Abstract
In the present work, ribosomes assembled in bacterial cells in the absence of essential ribosomal protein L5 were obtained. After arresting L5 synthesis, Escherichia coli cells divide a limited number of times. During this time, accumulation of defective large ribosomal subunits occurs. These 45S particles lack most of the central protuberance (CP) components (5S rRNA and proteins L5, L16, L18, L25, L27, L31, L33 and L35) and are not able to associate with the small ribosomal subunit. At the same time, 5S rRNA is found in the cytoplasm in complex with ribosomal proteins L18 and L25 at quantities equal to the amount of ribosomes. Thus, it is the first demonstration that protein L5 plays a key role in formation of the CP during assembly of the large ribosomal subunit in the bacterial cell. A possible model for the CP assembly in vivo is discussed in view of the data obtained.
INTRODUCTION
Reconstruction of functionally active bacterial ribosomal subunits was first performed successfully over 40 years ago (1–3), and since then a great body of experimental data regarding ribosome assembly in vitro has been accumulated (4–7). Crystal structures of ribosomes as well as that of functional ribosomal complexes from several bacteria are now available, providing insights into spatial organization of the assembled ribosome (8–13).
At the same time, these studies cannot take into account all of the factors driving the ribosome assembly in vivo (14–18). The present work is aimed at studying the role of the ribosomal protein (r-protein) L5 in the assembly of the large ribosomal subunit in the bacterial cell. The L5 protein is of particular interest for several reasons. First, it was shown earlier that two r-proteins, L5 and L18, are indispensable for the interaction between 5S rRNA and 23S rRNA (19,20). Recent crystallographic studies of bacterial ribosomes confirm the involvement of L5 in the conjunction of these rRNAs (8,10,12). Moreover, L5, which is conserved in Bacteria, Archaea and Eukarya, forms conserved intermolecular bonds with ribosomal RNAs (21–27). These observations may indicate that L5 performs a universal function in the formation of the ribosome. Second, protein L5 is known to be located on the ‘top’ of the central protuberance (CP) of the large ribosomal subunit (28) in the close vicinity of its functional centers (8,12,29–31). Crystallographic data show that L5 participates in formation of intersubunit bridge B1b and contacts the tRNA molecule in the ribosomal P-site (8–10,12). Taken together, these observations indicate the importance of protein L5 for the ribosome formation and functioning. However, the particular role of the protein in these processes still remains unknown. Earlier, we have shown that r-protein L5 is essential for the survival of Escherichia coli cells (32), but the reason for this has not been elucidated.
In this work we have studied the role of L5 in the ribosome assembly in vivo. We obtained an E. coli strain where the chromosomal gene encoding r-protein L5 (rplE) was inactivated in the presence of a complementing plasmid that provides inducible rplE expression. Thus, the synthesis of L5 could be stopped by removing the inducer from a growth medium. Analysis of the ribosomes from L5-depleted cells showed accumulation of large ribosomal subunits that lack most of the CP components. Our data demonstrate, for the first time, that r-protein L5 plays a key role in formation of this entire structural domain of the bacterial ribosome in vivo.
MATERIALS AND METHODS
Strains, plasmids and bacteriological techniques
All the strains and plasmids mentioned in the present work are listed in Supplementary Table S1. Cells were cultivated at 37°C in LB broth, and LB-agar plates, supplemented (where indicated) with 100 μg/ml ampicillin, 20 μg/ml chloramphenicol and 0.2% (wt/vol) l-arabinose. To obtain an rplE-encoding complementary plasmid pNK12, the NdeI-EcoRI fragment of pKAB122, carrying E. coli rplE open reading frame, was cloned into the plasmid pBADET. The latter is a gift from A. Kaliman. It is a derivative of pBAD18 (33), see Supplementary Table S1 for more information. Thus, pNK12 carries rplE under control of an arabinose-inducible promoter and confers resistance to ampicillin. The deletion of chromosomal rplE gene was made by transferring the ΔrplE::cat allele from KNB219 to the wild-type strain W3110, transformed with pNK12, via generalized transduction by phage P1 according to (34). Transductants were selected on LB agar, supplemented with chloramphenicol and arabinose. For simplicity, we call the resulting strain MS01, meaning that it contains the plasmid pNK12 at all times.
Cell cultivation conditions
To obtain E. coli cells depleted for protein L5, strain MS01 was grown at 37°C in LB medium supplemented with ampicillin and arabinose until A600 ∼ 0.4. Then, cells were washed and inoculated with 15–20 vol of pre-warmed at 37°C LB medium and cultivated until the growth curve reached a plateau (at A600 ∼ 0.8–1, within 6–8 h). During this time, aliquots of the cell culture were taken for the sedimentation analysis of ribosomes. The cells were collected, washed with buffer (30 mM Tris–HCl, pH 7.5, 200 mM NH4Cl, 10 mM MgCl2, 0.1 mM ethylenediaminetetraacetic acid (EDTA) and 3 mM 2-mercaptoethanol) and stored as described in (32).
