Abstract
Binding experiments with alkyl-transfer-active and -inactive mutants of human O6-alkylguanine DNA alkyltransferase (AGT) show that it forms an O6-methylguanine (6mG)-specific complex on duplex DNA that is distinct from non-specific assemblies previously studied. Specific complexes with duplex DNA have a 2:1 stoichiometry that is formed without accumulation of a 1:1 intermediate. This establishes a role for cooperative interactions in lesion binding. Similar specific complexes could not be detected with single-stranded DNA. The small difference between specific and non-specific binding affinities strongly limits the roles that specific binding can play in the lesion search process. Alkyl-transfer kinetics with a single-stranded substrate indicate that two or more AGT monomers participate in the rate-limiting step, showing for the first time a functional link between cooperative binding and the repair reaction. Alkyl-transfer kinetics with a duplex substrate suggest that two pathways contribute to the formation of the specific 6mG-complex; one at least first order in AGT, we interpret as direct lesion binding. The second, independent of [AGT], is likely to include AGT transfer from distal sites to the lesion in a relatively slow unimolecular step. We propose that transfer between distal and lesion sites is a critical step in the repair process.
INTRODUCTION
Many environmental and endogenous alkylating agents react with cellular DNA to produce base adducts (1). Among these, O6-alkylguanines are particularly mutagenic and carcinogenic because they base-pair with thymine in preference to cytosine during DNA replication (2). In human cells, O6-alkylguanines are repaired by the O6-alkylguanine DNA alkyltransferase (AGT, also known as methylguanine methyltransferase, MGMT) (3,4). This enzyme is of clinical interest because, in addition to its native roles, it also protects tumor cells against chemotherapeutic DNA alkylating drugs (4–7). AGT-inhibitors are currently in clinical trial with the goal of improving the efficacy of alkylating agents in cancer chemotherapy (8–10).
Human AGT is a small, monomeric protein (207 amino acids, Mr = 21 519) that is expressed constitutively in normal cells (6,11,12). It is overexpressed in some tumors that have been exposed to alkylating agents, and this is thought to be one mechanism by which tumor cells survive alkylating chemotherapy (4,6,13). AGT catalyzes DNA-repair reactions in which a single alkyl-group is transferred from the O6-position of guanine (or less efficiently, the O4-position of thymine (3,14)) to an active-site cysteine (C145 in the human protein). This returns the DNA base to its unmodified state, but the alkylated enzyme is permanently inactivated by this reaction and is rapidly degraded in vivo (15,16). Because repair by AGT is stoichiometric, the number of O6-alkylguanine and O4-alkylthymine adducts that can be repaired at one time depends on the cellular concentration of the un-alkylated form of AGT (6,17) and on its distribution between alkylated and competing unmodified sites throughout the genome. These facts motivate our study of AGT–DNA interactions.
AGT binds unmodified single-stranded and duplex DNAs with little sequence specificity and modest affinity but significant cooperativity (18–20). Cooperative DNA binding appears to be important for lesion search and/or repair, as mutations in the protein–protein interface that disrupt cooperativity also make cells highly sensitive to the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (21). Cooperative interactions allow AGT to bind available DNA sites to high densities (up to 1 AGT/4 bp (22)) and may speed the movement of AGT molecules between sites as part of the lesion-search mechanism (23).
Comparable binding studies using wild-type enzyme and alkylguanine-containing DNAs have proved difficult, because the alkyl-transfer reaction rapidly converts the enzyme-substrate mixture into alkyl-enzyme and unmodified DNA products (18,24). As a partial solution to this problem, alkyl-transfer-inactive forms of AGT have been studied, including active-site mutants, such as the C145S protein (24) and chemically modified forms such as the C145-methyl and C145-benzyl adducts (18,25). This strategy allowed DNase I mapping of a 6mG-specific complex, showing protection of 18 nt on the 6mG-strand and 14 nt on its complement (24), although the number of proteins and their distribution within the complex were not established. If binding densities like those found with unmodified DNA apply, this protected site might accommodate as many as 4 AGT molecules. Use of catalytically inactive forms of AGT and a related strategy using suicide substrates, allowed crystallization of AGT-substrate and substrate-analogue complexes, providing detailed views of structures involved in the alkyl-transfer reaction (3,26,27), including direct evidence for the stabilization of extrahelical bases in the active site of the enzyme. Intriguingly, all of these structures contained single AGT molecules bound to isolated DNA sites. If generalizable to free-solution conditions, this result would suggest that specific AGT-lesion complexes might have 1:1 stoichiometries and that lesion specific binding might not be cooperative. The contrast between the cooperative binding distributions observed with unmodified DNAs and the crystallography-based predictions for specific complexes calls for further examination. The results presented below are a first attempt to address this gap in our knowledge.
MATERIALS AND METHODS
Reagents
Agar, yeast extract and tryptone broth were obtained from Midwest Scientific. T4 polynucleotide kinase and endonuclease Nar I were purchased from New England Biolabs. [γ-32P]ATP was from ICN Radiochemicals. All other biochemicals were from Sigma.
Proteins
Human AGT, with wild-type sequence except for a C-terminal (His)6-tag replacing residues 202–207, was encoded on plasmid pQE-hAGT (26); active site mutants C145S, C145A as well as the alkyltransfer-active P140K were encoded on analogous plasmids pQE-hAGT-C145S, -C145A and -P140K. All were kindly provided by Dr A.E. Pegg (Penn State University). The P140K mutation reduces the size of the active site pocket and replaces an aliphatic surface within the pocket with a positively charged one (4). These changes do not abolish alkyltransferase activity, but they weaken binding of the inhibitor O6-benzylguanine (28). We anticipated that these changes might slow repair of 6mG in DNA enough to allow detection of a pre-repair complex by an active form of AGT. Experiments testing this possibility are presented below.
Wild-type and mutant sequences were confirmed by sequencing plasmid DNA from candidate clones (performed by Seqwright DNA Technology Services). Proteins were expressed in XL1-blue Escherichia coli (Stratagene), grown in Luria Broth supplemented with 100 µM ZnCl2 (29). They were purified by Talon® chromatography as described (26). Minor contaminants were removed and proteins were transferred into storage buffer (20 mM Tris (pH 8.0 at 20°C), 250 mM NaCl, 1 mM DTT (dithiothreitol)) by chromatography on Sephadex G-50. Protein solutions were stored frozen at −80°C until needed. AGT concentrations were measured spectrophotometrically using ε280 = 3.93 × 104 M−1 cm−1 (18). The samples of AGT used here were >95% active in DNA binding (20).
Nucleic acids
Oligonucleotides of 24 and 26 residues (sequences shown in Table 1) were purchased from The Midland Certified Reagent Company Inc. DNA samples for repair assay and Electrophoretic mobility shift assay (EMSA) experiments were labeled at 5′ termini with 32P as described by Maxam and Gilbert (30). Unincorporated [γ-32P]ATP was removed by buffer exchange using Sephadex G-10 mini-spin columns pre-equilibrated with 10 mM Tris (pH 8.0 at 21°C). If needed, labeled oligonucleotides were further purified by gel electrophoresis under denaturing conditions (30% polyacrylamide gel, 8 M Urea (30)) and recovered by the crush–soak method, precipitated with ethanol, resuspended and dialyzed against buffer consisting of 10 mM Tris, 0.1 mM EDTA, pH 8.0 at 20 ± 1°C. DNA duplexes were prepared by annealing purified 5′-labeled oligonucleotides with slight excess of the complementary unlabeled strands. Single-stranded DNA (ssDNA) concentrations were measured spectrophotometrically using extinction coefficients provided by the manufacturer.
Table 1.
Oligonucleotides used in this study
| Oligo | Sequence | Purpose |
|---|---|---|
| 1 | 5’-AGT CAG TCA GTC AGT CAG TCA GTC AG-3’ | 26mer binding substrate |
| 2 | 5’-AGT CAG TCA GTC AGT CAG TCA GTC AG-3’ | 6mG- 26mer binding substratea |
| 3 | 5’-CTG ACT GAC TGA CTG ACT GAC TGA CT-3’ | Complement to oligos 1 and 2 |
| 4 | 5’-GGG TCA TTT GGC GCC TTT CGA TCC-3’ | 24mer binding and repair substrateb |
| 123 456 | ||
| 5 | 5’-GGA TCG AAA GGC GCC AAA TGA CCC-3’ | 24mer binding and repair substrate, complement to oligo 4 |
Electrophoresis mobility shift assays
Two different binding buffers were used. Our standard buffer (used in previous work) is 10 mM Tris-HCl (pH 7.6 at 20°C), 100 mM KCl, 2 mM DTT. In a few cases where consistency with the alkyltransferase assay buffer was needed, the binding buffer was 20 mM Tris-acetate (pH 7.9 at 25°C), 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT. For each experiment the corresponding buffers are identified in text and figure captions. Samples were equilibrated at 20 ± 1°C for 1 h; tests with longer incubations gave indistinguishable results, indicating that equilibrium had been attained (results not shown). Electrophoresis was carried out as previously described (20) in 15% or 20% polyacrylamide gels, cast and run in 10 mM Tris-acetate (pH 8.0), 100 mM KCl. Autoradiographic images were captured on storage phosphor screens (type GP, GE Healthcare) detected with a Typhoon phosphorimager and bands were quantified with Image-Quant software (GE Healthcare).
Binding analyses
Stoichiometries were estimated using the method of Fried and Crothers (31). For the concerted binding of n protein molecules to a single DNA (nP + D ↔ PnD), the association constant K = [PnD]/[P]n[D] can be rearranged to give the linear relationship
| (1) |
Titration of DNA with protein varies the ratio [PnD]/[D], and [P] is calculated for each titration step using the conservation relation [P] = [P]tot − n[PnD], in which [P]tot is the total protein concentration. The linear dependence of ln [PnD]/[D] on ln [P] provides a new estimate of n and this value is fed back into the conservation relation. The cycle is iterated until values of n and K converge. This approach is particularly useful for analysis of mobility shift data like those in Figure 1, which allow direct measurement of [D] and [PnD] at every step (32). It is worthwhile to note that for n > 1, the equilibrium constants obtained in this analysis are composite values that include contributions from two or more protein–DNA interactions (for 6mG-containing DNAs these could be lesion-specific and non-specific interactions) as well as protein–protein contacts.
