Abstract
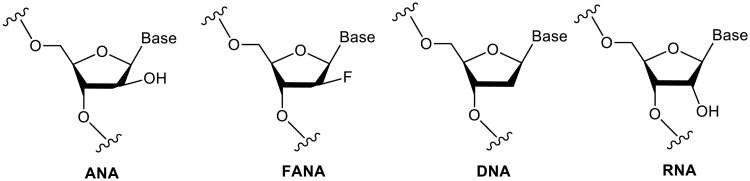
We report here the first structure of double helical arabino nucleic acid (ANA), the C2′-stereoisomer of RNA, and the 2′-fluoro-ANA analogue (2′F-ANA). A chimeric dodecamer based on the Dickerson sequence, containing a contiguous central segment of arabino nucleotides, flanked by two 2′-deoxy-2′F-ANA wings was studied. Our data show that this chimeric oligonucleotide can adopt two different structures of comparable thermal stabilities. One structure is a monomeric hairpin in which the stem is formed by base paired 2′F-ANA nucleotides and the loop by unpaired ANA nucleotides. The second structure is a bimolecular duplex, with all the nucleotides (2′F-ANA and ANA) forming Watson–Crick base pairs. The duplex structure is canonical B-form, with all arabinoses adopting a pure C2′-endo conformation. In the ANA:ANA segment, steric interactions involving the 2′-OH substituent provoke slight changes in the glycosidic angles and, therefore, in the ANA:ANA base pair geometry. These distortions are not present in the 2′F-ANA:2′F-ANA regions of the duplex, where the –OH substituent is replaced by a smaller fluorine atom. 2′F-ANA nucleotides adopt the C2′-endo sugar pucker and fit very well into the geometry of B-form duplex, allowing for favourable 2′F···H8 interactions. This interaction shares many features of pseudo-hydrogen bonds previously observed in 2′F-ANA:RNA hybrids and in single 2′F-ANA nucleotides.
INTRODUCTION
The increasing number of promising applications of modified nucleic acids ranges from new antisense/siRNA therapies to the design of DNA-based nanodevices. For this reason, understanding the structural effects of nucleic acids substitutions is a field of enormous interest. Among the many substitutions in the sugar moiety, arabino nucleic acid (ANA) and its 2′-fluorinated derivative (2′F-ANA; Figure 1) are particularly interesting (1–3). The attractiveness of arabinose modified oligonucleotides was first based on their nuclease resistance and their ability to bind to target mRNA and elicit enzymatic degradation of target mRNA through both RNase H and the RNA-induced silencing complex (1,4–7). More recently, arabino nucleotide derivatives have been used as enhanced in vivo cellular DNA labels (3). In today’s era of synthetic biology, ANA have become attractive systems for studying the evolution of functional biopolymers by in vitro selection (8,9). Recent work on ANA, 2′F-ANA and other nucleic acid analogues has been inspired by the desire to construct genetic systems based on alternative chemical platforms (10).
Figure 1.
Structures of ANA and 2′F-ANA in comparison with DNA and RNA.
Despite the close similarity between ANA and 2′F-ANA, their binding affinities for RNA are strikingly different (1,6,11–13). ANA binds to RNA with relatively low affinity, whereas 2′F-ANA forms thermally stable hybrids with RNA. The solution structure of 2′F-ANA:RNA and ANA:RNA hybrid duplexes, determined in a previous study, reveals that the different binding affinity between these two derivatives is related to several factors, among them a favourable inter-residual pseudo-hydrogen bond (2′F···purine H8) that contrasts with unfavourable inter 2′-OH···nucleobase steric interaction in the case of ANA:RNA hybrid duplexes (13). In 2′F-ANA:RNA hybrids, non-covalent pseudo-hydrogen bonding interactions occur between consecutive residues. Although the structures of these duplexes retain many features of the A-form family of double-stranded helices, 2′F-arabinoses adopt a south/east sugar pucker instead of a north-pucker, which is the common conformation for A-form duplexes. This particular south/east sugar pucker conformation provokes the close proximity between 2′F and H8 atoms of pyrimidine–purine steps as well as a favourable geometry for sequential pseudo-hydrogen bond formation (co-linearity of C8-H8-2′F). Interestingly, such interaction has been also observed in single 2′F-ANA nucleosides (14). This recent observation raises the question that whether the favourable pseudo-hydrogen bond formation could actually be a general feature of nucleic acids containing 2′F-ANA modifications.
Besides its high binding affinity toward RNA, it has also been reported that 2′F-ANA binds to itself to form B-like 2′F-ANA:2′F-ANA (FF) duplexes of high thermal stability (15,16). The structure of pure 2′F-ANA and pure ANA duplexes remains largely unexplored. We show here that ANA, unlike its C2′-epimer (RNA) or its 2′F cousin (2′F-ANA), forms ANA:ANA (AA) duplexes of poor thermal stability. However, the appropriate combination of 2′F-ANA and ANA nucleotides renders sufficient stability to an internal AA segment to permit its characterization by NMR for the first time. We also report on the first structural characterization of a FF duplex. Specifically, we focused on determination of the 3D structure of the chimeric dodecamer, 5′-fCfGfCfGaAaAaUaUfCfGfCfG-3′, by applying 2D NMR spectroscopy experiments and restrained molecular dynamics (MD).
MATERIALS AND METHODS
Oligonucleotide synthesis and purification
Oligonucleotides were synthesized from phosphoramidite precursors, using standard solid-phase methods (17). Purification of oligonucleotides was carried out using reverse phase high performance liquid chromatography. All masses were verified by electrospray ionization-mass spectrometry.
UV melting experiments and derivation of thermodynamic parameters
UV thermal denaturation data were obtained on a Varian Cary 5000 UV-visible spectrophotometer equipped with a Peltier temperature controller. Duplex concentration was 2 µM (total concentration of strands: 4 µM). After heating, samples were cooled to room temperature at a rate of 0.3°C/min and were then refrigerated overnight. Samples were transferred into cold cuvettes in the spectrophotometer and were kept under flowing nitrogen when below 15°C. Absorbance values were recorded after equilibration as the temperature was increased in 0.5°C steps at 1-minute intervals (buffer: 140 mM KCl, 1 mM MgCl2, and 5 mM Na2HPO4, pH 7.2). The optimal melting temperature (Tm) values were calculated using the baseline method, as assignment of baselines was clear in most cases.
NMR experiments
Sample gapmer design [gap(FA)] (Table 1) was suspended in 500 µL of either D2O or H2O/D2O 9:1 in 25 mM phosphate buffer, 100 mM NaCl, pH 7. NMR spectra were acquired in Bruker spectrometers operating at 600, 700 or 800 MHz and were processed with Topspin software. 19F and 1H 1D melting experiments, double quantum filtered correlation scpectroscopy (DQF-COSY), total correlation spectroscopy and nuclear overhauser effect spectroscopy (NOESY), were recorded in D2O and H2O/D2O 9:1. The NOESY spectra were acquired with mixing times of 50, 100, 150, 250 and 300 ms, and the total correlation spectroscopy spectra were recorded with standard MLEV-17 spin-lock sequence at 80 ms mixing time. For highly concentrated samples, NOESY spectra in H2O were acquired with 50 and 100 ms mixing times. For 2D experiments in H2O, water suppression was achieved by including a WATERGATE (18) module in the pulse sequence before acquisition. 2D experiments in D2O were carried out at temperatures ranging from 5°C to 35°C, whereas spectra in H2O were recorded at 5°C to reduce the exchange with water. 19F resonances were assigned from 19F detected using heteronuclear overhauser effect spectroscopy (HOESY) spectra (τm = 200 ms) (19). The spectral analysis program Sparky (20) was used for semiautomatic assignment of the NOESY cross-peaks and quantitative evaluation of the nuclear overhauser effect (NOE) intensities.