The same strategy was applied to obtain the control cells. However, in this case the inducer was always present in the growth medium, providing rplE expression in trans. The cells were harvested at A600 ∼ 0.8–1.
Preparation and analysis of ribosomes and ribosomal subunits
Ribosomes were obtained according to (35) with modifications published in (32). To obtain ribosomal subunits, the ribosomes from the L5-depleted cells were loaded onto 5–20% (wt/wt) sucrose gradients prepared in buffer A (10 mM Tris–HCl, pH 7.5, 100 mM NH4Cl, 10 mM MgCl2 and 3 mM 2-mercaptoethanol) and centrifuged at 64 000g and 4°C for 11–12 h. Under these ionic conditions intact 50S subunits remain associated with 30S subunits to form 70S ribosomes, which greatly facilitate purification of 45S particles. Ribosomal subunits from the control cells were prepared in the same manner but in the presence of 1 mM MgCl2. Lowering magnesium concentration to 1 mM and below is a conventional approach allowing complete 70S ribosomes dissociation onto 50S and 30S subunits (35). Separated subunits were collected by high-speed centrifugation, dissolved in cold buffer A with 10% (vol/vol) glycerol, frozen and stored at −70°C. Large ribosomal subunits from L5-depleted cells were additionally purified by a second centrifugation in 5–20% (wt/wt) sucrose gradients.
To analyze ribosome profiles, cells were lysed by freeze-thaw method with lysozyme as described earlier (32), with buffer (30 mM Tris–HCl, pH 7.5, 100 mM NH4Cl, 10 mM MgCl2, 0.1 mM EDTA and 3 mM 2-mercaptoethanol) being used at all the stages. Then, S30 fractions were loaded onto 5–20% (wt/wt) sucrose gradients made in buffer A and centrifuged at 210 000g and 4°C for 1.5 h. Experiments on the association of ribosomal subunits were performed as follows. Equal amounts of small and large ribosomal subunits were mixed in buffer A with varying concentrations of MgCl2 (5–20 mM) and incubated at 37°C for 20 min. Then, the mixtures were analyzed by centrifugation in 5–20% (wt/wt) sucrose gradients as described above. To determine the sedimentation coefficients, an Optima XL-I analytical ultracentrifuge (Beckman Coulter, USA) was used. Centrifugation was carried out in buffer A. Sedimentation data were analyzed with the program Sedfit (36). The integrity of high-molecular weight ribosomal RNA was tested using electrophoresis in composite 2.3% polyacrylamide/0.75% agarose gel as described (37). To estimate 5S rRNA distribution, low-molecular weight RNAs were extracted as described earlier (38) from equal amount (by A260) of S30 fractions from L5-depleted and control cells. Same volumes of the corresponding ribosome-free extracts were used for RNA extraction. An RNA content was analyzed by electrophoresis in 8% polyacrylamide gels containing 8 M urea (39). Band intensities were quantified using program ImageJ. In view of the fact that slow-growing L5-depleted cells contained reproducibly less ribosomes (per total A260), 5S rRNA band was equally less intense when analyzing L5-depleted S30 fraction as compared to the control. Ribosomal proteins were extracted from ribosomal subunits with acetic acid (40) and fractionated by 2D gel electrophoresis (41) using acidic–acidic system IV. A number of ribosomal proteins were eluted from the gel and identified by mass spectrometry on a Deca XP Plus ion trap mass spectrometer (Thermo Finnigan, USA).
Affinity purification of 5S rRNA from the cytoplasm of the L5-depleted cells
5S rRNA was isolated using affinity chromatography on L5-Sepharose. Ribosomal protein L5 was purified as previously described (42) and covalently attached to Sepharose-4B according to (43). Ribosome-free cytoplasmic fraction (S100) was incubated with L5-Sepharose in buffer (10 mM Tris–HCl, pH 7.5, 50 mM NH4Cl, 10 mM MgCl2 and 3 mM 2-mercaptoethanol) at room temperature for 30 min. Then, the resin was washed sequentially with the same buffer containing 50 and 400 mM NH4Cl. 5S rRNA and bound proteins were eluted with 3 M NaCl. RNA and protein content of the final wash was analyzed by electrophoresis in denaturing conditions, as described in (39) and (44), correspondingly. Protein bands were quantified using ImageJ software. Two proteins possessing the electrophoretic mobility similar to that of 5S rRNA-binding proteins L18 and L25 were eluted from the gel and identified by mass spectrometry.
