 2006;17:498–510. doi: 10.1091/mbc.E04-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.