Table 1.
Tm values for the self-complementary chimeric duplexes (buffer conditions: 140 mM KCl, 1 mM MgCl2 and 5 mM Na2HPO4; pH 7.2)
| Name | Sequence (5′-3′)a | Tm (°C) |
|---|---|---|
| DD | 5′-CGCGAATTCGCG-3′ | 58 |
| RR | 5′-CGCGAATTCGCG-3′ | 65 |
| FF | 5′-CGCGAATTCGCG-3′ | 76 |
| AA | 5′-CGCGAAUUCGCG-3′ | – |
| alt (AF) | 5′-CGCGAAUTCGCG-3′ | 36 |
| alt (DF) | 5′-CGCGAATTCGCG-3′ | 60 |
| gap (FA) | 5′-CGCGAAUUCGCG-3′ | 56 |
| gap (FD) | 5′-CGCGAATTCGCG-3′ | 58 |
a2′F-ANA, RNA, DNA and ANA.
NMR melting experiments
The dissociation of both structures in gap(FA), i.e. the double-stranded and the hairpin structure is described with the following scheme:

Evidence of unimolecular hairpin structure arises from NMR experiments. The equilibrium between the duplex and the hairpin is slow on the NMR timescale, so that resonances from both species can be observed simultaneously under the appropriate conditions. The equilibrium constants can be determined from the ratio of the areas of equivalent peaks. Thermodynamic parameters for the duplex-hairpin equilibrium are estimated using a van't Hoff analysis of the equilibrium constants at several temperatures well below the temperature when unfolded conformations become populated (21).
Experimental constraints
Quantitative distance constraints were obtained from NOESY experiments by using a complete relaxation matrix analysis with the program MARDIGRAS (22). Error bounds in the interprotonic distances were estimated by carrying out several MARDIGRAS calculations with different initial models, mixing times and correlation times. Standard A- and B-form duplexes were used as initial models, and three correlation times (1.0, 2.0 and 4.0 ns) were employed, assuming an isotropic motion for the molecule. Experimental intensities were recorded at three different mixing times (100, 150 and 250 ms) for non-exchangeable protons. Final constraints were obtained by averaging the upper and lower distance bounds in all the MARDIGRAS runs. Qualitative limits of 1.8 Å and 5 Å were set in those distances where no quantitative analysis could be carried out because of overlapping cross-peaks or peaks with weak intensities. In addition to these experimentally derived constraints, Watson–Crick hydrogen bond restraints were used as well. Target values for distances and angles related to hydrogen bonds were set as described from the crystallographic data. 19F-1H distance constraints from qualitative analysis of HOESY experiments were not used in these calculations. No backbone angle constraints were applied. Distance constraints with their corresponding error bounds were incorporated into the AMBER potential energy by defining a flat-well potential term.
1H-1H J-coupling constants could not be accurately measured because of the relatively broad line-widths of the sugar proton signals. However, sum of J-coupling constants were roughly estimated from DQF-COSY cross-peaks, and some 1H-19F J-coupling constants could be measured using NOESY and DQF-COSY spectra. Loose values were set for the sugar dihedral angles δ, ν1 and ν2 to constrain 2′F-arabinose and arabinose conformations to the east or south domain.
Structure determination of the duplex
Structures were calculated with the SANDER module of the MD package AMBER 7.0 (23). Starting models of the arabino duplexes were built in the A- and B- canonical structures using SYBYL. These structures were taken as starting points for the AMBER refinement, which started with a short run in vacuo (using hexahydrated Na+ counterions placed near the phosphates to neutralize the system). The resulting structures from in vacuo calculations were refined, including explicit solvent, periodic boundary conditions and the particle-mesh Ewald method, to evaluate long-range electrostatic interactions (24). Thus, the structures obtained in the previous step were placed in the center of a water-box with around 4000 water molecules and 20 sodium counterions to obtain electroneutral systems. We used the parmbsc0 (25) revision of the parm99 force field (26,27), including suitable parameters for the arabino and 2′F-arabino derivatives extracted from Noy et al. (28). The TIP3P model was used to describe water molecules (29). The protocol for the MD refinement consisted of an equilibration period of 160 ps using a standard equilibration process (30), followed by 10 independent 500 ps runs. Final structures were obtained by averaging the last ps of individual trajectories and further relaxation of the structure. Analysis of the representative structures and the MD trajectories was carried out with the programs CURVES V5.1 (31), MOLMOL (32), the analysis tools of AMBER and SYBYL and additional ‘in–house’ programs.
Molecular modeling of the hairpin structure
Initial structural models of the hairpin species were built with the program SYBYL on the basis of qualitative NMR information. Conformation of 2′F-ANA residues in the stem was set according to standard B-form parameters. Initial models were submitted to a MD calculation following the same protocols described previously. Only hydrogen bonds restraints for the Watson–Crick G-C base pairs in the stem were used.
RESULTS
Effect of ANA and 2′F-ANA modifications on duplex stability
Duplex formation and melting was monitored by ultraviolet (UV) spectroscopy. Besides the self-complementary sequences containing ANA modifications, isosequential RNA:RNA (RR) and DNA:DNA (DD) and 2′F-ANA:2′F-ANA (FF) control duplexes were also included in the UV melting experiments. Tm values are shown in Table 1 (see Supplementary Figure S1).
Although FF duplex, containing only 2′F-ANA nucleotides, exhibits the highest thermal stability, AA with only ANA nucleotides has a marginal stability and does not show any hypochromicity. Comparison between 1–1 altimer design, alt(AF), or gap(FA), with their corresponding control duplex FF also indicates that insertion of ANA nucleotides in the sequence is destabilizing. Oligonucleotide gap(FA) is especially interesting because although it contains a tract of destabilizing ANA modifications, insertion of 2′F-ANA nucleotides on flanks compensates this destabilization to the extent that permits the characterization of AA base pairs by NMR techniques.
Duplex-hairpin equilibrium
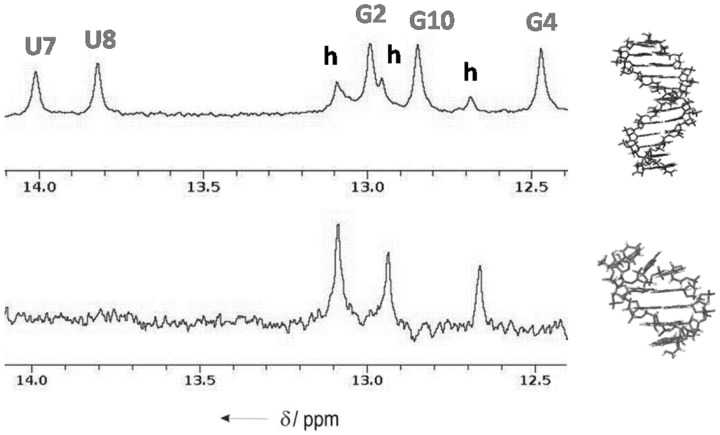
NMR spectra of gap(FA) recorded at different oligonucleotide concentrations are indicative of self-complementary equilibrium. The imino region, shown in Figure 2, is particularly informative. Signals corresponding to AU base pairs are not observed at low concentration, suggesting the formation of a hairpin structure in which the stems contain 2′F-ANA CG pairs, and the loop consists of four unpaired ANA nucleotides. However, at high oligonucleotide concentrations, the imino region is consistent with formation of a double-stranded duplex structure in which all nucleotides (ANA and 2′F-ANA) are base paired.