RESULTS AND DISCUSSION
Growth of the L5-depleted cells and the ribosome formation
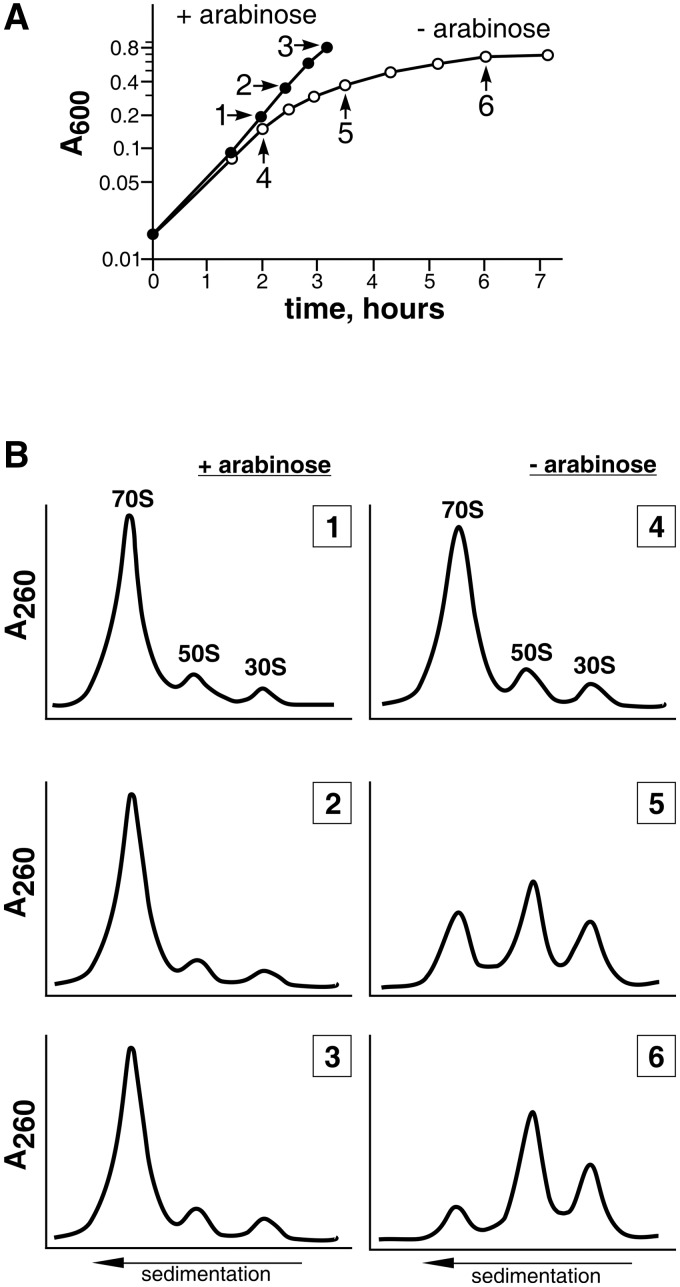
As we have shown earlier, the chromosomal gene rplE can be inactivated only in the presence of a complementing plasmid bearing the rplE gene (32). To study the role of the protein L5 in large ribosomal subunit assembly, we inactivated the chromosomal rplE gene in the presence of the plasmid, allowing arabinose-inducible expression of the rplE gene (strain MS01). Thus, we were able to stop L5 production by removing the inducer from the growth medium. To do this, cells were first grown in a medium with arabinose, then they were spun down and suspended in the medium with the inducer (control cells) or without it (L5-depleted cells). Figure 1A shows, that after removal of arabinose, when synthesis of L5 is stopped, cells start growing linearly and after 6 h the culture reaches a plateau. During this time the cells are only able to divide four to five times. This confirms our previous observation that r-protein L5 is essential for cell survival (32). During the linear growth, cells were harvested at several time points to analyze the ribosomal profile. While ribosomes from the control cells demonstrate a typical sedimentation profile (most of the subunits are associated to form 70S ribosome; Figure 1B, left panel), accumulation of free subunits can be seen in the cells grown without arabinose (Figure 1B, right panel). By the time the L5-depleted culture reaches the plateau, there are only 10–15% associated subunits. The large ribosomal subunits from the control and L5-depleted cells were isolated. The particles from the L5-depleted cells were unable to reassociate with 30S subunits to form the 70S ribosomes. Thus, the large ribosomal subunits assembled in cells in the absence of L5 are not able to form functional 70S ribosomes, which is lethal for the cells.
Figure 1.
(A) Growth curves of MS01 cells in the medium with (filled circle) or without (unfilled circle) an inducer. Arrows indicate the time points when aliquots were taken for ribosome analysis. The numerals (1–6) correspond to the ribosomal profiles in Section B. (B) Sedimentation analysis of ribosomes from of MS01 grown either in the presence (left panel) or absence (right panel) of the inducer. Components of cell cytoplasm were fractionated by centrifugation on a 5–20% (wt/wt) sucrose gradient.
