Abstract
The positive transcription elongation factor b (P-TEFb) regulates RNA polymerase II elongation. In cells, P-TEFb partitions between small active and larger inactive states. In the latter, HEXIM1 binds to 7SK snRNA and recruits as well as inactivates P-TEFb in the 7SK snRNP. Several stimuli can affect this P-TEFb equilibrium. In this study, we demonstrate that protein kinase C (PKC) phosphorylates the serine at position158 (S158) in HEXIM1. This phosphorylated HEXIM1 protein neither binds to 7SK snRNA nor inhibits P-TEFb. Phorbol esters or the engagement of the T cell antigen receptor, which activate PKC and the expression of the constitutively active (CA) PKCθ protein, which is found in T cells, inhibit the formation of the 7SK snRNP. All these stimuli increase P-TEFb-dependent transcription. In contrast, the kinase-negative PKCθ and the mutant HEXIM1 (S158A) proteins block effects of these PKC-activating stimuli. These results indicate that the phosphorylation of HEXIM1 by PKC represents a major regulatory step of P-TEFb activity in cells.
INTRODUCTION
Eukaryotic transcription by RNA polymerase II (RNAPII) is regulated at multiple steps including initiation, promoter clearance, elongation and cotranscriptional processing of nascent transcripts (1). Recent genome-wide analyses revealed that elongation is a critical step of transcription (2–4). The positive transcription elongation factor b (P-TEFb), which contains cyclins T1 or T2 (CycT1, CycT2; collectively, CycT) and cyclin-dependent kinase 9 (CDK9), plays a major stimulatory role in this process. P-TEFb phosphorylates serines at position 2 (S2) in the C-terminal domain (CTD) of RNAPII as well as DRB (5,6-dichloro-1 -β-d-ribofuranosylbenzimidazole) sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF) (5). In cells, P-TEFb exists in two major forms (5,6). The catalytically active P-TEFb binds bromodomain containing protein 4 (BRD4), subunits of the super elongation complex (SEC), or other DNA- or RNA-bound activators (7–10). In contrast, the 7SK snRNP is inactive and contains 7SK snRNA, hexamethylene bisacetamide-(HMBA)-induced mRNA-encoded proteins 1 or 2 (HEXIM1 or HEXIM2), La-related protein 7 (LARP7) and the methylphosphate capping enzyme (MePCE) (11). In this large complex, HEXIM proteins inhibit the kinase activity of CDK9 (5,12). Whereas the 7SK snRNP, which is loosely associated with chromatin, is extracted easily with low salt (10 mM), the P-TEFb that is engaged in transcription, is bound to chromatin, and thus requires a higher salt concentration (>0.15 M) for its extraction (13).
Depending on the cell type, up to 90% of P-TEFb is found in the 7SK snRNP, and the equilibrium between active and inactive complexes (P-TEFb equilibrium) determines the overall transcriptional activity of the cell (5). Many stresses such as UV light, heat, inhibition of transcription by Actinomycin D, DRB or flavopiridol, histone deacetylase inhibitors (HDACis) such as tricostatin A (TSA), suberoylanilide hydroxamic acid (SAHA) as well as specific intracellular signaling cascades can disrupt the 7SK snRNP and activate P-TEFb (6,14–17). Although precise molecular mechanisms leading to the disruption of 7SK snRNP and the release of P-TEFb remain to be elucidated, multiple post-transcriptional modifications of 7SK snRNP components are involved. For instance, HMBA and UV light activate PP2B (Ca++/Calmodulin-dependent protein phosphatase) and PP1a, which can dephosphorylate threonine at position 186 (T loop) in CDK9, and thus release P-TEFb (18,19). In a different cellular context, HMBA also activates the phosphatidylinositol-3-kinase (PI3K)/Akt-signaling pathway, which antagonizes the interaction between P-TEFb and HEXIM1 through phosphorylation of the threonine and serine at positions 270 and 278 of HEXIM1, respectively. T-cell antigen receptor (TCR) signaling also disrupts the 7SK snRNP by a signaling cascade that activates Erk, although its phosphorylation target remains unknown (20). In addition to these kinases and phosphatases, the acetylation of CycT1 contributes to this release, which could explain additional effects of HDACis on the activity of P-TEFb (21,22). Therefore, distinct molecular pathways target 7SK snRNP subunits to release the active free P-TEFb in cells.
Since P-TEFb also serves as the host cellular cofactor for HIV transcription and replication, studying its regulation is particularly important for the development of new antiviral therapies (16,23–27). Although the highly active antiretroviral therapy (HAART) reduces levels of HIV RNA below detection, persistence of latently infected cells prevents the cure of AIDS. To eradicate this reservoir, it is critical to reactivate viral replication and to eliminate these latently infected cells. Signals that activate NF-kB and P-TEFb, two critical complexes for HIV transcription, might accomplish this task. Indeed, protein kinase C (PKC) agonists activate both of them and can reactivate HIV replication (28–30). In this study, we found that PKC phosphorylates HEXIM1 on a specific serine residue, which increases not only levels of free P-TEFb but also transcription of a target gene. We focused on PKCθ, which is the major PKC isoform found in T cells (31–36).
MATERIALS AND METHODS
Cell lines, reagents and antibodies
Jurkat T cells were grown in RPMI containing 10% FCS at 37°C with 5% CO2. 293T and Hela cells were grown in DMEM containing 10% FCS at 37°C with 5% CO2. Chelerythrine chloride and phorbol myristyl acetate (PMA) were purchased from Sigma-Aldrich. Bisindolylmaleimide family compounds were purchased from Calbiochem. Flavopiridol was obtained from the AIDS Reference and Reagent Program (NIH). The anti-CycT1(sc-10750), anti-CDK9 (sc-484) antibodies were obtained from Santa Cruz Biotechnology. The anti-HEXIM and anti-tubulin antibodies were obtained from Abcam. The anti-PKCθ antibody was purchased from Cell Signaling Technologies. The anti-Flag M2 (F3165) antibody and the anti-Flag M2 agarose beads (A2220) were purchased from Sigma-Aldorich. The anti-CD3 and anti-CD28 antibodies were purchased from Pharmingen. Affinity purified anti-phosphoryated HEXIM1 antibody was raised in rabbits injected with peptide from HEXIM1 from positions 150 to 165, where the serine at 158 was replaced with phosphoserine, and affinity-purified using peptide antigen-conjugated columns.
Plasmids
Plasmids directing the expression of dominant negative (DN) and constitutively active (CA) PKCθ proteins were a generous gift from Art Weiss (33). The pG5TKLuc reporter plasmid was described previously (37). The pFLAG-CMV-2.HEXIM1 plasmid was described previously (38). To construct the plasmid coding for the mutant flag-HexS158A proteins, the pFLAG-CMV-2.HEXIM1 plasmid was subjected to site directed mutagenesis with the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene) (39).
In vitro kinase assays
An amount of 16 μl kinase reactions containing purified recombinant P-TEFb with human DSIF or RNA polymerase II (RNAPII) as substrates were carried out in 30 mM KCl, 20 mM HEPES pH 7.6, 7 mM MgCl2, 30 μM ATP, 1.3 μCi of [γ-32P]-ATP (Amersham), 1 μg BSA per reaction and the indicated amounts of HEXIM1 proteins. T7-transcribed 7SK snRNA or double-stranded (ds) RNA oligonucleotides were added last to the pre-incubation mixture. All reactions were incubated for 10 min at 23°C prior to the addition of ATP. The kinase reactions were then incubated for 20 min at 30°C and then stopped by the addition of sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) loading buffer. Reaction products were resolved by 9% SDS–PAGE. The dried gel was subjected to autoradiography and quantified with a Packard InstantImager. The PKC in vitro kinase assays (IVKAs) using wild-type or mutant HEXIM1 proteins as substrates were performed in 50 µl reactions in a buffer containing 25 mM HEPES (pH 7.5), 10 mM MgCl2, 200 µM ATP and 2 µCi of [γ-32P]ATP. All reactions were incubated for 15 min at 30°C, stopped by the addition of 2 × SDS–PAGE loading buffer. Fifteen microliters of each reaction mixture was subjected to 10% SDS–PAGE followed by autoradiography.