Figure 2.
Imino region of the 1H NMR spectra of 5′-fCfGfCfGaAaAaUaUfCfGfCfG-3′ at two different oligonucleotide concentrations: top, 0.9 mM and bottom, 0.2 mM (phosphate buffer 100 mM NaCl, pH = 7). Uppercase labels stands for duplex and lowercase labels for hairpin.
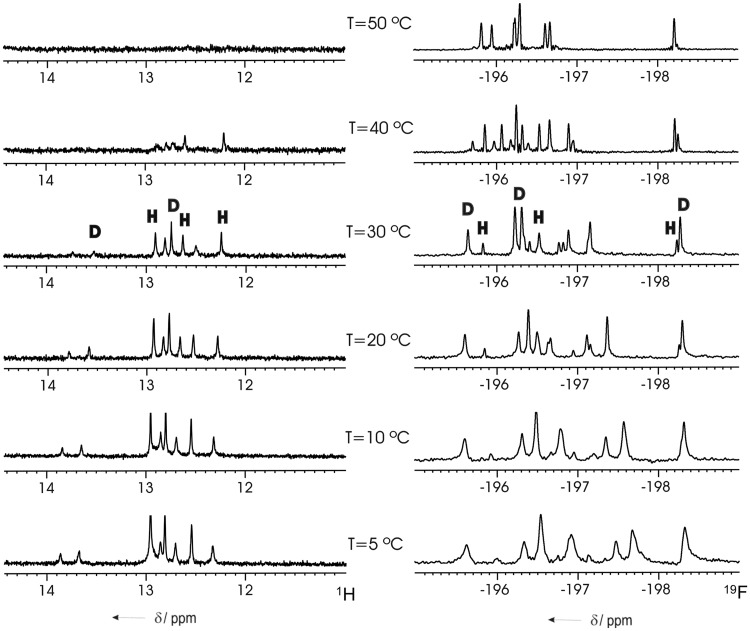
The observation of a hairpin species at low NMR concentrations suggests that the melting temperature of gap(FA) measured by UV at low oligonucleotide concentration corresponds, in fact, to hairpin denaturation and not to duplex dissociation as in the other oligonucleotides shown in Table 1 (the slightly different buffer conditions in UV and NMR experiments do not affect the NMR spectra, as shown in Supplementary Figure S2). Fortunately, hairpin and duplex melting processes can be studied independently by 1H or 19F NMR, as the equilibrium between duplex and hairpin species exhibits a slow kinetics on the NMR timescale (Figure 3). As previously observed by other authors, 19F NMR is particularly useful to monitor secondary structure transitions in nucleic acids (33,34). Both exchangeable 1H and 19F NMR spectra, recorded at different temperatures, indicate that these two species have similar melting temperatures at 0.8 mM oligonucleotide concentration. The thermodynamic parameters for the hairpin to duplex equilibrium were estimated from van't Hoff analysis of the equilibrium constants that were obtained from the relative intensities of the 19F NMR signals between equivalent protons in both forms (see Supplementary Figure S3). This analysis was carried out at temperatures somewhat below the Tm of both species, where the two-state approximation is valid. The resulting values for  , ΔH and TΔS are −0.7 kcal/mol, −6.6 kcal/mol and −5.9 kcal/mol·K, respectively. This ΔG value indicates that the duplex is slightly more stable than the hairpin under these experimental conditions. ΔS is negative as expected, but its value is surprisingly small, which could be attributed to better hydration of the hairpin or a less organized duplex structure. The small negative enthalpy change suggests that contribution of ANA:ANA base pairs to the stability of the duplex is only marginal.
, ΔH and TΔS are −0.7 kcal/mol, −6.6 kcal/mol and −5.9 kcal/mol·K, respectively. This ΔG value indicates that the duplex is slightly more stable than the hairpin under these experimental conditions. ΔS is negative as expected, but its value is surprisingly small, which could be attributed to better hydration of the hairpin or a less organized duplex structure. The small negative enthalpy change suggests that contribution of ANA:ANA base pairs to the stability of the duplex is only marginal.
Figure 3.
Imino region of the 1H (left) and complete 19F (right) NMR spectra of 5′-fCfGfCfGaAaAaUaUfCfGfCfG-3′ [gap(FA)] at different temperatures. Signals of duplex (D) and hairpin (H) species are indicated (oligonucleotide concentration of 0.8 mM, in the same buffer conditions as in Figure 2).
It is interesting to compare the NMR melting experiments of gap(FA) with that of the unmodified DNA control. As shown in Supplementary Figure S4, all the imino proton signals of the DNA control (DNA:DNA) disappear together at the same temperature, with the only exception being the terminal base pairs. However, for the gap(FA) sequence, ANA imino protons located in the central region of the duplex disappear more readily and at much lower temperatures compared with 2′F-ANA imino protons. Such a behaviour indicates that ANA:ANA base pairs are less stable than DNA:DNA base pairs.
NMR assignments
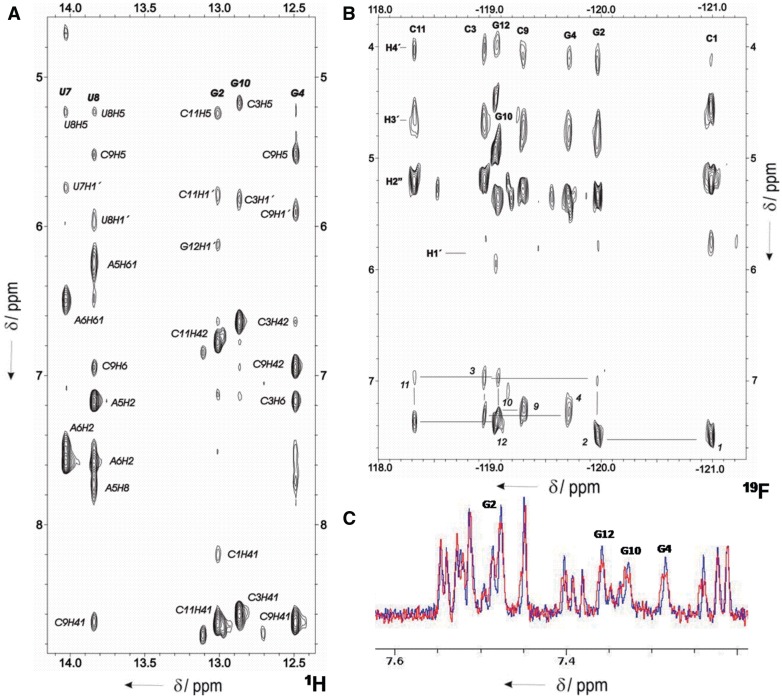
Sequential assignments of exchangeable and non-exchangeable proton resonances were performed following standard methods for right-handed double-stranded nucleic acids, using DQF-COSY, total correlation spectroscopy and 2D NOESY spectra. At high oligonucleotide concentrations, the assignment pathways could be followed in the base-H1′, and in the base-H2′′ regions (Supplementary Figure S5 and Supplementary Table S1). Assignment of the duplex was partially hindered by the low intensity of the intra-nucleotide signals base-H1′ and base-H2″, caused by the splitting of H2″ and H1′ signals by fluorine. The assignment pathways of the low-concentration species cannot be followed between residues A6 to C9 because of the loss of sequential connectivities. In spite of these problems, the complete assignment of 1H and 19F resonances of both species could be carried out by the analysis of experiments recorded at different temperatures and oligonucleotide concentrations. Except for some guanine and adenine amino resonances that could not be observed, most of the exchangeable protons of the duplex could be identified in the NOESY spectra in H2O (Figure 4A). The observed cross-peak patterns indicate that all bases in the double-stranded duplex structure are forming Watson–Crick base pairs. Only exchangeable protons corresponding to CG base pairs were observed in the low-concentration hairpin species.