Properties of large ribosomal subunits and the 5S rRNA–protein complex from L5-depleted cells
The sedimentation coefficient of large ribosomal subunits from the L5-depleted cells is 45S, which is less than that of intact subunits (50S) or even large subunits assembled in vitro in the absence of 5S rRNA (47S) (45,46). Such a change could be a result of a decompaction of rRNA or a loss of ∼10% (100–150 kDa) of the subunit mass. We therefore analyzed the composition of the 45S particles. 23S rRNA from these particles remains intact (fragmentation of 23S rRNA could affect compactness and shape of the particle). At the same time, as could be expected in view of published data (19,20,45,46) the absence of L5 greatly reduced the ability of 5S rRNA to incorporate into large ribosomal subunits. Only trace amount ( <10%) of 5S rRNA can be found in the 45S particles (Figure 2A, lanes 6 and 7). The presence of residual amounts of 5S rRNA and some proteins (see below) in preparations of the 45S particles is probably due to contamination with intact 50S subunits. As opposed to the control preparation (Figure 2A, lane 4), virtually all 5S rRNA is found in the ribosome-free cytoplasmic fraction of the L5-depleted cells (lane 5) in amounts corresponding to the amount of ribosomes. 5S rRNA from this fraction was purified using the affinity chromatography on L5-Sepharose. It was found that 5S rRNA copurifies with two proteins, whose electrophoretic mobility is similar to that of the 5S rRNA-binding proteins, L18 and L25 (Figure 2B). Subsequent mass spectrometry analysis has shown that the two major bands indeed correspond to the r-proteins L18 and L25 of E. coli. As a control, quadruple quantities of S100 from the control cells were treated with the affinity resin; neither 5S rRNA nor proteins L18 and L25 were detected in the high-salt wash. Thus, the components of the 5S rRNA–protein complex that are not incorporated into the ribosome in the absence of protein L5 are found in the cell cytoplasm as a stoichiometric complex. It has been shown previously that L5, L18 and L25 bind specifically to 5S rRNA (47,48), forming the 5S rRNA–protein complex (5S rRNP). These and other data concerning 5S rRNP properties in vitro suggest that its formation should precede incorporation into the large ribosomal subunit. Here, we provide evidences that in vivo 5S rRNP is formed independently of its association with the large ribosomal subunit, and that such an association is promoted by the r-protein L5.
Figure 2.
Distribution of 5S rRNA–protein complex components in L5-depleted cells. (A) Electrophoretic analysis of low-molecular weight RNAs (8% polyacrylamide gel, 8 M urea). 1—5S rRNA of E. coli (control); 2 and 3—S30 fraction from the control and L5-depleted cells, respectively; 4 and 5—S100 fraction from the control and L5-depleted cells, respectively; 6 and 7—large ribosomal subunits from the control and L5-depleted cells, respectively. Lanes 2 and 3 contain comparable amounts of the material and so do lanes 4 and 5. Lane 7 is loaded with twice as much material as lane 6. (B) Proteins that copurify with 5S rRNA by affinity chromatography on L5-Sepharose (15% sodium dodecyl sulphate–polyacrylamide gel electrophoresis). 1—S100 fraction of the L5-depleted cells; 2—3 M salt wash; 3 and 4—E. coli r-proteins L25 and L18, respectively.