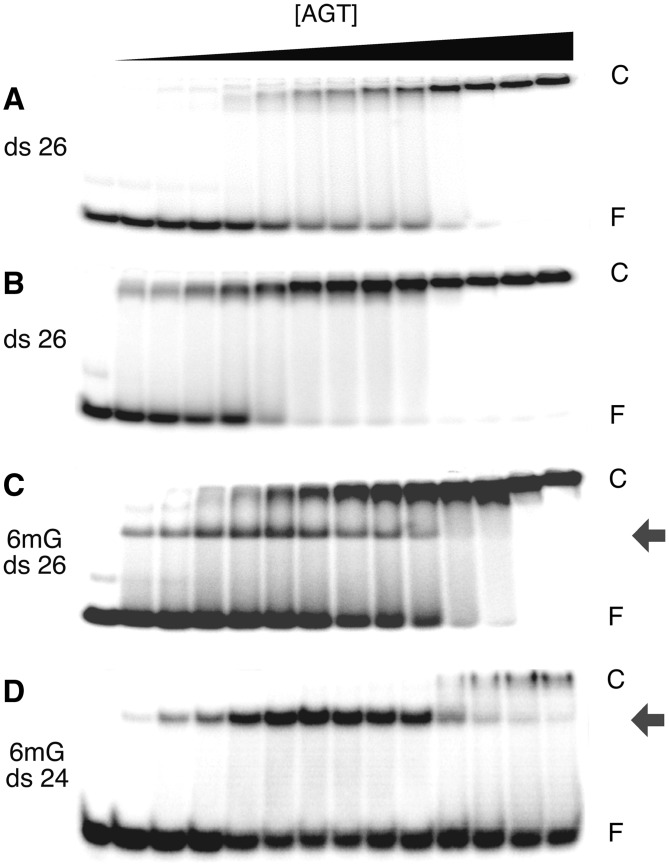
Figure 1.
Discrete, high-mobility complexes form when AGT binds 6mG-containing dsDNAs. EMSAs performed at 20 ±1°C. (A) Titration of unmodified double stranded 26 bp DNA (oligo 1 + 3 duplex; 0.17 µM) with wild-type AGT (0–9.6 µM) to form the low-mobility cooperative complex. The equilibration buffer was 10 mM Tris (pH 8.3 at 20°C), 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 5% glycerol, 0.1 mg/mL BSA (bovine serum albumin). Samples resolved in a 15% native polyacrylamide gel cast and run as described. (B) Titration of unmodified double stranded 26 bp DNA (oligo 1 + 3 duplex; 0.17 µM) with C145S-AGT (0–9.5 µM). Equilibration buffer and gel conditions as in (A). (C) Titration of 6mG-containing 26 bp duplex DNA (oligo 2 + 3 duplex; 0.15 µM) with C145S-AGT (0–5.1 µM). Equilibration buffer and gel conditions as in (A). (D) Titration of 6mG-containing 24 bp duplex DNA (oligo 4 + 5 duplex; 0.038 µM) with C145S-AGT (0–2.0 µM). Equilibration buffer was 40 mM Tris-acetate (pH 7.9 at 20°C), 100 mM potassium acetate, 20 mM magnesium acetate, 2 mM DTT, 0.5 mg/mL BSA. These samples resolved in a 20% native polyacrylamide gel cast and run as described. Band designations: F, free DNA; C, cooperative AGT-DNA complex; grey arrows, 6mG-specific complexes.
DNA alkyltransferase assays
NarI endonuclease is inactive when its substrate contains an O6-methylguanine (6mG) at residue 2 of its target sequence (33) (numbering shown in Table 1). Cleavage is restored when the methyl group is removed by AGT (25). To assay duplex DNA repair, oligonucleotides 4 and 5 containing the NarI sequence (Table 1) were annealed and transferred into reaction buffer (20 mM Tris-acetate (pH 7.9 at 25°C), 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol) by dialysis. Alkyltransferase reactions were carried out at 20 ± 0.1°C, with varying amounts of AGT for varying times (details given in figure captions). Reactions were stopped by addition of sodium dodecyl sulphate (SDS) to a final concentration 0.2% w/v, followed by two extractions with water-saturated phenol and three extractions with water-saturated diethylether. Samples were incubated at room temperature under vacuum to evaporate ether before digestion with a 10-fold unit excess of NarI endonuclease (3 h at 37°C). Samples were resolved by native electrophoresis on 20% polyacrylamide gels (34); electrophoretic distributions were recorded and quantified using a phosphorimager.
To assay ssDNA repair, oligo 4 was transferred into reaction buffer and incubated with AGT (concentrations and times given in figure captions). Reactions were stopped with SDS and AGT was extracted with phenol and ether as described above. Following evaporation of the ether, a 2-fold molar excess of oligo 5 was added and samples were heated to 80°C for 1 min and allowed to slowly cool to room temperature over 2 h. Samples were then digested with a 10-unit excess of NarI, and cleavage products were resolved by electrophoresis, as described above.
RESULTS
Single-step binding to unmodified duplex DNAs and two-step binding to 6mG-containing duplexes
AGT binds cooperatively to short double-stranded DNAs (dsDNAs) that contain no modified bases (18,23). This binding is characterized by a single-step transition from free DNA to a saturated complex, without significant accumulation of stoichiometric intermediates. Examples of this pattern for wild-type and alkyltransferase-inactive C145S mutant proteins binding to a 26 bp duplex (formed with unmodified oligos 1 and 2) are shown in the EMSAs of Figure 1A and B. The cooperative binding pattern is robust, appearing with single-stranded and duplex DNAs of varying sequence, base composition and length, over a range of solution conditions, and whether the protein is methylated at its active site (Cys145), or not (18–20). In contrast, when the 26mer duplex contained a single 6mG residue, titration with the alkyl-transfer-inactive C145S mutant protein gave an initial complex with greater gel-mobility than the saturated complex (Figure 1C). This initial complex is converted to one with the same gel-mobility as the saturated form, on further addition of AGT. A similar two-step binding pattern was also obtained by titrating the 6mG-duplex 26mer with the alkyltransfer-active P140K mutant (data not shown) and by titration of a 24 bp 6mG-duplex (containing oligos 4 and 5) with the C145A mutant protein (Figure 1D). Parallel binding experiments with wild-type AGT could not be performed. Under our conditions, intervals required for alkyl-transfer and binding equilibration were similar, so equilibrium could not be attained without alkyl-transfer. Since most experiments were carried out under conditions of AGT excess, the dominant species in such equilibrium mixtures were alkyl-AGT and lesion-free DNA.
The titrations of 6mG-26mer dsDNA were carried out in 10 mM Tris (pH 7.6 at 20°C), 1 mM EDTA, 50 mM sodium chloride, 5% v/v glycerol buffer, while those with the 6mG-duplex 24mer were carried out in 40 mM Tris-acetate (pH 7.9 at 25°C), 100 mM potassium acetate, 20 mM magnesium acetate, 2 mM dithiothreitol, 5% v/v glycerol. Together these results argue that the two-step pattern is not an artifact of enzyme preparation, identity of active-site mutation, DNA sequence or buffer conditions. Our current interpretation is that the first binding step is a 6mG-specific interaction of AGT. This interaction is characterized further below.
Stoichiometries of 6mG-dependent and non-specific AGT complexes differ
The electrophoretic mobilities of the first complexes formed with 6mG-containing dsDNAs are greater than those of the saturated complexes. This suggests that they might contain fewer molecules of protein than are present in the saturated complexes (31,35). To test this, we measured the stoichiometric differences between free dsDNA and the first binding state and between the first and second binding states. Species concentrations were calculated from EMSA band intensities and the dependence of ln ([PnD]/[D]) on ln [P] was determined using Equation (1), as described in the ‘Materials and Methods’ section. Graphs of these data are shown in Figure 2 and stoichiometries summarized in Table 2. Measured in this way, the single-step binding of wild-type and C145S AGTs to unmodified 26 bp dsDNA had stoichiometries of ∼6 in agreement with previous results (20,23). In contrast, the first binding steps of P140K and C145S AGTs with 6mG-26mer and 6mG-24mer dsDNAs had stoichiometries of ∼2. This result was unexpected because the repair of a single 6mG lesion in DNA consumes one active AGT molecule (4,36) and because the available crystal structures representing AGT bound to DNA lesions feature single protein molecules surrounded by free dsDNA (26,27). However, the small experimental uncertainties (<15% of the central value) and the facts that measurements were made with different protein preparations, different protein sequences, different DNA sequences and different buffer conditions, together argue that this stoichiometry is a bona fide property of AGT interaction with an isolated 6mG-containing site.
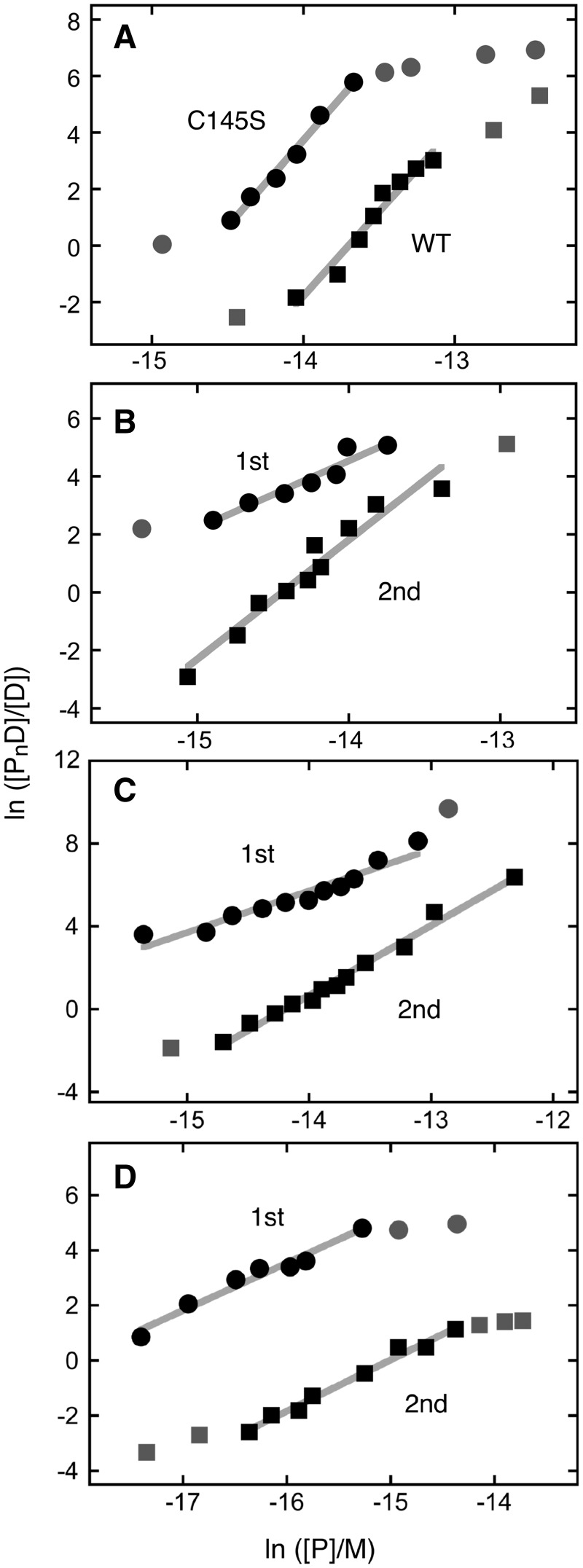
Figure 2.