Figure 4.
(A) Region of the NOESY spectrum of gap(FA) in H2O (mixing time: 150 ms). Watson–Crick base-pairs can be established for all the residues. (B) HOESY spectrum in D2O, showing the 19F-1H assignment pathway. (C) 19F-coupled (red) and 19F-decoupled (blue) 1H NMR spectra, demonstrating different signal intensity in H8 2′F-ANA purines. H8 of 2′F-ANA purines in the duplex are labelled. Oligonucleotide concentration of 0.8 mM, same buffer conditions as in Figure 2.
Assignment of 19F resonances was carried out through their heteronuclear correlations with the adjacent H2′′, H3′, H4′ and H1′ protons. Sequential and intra-residual 19F–1H6/1H8 cross-peaks were observed in the HOESY spectrum (Figure 4B).
Experimental constraints and structure calculations
Quantitative distance constraints were obtained from NOESY experiments by using a complete relaxation matrix analysis. A total of 194 structurally relevant experimental distance constraints were obtained (see Supplementary Table S2). The strong sequential H2″-H6/8 NOEs confirm a B-like conformation. In addition to the NOE-derived information, semi-quantitative analysis of the J-coupling constants obtained from DQF-COSY spectra was carried out. For 2′F-ANA nucleotides, J1′2″ are between 1 and 3 Hz, and J3′2″ are almost zero, whereas J1′2″ and J3′2″ values are both negligible for ANA nucleotides (see Supplementary Table S3). JF-H1' and JF-H3′ were estimated from the splitting in NOESY cross-peaks. Heteronuclear and homonuclear J-couplings are consistent with a south puckering in all arabinose nucleotides (35). Accordingly, sugar torsion angles were constrained to avoid north conformations (allowing east and south). Backbone dihedral angles were not constrained.
In the case of 2′F-ANA nucleotides, heteronuclear J-couplings were detected between 2′-fluorine and purine H8 of the same nucleotide (Figure 4C). These intra-residual couplings have been observed before in single 2′F-ANA modified nucleotides and in hybrids with RNA (11,13,14).
Distance and torsion angle constraints were used to calculate the structure by restrained MD as described in materials and methods section. Twenty final duplex structures were obtained from an explicit solvent refinement and are displayed in Figure 5A. The calculations converge to a well-defined structure, with a root-mean-square deviation (RMSD) of 0.9 Å (excluding the terminal residues). The final AMBER energies and NOE terms are reasonably low in all the structures, which do not exhibit significant constraint violations (see Supplementary Table S2). Both 2′F-ANA and ANA nucleotides are well defined. In the region of the 2′F-ANA-ANA junction, nucleotides A5 and A17 in strands 1 and 2, exhibit larger conformational deviations.
Figure 5.
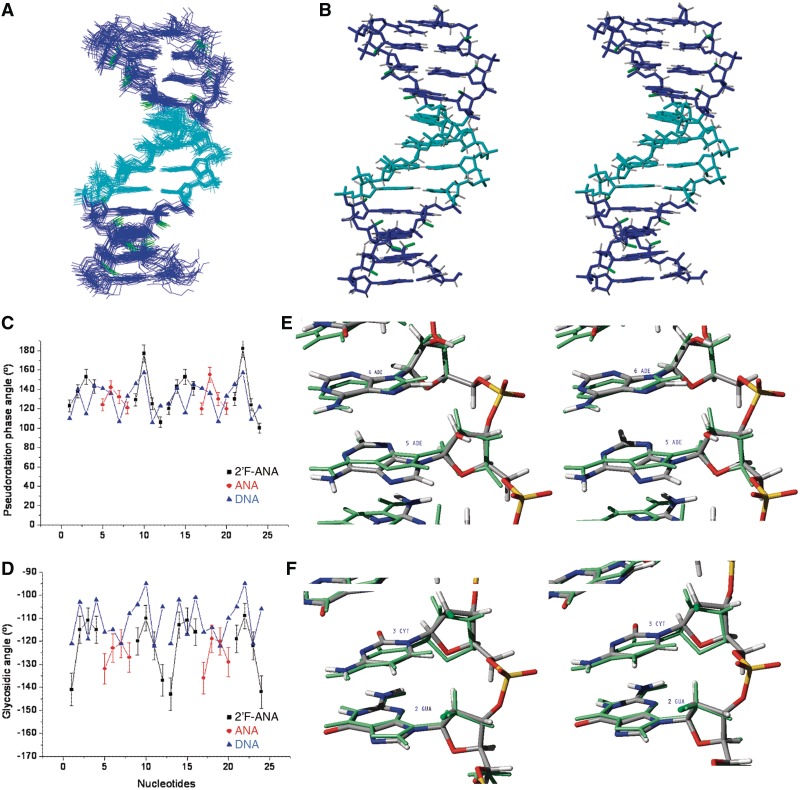
(A) Ensemble of the 20 refined structures of the duplex form of 5′-fCfGfCfGaAaAaUaUfCfGfCfG-3′. (B) Stereo view of the average structure. ANA nucleotides are shown in cyan, 2′F-ANA nucleotides in blue and fluorine atoms in green. (C) Pseudorotation phase angle and (D) glycosidic angle versus sequence. (E) Stereo view showing details of ANA residues, and (F) 2′F-ANA residues, superimposed to the unmodified DNA structure (in green colour, PDB: 1DUF).
Description of the duplex structure
The duplex structure of 5′-fCfGfCfGaAaAaUaUfCfGfCfG-3′, gap(FA), belongs to the B-form family (see Figure 5B). The RMSD between the average structure of gap(FA) and the canonical A-form structure of the same sequence is larger than the RMSD between gap(FA) and a canonical B-form (5.4 Å and 1.8 Å, respectively). In addition, helical parameters are characteristics of B-form helices, with an average rise of 3.3 Å and twist angles around 34°. A summary of helical parameters is shown in Supplementary Table S4.
The geometry of arabinoses and 2′F-arabinoses is similar; most sugar pseudorotation phase angles range from −120° to −170° and glycosidic angles from −110° to −135° (excluding terminal residues) (Figure 5C and D). Backbone torsion angles are well defined and within the usual range for double helical structures. The C2′-endo sugar pucker of arabinose residues is in agreement with the previous solution studies on DNA duplex with 2′F-ANA substitutions (36), and contrasts with the early crystallographic studies in which 2′F-ANA adopt an O4′-endo conformation (37).
An interesting observation was the splitting of the H8 proton signals because of heteronuclear 19F-1H J-couplings (Figure 4C and Supplementary Table S3). These couplings likely reflect pseudo-hydrogen bonds (13,14), as the intra-residual distances between the 2′–F and H8 protons are short (<2.7 Å). Interestingly, intra-residual distances between the arabinose 2′–OH group and H8 protons are also short (<2.6 Å), and we hypothesize that in this case, the 2′–OH/H8 interaction is destabilizing because of the larger size of the hydroxyl group (Figure 5E, F). It seems that the unfavourable steric contact between 2′–OH and H8 outweights their favourable electrostatic interaction, shifting the glycosidic angles of arabinoses toward high anti values (−135°) (Figure 5D) (12). Unfavourable van der Waals contacts also cause distortions in the AA base pairs, as shown in some base:base helical parameters, such as opening and inclination (see Supplementary Table S5). These distortions in ANA glycosidic angles partially hinder the co-planarity of A:U base pairs.