In vivo kinase assays
HeLa cells were seeded in 100-mm plates and transfected with 3 µg of flag-HEXIM1 expression plasmids. Twenty-four hours after transfection, cells were serum-deprived for 12 h and labeled with [32P] orthophosphate (300 µCi/ml) for 1.5 h at 37°C with or without a PKC inhibitor chelerythrine chloride (5 µM). Next, cells were washed with ice-cold PBS, resuspended in TNE buffer (10 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, complete EDTA-free protease inhibitor mixture (Roche Molecular Biochemicals), phosphatase inhibitor mixture (Sigma) containing 1% NP-40 and lysed for 1 h. Following centrifugation, flag-HEXIM1 proteins were immunoprecipitated using FLAG M2 agarose beads, separated by 10% SDS–PAGE and visualized by autoradiography.
Phosphorylation of HEXIM1 and repurification
Phosphorylation was carried out under identical conditions as in kinase assay described above, except that 1 mM ATP was used, and incubation time was at least 6 h at 37°C. After phosphorylation, His epitope-tagged HEXIM1 proteins were repurified by Ni–NTA chromatography.
Electrophoretic mobility shift assay
For electrophoretic mobility shift assay (EMSA) in Figure 2, 12 μl reactions were carried out in 25 mM HEPES, pH 7.6, 15% glycerol, 60 mM KCl, 0.1 mM EDTA, 5 mM DTT, 0.01% NP-40 and included 200 ng recombinant HEXIM1, P-TEFb and various RNA or DNA oligonucleotides as indicated. Reactions were incubated at room temperature for 15 min and resolved by 6% PAGE at 4°C for 1.5 h at 6 W. Proteins were visualized by silver staining. For EMSA in Figure 3, 12 μl reactions were carried out in 25 mM HEPES, pH 7.6, 15% glycerol, 60 mM KCl, 0.1 mM EDTA, 5 mM DTT, 0.01% NP-40 and included 200 ng recombinant HEXIM1, P-TEFb and α-32P-labeled 7SK snRNA. Reactions were incubated at room temperature for 15 min and resolved by 6% PAGE at 4°C for 1.5 h at 6 W. The free probe and RNA–protein complexes were visualized by autoradiography.
Figure 2.
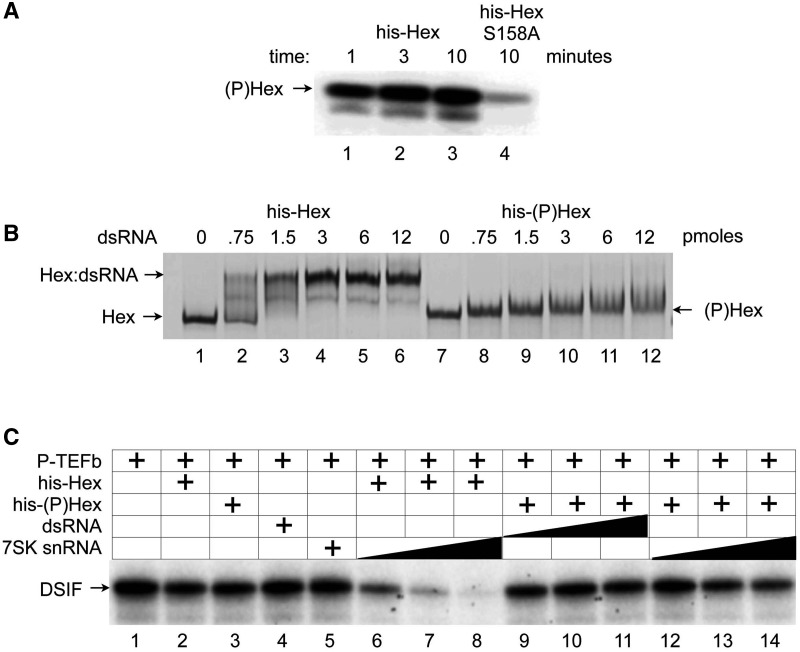
HEXIM1, which is phosphorylated on the serine at position 158, neither binds to RNA nor inhibits P-TEFb. (A) Only the WT HEXIM1 protein is phosphorylated by PKC in vitro. Purified His epitope-tagged WT HEXIM1 and mutant HEXIM1 (S158A) proteins (Hex and HexS158A) were incubated with native PKC catalytic subunit purified from rat brain in the presence of [γ-32P] ATP for indicated times at 30°C. The phosphorylated HEXIM1 protein ((P)Hex) was detected by autoradiography. (B) Only the unmodified HEXIM1 protein binds to RNA. Indicated amounts of WT and in vitro PKC-phosphorylated HEXIM1 proteins were incubated with dsRNA for 10 min. at room temperature. RNA:protein mixtures were then separated by non-denaturing gels, and visualized by silver staining. The positions for Hex:dsRNA complex and free Hex protein are indicated on the left of the gel. (C) The phosphorylated HEXIM1 protein no longer inhibits the kinase activity of CDK9. IVKAs were performed by incubating purified P-TEFb with DSIF as a substrate in the presence of WT HEXIM1 (lane 2, Hex) or the phosphorylated HEXIM1 protein alone (lane 3, (P)Hex) or in the presence of dsRNA or 7SK snRNA (lanes 4 and 5, dsRNA and 7SK) or with combinations of increasing amounts of HEXIM1 and 7SK snRNA (lanes 6–8, 7SK + Hex), phosphorylated HEXIM1 proteins (lanes 9–11, dsRNA + (P)Hex; or, lanes 12–14, 7SK + (P)Hex). Phosphorylated DSIF was visualized by autoradiography.
Figure 3.
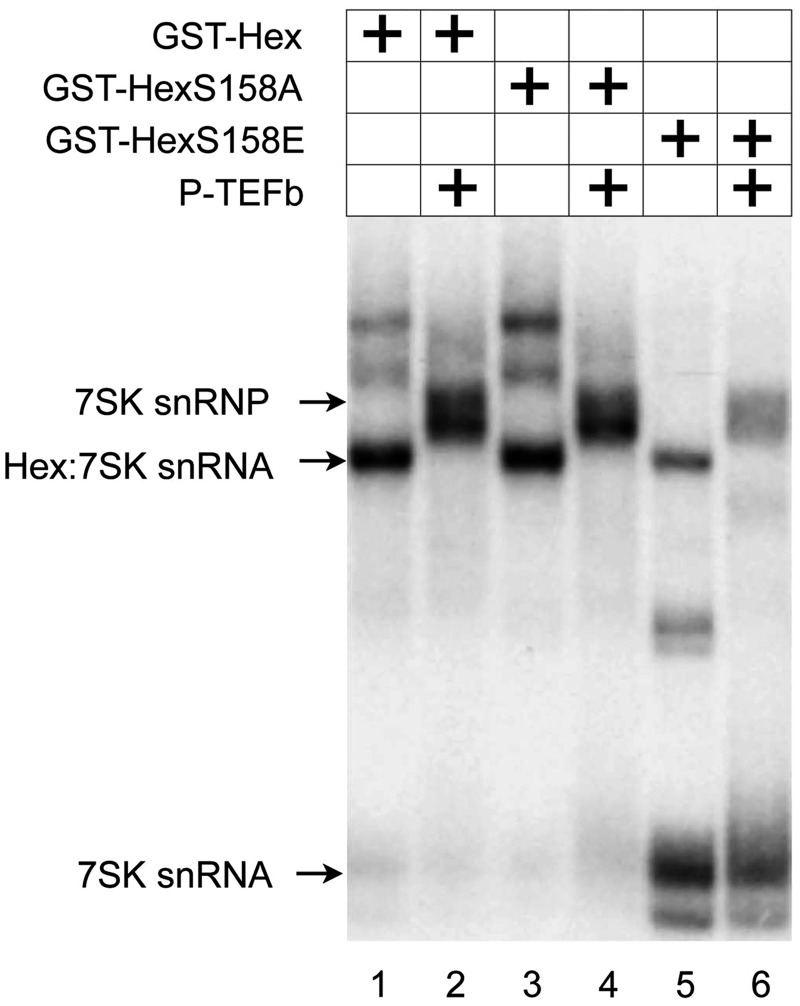
Changing S158 to glutamic acid reduces the binding of HEXIM1 and P-TEFb to 7SK snRNA in vitro. Recombinant GST-Hex, GST-HexS158A and GST-HexS158E proteins alone or with P-TEFb were incubated with α-32P-labeled 7SK snRNA. RNA–protein complexes were separated by non-denaturing PAGE and visualized by autoradiography. Arrows to the left indicate the free 7SK snRNA, HEXIM1-7SK snRNA complex or 7SK snRNP.