Participation of protein L5 in formation of the bacterial ribosome central protuberance
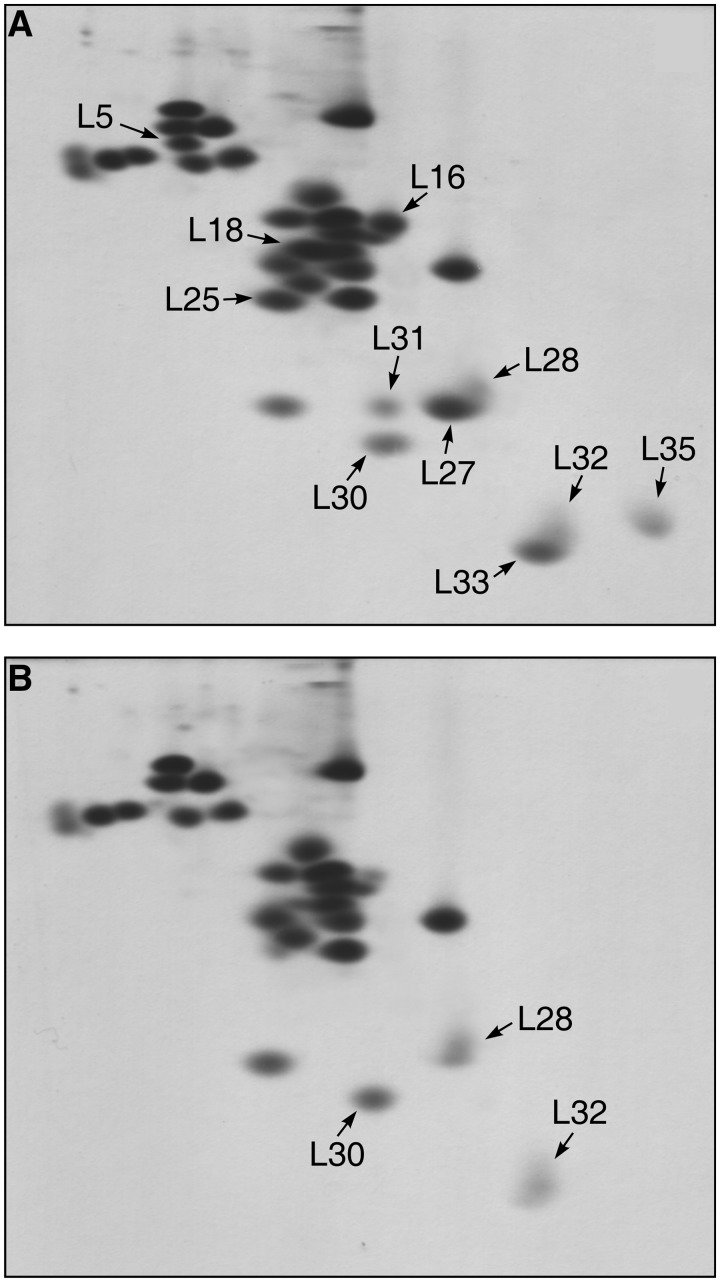
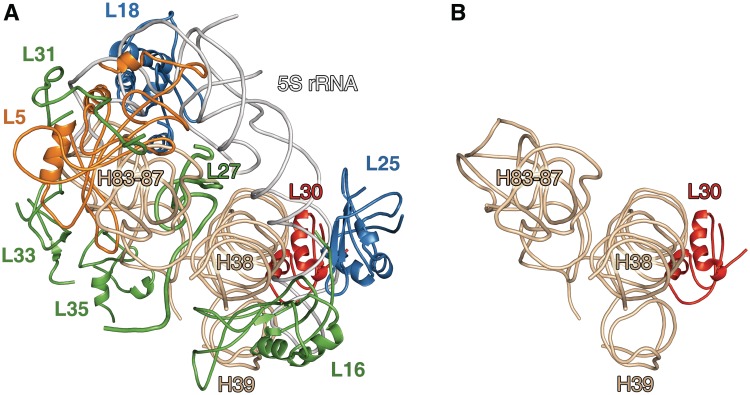
The protein composition of the 45S particles was determined using 2D gel electrophoresis with subsequent identification of some proteins by mass spectrometry. As seen in Figure 3B, the 45S particles are deprived almost completely of the 5S rRNA-binding proteins, L5, L18 and L25. The content of protein L16 is reduced to 10–15%. Furthermore, these particles contain only trace amounts (<10%) of several other r-proteins: L27, L31, L33 and L35. The proteins that partially overlap with proteins L27 and L33 on the gel have been identified as L28 and L32, respectively. These proteins (L28 and L32) are preserved in the 45S particles. The protein that migrates slower than L30 (Figure 3A) and which is missing from the 45S particles, has been identified by mass spectrometry as L31. This result is in agreement with the refined data on electrophoretic mobility of the full-length protein L31 (49). Thus, the 45S particles lose 5S rRNA and eight proteins: L5, L16, L18, L25, L27, L31, L33 and L35 (Figure 3B). The total mass of the components absent from the 45S particles in comparison with the intact 50S is ∼125 kDa. This correlates well with the difference in the sedimentation coefficients of these particles. According to the recent crystallographic data, the CP of the E. coli large ribosomal subunit comprises 5S rRNP, elements of domains II and V of 23S rRNA, and proteins L16, L27, L30, L33 and L35 (10) (Figure 4A). The only protein from this list that remains in the 45S particles is L30. Thus, the neighboring CP components (mentioned above) do not affect the incorporation of L30 into the large ribosomal subunit. In accordance with 50S ribosomal subunit in vitro assembly map, the other CP proteins do not influence the incorporation of protein L30 (5), which is consistent with our data. This allows to conclude that interaction of protein L30 with 23S rRNA occurs independently from the large ribosomal subunit CP formation. In the spatial structure of the Thermus thermophilus 70S ribosome (12) which was determined at a better resolution than that of the E. coli ribosome (10), the CP organization is similar to that of E. coli with only one exception. It contains r-protein L31, which is placed outside the CP in the E. coli ribosome model. It was suggested that L31 in the E. coli ribosome structure was confused with r-protein L28 due to insufficient resolution (12). Our data indicate that in the E. coli ribosome, protein L31 is most probably situated in the CP.