Hairpin structure
The hairpin structure is predominant only at low oligonucleotide concentrations, where conditions for obtaining structural constraints from NMR data are not optimal. Although most of the signals could be assigned, the number of distance constraints derived from NOESY experiments were low, impeding the determination of a high-resolution structure. Despite of this limitation, some features of this interesting unimolecular hairpin structure were deduced from the NMR data. The sequential sugar-base NOE connectivity could be traced for all 2′F-ANA residues and A5 residue in the 2′F-ANA-ANA interface, but not for other ANA residues. These data are consistent with a stem-loop hairpin structure comprising a four CG base pair stem, formed by 2′F-ANA nucleotides and a four-residue loop formed by ANA nucleotides. Furthermore, strong sequential H2″-H6/8 NOEs are indicative of a B-like conformation for the 2′F-ANA stem, which is similar to the FF segment of the duplex structure. In case of ANA nucleotides, the lack of NOEs involving aromatic protons suggests that the bases are somewhat disordered. This is also supported by the chemical shifts of ANA H6/H8 protons, which exhibit similar values as those of the fully denatured oligonucleotide observed at high temperatures. However, qualitative analysis of the J-coupling constants obtained from DQF-COSY spectra shows that J1′2″ and J2″3′ are small, with similar values as those found in 2′F-ANA and ANA nucleotides in the duplex (see Supplementary Figure S6). This observation indicates that arabino sugars have a similar rigidity in a duplex as in the single-stranded loop, retaining a south conformation in both cases.
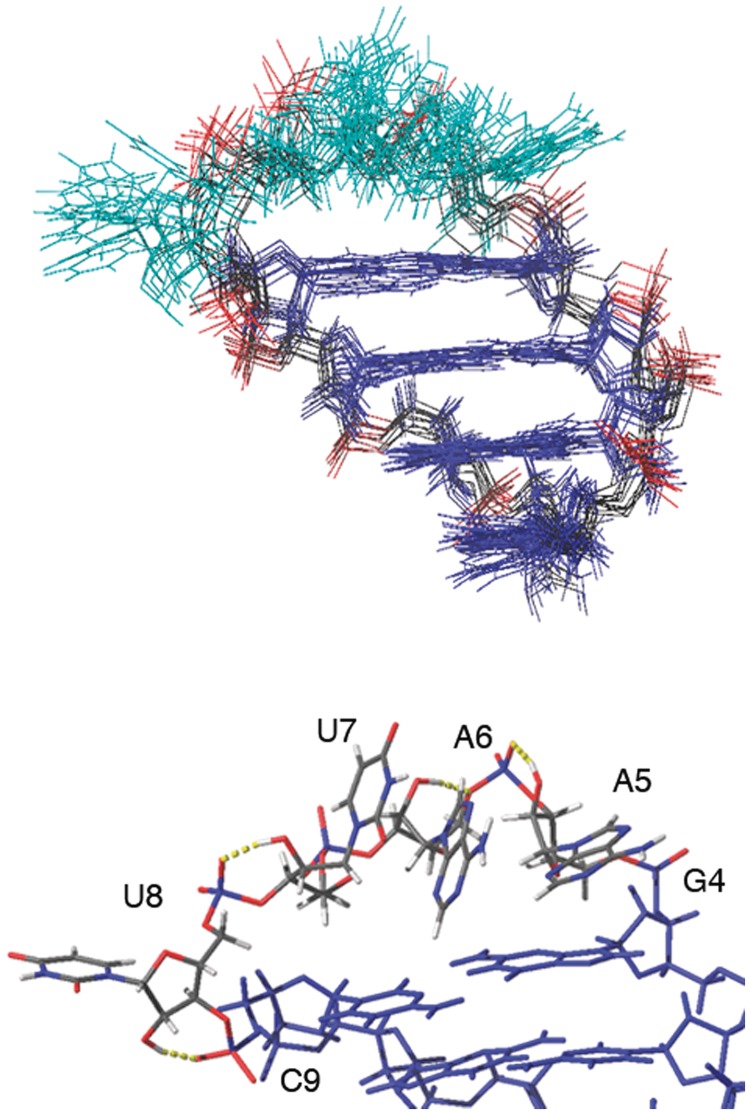
The structural features described previously were confirmed by the model calculation shown in Figure 6. Unrestrained MD trajectories show that the sugar-phosphate backbone of the loop and stem regions of the hairpin is well defined, whereas ANA nucleobases in the loop are disordered.
Figure 6.
Model of the hairpin structure of gap(FA). Top: ensemble of 10 structures from MD calculations. Bottom: detail of the ANA residues in the loop, showing hydrogen bonds between the 2′–OH and phosphate oxygens.
DISCUSSION
Comparison of ANA and 2′F-ANA with unmodified DNA structure
The Dickerson dodecamer is probably the most studied DNA duplex in the literature (38–41). Among the different structures deposited in the protein data bank (PDB), the structure obtained by Tjandra et al. (PDB: 1DUF) is the most appropriate model to use for comparison, as it was also determined by NMR methods, with a high resolution (40). Overall Tjandra’s structure is similar to the duplex conformation of gap(FA). However, some differences are observed. The RMSD between average structure of gap(FA) duplex and Tjandra’s average structure is 1.3 Å for heavy atoms and 0.9 Å for the bases. RMSD between ANA bases is 0.5 Å and between 2′F-ANA bases is ∼0.6 Å. The pseudo-rotation phase angles of ANA and 2′F-ANA sugars are more south relative to the DNA sugars (Figure 5C). These results suggest that arabinose sugars are more rigid compared with deoxyribose sugars, which are in dynamic equilibrium between the northern and southern conformations. In fact, observed J1′2″ coupling values (see Supplementary Table S3) convinced us that such north-south conformational transitions do not occur in ANA and 2′F-ANA nucleotides, as even a small population of north conformation would give rise to a larger value for J1′2″ couplings. Our findings are consistent with previous findings from NMR studies on ANA:RNA (PDB: 2KP3) and 2′F-ANA:RNA (PDB: 2KP4) hybrids, which also show rigid arabinose conformations (13).
Differences in glycosidic angles between gap(FA) and Tjandra’s structures are limited to the ANA residues mentioned previously. Backbone dihedral angles are also similar (Supplementary Tables S6 and S7). Exceptions are the zeta angles, which present smaller values than in the solution structures of unmodified DNA Dickerson dodecamer (40). This is caused by the repulsion between the 2′-substituent and the adjacent phosphate groups. Thus, sequential P-H2′ distances in Tjandra’s structure are smaller than 4 Å, whereas in gap(FA) these distances (sequential P-2′F and P-2′O) are around 5 Å. The minor groove width is narrower than in canonical B-form helices in solution (Supplementary Figure S7). Interestingly, a minor groove narrowing is observed in the central part of the sequence. A similar effect was described for the AATT moiety in the unmodified DNA helix (40).
ANA stabilizes stem-loop structures
All palindromic sequences can, in principle, form stem-loop hairpin structures, although such structures are mainly formed at much lower salt and oligonucleotide concentrations than those used in this study. Early spectroscopic studies on Dickerson sequence suggested the formation of alternative conformations (42,43). However, they only occur at much lower concentrations than those required in structural studies (44). In high-resolution NMR and crystallographic studies, unmodified DNA Dickerson dodecamer has been found as a duplex. In addition, most DNA analogues studied with this sequence adopt a predominant duplex structure. An exception is the recently reported crystallographic structure of Dickerson dodecamer containing N-methanocarba-T DNA analogues (45). However, this modification does not induce a detectable hairpin formation in solution (46,47). The pure RNA version of Dickerson sequence also adopts a duplex structure in solution (48). However, several modifications in the RNA sequence have been reported to induce hairpin formation (49–51).