Graphs of the dependence of log[PnD]/[D] on log[P] for AGT-complexes with duplex DNAs. Data are from the experiments shown in Figure 1 and others that provide additional [AGT] values. The lines represent least-squares fits to the data ensembles for the ranges about the midpoint of each reaction, with [AGT]free calculated as described in Experimental Procedures. Symbols: the points used in the fit are indicated by the black symbols; other points in the data sets are indicated by grey symbols. (A) Data from titrations of unmodified double stranded 26 bp DNA (oligos 1 + 3) with C145S-AGT (filled circle; displaced upward 3 units for clarity) and wild-type AGT (filled square). The slopes are 5.97 ± 0.49 and 6.09 ± 0.32, respectively. (B) Data from titration of 6mG-containing double stranded 26 bp DNA (oligos 2 + 3) with alkyltransfer-active P140K-AGT. First binding step (filled circle; points displaced upward 4 units for clarity); second binding step (filled square). The slopes are 2.33 ± 0.29 and 4.11 ± 0.34, respectively. (C) Data from titration of 6mG-containing double stranded 26 bp DNA (oligos 2 + 3) with C145S-AGT. First binding step (filled circle; points displaced upward 5 units for clarity); second binding step (filled square). The slopes are 2.01 ± 0.19 and 3.37 ± 0.12, respectively. (D) Data from titration of 6mG-containing double stranded 24 bp DNA (oligos 4 + 5) with C145S-AGT. First binding step (filled circle; points displaced upward 3 units for clarity); second binding step (filled square). The slopes are 1.72 ± 0.14 and 1.86 ± 0.09, respectively.
Table 2.
Stoichiometries and equilibrium constants for binding to normal and 6mG-containing DNAs
| DNA | Protein | Stoichiometry (n) |
Association constants |
|||
|---|---|---|---|---|---|---|
| 1st step | 2nd step | K1 | K2 | K1/K2 | ||
| Unmodified ds-26mer | Wild-type | 5.9 ± 0.5 | – | 9.1 ± 4.2 × 105 M−1 | – | – |
| Unmodified ds-26mer | C145S | 6.1 ± 0.3 | – | 2.2 ± 1.1 × 106 M−1 | – | – |
| 6mG ds-26mer | P140K | 2.3 ± 0.3 | 4.1 ± 0.3 | 1.7 ± 0.3 × 107 M−1 | 3.4 ± 0.8 × 106 M−1 | 5.0 ± 0.3 |
| 6mG ds-26mer | C145S | 2.0 ± 0.2 | 3.4 ± 0.1 | 6.2 ± 2.4 × 106 M−1 | 1.9 ± 0.4 × 106 M−1 | 3.3 ± 0.4 |
| 6mG ds-24mer | C145S | 1.8 ± 0.1 | 2.1 ± 0.1 | 2.5 ± 1.2 × 107 M−1 | 4.4 ± 2.8 × 106 M−1 | 5.7 ± 0.8 |
| Unmodified ss-26mer | C145S | 5.7 ± 0.4 | – | 2.2 ± 0.3 × 106 M−1 | – | – |
| 6mG ss-26mer | C145S | 6.3 ± 0.5 | – | 4.5 ± 0.5 × 106 M−1 | – | – |
For C145S- and P140K-mutant AGTs, the second binding steps with 6mG-26mer dsDNA had stoichiometries of 3–4 (Table 2) and the sum of first and second steps gave an overall stoichiometries of 5–6 in reasonable agreement with the saturating stoichiometry of 6 found for unmodified DNA. For C145S-AGT, the second binding step with the 6mG-24mer duplex had a stoichiometry of 2 and thus a saturating stoichiometry (∼4). This is less than the limiting value of 6 expected for cooperative non-specific binding to a 24-mer duplex that has no 6mG residues (20). In a previous analysis we found that AGT gave no detectable binding at partial sites containing 3 or fewer bp (22). It is possible that the initial binding to the 6mG-duplex 24mer gives an AGT distribution that leaves a smaller number of full-length sites for the second binding step than would be obtained if binding were unconstrained. The transition from the 2:1 6mG-specific complex to saturated assemblies containing at least 4 (and in some cases 6) proteins appears to take place in a single concerted step without formation of stoichiometric intermediates. This closely resembles the cooperative non-specific binding mode seen with unmodified DNAs and indicates that such cooperative binding operates in the presence of the lesion-specific AGT complex as well as in its absence.
Small differences in affinities for unmodified and 6mG-containing duplex DNAs
In addition to giving information about stoichiometry, the data in Figure 2 allow estimation of the equilibrium constants of the corresponding interactions. Equation (1) shows that at the midpoint of the binding transition, ln[PnD]/[D] = 0 and –n ln[P] = ln K. Here K is the association constant for the overall reaction, with stoichiometry n. A monomer-equivalent association constant, given by K1/n, allows easy comparison of the AGT-affinities of reactions with different stoichiometries. These values are given in Table 2. The association constants for wild-type and C145S-AGTs with unmodified 26mer duplex, ∼0.9 × 106 M−1 for wild-type enzyme and ∼2 × 106 M−1 for C145S-AGT, agree well with values found for short, unmodified dsDNAs, under similar conditions (20), and are 2–4 fold larger than affinities measured for linear and relaxed-circular pUC19 DNAs (2686 bp), under similar buffer conditions (23). Thus, they are fair representations of the lesion-non-specific interaction of AGT with duplex DNA.
The presence of a 6mG residue had a surprisingly small effect on binding affinities. In the first binding step, the C145S-AGT bound the 6mG-26mer duplex 3-fold more tightly than it did the unmodified 26mer DNA. Slightly larger differences (up to ∼10-fold) were found with C145S-AGT binding to the 6mG-24mer and P140K-AGT binding 6mG-26mer duplexes. Although these larger differences may reflect changes in DNA and protein sequences, they still barely reached a specificity ratio K6mG/Kunmodified of 10. Such weak specificity has important implications for the mechanism of lesion search, as will be discussed below. Analysis of the second binding step gave affinities (K = 2–4 × 106 M−1) that were within error the same as those for binding to unmodified dsDNAs. Thus, in spite of the presence of specifically bound AGT proteins, this second step appears to reflect an ensemble of non-specific interactions like that formed in the absence of 6mG residues.
Binding intermediates do not accumulate when DNA is single-stranded
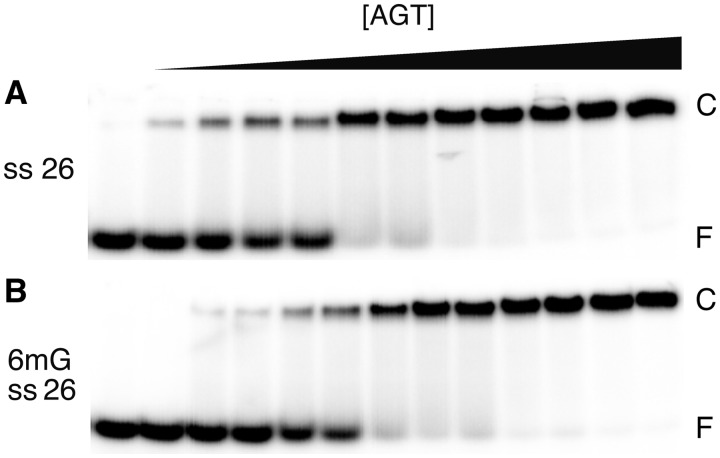
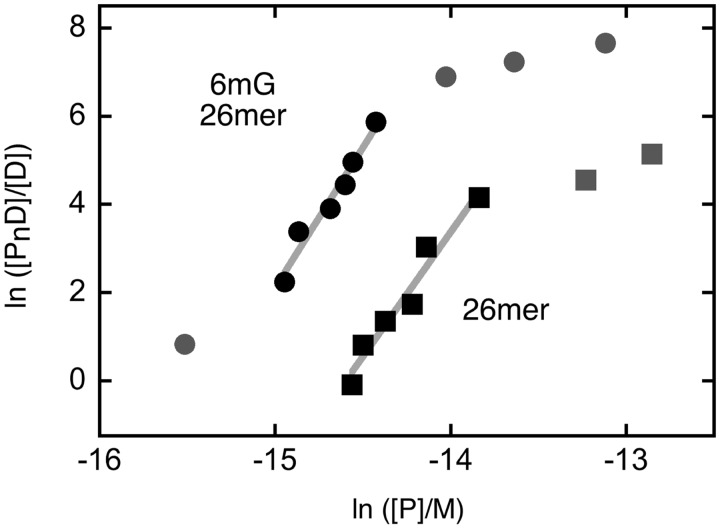
AGT binds both single-stranded and duplex forms of DNA (19,20), with a very modest (∼1.5-fold) preference for the duplex, under standard conditions (23). Such closely matched affinities suggest that AGT might function where DNA is single-stranded in vivo. It was of interest then, to determine whether the enzyme binds specifically to 6mG sites in ssDNAs. Shown in Figure 3 are titrations of unmodified and 6mG-containing single-stranded 26mers (oligos 1 and 2, respectively) with C145S-AGT. Both ssDNAs undergo single-step binding transitions to form saturated complexes with gel-mobilities similar to those formed with unmodified duplex 26mer. Graphs of the dependence of ln([PnD]/[D]) on ln[P] for these interactions are shown in Figure 4. Stoichiometries, estimated from the slopes near the binding midpoints, were 5.7 ± 0.4 and 6.3 ± 0.5 for unmodified and 6mG-ssDNAs, respectively. These values are within error the same as that measured for wild-type AGT with unmodified 26mer ssDNA (25). The monomer-equivalent association constants for unmodified single-stranded and duplex 26mers were indistinguishable in these assays (K ∼ 2.2 × 106 M−1), while that for the single-stranded 6mG-26mer was ∼2-fold larger (K ∼ 4.5 × 106 M−1; Table 2). This small stability difference (amounting to only ∼0.4 kcal/mol at 20°C) may account for the absence of a detectable 6mG-specific intermediate; our working hypothesis is that some interactions that generate a specific complex with a duplex substrate are missing or contribute substantially less to the ΔG° of binding, when the substrate is single-stranded.