Figure 3.
Protein composition of large ribosomal subunits from the control (A) and L5-depleted (B) cells by 2D gel electrophoresis. Arrows mark the positions of r-proteins that are discussed in the text. The gels contain all the proteins of large ribosomal subunits except for the proteins L34 and L36. Protein L34 was found in both control and 45S particles under different separation conditions. Protein L36 was not analyzed.
Figure 4.
(A) Model of the E. coli ribosome CP structure. Position of protein L31 is modeled in the structure of the CP according to its position in the ribosome of T. thermophilus. Positions of ribosomal proteins, 5S rRNA and helices of 23S rRNA are indicated. (B) A proposed model for the structure of the CP region of bacterial ribosome formed in the absence of protein L5. The models were generated using the crystal structures of the E. coli and T. thermophilus ribosomes (PDB entries 2AW4 and 2J01, respectively). Model building and refinement were carried out using the program Coot; the picture was made using PyMol.
Taken together, crystallographic data and our results suggest that bacterial large ribosomal subunits assembled in vivo in the absence of protein L5 lose 5S rRNP and all proteins (except L30) of the CP (Figure 4B). Thus, r-protein L5 affects the formation and (or) stability of the entire structural domain of the ribosome in the bacterial cell. In early experiments it was demonstrated that large ribosomal subunits assembled in vitro in the absence of 5S rRNA are functionally inactive (45,50). These subunits contained all the r-proteins except for L5, L16, L18 and L25 (45,46,51). Differences in composition of these ribosomal subunits, reconstructed in vitro, and those obtained in the present work can be explained by different assembly conditions. Reconstitution of the 50S ribosomal subunits requires the conditions (high salt concentrations and prolonged incubation at elevated temperatures) that are far from physiological environment (3,5,17). Thereby, the results of the present work contribute significantly to a better understanding of the large ribosomal subunit assembly in bacteria. On the base of our data along with the data on structural organization of the CP (8,10,12,28,46,52) we suggest a model for the assembly of this 50S subunit structural domain in the bacterial cell. Incorporation of the preformed 5S rRNP into the large ribosomal subunit takes place at the beginning of CP formation. According to our results, only r-protein L30 (of the CP proteins) may be already present in the subunit during this stage (Figure 4B). The position of the 5S rRNP in the ribosome is mainly determined by two conserved contacts with 23S rRNA (8,10,12). Firstly, 5S rRNA interacts directly with helix 38 of 23S rRNA domain II (A-site finger, ASF). Secondly, 5S rRNP contacts helices H83–H85 of 23S rRNA domain V through proteins L5 and L18. These intermolecular contacts probably induce local rearrangements in 23S rRNA that, in turn, allow incorporation of the other CP proteins. This model is in accordance with the data on protein composition of an early large ribosomal subunit precursor in E. coli (53). It was shown that these particles (p150S) contain 5S rRNP but do not carry many r-proteins, inter alia L16, L31 and L33 (protein L35 was not analyzed).
Cooperativity and independence of the CP assembly suggests that it appears as a single structural unit not only during the ribosome assembly but also in ribosome functioning. On the basis of intermolecular cross-links in the ribosome, 5S rRNA was proposed to be a mediator between several ribosomal functional centers (31). According to the recent data on the ribosome structural organization (9–13), the CP components interact with each other, contacting with more than one component simultaneously (Figure 4A). Thereby, they form a complex self-contained system within the 50S subunit, where changes in one component can be transmitted to the others. Most of the components contact directly 5S rRNP, what explains its importance for CP formation and suggests that it plays a key role in CP function. At the same time, CP components contact directly tRNAs in all the three ribosomal sites (L16 and ASF in A-site, L5 in P-site and L33 in E-site); L27 protrudes into the 50S subunit, reaching the peptidyltransferase center; L25 is situated in close vicinity of the GTPase-associated center. In view of this, we propose that the CP (as a whole) serves as a coordinator of the functioning of ribosomal active centers.