In the case of ANA modification, the unfavourable steric interactions because of short –OH to aromatic proton distances observed in the duplex are partially alleviated in the loop structure. Such a reduction of unfavourable steric clashes has an effect in the energetic balance between the duplex and the hairpin forms, and is partially responsible for the similar stability of the two forms. Furthermore, several intra-residual hydrogen bonds between 2′-OH and phosphate oxygens were observed for the arabinose loop residues during the unrestrained MD calculations (Figure 6). Intra O2′-H2′… O5′ hydrogen bonds have been previously observed in ANA nucleosides (12,52) and have stabilized the south sugar conformation. It must be noticed that these interactions are not possible in a B-form helical conformation.
Furthermore, arabinose 2′-OH groups are more solvent exposed in the hairpin loop than in the duplex species. Therefore, dehydration of these groups is required for duplex formation, but not for hairpin folding. This effect may be responsible for the small entropy difference between the two conformations. Solvation of 2′-OH groups is an important contribution to the stability of RNA hairpins. Stem-loop hairpins are more common in RNA than in DNA (53). It is not uncommon that short oligoribonucleotides adopt duplex or hairpin structures of comparable stabilities, and transitions between them are related to different biological processes (54). This observation raises the question whether ANA is a DNA or an RNA mimic. In the context of double helical structures, ANA is clearly a DNA analogue (i.e. C2′-endo sugar conformations, B-type duplex). However, ANA promotes formation of stem-loop structures and, in this sense, behaves as an RNA analogue. This tendency of ANA nucleotide tracts to form loop structures may be useful for the rational design of nucleic acid hairpins or in applications where a duplex/hairpin conformational switch is desirable.
ANA versus 2′F-ANA duplexes—why such a difference in stability?
ANA and 2′F-ANA segments of gap(FA) duplex exhibit similar structures. Sugar and glycosidic angle conformations provoke a systematic steric clash between the 2′-OH and their own H6/H8 base protons in ANA nucleotides. However, the same geometry provokes favourable electrostatic interactions with 2′F-ANA nucleotides, because of the smaller radii of fluorine and its higher electronegativity, compared with the 2′-OH group. Similar effects have been observed previously for hybrids of 2′F-ANA and ANA with RNA (13). In 2′F-ANA:RNA hybrids, the geometry of the C2′-F/H8 network is almost co-linear, facilitating the formation of sequential pseudo-hydrogen bonds. The 2′F-H8 contacts are particularly favourable in pyrimidine-purine steps where the base stacking geometry can adjust to optimize the interaction without incurring a steric penalty. In the duplex structure of gap(FA), 2′F..H8 interactions are intra- not inter-residual. Whereas this geometry is not so favourable (the C2′-F..H6/8 angle is around 100–110°), a similar geometry has been found in solution and the crystal for N-H..F-C hydrogen bonds in substituted benzamide derivatives (55).
Organic fluorine is considered to be a poor hydrogen bond acceptor (56–62); however, there is evidence that fluorine-mediated pseudo-hydrogen bonding is important in duplex thermal stability (13,63–66). Because in gap(FA) these interactions are intra-residual, they can occur in the duplex and in the single-stranded denatured states and, consequently, their contribution to duplex stability is not direct. Most probably, such favourable non-covalent interactions act by pre-organizing the bases and, therefore, reducing the entropic penalty of duplex formation. Alternatively, duplex formation may facilitate intra-residual pseudo-hydrogen bonding, and thus, making duplexation enthalpically favourable. Finally, it has been suggested that 2′-substituents in riboses may modulate base pairing strength and π-π stacking interactions in 2′F-RNA structures (67,68). Although this hypothesis is difficult to probe experimentally, fluorine substitution in arabinoses may also polarize their nucleobases and affect the strength of Watson–Crick hydrogen bonds in FF base pairs (67,68). Consistent with this notion, the 2′-fluorine in the nucleoside araF-A increases the polarity of the nucleobase, and thus, increases the acidity of the H8 proton as seen by the faster deuterium exchange (14).
Alternative genetic systems based on arabinose?
Recent work on nucleic acid analogues have been inspired by the desire to construct genetic systems based on alternative chemical platforms (8–10). Two groups have recently reported that DNA polymerases can catalyze DNA synthesis on a 2′F-ANA template strand (8–10). Furthermore, polymerases are able to synthesize ANA (10), 2′F-ANA (8,9) or chimeric 2′F-ANA-DNA (8,9) strands on template DNA strands. Since ANA and 2′F-ANA polymers can be copied to and from DNA with good fidelity (8–10), this attribute is in principle sufficient to carry out the directed evolution of functional ANA and 2′F-ANA molecules.
It is generally assumed that stable duplex formation is a crucial aspect of directed evolution of functional biopolymers based on a synthetic nucleic acid system. Although the poor stability of ANA duplexes would suggest that ANA may not be used to store or propagate information, there is ample evidence that polymerases are able to catalyze oligonucleotide synthesis in the absence of a stable product-template duplex (69). Consistent with this observation, an engineered D4K polymerase is able to carry out template-dependent DNA synthesis on an ANA template (10), despite the poor stability of ANA:DNA duplexes (1). 2′F-ANA has been copied to its own complement (10), but this process is less efficient than copying information between 2′F-ANA to and from DNA (8–10), pointing to subtle structural differences between DNA and 2′F-ANA duplexes (e.g., rigidity of fluorinated sugar) as revealed in this study.
CONCLUSION
ANA can adopt duplex structures in solution similar to standard B-form DNA duplexes. Arabinoses and their fluorine-substituted analogue, 2′-fluoroarabinose, adopt pure C2′-endo conformations, with little variation along the sequence. Despite of these structural similarities, the thermal stability of ANA and 2′F-ANA duplexes is different. Pure 2′F-ANA duplexes are substantially more stable than RNA duplexes of the same sequence, whereas AA duplexes are weak at best (duplex thermal stability increasing from ANA < DNA < RNA < 2′F-ANA).
The electronegativity of fluorine provokes the polarization of the C-H8 bonds, favouring the formation of intra-residue C-2′F…H8-C pseudo-hydrogen bonds. The chimeric oligonucleotide sequence, gap(FA), adopts a duplex and a hairpin structure of surprisingly similar thermal stability. The co-existence of these two species, as observed independently by NMR methods, offered a good opportunity to study the structure of the AA duplex. Analysis of the ANA residues in the loop or duplex structure indicates that they are rigid and pre-organized in a conformation that cannot adjust to optimize the base stacking and Watson–Crick hydrogen bond formation without incurring a steric penalty. NMR studies of other single-stranded arabino and fluoro-arabino oligonucleotides, currently underway in our laboratories, will help test and refine this hypothesis.
ACCESSION NUMBERS
Atomic coordinates have been deposited in the PDB (accession numbers 2LSC).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–7 and Supplementary Figures 1–7.
FUNDING
MICINN [CTQ2010-21567-C02-02 and I-LINK-0216 to C.G. and M.J.D.]; CIHR (operating grant to M.J.D.); CIHR DDTP fellowship (to M.Y.A.); JAE-CSIC pre-doctoral fellowship (to N.M.P.). Funding for open access charge: MICINN.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Dr. J.K. Watts for useful comments and editorial review.