Glycerol gradients
Glycerol gradients (10–30%) were established by pipeting 2 ml of each glycerol fraction (10, 15, 20, 25 and 30% v/v) in buffer A (20 mM HEPES (pH7.9), 0.3 M KCl, 0.2 mM EDTA, 0.1% NP40) into centrifugation tubes (Beckman 331372). Gradients were formed by standing for 6 h at 4°C. Stimulated or unstimulated Jurkat T cells transfected or not with corresponding plasmids were lysed in 0.6 ml of buffer A containing protease inhibitor cocktail (Fermentas) and an RNase inhibitor (Roche) for 30 min at 4°C. Lysates were centrifuged at 10 000g 14 000 rpm for 10 min and supernatants were loaded into tubes with preformed glycerol gradients. Protein complexes were then fractionated by centrifugation in an SW4T1 rotor (Beckman) at 38 000 rpm<AQ10> for 21 h. Ten fractions (1 ml) were collected, precipitated with trichloracetic acid and analyzed with the indicated antibodies by western blotting. Fractions with detectable bands are presented in Figures 5 and 6. Band intensities were quantified by LI-COR Odyssey Imaging System and plotted as indicated in Figures 5 and 6.
Figure 5.
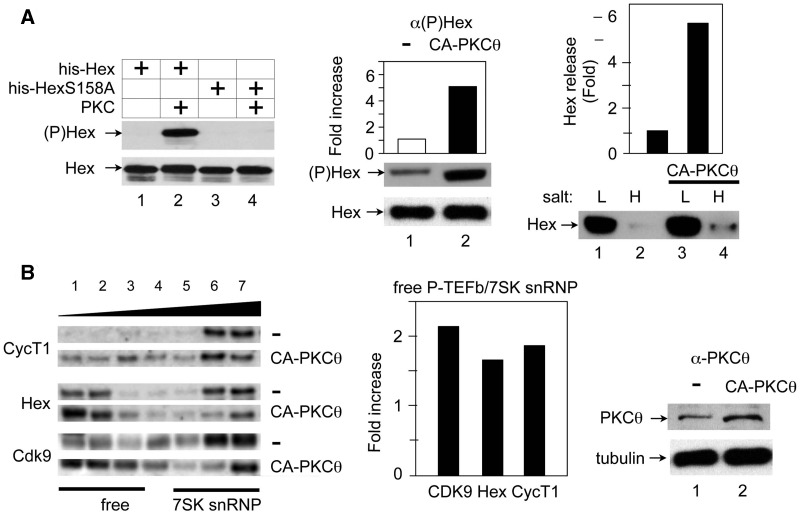
The CA-PKCθ protein phosphorylates HEXIM1, which releases free P-TEFb. (A) The CA PKCθ protein (CA-PKCθ) phosphorylates HEXIM1 and leads to its release in cells. Left panel: Evaluation of antibodies against HEXIM1, which is phosphorylated on the serine at position 158 ((P)Hex). IVKAs were performed with purified PKC and WT HEXIM1 or mutant HEXIM1 (S158A) proteins (Hex or HexS158A) and non-radioactive ATP as the substrate. Protein complexes were then separated by SDS–PAGE, followed by western blotting with anti-phospho-HEXIM1 ((P)Hex, upper panel) and anti-HEXIM1 (Hex, lower panel) antibodies. Center and right panels: Cell fractionation analyses were performed with Jurkat T cells, which expressed the empty plasmid vector (−) or the CA PKCθ protein (CA-PKCθ). Complexes, which were bound loosely or tightly to chromatin were extracted with buffers containing low (L,10 mM) and high (H, 300 mM) salt concentrations. Center panel: Phosphorylated and unphosphorylated HEXIM1 proteins (middle panel, (P)Hex; bottom panel, Hex) in the high salt (H) fraction was detected with anti-phospho-HEXIM1 and anti-HEXIM1 antibodies, respectively. Band intensities were quantified (top panel) and are presented as fold increase over the value obtained with Jurkat T cells, which expressed only the empty plasmid vector. Right panel: After fractionation, HEXIM1 was detected with anti-HEXIM1 antibodies by western blotting (lower panel, Hex). The release of HEXIM1 to chromatin (H, high salt) was quantified relative to the value obtained with Jurkat T cells, which expressed the empty plasmid vector only (Hex release). (B) The CA PKCθ protein (CA-PKCθ) releases of P-TEFb and HEXIM1 from 7SK snRNP. Left panel: Glycerol gradient sedimentation was performed with Jurkat T cells, which expressed the empty plasmid vector (upper panels, −) or CA-PKCθ (lower panels, CA-PKCθ). Twenty-four hours after transfection, cell lysates were subjected to glycerol gradient (10–30%) sedimentation. Fractions were separated by SDS–PAGE and analyzed with anti-CycT1 (top panels), anti-HEXIM1 (middle panels) and anti-CDK9 (bottom panels) antibodies by western blotting. Center Panel: Band intensities in fractions corresponding to free P-TEFb (lanes 1–3) and 7SK snRNP (lanes 5–7) were quantified and ratios between these two groups were calculated and plotted as a fold-increase over values obtained with untransfected Jurkat cells. Right panel: The expression of CA-PKCθ was confirmed with anti-PKCθ antibodies by western blotting.
Cell fractionation
Jurkat T cells, confluent in 12-well plates, were pelletted and washed once in ice-cold PBS, 0.1% PMSF. Cells were resuspended in 70 μl of cold buffer A (10 mM HEPES, 15 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.1% PMSF, 40 U/ml RNaseOUT (Invitrogen)) and incubated on ice with occasional vortexing for 10 min. Cell lysates were spun for 5 min at 4000 rpm in a cold table microcentrifuge to isolate the 7SK snRNP. Nuclei were resuspended with 70 μl of cold buffer B (10 mM HEPES, 2 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 420 mM NaCl, 0.5% NP-40, 0.1% PMSF, 40 U/ml RNaseOUT (Invitrogen)) and incubated on ice with occasional vortexing for 10 min. Cell lysates were spun for 5 min at 4000 rpm in a cold table microcentrifuge to isolate the fraction containing the free P-TEFb.
Transient transfection and reporter gene assays
293T or HeLa cells were seeded into 24-well plates ∼12 h prior to transfection using Extreme (Roche). Luciferase enzymatic assays were performed as described previously (15,40). Jurkat T cells were transiently transfected with mutant PKCθ expressing plasmids using Extreme (Figure 5) or by electroporation (Biorad, Figure 6) at 250 V/960 µF as described previously (33).
Figure 6.