Figure 3.
Single-step binding to ssDNAs. EMSAs performed at 20 ± 1°C. (A) Titration of unmodified single stranded 26 nt DNA (oligo 1, 0.17 µM) with C145S-AGT (0–2.2 µM) to form the low-mobility cooperative complex. The equilibration buffer was 10 mM Tris (pH 8.3 at 20°C), 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 5% glycerol, 0.1 mg/ml BSA. Samples resolved in 20% native polyacrylamide gels cast and run as described. (B) Titration of 6mG containing 26 nt DNA (oligo 2; 0.19 µM) with C145S-AGT (0–3.4 µM). The equilibration buffer and electrophoresis conditions were as in (A).
Figure 4.
Graphs of the dependence of log[PnD]/[D] on log[P] for C145S-AGT-complexes with ssDNAs. Data are from the experiments shown in Figure 3. The lines represent least-squares fits to the data ensemble for the range about the midpoint of each reaction, with [AGT]free calculated as described in Experimental Procedures. Symbols: the points used in the fit are indicated by the black symbols; other points in the data sets are indicated by grey symbols. Titration of 6mG-containing 26mer (oligo 2, filled circle, displaced upward 4 units for clarity) and unmodified 26mer (oligo 1, filled square). The slopes are 6.35 ± 0.64 and 5.64 ± 0.62, respectively.
Quantitative DNA repair
If specific complex formation is the rate-limiting step in the repair reactions of AGT, the data shown above predict that repair rates will have a second-order dependence on [AGT] when the substrate is duplex, and possibly a higher-order dependence when the substrate is single-stranded. As a first step in a kinetic analysis, we determined the repair activities of our AGT preparations when reaction time was not limiting. Our assay takes advantage of the fact that NarI endonuclease is inactive when substrate DNA contains an 6mG at position 2 of its target sequence (33) (Table 1), but is restored by DNA alkyltransferase activity provided by AGT. In addition, the assay relies on the fact that 6mG repair is stoichiometric, i.e. that one active AGT molecule is consumed for each 6mG converted to guanine (3,4). Shown in Figure 5 are continuous variation assays (37,38) in which AGT and dsDNA solutions of equimolar concentration were combined in a range of volume ratios, giving samples in which the total macromolecular concentration was constant but the AGT:DNA molar ratios varied. The greatest amount of NarI cleavage occurs where AGT and DNA combine in the optimal ratio for repair. Linear fits to flanking data subsets intersect at optima predicted by the entire dataset (Figure 6). For wild-type AGT, the optimal combining ratio was reached when the AGT mole-fraction was 0.503 (i.e. [AGT]/[DNA] = 1.01, corresponding to an active fraction of 0.99); for P140K-AGT, the optimal mole fraction was 0.51, corresponding to [AGT]/[DNA] = 1.04 and an active fraction of 0.96. When wild-type AGT was mixed in 1:1 molar ratio with alkyl-transfer-inactive C145S mutant protein, the optimal mole fraction was 0.65, corresponding to [AGT]/[DNA] = 1.86 and an active fraction of 0.53. If one molecule of AGT is inactivated for each DNA molecule repaired (as previously shown (39)), these results indicate that the DNA is fully competent for repair and subsequent NarI cleavage, that the active fractions of wild-type and P140K AGTs are near unity, and that the C145S mutant is devoid of alkyltransferase activity.
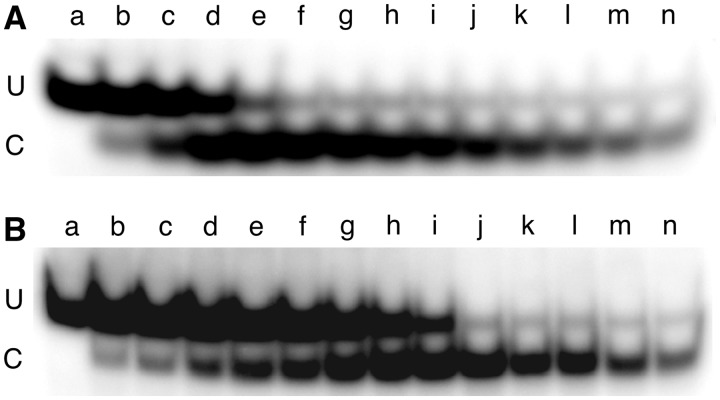
Figure 5.
Continuous-variation NarI DNA repair assays. Images of 20% polyacrylamide gels resolving NarI cleaved fragment (C) from uncleaved DNA (U). The DNA used was the double-stranded 6mG-containing 24mer (oligos 5 + 6) and oligo 6 carried a 5′-32P label. The reaction buffer was 20 mM Tris acetate (pH 7.9), 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT. (A) DNA and wild-type AGT solutions (each 5.8 µM) were mixed in volume ratios giving mole-fractions of AGT ranging from 0.1 to 0.9. For samples a–n, these were 0, 0.1, 0.15, 0.2, 0.36, 0.42, 0.5, 0.55, 0.6, 0.66, 0.72, 0.82, 0.86 and 0.92, respectively. Incubation was for 3 h at 20 ± 0.1°C. Repair reactions were quenched by addition of SDS to a final concentration of 0.2%, and samples were extracted with phenol, then ether, as described, then digested with 10U of NarI for 2 h at 37°C, prior to electrophoresis. (B) Effect of mixing wild-type and C145S-AGTs in 1:1 molar ratio. Assays carried out as described above. For samples a–n, the mole fractions of AGT (wild-type plus C145A) were 0, 0.1, 0.15, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.8, 0.85 and 0.9, respectively.
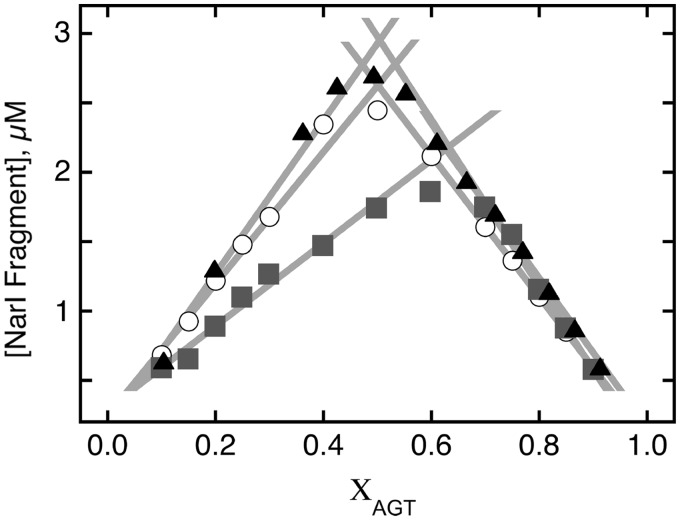
Figure 6.
Continuous variation analyses of dsDNA-repair reactions. The data are from the experiments shown in Figure 5, plus an additional one carried out with P140K-AGT. Symbols: wild-type AGT (filled triangle); P140K mutant (open circle); wild-type- and C145S-AGTs in 1:1 ratio (filled square). The solid lines are linear fits to rising and falling subsets of the data. Optimal combining ratios are given by their intersection. For wild-type- and P140K-AGTs and the wild-type plus C145S-AGT mixture, these were 0.503, 0.51 and 0.65, respectively.
Kinetic orders of reaction and equilibrium binding stoichiometries differ
We used a variant of the NarI assay to measure the rates of alkyl-transferase reactions. In these measurements, dsDNA, labeled with 32P at one 5′-end, was combined with AGT to start the reaction, and aliquots were withdrawn as a function of time and quenched with 0.2% SDS. Samples were processed with solvent extraction and NarI digestion as before. Shown in Figure 7 is a representative gel image showing the change in NarI cleavage as a function of the AGT-DNA incubation time. Band intensities, converted to mole fractions of DNA repaired, were plotted as functions of time for representative AGT concentrations (Figure 8A); the smooth curves correspond to fits of Equation (2) to the data. This expression describes the time course of product formation, P(t), expected for a one-step reaction, in which F is the mole fraction of repair-competent DNA, kobs is the apparent rate constant and t is time (s) (40).
| (2) |
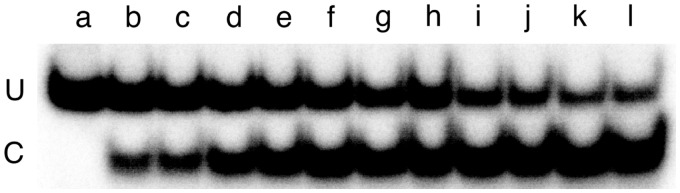
Figure 7.
Time-dependence of dsDNA repair detected by NarI sensitivity. The 6mG-containing 24mer duplex (oligos 4 and 5) 0.037 µM, was dissolved in 40 mM Tris acetate (pH 7.9 at 20°C), 100 mM potassium phosphate, 20 mM magnesium acetate, 2 mM DTT, containing 0.5 mg/ml BSA. To start the reaction, AGT was added to a final concentration of 0.11 µM. Aliquots were withdrawn and quenched in 0.2% SDS after 15 s, 30 s, 45 s, 60 s, 120 s, 180 s, 240 s, 360 s, 900 s, 2400 s and 3600 s (samples b–l, respectively); unrepaired DNA is shown in sample a. Samples were deproteinized by phenol extraction followed by ether extractions as described; they were then digested with NarI (15 U) for 2 h at 37°C and resolved by electrophoresis on a 20% gel.