REFERENCES
- 1.Noronha AM, Wilds CJ, Lok CN, et al. Synthesis and biophysical properties of arabinonucleic acids (ANA): circular dichroic spectra, melting temperatures, and ribonuclease H susceptibility of ANA·RNA hybrid duplexes. Biochemistry. 2000;39:7050–7062. doi: 10.1021/bi000280v. [DOI] [PubMed] [Google Scholar]
- 2.Watts JK, Damha MJ. 2′F-arabinonucleic acids (2′F-ANA)-history, properties, and new frontiers. Can. J. Chem. 2008;86:641–656. [Google Scholar]
- 3.Neef AB, Luedtke NW. Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides. Proc. Natl. Acad. Sci. USA. 2011;108:20404–20409. doi: 10.1073/pnas.1101126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowler T, Bergeron D, Tedeschi AL, et al. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-β-d-arabinonucleic acid (FANA) Nucleic Acids Res. 2006;34:1669–1675. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalota A, Karabon L, Swider CR, et al. 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid (2′F-ANA) modified oligonucleotides (ON) effect highly efficient, and persistent, gene silencing. Nucleic Acids Res. 2006;34:451–461. doi: 10.1093/nar/gkj455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, Sarkhel S, Wilds CJ, et al. 2′-fluoroarabino- and arabinonucleic acid show different conformations, resulting in deviating RNA affinities and processing of their heteroduplexes with RNA by RNase H. Biochemistry. 2006;45:4141–4152. doi: 10.1021/bi052322r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denisov AY, Noronha AM, Wilds CJ, et al. Solution structure of an arabinonucleic acid (ANA)/RNA duplex in a chimeric hairpin: comparison with 2′-fluoro-ANA/RNA and DNA/RNA hybrids. Nucleic Acids Res. 2001;29:4284–4293. doi: 10.1093/nar/29.21.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng CG, Damha MJ. Probing DNA polymerase activity with stereoisomeric 2′-fluoro-β-D-arabinose (2′F-araNTPs) and 2′-fluoro-β-D-ribose (2′F-rNTPs) nucleoside 5′-triphosphates. Can. J. Chem. 2008;86:881–891. [Google Scholar]
- 9.Peng CG, Damha MJ. Polymerase-directed synthesis of 2′-deoxy-2′-fluoro-β-D-arabinonucleic acids. J. Am. Chem. Soc. 2007;129:5310–5311. doi: 10.1021/ja069100g. [DOI] [PubMed] [Google Scholar]
- 10.Pinheiro VB, Taylor AI, Cozens C, et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilds CJ, Damha MJ. 2′-deoxy-2′-fluoro-β-d-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao YG, Van der Marel GA, Van Boom JH, Wang AH. Molecular structure of a DNA decamer containing an anticancer nucleoside arabinosylcytosine: conformational perturbation by arabinosylcytosine in B-DNA. Biochemistry. 1991;30:9922–9931. doi: 10.1021/bi00105a016. [DOI] [PubMed] [Google Scholar]
- 13.Watts JK, Martín-Pintado N, Gómez-Pinto I, et al. Differential stability of 2′F-ANA*RNA and ANA*RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 2010;38:2498–2511. doi: 10.1093/nar/gkp1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yahyaee-Anzahaee M, Watts JK, Alla NR, Nicholson AW, Damha MJ. Energetically important C−H···F−C pseudohydrogen bonding in water: evidence and application to rational design of oligonucleotides with high binding affinity. J. Am. Chem. Soc. 2010;133:728–731. doi: 10.1021/ja109817p. [DOI] [PubMed] [Google Scholar]
- 15.Deleavey GF, Watts JK, Alain T, et al. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 2010;38:4547–4557. doi: 10.1093/nar/gkq181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilds CJ. 2000. Synthesis, Physicochemical and Biological Properties of Oligonucleotides Containing 2-fluoro-2-deoxy-b-D-arabinose. Ph.D. Thesis. McGill University. [Google Scholar]
- 17.Damha MJ, Ogilvie KK. Oligoribonucleotide synthesis. The silyl-phosphoramidite method. Methods Mol. Biol. 1993;20:81–114. doi: 10.1385/0-89603-281-7:81. [DOI] [PubMed] [Google Scholar]
- 18.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 19.Yu C, Levy GC. Two-dimensional heteronuclear NOE (HOESY) experiments: investigation of dipolar interactions between heteronuclei and nearby protons. J. Am. Chem. Soc. 1984;106:6533–6537. [Google Scholar]
- 20.Goddard DT, Kneller DG. 3rd edn. San Francisco: University of California; SPARKY. [Google Scholar]
- 21.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 22.Borgias BA, James TL. MARDIGRAS, a procedure for matrix analysis of relaxation for discerning geometry of an aqueous structure. J. Magn. Reson. 1990;87:475–487. [Google Scholar]
- 23.Case DA, Pearlman DA, Caldwell JW, III, et al. San Francisco: University of California; 2007. AMBER7. [Google Scholar]
- 24.Darden TE, York D, Pedersen L. Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 25.Pérez A, Marchán I, Svozil D, et al. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dynamics. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 27.Cornell WD, Cieplak P, Bayly CI, et al. A 2nd generation force field for the simulation of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 28.Noy A, Luque FJ, Orozco M. Theoretical analysis of antisense duplexes: determinants of the RNase H susceptibility. J. Am. Chem. Soc. 2008;130:3486–3496. doi: 10.1021/ja076734u. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 30.Shields GC, Laughton CA, Orozco M. Molecular dynamics simulations of the d(T.A.T) triple helix. J. Am. Chem. Soc. 1997;119:7463–7469. [Google Scholar]
- 31.Lavery R, Sklenar H. 1990. CURVES. helical analysis of irregular nucleic acids. Laboratory of Theoretical Biochemistry CNRS, Paris. [Google Scholar]
- 32.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55, 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 33.Graber D, Moroder H, Micura R. 19F NMR spectroscopy for the analysis of RNA secondary structure populations. J. Am. Chem. Soc. 2008;130:17230–17231. doi: 10.1021/ja806716s. [DOI] [PubMed] [Google Scholar]
- 34.Kreutz C, Kahlig H, Konrat R, Micura R. Ribose 2′-F labeling: a simple tool for the characterization of RNA secondary structure equilibria by 19F NMR spectroscopy. J. Am. Chem. Soc. 2005;127:11558–11559. doi: 10.1021/ja052844u. [DOI] [PubMed] [Google Scholar]
- 35.Thibaudeau C, Plavec J, Chattopadhyaya J. A new generalized karplus-type equation relating vicinal proton-fluorine coupling constants to H−C−C−F torsion angles. J. Org. Chem. 1998;63:4967–4984. [Google Scholar]
- 36.Ikeda H, Fernandez R, Barchi JJ, Huang X, Marquez VE, Wilk A. The effect of two antipodal fluorine-induced sugar puckers on the conformation and stability of the Dickerson–Drew dodecamer duplex [d(CGCGAATTCGCG)]2. Nucleic Acids Res. 1998;26:2237–2244. doi: 10.1093/nar/26.9.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger I, Tereshko V, Ikeda H, Marquez VE, Egli M. Crystal structures of B-DNA with incorporated 2′-deoxy-2′-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability. Nucleic Acids Res. 1998;26:2473–2480. doi: 10.1093/nar/26.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drew HR, Wing RM, Takano T, et al. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tereshko V, Minasov G, Egli M. The Dickerson–Drew B-DNA dodecamer revisited at atomic resolution. J. Am. Chem. Soc. 1999;121:470–471. [Google Scholar]