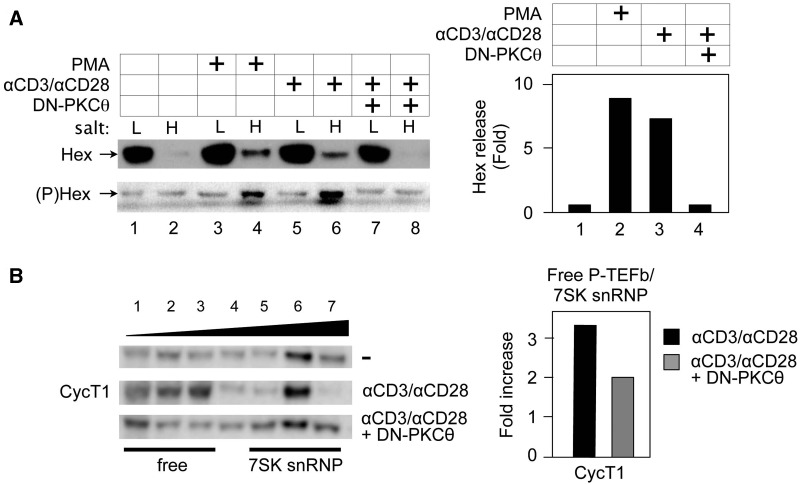
Activation of PKC via phorbol ester or the TCR releases HEXIM1 in Jurkat T cells, which is blocked by the DN PKCθ protein. (A) PMA and anti-CD3/anti-CD28 antibodies release phosphorylated HEXIM1 protein from 7SK snRNP. Jurkat T cells expressed the empty plasmid vector (lanes 1–6) or DN-PKCθ (lanes 7 and 8). Forty-eight hours after the transfection, cells were untreated (lanes 1 and 2), or treated with PMA (20 nM, lanes 3 and 4), or anti-CD3/anti-CD28 antibodies (lanes 4 and 8) for 30min. High and low salt fractions were subjected to western blotting with anti-HEXIM1 (upper panel, Hex) and anti-phosphoylated HEXIM1 (lower panel, (P)Hex) antibodies. Hex release was determined as in Figure 5A. (B) The DN PKCθ protein inhibits the release of CycT1 from the 7SK snRNP. Left panel: Glycerol gradient sedimentation was performed as above (top panels, CycT1) and in the presence of DN-PKCθ (bottom panel). CycT1 of each fraction was detected with anti-CycT1 antibodies by western blotting. Whereas CycT1 in lanes 1–3 represents free P-TEFb, that in lanes 5–7 is in the 7SK snRNP. Right panel: Band intensities in fractions corresponding to free P-TEFb and 7SK snRNP were quantified and the ratio between these two groups was calculated and plotted as a fold-increase over the value obtained with unstimulated Jurkat cells.
RESULTS
PKC phosphorylates HEXIM1 on the serine at position 158 (S158)
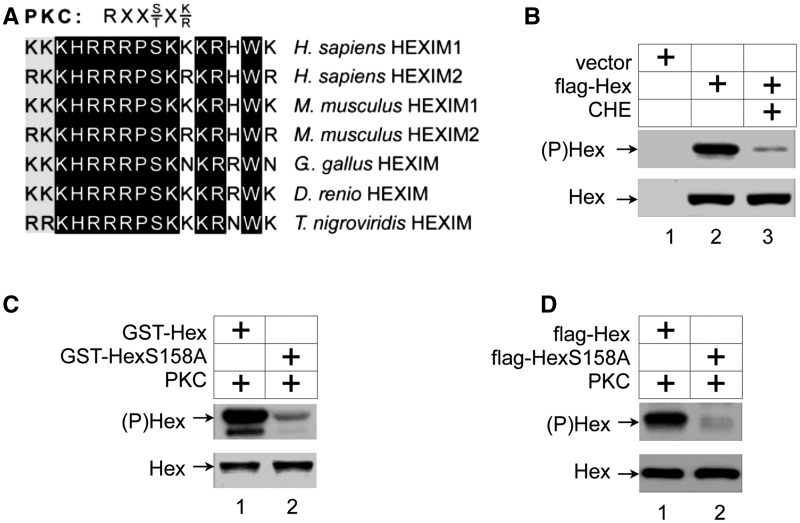
It is well established that activation of cellular-signaling pathways induces a disruption of the 7SK snRNP via post-transcriptional modifications of its individual components (14,17,18,20,22). In particular, phosphorylation and acetylation play important roles in the removal and activation of P-TEFb from the 7SK snRNP. To investigate potential additional phosphorylation targets, we aligned primary amino acid sequences of HEXIM proteins from human, mouse, chicken and fish. This comparison revealed that there is an evolutionally conserved region from positions 150 to 165, in which the serine at position 158 (S158) lies in a canonical PKC phosphorylation site (RxxS/TxK/R, Figure 1A) (41). Since this highly basic region also interacts with 7SK snRNA (42–45), the addition of a negative charge by phosphorylation of S158 should decrease the ability of HEXIM1 to bind to RNA.
Figure 1.
HEXIM1 is phosphorylated in vivo and is a substrate of PKC in vitro. (A) Serine at position 158 in HEXIM1 is an evolutionary conserved residue in a consensus PKC phosphorylation sequence. Primary sequence of the human HEXIM1 protein from positions 150 to 165 is aligned with the corresponding positively charged regions of HEXIM proteins from mouse, chicken, zebrafish and puffer fish, respectively. Whereas white letters in a black background indicate amino acid identity, black letters in a gray background indicate amino acid similarity. The consensus PKC phosphorylation sequence is presented above the alignment. (B) The PKC inhibitor chelerythrine chloride reduces the phosphorylation of HEXIM1 in vivo. Flag epitope-tagged HEXIM1 proteins (Hex) were expressed and labeled with [γ-32P]-ATP in HeLa cells and immunoprecipitated with anti-flag antibodies. Cells were also treated with chelerythrine chloride (PKC inhibitor). Upper and lower panels contain phosphorylated chimeras (autoradiograph, (P)Hex) and input (25%) of proteins (western blot, Hex), respectively. (C) PKC phosphorylates serine at position 158 in HEXIM1, which was purified from E. coli, in vitro. WT GST-HEXIM1 and mutant GST-HEXIM1 (S158A) chimeras (GST-Hex and GST-HexS158A, respectively), which were extensively purified from E. coli, were subjected to IVKAs by PKC as indicated. Upper and lower panels contain phosphorylated chimeras (autoradiograph, (P)Hex) and the input (25%) of proteins (western blot, Hex), respectively. (D) PKC phosphorylates serine at position 158 in HEXIM1, which was purified from HeLa cells, in vitro. Flag epitope-tagged WT HEXIM1 and mutant HEXIM1 (S158A) proteins (Hex and HexS158A, respectively), which were expressed in HeLa cells were immunoprecipitated with anti-flag antibodies, and subjected to IVKAs by PKC. Upper and lower panels contain phosphorylated chimeras (autoradiograph, (P)Hex) and the input (25%) of proteins (western blot, Hex), respectively.
To this end, we examined whether HEXIM1 can be phosphorylated by PKC in vitro and in vivo. First, flag epitope-tagged wild-type (WT) HEXIM1 (flag-Hex) protein was expressed in HeLa cells. Cells were labeled with [32P] orthophosphate with or without the PKC-inhibitor chelerythrine chloride (5 μM) for 1.5 h prior to cell lysis followed by immunoprecipitation with anti-flag antibodies and autoradiography. As presented in Figure 1B, flag-Hex was heavily phosphorylated ((P)Hex, lane 2, upper panel). The amount of phosphorylated HEXIM1 protein was dramatically reduced when cells were incubated with this PKC inhibitor (Figure 1B, lane 3, upper panel). The specificity and the efficiency of immunoprecipitations were confirmed by western blotting (Figure 1B, bottom panel, Hex). IVKAs using purified PKC and HEXIM1 proteins expressed and purified from Escherichia coli as GST-fusion proteins (Figure 1C) or the flag-Hex protein expressed in HeLa cells and immunoprecipitated with anti-flag antibodies (Figure 1D) further confirmed that HEXIM1 can be phosphorylated by PKC (Figure 1C and D, upper panels, lanes 1, (P)Hex). Of note, under these conditions P-TEFb cannot phosphorylate HEXIM1 and PKC did not phosphorylate CDK9 or CycT1 (data not presented). Changing the S158 in HEXIM1 to alanine (HexS158A) almost completely abolished this phosphorylation by PKC (Figure 1C and D, upper panels, lanes 2, (P)Hex), indicating that no other residues in HEXIM1 were phosphorylated by PKC. We conclude that PKC phosphorylates S158 in HEXIM1.