Considering the fact that 45S particles are not able to associate with 30S subunits a few words should be said about the role of the CP in the subunit association. As shown by recent crystallographic data, the ASF and the protein L5 form intersubunit bridges 1a and 1b, respectively, interacting with protein S13 of the small ribosomal subunit (8,10,12). Moreover, it has recently been shown that a knockout of the S13 chromosomal gene (rpsM) as well as certain mutations in the S13 or ASF, impair association of ribosomal subunits (54,55). Protein L5 is absent from the 45S particles, thereby, excluding the formation of bridge 1b. However, earlier it was demonstrated that large ribosomal subunits lacking 5S rRNP (including protein L5) but containing the other proteins of the CP were not completely incapable of associating with the small ribosomal subunit (45,46). The position of ASF, forming bridge 1a, in the ribosomal subunit is stabilized by the contacts with 5S rRNA and proteins L16, L27 and L30 (Figure 4A). One can suggest that the absence of most of the CP components in the 45S particles destabilizes position of the ASF and, as a consequence, impedes the formation of bridge 1a. Thereby, according to our data and results of (54) it seems that together these bridges contribute significantly to association of the ribosomal subunits.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
The Russian Academy of Sciences, the Russian Foundation for Basic Research; the Program of RAS on Molecular and Cellular Biology. Funding for open access charge: Program of RAS on Molecular and Cellular Biology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Alexander Kaliman for the plasmid pBADET, Alexei Surin for mass spectrometry analysis, Oleg Nikonov for his assistance with figure preparation, Ciaran Condon and Mathias Springer for careful reading of this article and Mikhail Bubunenko for helpful discussion.
REFERENCES
- 1.Traub P, Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl Acad. Sci. USA. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nomura M, Erdmann VA. Reconstitution of 50S ribosomal subunits from dissociated molecular components. Nature. 1970;228:744–748. doi: 10.1038/228744a0. [DOI] [PubMed] [Google Scholar]
- 3.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl Acad. Sci. USA. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 5.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 6.Talkington MWT, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 9.Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HA, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 10.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 11.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 12.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 13.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 15.Williamson JR. After the ribosome structures: how are the subunits assembled? RNA. 2003;9:165–167. doi: 10.1261/rna.2164903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaczanowska M, Rydén-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 18.Sykes MT, Shajani Z, Sperling E, Beck AH, Williamson JR. Quantitative proteomic analysis of ribosome assembly and turnover in vivo. J. Mol. Biol. 2010;403:331–345. doi: 10.1016/j.jmb.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spierer P, Wang CC, Marsh TL, Zimmermann RA. Cooperative interactions among protein and RNA components of the 50S ribosomal subunit of Escherichia coli. Nucleic Acids Res. 1979;6:1669–1682. doi: 10.1093/nar/6.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Röhl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wool IG, Chan YL, Glück A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima T, Yao M, Kawamura S, Iwasaki K, Kimura M, Tanaka I. Ribosomal protein L5 has a highly twisted concave surface and flexible arms responsible for rRNA binding. RNA. 2001;7:692–701. doi: 10.1017/s1355838201002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perederina A, Nevskaya N, Nikonov O, Nikulin A, Dumas P, Yao M, Tanaka I, Garber M, Gongadze G, Nikonov S. Detailed analysis of RNA-protein interactions within the bacterial ribosomal protein L5/5S rRNA complex. RNA. 2002;8:1548–1557. [PMC free article] [PubMed] [Google Scholar]
- 24.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 26.Nevskaia NA, Nikonov OS, Revtovich SV, Garber MB, Nikonov SV. Identification of RNA-recognizing modules on the surface of ribosomal proteins. Mol. Biol. (Mosk) 2004;38:926–936. [PubMed] [Google Scholar]
- 27.Hartman H, Favaretto P, Smith TF. The archaeal origins of the eukaryotic translational system. Archaea. 2006;2:1–9. doi: 10.1155/2006/431618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lotti M, Noah M, Stöffler-Meilicke M, Stöffler G. Localization of proteins L4, L5, L20 and L25 on the ribosomal surface by immuno-electron microscopy. Mol. Gen. Genet. 1989;216:245–253. doi: 10.1007/BF00334363. [DOI] [PubMed] [Google Scholar]
- 29.Spirin AS. Location of tRNA on the ribosome. FEBS Lett. 1983;156:217–221. doi: 10.1016/0014-5793(83)80499-2. [DOI] [PubMed] [Google Scholar]