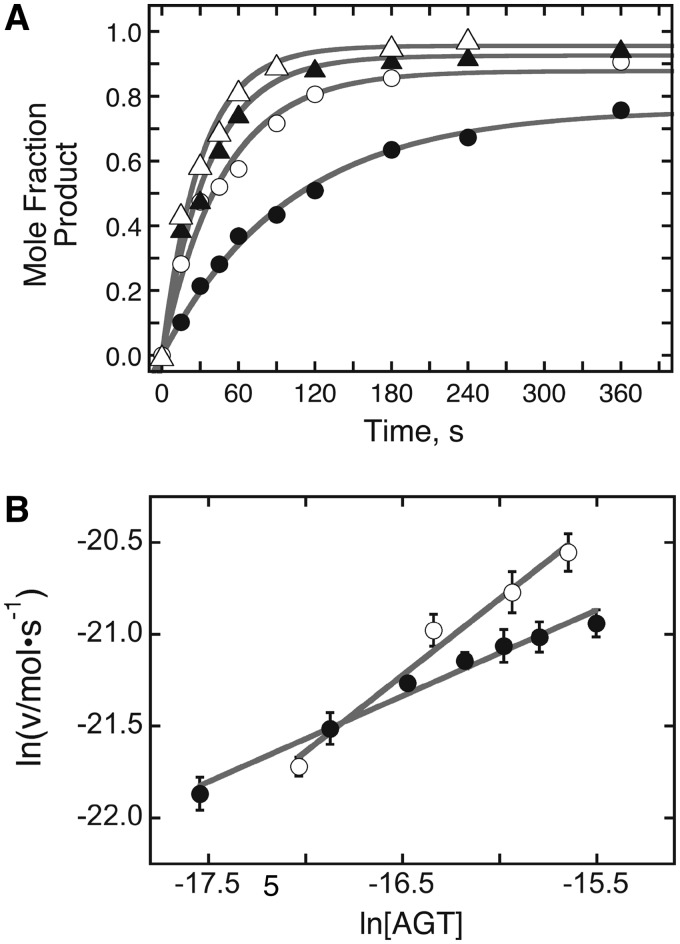
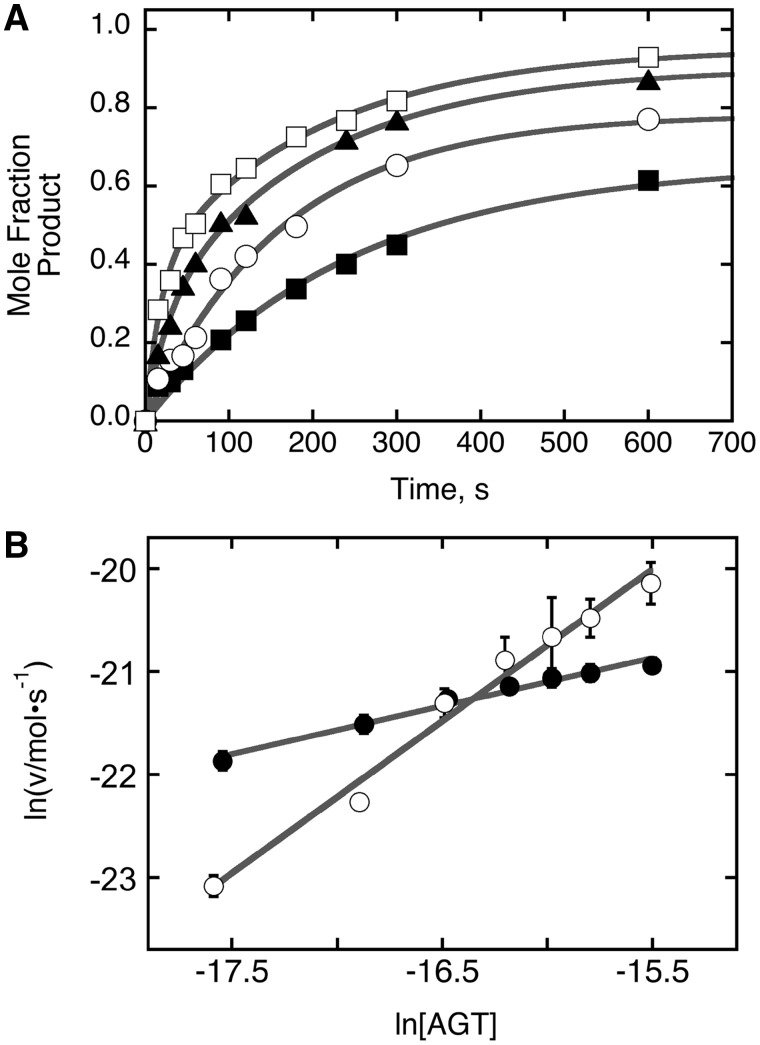
Figure 8.
Analysis of repair reactions carried out on duplex DNA at several AGT concentrations. (A) Time profiles for repair reactions. Data were obtained as shown in Figure 7, with [6mG DNA] = 0.037 µM and [AGT] = 0.037 µM (filled circle); [AGT] = 0.074 µM (open square); [AGT] = 0.11 µM (filled triangle); [AGT] = 0.148 µM (open triangle). The smooth curves are fits of Equation (2) to the data. (B) Graphs of the dependence of ln v as functions of ln [AGT] for repair reactions run with [DNA] = 0.037 µM (open circle) and [DNA] = 0.023 µM (filled circle). Here v is the initial reaction rate; error bars correspond to the 95% confidence intervals on v determined from fits like those shown in Panel A. The solid lines are linear fits to the data as described for Equation (3); the slopes provide estimates of the order of the reaction in AGT. For [DNA] = 0.037 µM, α = 0.83 ± 0.08; for [DNA] = 0.023 µM α = 0.47 ± 0.03.
These fits show that a single-step model accounts for the time course of each reaction, but do not establish the number of protein molecules contributing to its rate-limiting step. However, agreement between fits and data show that this model accounts well for initial reaction rates, allowing us to determine reaction orders using a version of the method of initial rates (41). For a reaction step in which αAGT molecules participate with one DNA, the rate equation v = k[AGT]α[DNA] can be linearized as:
| (3) |
where v is the initial rate of the reaction and k′ = k [DNA]. A graph of ln v as a function of ln [AGT] will have a slope equal to α, the order of the reaction in AGT. Shown in Figure 8B is this graph for dsDNA concentrations of 23 nM and 37 nM; linear fits to these data returned reaction orders of 0.47 ± 0.03 and 0.83 ± 0.08, respectively. As the enzyme preparation used in these measurements was fully active (Figure 6), this difference in reaction order is not easily attributable to a difference in time-independent alkyltransferase activity. These results are striking in several ways. First, they indicate a change in the rate-limiting reaction mechanism with change in [DNA]. Second, both values are significantly less than 2 (the order predicted on the basis of equilibrium binding stoichiometries) indeed, they are significantly less than 1, the smallest value consistent with direct binding of 6mG sites by a free protein molecule. We interpret reaction orders between 0 and 1 as evidence for a mixed mechanism in which some repair reactions are independent of free AGT concentration (e.g. through transfer of protein(s) to the 6mG site from one or more sites elsewhere on the DNA) while some reactions depend on the binding of AGT from free solution. The fact that the order in AGT is smaller at lower DNA concentration is consistent with this view, since bimolecular encounters with the 6mG site will be slower at low [DNA], increasing the time available for AGT transfer from distal sites. Such transfer mechanisms are a feature of many sequence or structure-specific protein–DNA interactions [c.f. (42–44)].
Alkyl-transfer-inactive C145S-AGT competes with wild-type enzyme
The C145S-AGT binds unmodified dsDNA cooperatively, forming complexes with gel mobilities, affinities and stoichiometries that are indistinguishable from those of the wild-type enzyme (Figures 1 and 2). In addition, it forms a specific complex with 6mG-containing dsDNA (Figure 1), but is, itself, fully alkyl-transfer-inactive. Finally, apart from the effect of diluting active enzyme, it has no discernable effect on ‘time-independent’ alkyl-transfer activity (Figures 5 and 6). These results suggest that its presence in a mixture might affect the alkyl-transfer kinetics of the wild-type enzyme. Since the C145-methyl enzyme also binds DNA cooperatively and with nearly wild-type affinity (18), mixtures of C145S-mutant and wild-type AGTs are a useful model for the mixtures of alkyltransferase-active and -inactive AGTs in cells exposed to chemotherapeutic methylating agents such as temozolomide (45).
We envisioned two possibilities. Since C145S-AGT is unimpaired in its ability to form cooperative complexes (Figure 1 and Melikishvili, unpublished result), addition of this protein to a solution containing wild-type AGT and DNA might increase the concentration of AGT-DNA complexes by mass action, and thus increase the repair rate of the wild-type protein in the mixture. On the other hand, mixing C145S and wild-type AGTs will increase the probability that the 6mG site will be occupied by an inactive monomer, and if exchange of proteins between distal and 6mG sites is rate limiting, this effect could slow repair. These possibilities were tested by measuring the initial repair rates of mixtures in which the concentrations of wild-type (active) enzyme were held constant and those of the C145S (inactive) enzyme were varied. As shown in Figure 9, even though the ‘total’, ‘time-independent’ repair activity of the solutions remained constant, the relative rate (v/v0) fell as the [C145S]/[wild-type] ratio increased. This reflects a competition between wild-type and mutant enzymes. However, the proportionality constant (equal to the slope, −0.17 ± 0.01) indicates that the change in activity was not equal to the ratio of the two proteins, as would be the case if initial 6mG-binding were irreversible on the time scale of the assay. This result is most simply accounted for by mechanisms in which C145S and wild-type enzymes exchange at the 6mG site during the assay interval. A similar result has been reported for a mixture of AGT and an analogous protein, alkyltransferase-like (ATL), that lacks alkyl-transfer activity (46). Our current view is that, in mixed solutions of active and inactive AGTs, repair rates are influenced by a combination of positive effects (such as mass action-driven binding and protein exchange) and negative effects (such as competition for 6mG sites).
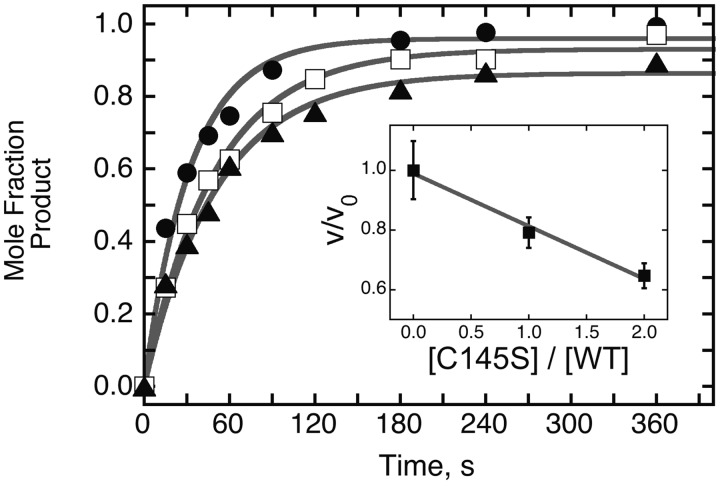
Figure 9.