Phosphorylated HEXIM1 fails to bind to 7SK snRNA and to inhibit P-TEFb
Next, we examined effects of HEXIM1 phosphorylation on its ability to interact with RNA and to inhibit P-TEFb activity. His epitope-tagged WT HEXIM1 and mutant HEXIM1 (S158A) proteins (his-Hex and his-HexS158A) were expressed and purified from E. coli. Recombinant proteins were phosphorylated with the purified PKC protein in an in vitro reaction with cold ATP, and then repurified by Ni–NTA affinity chromatography. A parallel phosphorylation experiment with [γ-32P]-ATP confirmed that the WT his-Hex but not mutant his-HexS158A proteins were phosphorylated by PKC under these conditions (Figure 2A, lanes 1–4, (P)Hex).
To examine interactions between HEXIM1 and RNA, the in vitro phosphorylated and WT HEXIM1 proteins were tested for RNA binding in EMSAs. Increasing amounts of unlabeled dsRNA were incubated with 200 ng of WT (Figure 2B, lanes 2–6) or phosphorylated (P) (Figure 2A, lanes 8–12) HEXIM1 (Hex and (P)Hex) proteins for 15 min at room temperature. RNA–protein complexes were separated by a native PAGE followed by silver staining to detect free HEXIM1 proteins (Hex) and HEXIM1 associated with dsRNA (Hex:dsRNA). As presented in Figure 2B, the WT his-Hex protein exhibited a mobility shift when incubated with dsRNA (lanes 2–6, Hex:dsRNA) whereas no apparent mobility shifts were observed when dsRNA was added to the phosphorylated his-(P)Hex protein (lanes 8–12, (P)Hex). A parallel experiment with the mutant his-HexS158A protein demonstrated that it interacted with dsRNA and there was no difference in this mobility shift whether or not the proteins had been pre-incubated with PKC and ATP (data not presented).
An EMSA using recombinant GST-Hex fusion proteins and α-32P-labeled 7SK snRNA gave similar results (Figure 3). Whereas the WT GST-Hex and mutant GST-HexS158A proteins bound to 7SK snRNA in the presence or absence of P-TEFb, changing the S158 to glutamic acid (S158E) diminished the ability of HEXIM1 to interact with 7SK snRNA (Figure 3, lanes 1–6). The slight residual binding of the mutant GST-HexS158E protein to 7SK snRNA could be due to differences in the negative charge between the substituted glutamic acid and the phospho-serine in the phosphorylated HEXIM1 protein. These results indicate that HEXIM1, which is phosphorylated at S158 loses its ability to interact with RNA.
In addition, the WT and phosphorylated HEXIM1 proteins were examined for their ability to inhibit CDK9 kinase activity in the presence of 7SK snRNA. We performed P-TEFb kinase assays by incubating the purified recombinant P-TEFb complex (CDK9 and CycT1) with DSIF and [γ-32P]-ATP in the absence (Figure 2C, lane 1, DSIF) or presence of HEXIM1 and/or 7SK snRNA (Figure 2C, lanes 2–14, DSIF). Indeed, whereas CDK9 kinase activity was inhibited by the addition of increasing amounts of WT his-Hex protein and 7SK snRNA (Figure 2C, lanes 7–9, DSIF), the phosphorylated his-(P)Hex protein failed to inhibit P-TEFb even in the presence of dsRNA or 7SK snRNA (Figure 2C, lanes 9–14, DSIF). HEXIM1 or 7SK snRNA alone had no inhibitory effect on P-TEFb (Figure 2C, lanes 2–4, DSIF). We conclude that the phosphorylated HEXIM1 protein neither binds to RNA nor inhibits P-TEFb.
PKCθ stimulates the activity of P-TEFb and NF-kB
The above in vitro data implicated PKC in the phosphorylation of S158 in HEXIM1, which inactivated its ability to bind to 7SK snRNA and to inhibit P-TEFb. Next, we sought to determine the functional consequence of this HEXIM1 phosphorylation in vivo. As presented in Figure 1B, the treatment of cells with the pan-PKC inhibitor chelerythrine chloride inhibited HEXIM1 phosphorylation. Surprisingly, it also inhibited the phosphorylation of RNAPII (data not presented), which prompted us to examine if specific PKC inhibitors could also inhibit CDK9. Indeed, Ro-31-8220, one of the commonly used pan-PKC inhibitors exhibited strong inhibitory effects on PKC and CDK9 with similar IC50s (between 1 and 10 nM), when used in PKC- and P-TEFb-specific kinase assays using HEXIM1 and RNAPII as substrates, respectively (Supplementary Figure S1A and B). Indeed, other bisindolylmaleimide family of PKC inhibitors also inhibited CDK9 with IC50s comparable to those for PKC (Supplementary Figure S1C and D). In contrast, flavopiridol, a highly potent CDK9 inhibitor, did not inhibit PKC within the range of concentration required to inhibit CDK9 (Supplementary Figure S1C).
Since PKC inhibitors inhibit CDK9, we could not use them for the analysis of P-TEFb-dependent transcription in cells. Therefore, we examined directly effects of PKC on the activity of a reporter gene. To this end, we employed a thymidine kinase promoter-luciferase plasmid target that also contained five repeats of the Gal4 DNA-binding sequence (pTG5Luc), and a plasmid effector encoding the p65 (RelA) subunit of NF-kB linked to the Gal4 DNA-binding motif (Gal.p65). Previously, we demonstrated that this Gal.p65 chimera depended entirely on P-TEFb for its activity (46).
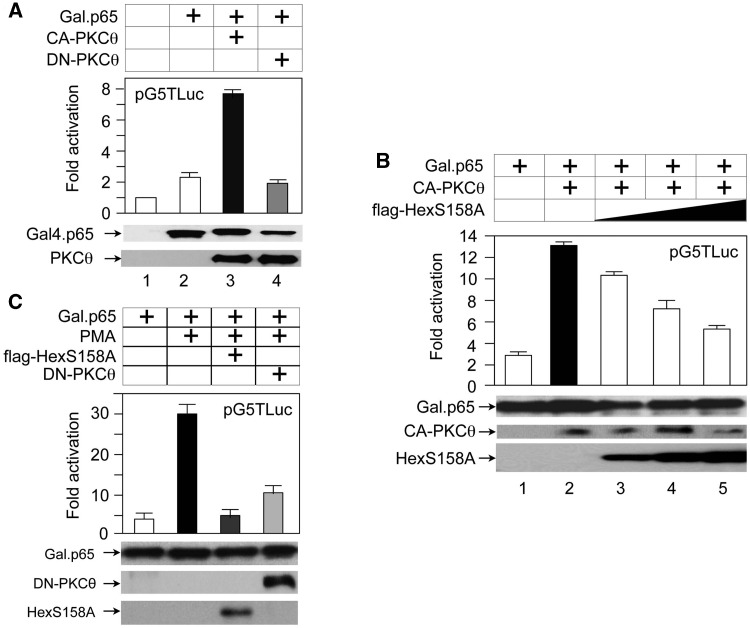
Although there are at least 15 isoforms of PKC in humans, we focused on PKCθ, which is expressed predominantly in T cells where HIV replicates (33–36). In addition, PKCθ can be found in the nucleus where it functions in transcriptional regulation (47). To this end, we coexpressed pTG5Luc and the Gal.p65 chimera in the presence or absence of CA or kinase-negative (DN) mutant PKCθ proteins (CA-PKCθ and DN-PKCθ) in 293 T cells. Whereas the expression of CA-PKCθ stimulated the activity of the Gal.p65 chimera 3-fold (Figure 4A, bars 2 and 3), the expression of DA-PKCθ had no or slightly negative effects (Figure 4A, bars 2 and 4). Next, we examined whether the exogenous expression of the WT HEXIM1 or mutant HEXIM1 (S158A) proteins blocks this PKCθ-dependent activation. Indeed, the WT flag-Hex and mutant flag-HexS158A proteins inhibited the CA-PKCθ-dependent activation of the Gal.p65 chimera on pG5Luc to a lesser and greater extent, respectively (Supplementary Figure S2, bars 3 and 4 and Figure 4B, bars 2–5). In the resting state, the WT HEXIM1 and mutant HEXIM1 (S158A) proteins bind to 7SK snRNA. Whereas CA-PKCθ can phosphorylate the WT protein at S158, it cannot phosphorylate the mutant HEXIM1 (S158A) protein. Thus, the WT HEXIM1 dissociates from 7SK snRNA, but the mutant HEXIM1 (S158A) protein remains bound to 7SK snRNA, thus exhibiting a more potent inhibitory effect on CA-PKCθ-mediated stimulation of p65 activity. Neither mutant PKCθ nor HEXIM1 proteins altered the expression of our protein effectors (Figures 4A and B, lower panels, Gal.p65, PKCθ, Hex). Thus, PKCθ increases the activity of NF-kB by targeting the 7SK snRNP.