- 30.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 31.Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN. Structure and function of 5S rRNA in the ribosome. Biochem. Cell Biol. 1995;73:869–876. doi: 10.1139/o95-094. [DOI] [PubMed] [Google Scholar]
- 32.Korepanov AP, Gongadze GM, Garber MB, Court DL, Bubunenko MG. Importance of the 5 S rRNA-binding ribosomal proteins for cell viability and translation in Escherichia coli. J Mol. Biol. 2007;366:1199–1208. doi: 10.1016/j.jmb.2006.11.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 35.Staehelin T, Maglott DM, Monro RE. On the catalytic center of peptidyl transfer: a part of the 50 S ribosome structure. Cold Spring Harb. Symp. Quant. Biol. 1969;34:39–48. doi: 10.1101/sqb.1969.034.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Schuck P, Millar DB. Rapid determination of molar mass in modified archibald experiments using direct fitting of the Lamm equation. Anal. Biochem. 1998;259:48–53. doi: 10.1006/abio.1998.2638. [DOI] [PubMed] [Google Scholar]
- 37.Peacock AC, Dingman CW. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968;7:668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- 38.Zubay G. The isolation and fractionation of soluble ribonucleic acid. J. Mol. Biol. 1962;4:347–356. [Google Scholar]
- 39.Meshcheryakov VA, Gryaznova OI, Davydova NL, Mudrik ES, Perederina AA, Vasilenko NL, Gongadze GM, Garber MB. RNA-binding properties of an unusual ribosomal protein TL5 from Thermus thermophilus. Biochemistry (Mosc) 1997;62:537–542. [PubMed] [Google Scholar]
- 40.Hardy SJ, Kurland CG, Voynow P, Mora G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 41.Madjar JJ, Michel S, Cozzone AJ, Reboud JP. A method to identify individual proteins in four different two-dimensional gel electrophoresis systems: application to Escherichia coli ribosomal proteins. Anal. Biochem. 1979;92:174–182. doi: 10.1016/0003-2697(79)90641-9. [DOI] [PubMed] [Google Scholar]
- 42.Gongadze GM, Perederina AA, Meshcheriakov VA, Fedorov RV, Moskalenko SE, Rak AV, Serganov AA, Shcherbakov DV, Nikonov SV, Garber MB. The Thermus thermophilus 5S rRNA-protein complex: identifications of specific binding sites for proteins L5 and L18 in 5S rRNA. Mol. Biol. (Mosk) 2001;35:610–616. [PubMed] [Google Scholar]
- 43.Harrison LC, Itin A. Purification of the insulin receptor from human placenta by chromatography on immobilized wheat germ lectin and receptor antibody. J. Biol. Chem. 1980;255:12066–12072. [PubMed] [Google Scholar]
- 44.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Dohme F, Nierhaus KH. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc. Natl Acad. Sci. USA. 1976;73:2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selivanova OM, Gongadze GM, Gudkov AT, Vasiliev VD. Structure of protein-deficient 50 S ribosomal subunits. Particles without 5 S RNA-protein complex retain the L7/L12 stalk and associate with 30 S subunits. FEBS Lett. 1986;197:79–83. doi: 10.1016/0014-5793(86)80302-7. [DOI] [PubMed] [Google Scholar]
- 47.Horne JR, Erdmann VA. Isolation and characterization of 5S RNA-protein complexes from Bacillus stearothermophilus and Escherichia coli ribosomes. Mol. Gen. Genet. 1972;119:337–344. doi: 10.1007/BF00272091. [DOI] [PubMed] [Google Scholar]
- 48.Chen-Schmeisser U, Garrett RA. A new method for the isolation of a 5 S RNA complex with proteins L5, L18 and L25 from Escherichia coli ribosomes. FEBS Lett. 1977;74:287–291. doi: 10.1016/0014-5793(77)80866-1. [DOI] [PubMed] [Google Scholar]
- 49.Eistetter AJ, Butler PD, Traut RR, Fanning TG. Characterization of Escherichia coli 50S ribosomal protein L31. FEMS Microbiol. Lett. 1999;180:345–349. doi: 10.1111/j.1574-6968.1999.tb08816.x. [DOI] [PubMed] [Google Scholar]
- 50.Erdmann VA, Fahnestock S, Higo K, Nomura M. Role of 5S RNA in the functions of 50S ribosomal subunits. Proc. Natl Acad. Sci. USA. 1971;68:2932–2936. doi: 10.1073/pnas.68.12.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dohme F, Nierhaus KH. Total reconstitution and assembly of 50 S subunits from Escherichia coli Ribosomes in vitro. J. Mol. Biol. 1976;107:585–599. doi: 10.1016/s0022-2836(76)80085-x. [DOI] [PubMed] [Google Scholar]
- 52.Shatsky IN, Evstafieva AG, Bystrova TF, Bogdanov AA, Vasiliev VD. Topography of RNA in the ribosome: location of the 3′-end of 5 S RNA on the central protuberance of the 50 S subunit. FEBS Lett. 1980;121:97–100. doi: 10.1016/0014-5793(80)81274-9. [DOI] [PubMed] [Google Scholar]
- 53.Nierhaus KH, Bordasch K, Homann HE. Ribosomal proteins. 43. In vivo assembly of Escherichia coli ribosomal proteins. J. Mol. Biol. 1973;74:587–597. doi: 10.1016/0022-2836(73)90049-1. [DOI] [PubMed] [Google Scholar]
- 54.Cukras AR, Green R. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J. Mol. Biol. 2005;349:47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sergiev PV, Kiparisov SV, Burakovsky DE, Lesnyak DV, Leonov AA, Bogdanov AA, Dontsova OA. The conserved A-site finger of the 23 S rRNA: just one of the intersubunit bridges or a part of the allosteric communication pathway? J. Mol. Biol. 2005;353:116–123. doi: 10.1016/j.jmb.2005.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.