Effect of alkyl-transfer-inactive AGT on dsDNA repair. All samples contained 32P-labeled 6mG-duplex 24mer DNA (0.037 µM) and wild-type AGT (0.111 µM); samples contained, in addition, C145S-AGT at 0 µM (filled circle), 0.111 µM (open square) or 0.222 µM (filled triangle). The time course of repair was measured as described for Figures 7 and 8. Inset: graph of relative velocity in the presence of C145S-AGT (v) with respect to velocity in its absence (v0) as a function of the molar ratio [C145S]/[WT] in each assay. The solid line is a linear fit to the data.
For a single-stranded substrate, the kinetic order of reaction differs from binding stoichiometry
AGT repairs 6mG-lesions in single-stranded as well as duplex substrates (47). On the other hand, the equilibrium binding data indicate that the stabilities of 6mG-specific complexes must be different on single- and double-stranded substrates (compare Figures 1C and 3B). To learn how these differences in DNA secondary structure and complex stability affect repair kinetics, we used a variant of the NarI assay in which single stranded DNA (oligo 4) was incubated with AGT for various times. Reactions were stopped and samples deproteinized as described for dsDNAs (see above) and the single-stranded products were annealed with an excess of complementary strand (oligo 5), before digestion with NarI and analysis by gel electrophoresis (Figure 10). Mole fractions of DNA repaired were graphed as functions of time for representative AGT concentrations (Figure 11A), together with fits of Equation (2) to the data. These fits show that the early stages of each repair reaction are kinetically homogeneous (i.e. they can be described by a single rate constant) and they give reaction rates that are similar to those obtained with duplex substrates (see below). The reaction rates depended on [AGT], allowing us to determine the reaction-order in AGT using the initial rates method described above. Shown in Figure 11B is a graph of ln v against ln [AGT] for single-stranded [DNA] = 23 nM; included for comparison are the data for the corresponding duplex, replotted from Figure 8. As described for Equation (3), the slopes of these graphs give the kinetic orders of the reactions in [AGT]. A linear fit to the single-stranded data returned a reaction order α = 1.48 ± 0.08. For this single-stranded substrate, the order of reaction in AGT is greater than that found for duplex DNA (αduplex = 0.47 ± 0.03), consistent with the notion that the pathways contributing to the repair of these substrates are different. Time-independent alkyl-transferase assays like those shown in Figure 6, established that the preparations compared here were of equal activity (results not shown). Thus, the difference in reaction order does not simply reflect a difference in enzymatic activity. As above, we interpret the non-integral reaction orders as evidence for mixed mechanisms. For repair of ssDNA, the result suggests the contributions of at least two repair pathways; one with a rate-limiting step that requires two or more AGT monomers, and a second pathway, involving one or fewer AGT molecules. Finally, the reaction order in AGT is much less than the equilibrium binding stoichiometry (∼6) measured by EMSA (Figures 3 and 4). This is not surprising in view of the fact that a single AGT molecule acts as alkyl-acceptor in each repair reaction (Figure 6), and it argues against models in which the binding of 6 AGT monomers, attaining binding saturation on this DNA, is the ‘rate-limiting step’ in repair.
Figure 10.
Time course of repair for a ssDNA. The single-stranded 6mG-24mer (oligo 4) was 5′-labeled with 32P. Repair was carried out as described for Figure 7 in a solution containing 0.023 µM DNA and 0.092 µM AGT. Samples b–k correspond to reactions stopped by addition of 0.2% SDS after 15 s, 30 s, 45 s, 60 s, 90 s, 120 s, 180 s, 240 s, 360 s and 600 s, respectively. Samples were deproteinized as described and the substrate DNA was annealed with 1.1 equivalents of complementary strand (oligo 5), prior to digestion with NarI. Digestion products were resolved on a 20% polyacrylamide gel. Sample a contains ssDNA that has not been subjected to repair or annealing to its complement, or digestion with NarI. Band designations: U, uncut; C, cut with NarI.
Figure 11.
Analysis of repair reactions carried out on ssDNA at several AGT concentrations. (A) Time profiles for repair reactions. Data were obtained as shown in Figure 10, with [6mG DNA] = 0.023 µM and [AGT] = 0.023 µM (filled square); [AGT] = 0.046 µM (open circle); [AGT] = 0.69 µM (filled triangle); [AGT] = 0.092 µM (open square). The smooth curves are fits of Equation (2) to the data. (B) Graphs of the dependence of ln v as functions of ln [AGT] for repair reactions run with ssDNA (0.023 µM; open circle); corresponding data for dsDNA, from Figure 8, is shown for comparison (filled circle). Here v is the initial reaction rate; error bars correspond to the 95% confidence intervals on v determined from fits like those shown in Panel A. The solid lines are linear fits to the data as described for Equation (3); the slopes provide estimates of the order of each reaction in AGT. For single stranded DNA, α = 1.48 ± 0.08; for double stranded DNA, α = 0.47 ± 0.03.
DISCUSSION
Our current understanding of the mechanisms by which AGT performs its surveillance and repair functions offers intriguing contrasts. For example, AGT binds DNA to form cooperative clusters (20) but the alkyl-transfer reaction is stoichiometric (3,12). In addition, it performs repair efficiently enough to protect cells against guanine alkylation, but its affinity for 6mG-containing DNA is only marginally greater than that for the unmodified homologous sequence ((18); Table 2). To learn how these disparate properties combine in the surveillance and repair mechanisms of AGT, we have characterized its activities on a series of short single-and double-stranded substrates. We were initially surprised to be able to detect discrete 6mG-specific complexes using EMSA, because previous attempts with wild-type, C145A and C145-alkyl forms of AGT gave only single-step cooperative binding that was qualitatively indistinguishable from the pattern found with unmodified DNA (18). However, better stabilization of complexes during electrophoresis through use of higher percentage polyacrylamide gels (15% and 20% as opposed to 10% (48)) allowed us to resolve 6mG-dependent complexes for the first time.
A survey of proteins and binding conditions revealed that mixtures containing the alkyl-transfer-active P140K mutant of AGT or the alkyl-transfer-inactive C145S or C145A mutants formed detectable 6mG-specific complexes with duplex DNA (Figure 1), but wild-type AGT did not. The result with wild-type enzyme is understandable, because repair reactions with similar concentrations of wild-type AGT (Figure 8) were complete within 6 min (just 10% of the incubation time used in our equilibrium experiments). Since the products of repair are C145-methyl AGT and unmodified DNA, and since [AGT]>>[DNA], the dominant pattern at binding equilibrium would be that of unmodified AGT binding unmodified DNA. We believe that we were able to detect specific complexes with the alkyl-transfer active P140K mutant, because its repair reaction is slower than that of the wild-type enzyme (results not shown). Stoichiometry analyses returned values of ∼2 for 6mG-specific complexes formed with three different protein preparations, in two dsDNA sequence contexts, and under two different buffer conditions. The consistency of these results suggests that this stoichiometry is a bona-fide property of 6mG-specific complexes.
The 2:1 stoichiometry of 6mG-specific complexes is striking, because it differs from expectation based on the 1:1 crystalline complexes currently available (26,27) and because AGT repair reactions have obligate 1 alkyl-transfer/protein stoichiometries (4). However, individual AGT molecules occupy 8 bp centered on the minor groove face of the DNA (26,27), so a 2:1 stoichiometry may account for DNase I footprints spanning ∼18 nt on the 6mG-strand and ∼14 nt on its complement (24). The 2:1 stoichiometry is also striking because the formation of such a complex without the accumulation of detectable concentrations of 1:1 intermediates indicates that specific binding is cooperative. While the phenotypes of cells expressing mutant AGT proteins with reduced binding cooperativity suggested a role for cooperative binding in DNA repair (21), the stoichiometry data presented here provides the first direct evidence that lesion binding is, itself, cooperative. Although this result shows that cooperative binding is a feature of both specific and non-specific interactions, the fact that the specific stoichiometry is limited to 2:1 while non-specific complexes are often larger (20), suggests that the 6mG-specific complex may be structurally distinct from those formed on lesion-free DNA. An additional striking feature of the 6mG complexes is their low stability relative to non-specific interactions. The resolved complexes have equilibrium constants a factor of 3–6 greater than those of the background, non-specific interactions available on the DNA substrates (Table 2). This difference corresponds to ≤1 kcal/mol at 20°C. If this outcome is generalizable to cellular conditions, the low relative affinity for 6mG sites must greatly limit the role of sequence-specific binding in the lesion-search mechanism.
Single-step, cooperative transitions from free DNA to saturated 6:1 complexes were obtained with ssDNAs, whether the sequence contained a 6mG residue, or not (Figure 3). While this qualitative pattern contrasts with that found for duplex DNAs, the quantitative theme is maintained: binding to 6mG-containing DNA is only marginally (2-fold) stronger than that for the unmodified homologue. Thus, if AGT repairs single-stranded substrates in vivo, it seems unlikely that binding specificity plays an important role in lesion search in that context, either. In addition, AGT’s affinities for single-stranded and duplex DNAs are within error the same (Table 2). This indicates that AGT is not an effective DNA-melting protein and that it does not partition strongly in favor of single-stranded or duplex DNA sites. Thus, neither DNA secondary state benefits disproportionately from AGT activity, or acts as a sink for it.
The kinetics of DNA repair by AGT have been studied several times, using different protein preparations, DNA substrates, buffer compositions, methods for the detection of alkyl-transfer and reaction models (49–54). Unsurprisingly, the reported bimolecular rate constants (krepair) vary widely (1.4 × 105 M−1s−1 ≤ krepair ≤ 1 × 109 M−1s−1). One potential source of variation is difference in reaction mechanism under different reaction conditions or with different substrates. As shown above, single-stranded and duplex 6mG-containing DNAs form complexes that differ in stoichiometry and structure (compare Figure 1, panels C and D with Figure 3). If assembly of the complex at the 6mG site is rate-limiting, the dependence of repair rate on [AGT] will itself depend on the secondary structure of the substrate. The repair kinetics experiments (Figures 7–11) give evidence for different reaction orders in AGT (αdouble stranded = 0.47 ± 0.03; αsingle stranded = 1.48 ± 0.08 at [DNA] = 23 nM) that strongly supports this idea.