Figure 4.
PKCθ stimulates NF-kB activity, which is blocked by mutant HEXIM1 (S158A) and/or a DN PKCθ proteins. (A) CA and DN PKCθ proteins (CA-PKCθ and DN-PKCθ) affect the activity of NF-kB (Gal.p65). These proteins were coexpressed with p5GTLuc in 293T cells. Luciferase activities were measured 48 h after the transfection. They are presented as fold activation over the value obtained with the plasmid target and equivalent amounts of the empty plasmid vector (lane 1). The expression of the Gal.p65 chimera (middle panel, Gal.p65) and PKCθ (bottom panel, PKCθ) was confirmed with anti-Gal4 and anti-PKCθ antibodies by western blotting. Error bars represent standard errors from three experiments performed in duplicate. (B) Increasing amounts of mutant HEXIM1 (S158A) inhibit effects of CA-PKCθ. These proteins, with increasing amounts of the mutant HEXIM1 (S158A), were coexpressed with p5GTLuc in 293T cells. Luciferase assays were performed as described above. The expression of Gal.p65, PKCθ and the mutant HEXIM1 (S158A) proteins was confirmed with anti-Gal4, anti-PKCθ, anti-Flag antibodies by western blotting, respectively. Error bars are as in (A). (C) Phorbol esters also activate NF-kB, which is blocked by mutant HEXIM1 (S158A) and/or a DN-PKCθ proteins. p5GTLuc was expressed alone or with these proteins in 293 T cells. Twenty-four hours after the transfection, cells were untreated (lane 1) or treated with PMA (20 nM)(lanes 2–4) for additional 24 h before performing luciferase assays. The expression of proteins was confirmed with anti-Gal4, anti-PKCθ, anti-flag antibodies by western blotting, respectively. Errors are as in (A).
Finally, we examined if the activation of endogenous PKC-signaling pathways can also stimulate P-TEFb. First, 293 T cells, which coexpressed pGT5luc and the Gal.p65 chimera, were treated with the PKC agonist PMA (10 nM) for 24 h prior to our luciferase assays. PMA increased Gal.p65-dependent luciferase activity 6-fold (Figure 4C, bars 1 and 2). Expressing the mutant flag-HexS158A (bar 3) or DN-PKCθ (lane 4) proteins abolished this stimulatory effect of PMA. Similar results were observed when the Gal4–CycT1 fusion protein was used as the protein effector instead of the Gal.p65 chimera (data not presented). We conclude that the phosphorylation of HEXIM1 at S158 by PKCθ activates P-TEFb in vivo.
CA-PKCθ shifts the P-TEFb equilibrium
Experiments presented in Figure 2 indicated that the phosphorylated HEXIM1 protein not only fails to bind to 7SK snRNA but loses its ability to inhibit the kinase activity of P-TEFb, which stimulates transcription. To investigate functional consequence of this HEXIM1 phosphorylation, we raised phospho-specific polyclonal antibodies against a HEXIM1 peptide containing the phosphorylated serine at position 158. These anti-phospho-HEXIM1 ((P)Hex) antibodies were further purified from serum by phosphopeptide-affinity chromatography. Their specificity was confirmed with purified GST-Hex and GST-HexS158A chimeras, which were pre-incubated with PKC and unlabeled ATP, by western blotting (Figure 5A, lane 2, (P)Hex). Next, we determined if PKCθ can phosphorylate HEXIM1 and release P-TEFb from the 7SK snRNP. Indeed, expressing the CA-PKCθ in Jurkat T cells, levels of phosphorylated HEXIM1 protein increased 5-fold, as determined with anti-(P)Hex antibodies by western blotting (Figure 5A, center, middle panel). We conclude that PKCθ can phosphorylate S158 in HEXIM1.
Previously, we established a method to separate quickly the 7SK snRNP from the free P-TEFb using step-wise fractionation of cell lysates (13). Using this method, we analyzed P-TEFb and HEXIM1 in the presence and absence of CA-PKCθ in Jurkat T cells. Cells were lysed first in low salt (10 mM KCl) to extract the 7SK snRNP (free nuclear fraction), then in a high salt (0.3 M KCl) buffer to extract free P-TEFb (chromatin bound fraction). Since up to 6-fold more HEXIM1 was detected in the high salt fraction, the expression of CA-PKCθ increased levels of free P-TEFb (Figure 5A, right panel, lanes 2 and 4, H). Glycerol gradient sedimentation also indicated that more CycT1, CDK9 and HEXIM1 were removed from the 7SK snRNP when CA-PKCθ was expressed in cells (Figure 5B, left panel, lanes 1–4, CA-PKCθ, free). The quantification of CycT1, HEXIM1 and Cdk9 in fractions corresponding to free P-TEFb (lanes 1–3) and 7SK snRNP (lanes 5–7) revealed that the expression of CA-PKCθ increased ratios between free P-TEFb and 7SK snRNP 2-, 1.7- and 1.8-fold, respectively (Figure 5B, center panel). These results indicate that CA-PKCθ phosphorylates HEXIM1, which shifts the P-TEFb equilibrium towards increased levels of free P-TEFb in cells.
T-cell receptor engagement also shifts the P-TEFb equilibrium in cells
To confirm that cellular-signaling pathways that activate endogenous PKCθ also activate P-TEFb via the phosphorylation of HEXIM1 and disruption of the 7SK snRNP, we analyzed changes in the P-TEFb equilibrium by stimulating Jurkat T cells with PMA or by engaging the TCR with anti-CD3 and anti-CD28 antibodies. Indeed, cell fractionation analyses indicated that PMA and TCR signaling increased the phosphorylation of HEXIM1 ((P)Hex, Figure 6A, left, lower panel, lanes 3 and 6) and the removal of HEXIM1 from the 7SK snRNP up to 8-fold (Hex, Figure 6A, left, upper panel, lanes 4 and 6, Hex and (P)Hex and right panel, bars 1–3). The expression of DN-PKCθ inhibited this shift of HEXIM1, further confirming the involvement of PKCθ in this process (Figure 6A, left panels, lanes 7 and 8, Hex and (P)Hex and right panel, bar 4). Glycerol gradient analyses also supported this finding (Figure 6B, left panels, lanes 1–4, free). Indeed, just 30 min after the addition of anti-CD3 and anti-CD28 antibodies to Jurkat cells, more CycT1 was detected in earlier fractions corresponding to free P-TEFb (Figure 6B, left, top and middle panels, lanes 1–4, CycT1, free). The expression of DN-PKCθ also inhibited this shift (Figure 6B, left bottom panel). Indeed, the addition of anti-CD3 and anti-CD28 antibodies increased the ratio between free P-TEFb and 7SK snRNP 3.3-fold, which was decreased to 1.9-fold by DN-PKCθ (Figure 6, right panel). HEXIM1 and Cdk9 exhibited similar patterns (data not presented). A similar result was obtained with another PKC agonist bryostatin-1 (data not presented). We conclude that the induction of PKC signaling via phorbol esters or TCR engagement leads to the phosphorylation of HEXIM1, which shifts the P-TEFb equilibrium towards more free P-TEFb in cells.