- 40.Tjandra N, Tate S-I, Ono A, Kainosho M, Bax A. The NMR structure of a DNA dodecamer in an aqueous dilute liquid crystalline phase. J. Am. Chem. Soc. 2000;122:6190–6200. [Google Scholar]
- 41.Kuszewski J, Schwieters C, Clore GM. Improving the accuracy of NMR structures of DNA by means of a database potential of mean force describing base−base positional interactions. J. Am. Chem. Soc. 2001;123:3903–3918. doi: 10.1021/ja010033u. [DOI] [PubMed] [Google Scholar]
- 42.Marky LA, Blumenfeld KS, Kozlowski S, Breslauer KJ. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983;22:1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- 43.Miller M, Kirchhoff W, Schwarz F, et al. Conformational transitions of synthetic DNA sequences with inserted bases, related to the dodecamer d(CGCGAATTCGCG) Nucleic Acids Res. 1987;15:3877–3890. doi: 10.1093/nar/15.9.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hald M, Pedersen JB, Stein PC, Kirpekar F, Jacobsen JP. A comparison of the hairpin stability of the palindromic d(CGCG(A/T)4CGCG) oligonucleotides. Nucleic Acids Res. 1995;23:4576–4582. doi: 10.1093/nar/23.22.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pallan PS, Marquez VE, Egli M. The conformationally constrained N-methanocarba-dT analogue adopts an unexpected C4′-exo sugar pucker in the structure of a DNA hairpin. Biochemistry. 2012;51:2639–2641. doi: 10.1021/bi300215k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Z, Maderia M, Barchi JJ, Marquez VE, Bax A. Changes in DNA bending induced by restricting nucleotide ring pucker studied by weak alignment NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2005;102:24–28. doi: 10.1073/pnas.0408498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maderia M, Shenoy S, Van QN, Marquez VE, Barchi JJ. Biophysical studies of DNA modified with conformationally constrained nucleotides: comparison of 2′-exo (north) and 3′-exo (south) ‘locked’ templates. Nucleic Acids Res. 2007;35:1978–1991. doi: 10.1093/nar/gkm025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chou SH, Flynn P, Reid B. Solid-phase synthesis and high-resolution NMR studies of two synthetic double-helical RNA dodecamers: r(CGCGAAUUCGCG) and r(CGCGUAUACGCG) Biochemistry. 1989;28:2422–2435. doi: 10.1021/bi00432a013. [DOI] [PubMed] [Google Scholar]
- 49.Micura R, Pils W, Hobartner C, et al. Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic Acids Res. 2001;29:3997–4005. doi: 10.1093/nar/29.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plevnik M, Gdaniec Z, Plavec J. Solution structure of a modified 2′,5′-linked RNA hairpin involved in an equilibrium with duplex. Nucleic Acids Res. 2005;33:1749–1759. doi: 10.1093/nar/gki318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Höbartner C, Ebert MO, Jaun B, Micura R. RNA two-state conformation equilibria and the effect of nucleobase methylation. Angew. Chem. Int. Ed. Engl. 2002;41:605–609. [Google Scholar]
- 52.Venkateswarlu D, Ferguson DM. Effects of C2′-substitution on arabinonucleic acid structure and conformation. J. Am. Chem. Soc. 1999;121:5609–5610. [Google Scholar]
- 53.Varani G. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- 54.Bernacchi S, Ennifar E, Toth K, et al. Mechanism of hairpin-duplex conversion for the HIV-1 dimerization initiation site. J. Biol. Chem. 2005;280:40112–40121. doi: 10.1074/jbc.M503230200. [DOI] [PubMed] [Google Scholar]
- 55.Hennig L, Ayala-Leon K, Angulo-Cornejo J, Richter R, Beyer L. Fluorine hydrogen short contacts and hydrogen bonds in substituted benzamides. J. Fluorine Chem. 2009;130:453–460. [Google Scholar]
- 56.Zhou P, Zou J, Tian F, Shang Z. Fluorine bonding — how does it work in protein−ligand interactions? J. Chem. Inf. Model. 2009;49:2344–2355. doi: 10.1021/ci9002393. [DOI] [PubMed] [Google Scholar]
- 57.Dunitz JD. Organic fluorine: odd man out. Chembiochem. 2004;5:614–621. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- 58.Böhm H-J, Banner D, Bendels S, et al. Fluorine in medicinal chemistry. Chembiochem. 2004;5:637–643. doi: 10.1002/cbic.200301023. [DOI] [PubMed] [Google Scholar]
- 59.Ojala WH, Skrypek TM, MacQueen BC, Ojala CR. Intermolecular C-F…H-C contacts in the molecular packing of three isostructural N-(fluorophenyl)mannopyranosylamines. Acta Crystallogr. C. 2010;66:o565–o570. doi: 10.1107/S0108270110043556. [DOI] [PubMed] [Google Scholar]
- 60.Carosati E, Sciabola S, Cruciani G. Hydrogen bonding interactions of covalently bonded fluorine atoms: from crystallographic data to a new angular function in the GRID force field. J. Med. Chem. 2004;47:5114–5125. doi: 10.1021/jm0498349. [DOI] [PubMed] [Google Scholar]
- 61.Mehta G, Sen S. Probing fluorine interactions in a polyhydroxylated environment: conservation of a C–F···H–C recognition motif in presence of O–H···O hydrogen bonds. Eur. J. Org. Chem. 2010;2010:3387–3394. [Google Scholar]
- 62.Mele A, Vergani B, Viani F, et al. Experimental evidence for intramolecular attractive nonbonded C–F···H–C interactions in 2′,3′-dideoxy-4′-(fluoromethyl)nucleosides — through-space JCF and JHF NMR coupling constants, correlation with empirical parameters of solvent polarity and single-crystal X-ray structures. Eur. J. Org. Chem. 1999;1999:187–196. [Google Scholar]
- 63.Sun Z, McLaughlin LW. Probing the nature of three-centered hydrogen bonds in minor-groove ligand−DNA interactions: the contribution of fluorine hydrogen bonds to complex stability. J. Am. Chem. Soc. 2007;129:12531–12536. doi: 10.1021/ja073981p. [DOI] [PubMed] [Google Scholar]
- 64.Pallan PS, Egli M. Pairing geometry of the hydrophobic thymine analogue 2,4-difluorotoluene in duplex DNA as analyzed by X-ray crystallography. J. Am. Chem. Soc. 2009;131:12548–12549. doi: 10.1021/ja905739j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parsch J, Engels JW. C−F···H−C hydrogen bonds in ribonucleic acids. J. Am. Chem. Soc. 2002;124:5664–5672. doi: 10.1021/ja012116g. [DOI] [PubMed] [Google Scholar]
- 66.Egli M, Pallan PS, Allerson CR, et al. Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3′-fluoro hexitol nucleic acid (FHNA and Ara-FHNA) modified oligonucleotides. J. Am. Chem. Soc. 2011;133:16642–16649. doi: 10.1021/ja207086x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manoharan M, Akinc A, Pandey RK, et al. Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew. Chem. Int. Ed. Engl. 2011;50:2284–2288. doi: 10.1002/anie.201006519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pallan PS, Greene EM, Jicman PA, et al. Unexpected origins of the enhanced pairing affinity of 2′-fluoro-modified RNA. Nucleic Acids Res. 2011;39:3482–3495. doi: 10.1093/nar/gkq1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai C-H, Chen J, Szostak JW. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc. Natl Acad. Sci. USA. 2007;104:14598–14603. doi: 10.1073/pnas.0704211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.