A previous report indicated that AGT repaired a duplex DNA significantly more rapidly than a corresponding single stranded form (55). Data shown in Figure 11B are consistent with this conclusion for low AGT concentrations. However, these data also show differences in kinetic order in AGT for single-stranded and duplex substrates that result in more rapid repair for single-stranded substrates than for duplex at high [AGT]. Under our conditions the cross-over point occurs at [AGT] ∼10−7 M, a value that is close to its Kd value for duplex 6mG sites (Kd ∼ 2.2 × 10−7 M, corresponding to K ∼ 4.5 × 106 M−1 (Table 2)).
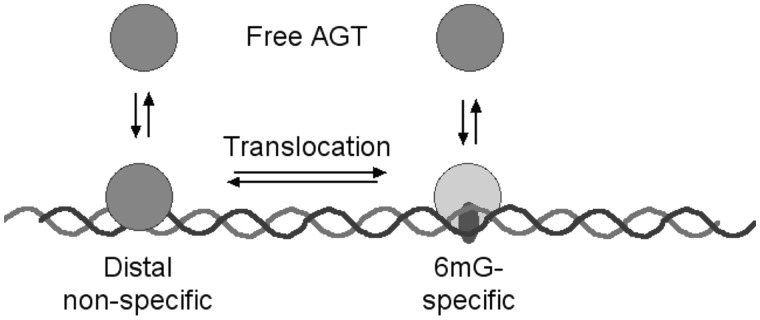
Preliminary experiments indicated that reaction rates increased with increasing [dsDNA] (results not shown). This was expected for a process in which the initial bimolecular collision is rate limiting. However, a more detailed analysis (Figure 8B) showed that changing the [dsDNA] also changed the order of the reaction in AGT (α = 0.47 ± 0.03 at [dsDNA] = 23 nM, α = 0.83 ± 0.08 at [dsDNA] = 37 nM). This effect is incompatible with models in which the rate-limiting step is a simple bimolecular reaction. In addition, the non-integral values of α are most-simply explained by a mixed mechanism in which two or more reactions with different [AGT] dependence contribute to the rate-limiting step. Our working model is that formation of the 6mG-specific complex is rate limiting and that two pathways contribute to the observed rate (Figure 12). In one, AGT binds first to a distal site. Translocation to the 6mG site is rate-limiting for that branch of the path. Since the translocation step is unimolecular, that branch is of zero-order in AGT. In the second, parallel branch, AGT binds directly to the 6mG site, and the AGT-order of this branch is equal to the stoichiometry. The AGT-order observed for the entire pathway will be a weighted average of the orders of the two branches. This parallel-path model accounts for the increase in the AGT reaction order with increasing [DNA] (Figure 8B), because increasing [DNA] will increase the rate of the direct association (bimolecular) branch of the reaction path and thus its proportional contribution to the observed order. The model also makes testable predictions. It predicts that increasing the translocation time by increasing the length of distal DNA should favor the direct-association branch of the pathway, increasing the observed order in AGT. It also predicts that barriers to translocation (for instance, other proteins), located near but not in the lesion site, should also increase reaction order. Experiments to test these predictions are currently underway.
Figure 12.
Diagram of a reaction pathway in which two branches lead to formation of the specific 6mG complex. In the left-hand branch, the protein binds first to distal sites and then transfers in a unimolecular step to the 6mG site. In the right-hand branch, the protein binds the 6mG-site directly from solution. For simplicity, a single oval is used to represent AGT, however this is not intended to indicate the stoichiometry of any step.
How do alkyl-transfer-inactive forms of AGT affect the repair process? This question is relevant to chemotherapy, because the inactive C145-methyl-AGT is produced in cells exposed to methylating drugs such as temozolomide, while C145-benzyl-AGT is produced when the enzyme is exposed to the chemotherapy-enhancing drug O6-benzylguanine (56,57). Alkylated forms of AGT are ultimately ubiquitinated and degraded (16), but these processes are not instantaneous, so at least some alkyl-AGT must be present at the sites of DNA repair in vivo. Further, the C145-methyl- and C145-benzyl- forms bind unmodified- and 6mG-containing DNAs with stoichiometries and affinities like those of the wild-type enzyme (18), so competitive effects might be expected. We tested alkyl-transfer-inactive C145S-AGT in repair reactions with wild-type AGT. Under conditions in which the time-independent repair activity was constant, we found that C145S-AGT slowed repair rates, but not in direct proportion to its mole fraction. The simplest model consistent with this result is one in which the life-time of the specific AGT-6mG complex is short when compared to the time intervals in our assays, and cycles of dissociation and binding eventually bring active AGT molecules to 6mG sites that were initially occupied by the C145S-protein. Such protein exchange between DNA sites may be part of the lesion-search mechanism, and it may also reduce the risk that a mutagenic 6mG-residue will go un-repaired because it is obscured by an alkyl-transfer-inactive AGT molecule.
How, then does AGT conduct its lesion search? Even though we are now able to isolate a 6mG-specific complex, its stability relative to non-specific interactions (less than a factor of 10) is not great enough to drive an efficient lesion search in a large genome. It is possible that AGT interacts with a protein or proteins that provide the necessary 6mG-specificity, but none have been identified, to date. Thus, our working model requires no additional specificity-generating components. AGT has been shown to interact in vitro with proteins involved in DNA-replication and repair (58). These include MCM2 (59), PCNA (60) and MSH2 (61). Such interactions may concentrate AGT where access to DNA is provided by chromatin remodeling (21,23,62). In addition, remodeling may provide segments of open DNA suitable for cooperative AGT binding, which should further increase the local AGT concentration (20,21,23). Searching open DNA segments may not require highly specific lesion binding, since open regions make up only a small subset of the genome at any one time. Cooperative interactions have the potential to speed exchange between available sites, making the lesion-search time-efficient. In this process, AGT exchange between non-specific and specific sites (as inferred from the kinetic analysis above) is likely to play a key role. AGT-function near the replication fork may require an ability to bind and repair ssDNA, like that shown here. The movement of open DNA regions with the advance of the replication machinery would make possible a processive search for alkylated sites that spans nearly the entire genome. This model predicts that AGT will colocalize with chromatin-remodeling enzymes in the cell and that AGT-mediated repair may be especially concentrated in chromatin regions undergoing DNA replication. Experiments to test these predictions are underway.
FUNDING
National Institutes of Health (NIH) [GM-070662 to M.G.F.]; National Center for Research Resources [2P20 RR020171]. Funding for open access charge: NIH [GM-070662].
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the NCRR. We thank Drs Geoffrey Margison (Paterson Institute for Cancer Research, Manchester, UK) and Ingrid Tessmer (Rudolf-Virchow Center for Experimental Biomedicine, Würzburg, Germany) for valuable discussions.
REFERENCES
- 1.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyrtopoulos SA, Anderson LM, Chhabra SK, Souliotis VL, Pletsa V, Valavanis C, Georgiadis P. DNA adducts and the mechanism of carcinogenesis and cytotoxicity of methylating agents of environmental and clinical significance. Cancer Detect. Prev. 1997;21:391–405. [PubMed] [Google Scholar]
- 3.Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 2007;6:1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pegg AE. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy and research tools. Chem. Res. Toxicol. 2011;24:618–639. doi: 10.1021/tx200031q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J. Clin. Oncol. 2002;20:2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 6.Margison GP, Santibáñez-Koref MF. O6-Alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. BioEssays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- 7.Kaina B, Margison GP, Christmann M. Targeting O6-methylguanine-DNA methyltransferase with specific inhibitors as a strategy in cancer therapy. Cell Mol. Life Sci. 2010;67:3663–3681. doi: 10.1007/s00018-010-0491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verbeek B, Southgate TD, Gilham DE, Margison GP. O6-Methylguanine-DNA methyltransferase inactivation and chemotherapy. Br. Med. Bull. 2008;85:17–33. doi: 10.1093/bmb/ldm036. [DOI] [PubMed] [Google Scholar]
- 9.Watson AJ, Sabharwal A, Thorncroft M, McGown G, Kerr R, Bojanic S, Soonawalla Z, King A, Miller A, Waller S, et al. Tumor O6-methylguanine-DNA methyltransferase inactivation by oral lomeguatrib. Clin. Cancer Res. 2010;16:743–749. doi: 10.1158/1078-0432.CCR-09-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tawbi H, Villaruz L, Tarhini A, Moschos S, Sulecki M, Viverette F, Shipe-Spotloe J, Radkowski R, Kirkwood JM. Inhibition of DNA repair with MGMT pseudosubstrates: phase I study of lomeguatrib in combination with dacarbazine in patients with advanced melanoma and other solid tumours. Br. J. Cancer. 2011;105:773–777. doi: 10.1038/bjc.2011.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pegg AE, Dolan ME, Moschel RC. Structure, function and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog. Nucl. Acid Res. and Mol. Biol. 1995;51:167–223. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]
- 12.Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 13.Tano K, Dunn WC, Darroudi F, Shiota S, Preston RJ, Natarajan AT, Mitra S. Amplification of the DNA repair gene O6-methylguanine-DNA methyltransferase associated with resistance to alkylating drugs in a mammalian cell line. J. Biol. Chem. 1997;272:13250–13254. doi: 10.1074/jbc.272.20.13250. [DOI] [PubMed] [Google Scholar]
- 14.Fang Q, Kanugula S, Tubbs JL, Tainer JA, Pegg AE. Repair of O4-alkylthymine by O6-alkylguanine-DNA alkyltransferases. J. Biol. Chem. 2010;285:8185–8195. doi: 10.1074/jbc.M109.045518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivenugopal KS, Yuan XH, Friedman HS, Ali-Osman F. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA alkyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3 bis(2-chloroethyl)-1-nitrosourea. Biochemistry. 1996;35:1328–1334. doi: 10.1021/bi9518205. [DOI] [PubMed] [Google Scholar]