DISCUSSION
Our results demonstrate that the phosphorylation of HEXIM1 at S158 by PKCθ shifts the P-TEFb equilibrium away from the 7SK snRNP. Although we presented data on PKCθ, we also performed limited experiments with other PKC isoforms, namely PKCα, PKCδ and PKCι. In vitro, all of these three PKC isoforms and possibly others can phosphorylate S158 in HEXIM1. Of these, PKCι is found predominantly in the nucleus, where it also activates the activity of NF-kB (48,49). Thus, it is very likely that many PKC isoform that can access 7SK snRNP in the nucleus will activate P-TEFb by this mechanism. In this study, we concentrated on PKCθ because of previous reports of its involvement in TCR signaling, nuclear localization and transcriptional activity in T cells (33,34,36,47). While we were impressed by how conserved this consensus PKC target site is during evolution of HEXIM1 and is also present in HEXIM2, we found no other target sequence for PKC phosphorylation on other subunits of 7SK snRNP, which include CycT1 and CDK9. We were also surprised that thus phosphorylated, (P)Hex, like free P-TEFb, moved to chromatin from the soluble nuclear fraction. This movement could represent a conformational change in the protein and/or association with a new set of proteins that interact with chromatin. Of note, others had also noted this association between HEXIM1 with chromatin (50,51). Indeed, this chromatin-associated HEXIM1 protein, when dephosphorylated, could play an active role by helping to remove P-TEFb from abortive or arrested transcription as well as after termination and 3′-end processing events.
Changes in the P-TEFb equilibrium via intracellular signaling represent important steps in the global control of transcription elongation. While no unified mechanisms for the sequestration of P-TEFb exist, distinct post-translational modification of 7SK snRNP components, such as phosphorylation and acetylation play important roles in this process. For instance, our previous studies indicated that HBMA stimulates PI3K/Akt kinase pathway and phosphorylates HEXIM1 on the threonine and serine at positions 270 and 278 (14). Karn and colleague demonstrated that P-TEFb release via TCR signaling also involves the Erk kinase, although its exact target on the 7SK snRNP was not identified (20). In the study presented here, we observed a quantitatively similar release of P-TEFb via small molecule PKC agonists (i.e. PMA, bryostatin-1) and TCR signaling. However, we did not detect any phosphorylation of HEXIM1 by the Erk kinase (data not presented). This finding is not surprising, as none of our phosphorylation sites represents a canonical Erk phosphorylation motif (SPKTP) (52). Nevertheless, our results are in agreement with their observation that these agonists increase greatly the transcription from the HIV LTR (20). In addition to these phosphorylation events, PP2B and PP1a can disrupt the 7SK snRNP by dephosphorylating the threonine at position 186 (T loop) of CDK9 (18). Since some HDACis activate P-TEFb, the acetylation of CycT1 also plays an important role in the release of free P-TEFb. Indeed, thus acetylated, CycT1 preferentially associates with BRD4 (21,22). Thus, different stimuli disrupt the 7SK snRNP for signal-dependent stimulation of transcription elongation. Most likely, these stimuli set up new P-TEFb equilibria, each of which has small effects on global transcription. However, separate and convergent pathways should act synergistically with far greater effects on levels of free P-TEFb, thereby increasing transcription globally, which may result in cell growth and proliferation of target cells.
Besides leading to increased levels of free P-TEFb and activation of NF-kB-dependent transcription, there exist many publications on the role of PKC in biology and HIV replication, which include positive and negative effects (53–60). However, studies using specific PKC inhibitors must be reexamined in light of our findings because these agents, particularly bisindolylmaleimide group of compounds, exhibit very potent inhibitory effects on CDK9 (Supplementary Figure S1). In the case of Ro-31-8220, its IC50 for P-TEFb was similar to that for PKC. The inhibition of P-TEFb by Ro-31-8220 was also independent of the concentration of ATP, suggesting that it is a non-competitive inhibitor (data not presented). Importantly, the converse was not the case as flavopiridol, a CDK9 inhibitor, did not inhibit PKC. In summary, some transcriptional effects of PKC inhibitors must be reinterpreted in this new light.
Recent development in genome-wide analyses and bioinformatics revealed that the elongation –step of transcription plays a central role in the regulation of gene expression (2–4). Particularly, P-TEFb plays a critical role in this process. Thus, its function must be tightly controlled at multiple steps, which include levels of expression of its subunits and those of 7SK snRNP, their post-translational modifications, the CDK9 kinase activity and the recruitment of P-TEFb to target genes (17,23,61,62). In this study, we elucidated a new mechanism by which cellular signaling affects directly the integrity of the 7SK snRNP. Further studies of 7SK snRNP, its assembly and disassembly via discrete and convergent signaling pathways will shed light onto this complex regulation that likely controls the transcription of most cellular genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
The Academy of Finland [137077 and 140996 to M.B.]; Sigrid Juselius Foundation [4702687 to M.B.]; AmFAR [108241-51-RGRL to K.F. in part]; National Basic Research Program of China/973 Program [2011CB504200 and 2012CB910700]; National Natural Science Foundation of China [30970625 and 31171260]; Program for New Century Excellent Talents in University of Ministry of Education of China [NCET-10-0565 to Q.L.]; National Institutes of Health (NIH) [P50 GM082250 and R01 AI049104]. Funding for open access charge: NIH [R01 AI049104].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Art Weiss, Warner Greene and Alan Fields for reagents, and Dr Paul Luciw and the members of Peterlin laboratory for helpful discussions.
REFERENCES
- 1.Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 2.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterlin BM. Transcription elongation takes central stage: the P-TEFb connection. Cell Cycle. 2010;9:2933–2934. doi: 10.4161/cc.9.15.12698. [DOI] [PubMed] [Google Scholar]
- 4.Price DH. Poised polymerases: on your mark…get set…go! Mol. Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 11.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biglione S, Byers SA, Price JP, Nguyen VT, Bensaude O, Price DH, Maury W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contreras X, Schweneker M, Chen CS, McCune JM, Deeks SG, Martin J, Peterlin BM. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 2009;284:6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karn J. The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr. Opin. HIV AIDS. 2011;6:4–11. doi: 10.1097/COH.0b013e328340ffbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA. 2011;3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liou LY, Herrmann CH, Rice AP. Transient induction of cyclin T1 during human macrophage differentiation regulates human immunodeficiency virus type 1 Tat transactivation function. J. Virol. 2002;76:10579–10587. doi: 10.1128/JVI.76.21.10579-10587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YK, Mbonye U, Hokello J, Karn J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 2011;410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho S, Schroeder S, Kaehlcke K, Kwon HS, Pedal A, Herker E, Schnoelzer M, Ott M. Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 2009;28:1407–1417. doi: 10.1038/emboj.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, Lau J, Bisgrove D, Schnolzer M, Verdin E, et al. Two-pronged binding with bromodomain-containing Protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J. Biol. Chem. 2011;287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott M, Geyer M, Zhou Q. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe. 2011;10:426–435. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margolis DM. Confronting proviral HIV infection. Curr. HIV/AIDS Rep. 2007;4:60–64. doi: 10.1007/s11904-007-0009-6. [DOI] [PubMed] [Google Scholar]