- 16.Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23:823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 17.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogens and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 18.Rasimas JJ, Pegg AE, Fried MG. DNA-binding mechanism of O6-alkylguanine-DNA alkyltransferase. Effects of protein and DNA alkylation on complex stability. J. Biol. Chem. 2003;278:7973–7980. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- 19.Rasimas JJ, Kar SR, Pegg AE, Fried MG. Interactions Of human O6-alkylguanine-DNA alkyltransferase (AGT) with short single-stranded DNAs. J. Biol. Chem. 2007;282:3357–3366. doi: 10.1074/jbc.M608876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melikishvili M, Rasimas JJ, Pegg AE, Fried MG. Interactions of human O6-alkylguanine-DNA alkyltransferase (AGT) with short double-stranded DNAs. Biochemistry. 2008;47:13754–13763. doi: 10.1021/bi801666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams CA, Fried MG. Mutations that probe the cooperative assembly of O6-alkylguanine-DNA alkyltransferase complexes. Biochemistry. 2011;50:1590–1598. doi: 10.1021/bi101970d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melikishvili M, Hellman LM, Fried MG. Use of DNA length varition to detect periodicities in positively cooperative, nonspecific binding. Methods Enzymol. 2009;466:66–82. doi: 10.1016/S0076-6879(09)66004-3. [DOI] [PubMed] [Google Scholar]
- 23.Adams CA, Melikishvili M, Rodgers DW, Rasimas JJ, Pegg AE, Fried MG. Topologies of complexes containing O6-alkylguanine-DNA alkyltransferase and DNA. J. Mol. Biol. 2009;389:248–263. doi: 10.1016/j.jmb.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazra TK, Roy R, Biswas T, Grabowski DT, Pegg AE, Mitra S. Specific recognition of O6-methylguanine in DNA by active site mutants of human O6-methylguanine-DNA methyltransferase. Biochemistry. 1997;36:5769–5776. doi: 10.1021/bi963085i. [DOI] [PubMed] [Google Scholar]
- 25.Melikishvili M, Rodgers DW, Fried MG. 6-Carboxyfluorescein and structurally-similar molecules inhibit DNA binding and repair by O6-alkylguanine DNA alkyltransferase. DNA Repair. 2011;10:1193–1202. doi: 10.1016/j.dnarep.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 27.Duguid EM, Rice PA, He C. The Structure of the human AGT protein bound to DNA and its implications for damage detection. J. Mol. Biol. 2005;350:657–666. doi: 10.1016/j.jmb.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 28.Xu-Welliver M, Kanugula S, Pegg AE. Isolation of human O6-alkylguanine-DNA alkyltransferase mutants highly resistant to inactivation by O6-benzylguanine. Cancer Res. 1998;58:1936–1945. [PubMed] [Google Scholar]
- 29.Rasimas JJ, Kanugula S, Dalessio PM, Ropson IJ, Fried MG, Pegg AE. Effects of zinc occupancy on human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2003;42:980–990. doi: 10.1021/bi026970b. [DOI] [PubMed] [Google Scholar]
- 30.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:449–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 31.Fried MG, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams C, Fried MG. In: Protein Interactions: Biophysical Approaches For The Study of Multicomponent Systems. Schuck P, editor. New York: Academic Press; 2007. pp. 417–446. [Google Scholar]
- 33.Voigt JM, Topal MD. O6-methylguanine in place of guanine causes asymmetric single-strand cleavage of DNA by some restriction enzymes. Biochemistry. 1990;29:1632–1637. doi: 10.1021/bi00458a039. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis T, Efstratiadis A. Fractionation of low molecular weight DNA or RNA in polyacrylamide gels containing 98% formamide or 7M urea. Method Enzymol. 1980;65:299–305. doi: 10.1016/s0076-6879(80)65040-x. [DOI] [PubMed] [Google Scholar]
- 35.Bading H. Determination of the molecular weight of DNA-bound protein(s) responsible for gel electrophoretic mobility shift of linear DNA fragments examplified with purified viral myb protein. Nucleic Acids Res. 1988;16:5241–5248. doi: 10.1093/nar/16.12.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coulter R, Blandino M, Tomlinson JM, Pauly GT, Krajewska M, Moschel RC, Peterson LA, Pegg AE, Spratt TE. Differences in the rate of repair of O6-alkylguanines in different sequence contexts by O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 2007;20:1966–1971. doi: 10.1021/tx700271j. [DOI] [PubMed] [Google Scholar]
- 37.Job P. Formation and stability of inorganic complexes in solution. Ann. Chim. (Paris) 1928;9:113–203. [Google Scholar]
- 38.Huang CY. Determination of binding stoichiometry by the continuous variation method: the Job plot. Methods Enzymol. 1982;87:509–525. doi: 10.1016/s0076-6879(82)87029-8. [DOI] [PubMed] [Google Scholar]
- 39.Pegg AE, Dolan ME. Properties and assay of mammalian O6-alkylguanine-DNA alkyltransferase. Pharmacol. Ther. 1987;34:167–179. doi: 10.1016/0163-7258(87)90010-6. [DOI] [PubMed] [Google Scholar]
- 40.Connors KA. Chemical Kinetics. New York: John Wiley and Sons; 1990. pp. 17–58. [Google Scholar]
- 41.Gardiner WC., Jr . In: Rates and Mechanisms of Chemical Reactions. Benjamin WA, editor. Menlo Park; 1972. pp. 41–74. [Google Scholar]
- 42.Fried MG, Crothers DM. Kinetics and mechanism in the reactions of gene regulatory proteins with DNA. J. Mol. Biol. 1984;172:263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- 43.Fickert T, Muller-Hill B. How lac repressor finds lac operator in vitro. J. Mol. Biol. 1992;226:59–68. doi: 10.1016/0022-2836(92)90124-3. [DOI] [PubMed] [Google Scholar]
- 44.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watson AJ, Middleton MR, McGown G, Thorncroft M, Ranson M, Hersey P, McArthur G, Davis ID, Thomson D, Beith J, et al. O6-methylguanine-DNA methyltransferase depletion and DNA damage in patients with melanoma treated with temozolomide alone or with lomeguatrib. Br. J. Cancer. 2009;100:1250–1256. doi: 10.1038/sj.bjc.6605015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson SJ, Wharton S, Watson AJ, Begum G, Butt A, Glynn N, Williams DM, Shibata T, Santibanez-Koref MF, Margison GP. A novel DNA damage recognition protein in Schizosaccharomyces pombe. Nucleic Acids Res. 2006;34:2347–2354. doi: 10.1093/nar/gkl270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luu KX, Kanugula S, Pegg AE, Pauly GT, Moschel RC. Repair of oligodeoxyribonucleotides by O6-alkylguanine-DNA alkyltransferase. Biochemistry. 2002;41:8689–8697. doi: 10.1021/bi025857i. [DOI] [PubMed] [Google Scholar]
- 48.Fried MG, Bromberg JL. Factors that affect the stability of protein-DNA complexes during gel electrophoresis. Electrophoresis. 1997;18:6–11. doi: 10.1002/elps.1150180103. [DOI] [PubMed] [Google Scholar]
- 49.Bhattacharyya D, Foote RS, Boulden AM, Mitra S. Physicochemical studies of human O6-methylguanine-DNA methyltransferase. Eur. J. Biochem. 1990;193:337–343. doi: 10.1111/j.1432-1033.1990.tb19343.x. [DOI] [PubMed] [Google Scholar]
- 50.Chan CL, Wu Z, Ciardelli T, Eastman A, Bresnick E. Kinetic and DNA-binding properties of recombinant human O6-methylguanine-DNA methyltransferase. Arch. Biochem. Biophys. 1993;300:193–200. doi: 10.1006/abbi.1993.1027. [DOI] [PubMed] [Google Scholar]
- 51.Bender K, Federwisch M, Loggen U, Nehls P, Rajewsky MF. Binding and repair of O6-ethylguanine in double-stranded oligodeoxynucleotides by recombinant human O6-alkylguanine-DNA alkyltransferase do not exhibit significant dependence on sequence context. Nucleic Acids Res. 1996;24:2087–2094. doi: 10.1093/nar/24.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spratt TE, Wu JD, Levy DE, Kanugula S, Pegg AE. Reaction and binding of oligodeoxynucleotides containing analogs of O6-methylguanine with wild type and mutant humanO6-alkylguanine-DNA alkyltransferase. Biochemistry. 1999;38:6801–6806. doi: 10.1021/bi982908w. [DOI] [PubMed] [Google Scholar]
- 53.Meyer AS, McCain MD, Fang Q, Pegg AE, Spratt TE. O6-alkylguanine-DNA alkyltransferases repair O6-methylguanine in DNA with Michaelis-Menten-like kinetics. Chem Res Toxicol. 2003;16:1405–1409. doi: 10.1021/tx0341254. [DOI] [PubMed] [Google Scholar]
- 54.Zang H, Fang Q, Pegg AE, Guengerich FP. Kinetic analysis of steps in the repair of damaged DNA by human O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 2005;280:30873–30881. doi: 10.1074/jbc.M505283200. [DOI] [PubMed] [Google Scholar]
- 55.Guza R, Ma L, Fang Q, Pegg AE, Tretyakova N. Cytosine methylation effects on the repair of O6-methylguanines within CG dinucleotides. J. Biol. Chem. 2009;284:22601–22610. doi: 10.1074/jbc.M109.000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pegg AE, Boosalis M, Samson L, Moschel RC, Byers TL, Swenn K, Dolan ME. Mechanism of inactivation of human O6-alkylguanine-DNA alkyltransferase by O6-benzylguanine. Biochemistry. 1993;32:11998–12006. doi: 10.1021/bi00096a009. [DOI] [PubMed] [Google Scholar]
- 57.Dolan ME, Pegg AE. O6-Benzylguanine and its role in chemotherapy. Clinical Cancer Res. 1997;3:837–847. [PubMed] [Google Scholar]
- 58.Niture SK, Doneanu CE, Velu CS, Bailey NI, Srivenugopal KS. Proteomic analysis of human O6-methylguanine-DNA methyltransferase by affinity chromatography and tandem mass spectrometry. Biochem. Biophys. Res. Commun. 2005;337:1176–1184. doi: 10.1016/j.bbrc.2005.09.177. [DOI] [PubMed] [Google Scholar]
- 59.Ishimi Y, Komamura-Kohno Y, Arai K, Masai H. Biochemical activities associated with mouse MCM2 protein. J. Biol. Chem. 2001;276:42744–42752. doi: 10.1074/jbc.M106861200. [DOI] [PubMed] [Google Scholar]
- 60.Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geng H, Sakato M, DeRocco V, Yamane K, Du C, Erie DA, Hingorani M, Hsieh P. Biochemical analysis of the human mismatch repair proteins hMutSα MSH2G674A-MSH6 and MSH2-MSH6T1219D. J. Biol. Chem. 2012;287:9777–9791. doi: 10.1074/jbc.M111.316919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hübscher U. DNA replication fork proteins. Methods Mol. Biol. 2009;521:19–33. doi: 10.1007/978-1-60327-815-7_2. [DOI] [PubMed] [Google Scholar]