- 25.Peterson S, Reid AP, Kim S, Siliciano RF. Treatment implications of the latent reservoir for HIV-1. Adv. Pharmacol. 2007;55:411–425. doi: 10.1016/S1054-3589(07)55012-X. [DOI] [PubMed] [Google Scholar]
- 26.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 27.Shen L, Siliciano RF. Viral reservoirs, residual viremia, and the potential of highly active antiretroviral therapy to eradicate HIV infection. J. Allergy Clin. Immunol. 2008;122:22–28. doi: 10.1016/j.jaci.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehla R, Bivalkar-Mehla S, Zhang R, Handy I, Albrecht H, Giri S, Nagarkatti P, Nagarkatti M, Chauhan A. Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PLoS One. 2010;5:e11160. doi: 10.1371/journal.pone.0011160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez M, de Vinuesa AG, Sanchez-Duffhues G, Marquez N, Bellido ML, Munoz-Fernandez MA, Moreno S, Castor TP, Calzado MA, Munoz E. Bryostatin-1 synergizes with histone deacetylase inhibitors to reactivate HIV-1 from latency. Curr. HIV Res. 2010;8:418–429. doi: 10.2174/157016210793499312. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez-Duffhues G, Vo MQ, Perez M, Calzado MA, Moreno S, Appendino G, Munoz E. Activation of latent HIV-1 expression by protein kinase C agonists. A novel therapeutic approach to eradicate HIV-1 reservoirs. Curr. Drug Targets. 2010;12:348–356. doi: 10.2174/138945011794815266. [DOI] [PubMed] [Google Scholar]
- 31.Barouch-Bentov R, Altman A. Protein kinase C-theta (PKCtheta): new perspectives on its functions in T cell biology. Adv. Exp. Med. Biol. 2006;584:1–13. doi: 10.1007/0-387-34132-3_1. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi K, Altman A. Protein kinase C theta (PKCtheta): a key player in T cell life and death. Pharmacol. Res. 2007;55:537–544. doi: 10.1016/j.phrs.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kane LP, Andres PG, Howland KC, Abbas AK, Weiss A. Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-gamma but not TH2 cytokines. Nat. Immunol. 2001;2:37–44. doi: 10.1038/83144. [DOI] [PubMed] [Google Scholar]
- 34.Lin X, O'Mahony A, Mu Y, Geleziunas R, Greene WC. Protein kinase C-theta participates in NF-kappaB activation induced by CD3-CD28 costimulation through selective activation of IkappaB kinase beta. Mol. Cell. Biol. 2000;20:2933–2940. doi: 10.1128/mcb.20.8.2933-2940.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Matsumoto R, You Y, Che T, Lin XY, Gaffen SL, Lin X. CD3/CD28 costimulation-induced NF-kappaB activation is mediated by recruitment of protein kinase C-theta, Bcl10, and IkappaB kinase beta to the immunological synapse through CARMA1. Mol. Cell. Biol. 2004;24:164–171. doi: 10.1128/MCB.24.1.164-171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witte V, Laffert B, Gintschel P, Krautkramer E, Blume K, Fackler OT, Baur AS. Induction of HIV transcription by Nef involves Lck activation and protein kinase C theta raft recruitment leading to activation of ERK1/2 but not NF kappa B. J. Immunol. 2008;181:8425–8432. doi: 10.4049/jimmunol.181.12.8425. [DOI] [PubMed] [Google Scholar]
- 37.Nojima M, Huang Y, Tyagi M, Kao HY, Fujinaga K. The positive transcription elongation factor b is an essential cofactor for the activation of transcription by myocyte enhancer factor 2. J. Mol. Biol. 2008;382:275–287. doi: 10.1016/j.jmb.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 39.Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res. 2005;33:7000–7010. doi: 10.1093/nar/gki997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija Peterlin B, Zhou Q. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loog M, Toomik R, Sak K, Muszynska G, Jarv J, Ek P. Peptide phosphorylation by calcium-dependent protein kinase from maize seedlings. Eur. J. Biochem. 2000;267:337–343. doi: 10.1046/j.1432-1327.2000.01002.x. [DOI] [PubMed] [Google Scholar]
- 42.Li Q, Cooper JJ, Altwerger GH, Feldkamp MD, Shea MA, Price DH. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 2007;35:2503–2512. doi: 10.1093/nar/gkm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yik JH, Chen R, Pezda AC, Samford CS, Zhou Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 47.Sutcliffe EL, Bunting KL, He YQ, Li J, Phetsouphanh C, Seddiki N, Zafar A, Hindmarsh EJ, Parish CR, Kelleher AD, et al. Chromatin-associated protein kinase C-theta regulates an inducible gene expression program and microRNAs in human T lymphocytes. Mol. Cell. 2011;41:704–719. doi: 10.1016/j.molcel.2011.02.030. [DOI] [PubMed] [Google Scholar]
- 48.Fields AP, Regala RP. Protein kinase C iota: human oncogene, prognostic marker and therapeutic target. Pharmacol. Res. 2007;55:487–497. doi: 10.1016/j.phrs.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Y, Jamieson L, Brasier AR, Fields AP. NF-kappaB/RelA transactivation is required for atypical protein kinase C iota-mediated cell survival. Oncogene. 2001;20:4777–4792. doi: 10.1038/sj.onc.1204607. [DOI] [PubMed] [Google Scholar]
- 50.D'Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010;17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wittmann BM, Fujinaga K, Deng H, Ogba N, Montano MM. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene. 2005;24:5576–5588. doi: 10.1038/sj.onc.1208728. [DOI] [PubMed] [Google Scholar]
- 52.Makagiansar IT, Williams S, Mustelin T, Stallcup WB. Differential phosphorylation of NG2 proteoglycan by ERK and PKCalpha helps balance cell proliferation and migration. J Cell. Biol. 2007;178:155–165. doi: 10.1083/jcb.200612084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bocklandt S, Blumberg PM, Hamer DH. Activation of latent HIV-1 expression by the potent anti-tumor promoter 12-deoxyphorbol 13-phenylacetate. Antiviral Res. 2003;59:89–98. doi: 10.1016/s0166-3542(03)00034-2. [DOI] [PubMed] [Google Scholar]
- 54.Folgueira L, McElhinny JA, Bren GD, MacMorran WS, Diaz-Meco MT, Moscat J, Paya CV. Protein kinase C-zeta mediates NF-kappa B activation in human immunodeficiency virus-infected monocytes. J. Virol. 1996;70:223–231. doi: 10.1128/jvi.70.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujiwara M, Okamoto M, Ijichi K, Tokuhisa K, Hanasaki Y, Katsuura K, Uemura D, Shigeta S, Konno K, Yokota T, et al. Upregulation of HIV-1 replication in chronically infected cells by ingenol derivatives. Arch. Virol. 1998;143:2003–2010. doi: 10.1007/s007050050436. [DOI] [PubMed] [Google Scholar]
- 56.Kim CH, Gollapudi S, Kim A, Lee T, Gupta S. Role of protein kinase C-beta isozyme in activation of latent human immunodeficiency virus type 1 in promonocytic U1 cells by phorbol-12-myristate acetate. AIDS Res. Hum. Retroviruses. 1996;12:1361–1366. doi: 10.1089/aid.1996.12.1361. [DOI] [PubMed] [Google Scholar]
- 57.Kim CH, Lim SJ, Gollapudi S, Gupta S. Role of protein kinase C isozymes in activation of human immunodeficiency virus type 1 in chronically infected promonocytic cells: evidence against a role of PKC beta 1. Biochem. Biophys. Res. Commun. 1994;199:292–297. doi: 10.1006/bbrc.1994.1227. [DOI] [PubMed] [Google Scholar]
- 58.Lopez-Huertas MR, Mateos E, Diaz-Gil G, Gomez-Esquer F, Sanchez del Cojo M, Alcami J, Coiras M. Protein kinase Ctheta is a specific target for inhibition of the HIV type 1 replication in CD4+ T lymphocytes. J. Biol. Chem. 2011;286:27363–27377. doi: 10.1074/jbc.M110.210443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qatsha KA, Rudolph C, Marme D, Schachtele C, May WS. Go 6976, a selective inhibitor of protein kinase C, is a potent antagonist of human immunodeficiency virus 1 induction from latent/low-level-producing reservoir cells in vitro. Proc. Natl Acad. Sci. USA. 1993;90:4674–4678. doi: 10.1073/pnas.90.10.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warrilow D, Gardner J, Darnell GA, Suhrbier A, Harrich D. HIV type 1 inhibition by protein kinase C modulatory compounds. AIDS Res. Hum. Retroviruses. 2006;22:854–864. doi: 10.1089/aid.2006.22.854. [DOI] [PubMed] [Google Scholar]
- 61.He N, Zhou Q. New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC) J. Neuroimmune Pharmacol. 2011;6:260–268. doi: 10.1007/s11481-011-9267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2011;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.