Abstract
Transcriptional repression of pathogen defense-related genes is essential for plant growth and development. Several proteins are known to be involved in the transcriptional regulation of plant defense responses. However, mechanisms by which expression of defense-related genes are regulated by repressor proteins are poorly characterized. Here, we describe the in planta function of CBNAC, a calmodulin-regulated NAC transcriptional repressor in Arabidopsis. A T-DNA insertional mutant (cbnac1) displayed enhanced resistance to a virulent strain of the bacterial pathogen Pseudomonas syringae DC3000 (PstDC3000), whereas resistance was reduced in transgenic CBNAC overexpression lines. The observed changes in disease resistance were correlated with alterations in pathogenesis-related protein 1 (PR1) gene expression. CBNAC bound directly to the PR1 promoter. SNI1 (suppressor of nonexpressor of PR genes1, inducible 1) was identified as a CBNAC-binding protein. Basal resistance to PstDC3000 and derepression of PR1 expression was greater in the cbnac1 sni1 double mutant than in either cbnac1 or sni1 mutants. SNI1 enhanced binding of CBNAC to its cognate PR1 promoter element. CBNAC and SNI1 are hypothesized to work as repressor proteins in the cooperative suppression of plant basal defense.
INTRODUCTION
Plants, unlike animals, do not possess specialized cells for protection against invading pathogens. When a pathogen challenge is detected, plant defense responses occur through the activation of cellular signal transduction pathways leading to global transcriptional reprogramming. These changes favor immune responses over normal cellular functions (1,2). Equally important is the suppression of immune responses in the absence of a pathogen threat that is necessary for proper plant growth and development. Thus, the induction of defense response to a specific pathogen occurs by a complex signaling network interconnected by crosstalk with networks that regulate response to other stressors, growth and development (3). Research on Arabidopsis thaliana has demonstrated that local and systemic resistance responses to biotrophic pathogens such as Pseudomonas syringae are mediated by the plant hormone salicylic acid (SA). Accumulation of SA leads to reduction of the oligomeric cytoplasmic form of the transcriptional co-activator NPR1/NIM1 (nonexpressor of PR genes1) to a monomeric form that translocates to the nucleus (4–8). Once there, NPR1 interacts with three redundant transcription factors, TGA2, TGA5 and TGA6 to activate expression of defense genes such as pathogenesis-related protein 1 (PR1) (9–13).
Constitutive activation of defense is detrimental to the normal growth of plants (14–17). Therefore, negative regulation of defense responses is very important. Negative regulators of the PR1 expression and resistance to P. syringae include SNI1 (suppressor of npr1-1, inducible1), NIM1-interacting1 and several WRKY transcription factors such as WRKY7, WRKY11 and WRKY17 (18–21). SNI1 was identified in a screen for suppressors of npr1 (18). SNI1 encodes a protein with structural similarity to Armadillo-repeat proteins that are involved in scaffolding or protein–protein interactions. The mechanism by which NPR1 and SNI1 interact to control PR1 expression is not clear. SA-inducible PR gene expression and resistance are restored in the npr1 sni1 double mutant suggesting that there is an NPR1-independent pathway of SA activation of PR1 transcription and that NPR1 blocks SNI1 activity. The deoxyribonucleic acid (DNA) recombination protein RAD15 seems to be involved in the regulation of PR1 expression by the NPR1-independent pathway (22). Both SNI1 and RAD51D were found to play roles in PR gene transcription and DNA recombination (22). Histone modifications are involved in SNI1-mediated repression (23).
Calcium signaling is another component of the defense response. Calcium signals are transduced in many ways including the binding of calcium to calmodulins (CaMs) or CaM-like proteins (24). The Ca2+/CaM complex modulates immune responses by repressing or activating transcription. Transcription of genes involved in SA biosynthesis is modulated by Ca2+/CaM (25). TGA and WRKY transcription factors are involved in controlling PR1 expression, and some members of these families of transcription factors are known to bind Ca2+/CaM. Details of CaM regulation of defense gene expression are not well-understood.
We show here a novel connection between SNI1 and Ca2+/CaM control of PR1 expression. We demonstrate that a previously identified CaM-binding NAC transcription repressor designated CBNAC (26) binds to cis-elements on the PR1 promoter that contain a GCTT core sequence and also interacts physically with SNI1. Genetic analyses showed that CBNAC functions as a negative regulator of pathogen-induced PR1 expression and basal resistance to a virulent strain of P. syringae. CBNAC and SNI1 were found to function synergistically as negative regulators of both PR1 expression and disease resistance.
MATERIALS AND METHODS
Plant and bacterial materials
All Arabidopsis plants used in this study were of the Columbia (Col-0) ecotype. The virulent bacterial pathogen, P. syringae pv. tomato (Pst) DC3000 was used for disease response tests. Escherichia coli BL21 (DE3) pLysS was used to express and produce recombinant GST-CBNAC protein. Arabidopsis transformation was performed as described previously (27).
Generation of transgenic plants
To generate transgenic plants, CBNAC complementary DNA (cDNA) with or without the FLAG tag was placed under the control of the CaMV 35S promoter. These constructs were cloned into pCAMBIA 1300 and transformed into Agrobacterium tumefaciens GV3101. Arabidopsis wild-type plants were transformed with the 35S:Flag-CBNAC construct according to a published protocol (27), and T3 progeny lines overexpressing CBNAC were selected for experiments. The 35S:CBNAC construct was used to transform cbnac1 plants and T3 progeny lines (cbnac1/CBNAC) expressing approximately the same level of CBNAC as wild-type plants in 1 mM SA-treated leaves were selected for experiments. MS medium containing 40 μg/ml hygromycin was used for selection of transformants.
Plant growth conditions
Arabidopsis thaliana plants were grown in growth chambers at 22°C and under 120 μEm−2 s−1 light intensity and 16-h-light/8-h-dark photoperiod.
Isolation of the cbnac1 and cbnac1 sni1 mutant lines
The cbnac1 (Salk_065051) T-DNA insertion mutant was identified from the Salk Arabidopsis T-DNA population (28). The T-DNA insertion was confirmed by polymerase chain reaction (PCR) using a T-DNA-specific primer (T-DNA) and a CBNAC-specific primer (CBNAC-S). A homozygous cbnac1 line was identified by PCR using a pair of primers corresponding to T-DNA flanking sequences (F1 and F2). The sni1 mutant was provided by Dr Xinnian Dong. The cbnac1 sni1 double mutant was obtained by crossing cbnac1 and sni1, selfing the F1 progeny and screening of the F2 population. The F2 population was screened for an absence of both genes by gene-specific PCR using the following primers: SNI1-specific primers (SNI1-S1 and SNI1-S2) and CBNAC-specific primers (F1 and F2). The primers used for PCR are listed in Supplementary Table S1.
Pseudomonas infection
Pseudomonas infection was carried out as described previously (29). Pseudomonas syringae DC3000 (PstDC3000) carrying empty vector (pVSP61) was grown at 28°C on King’s agar plates supplemented with 50 μg/ml rifampicin and 50 µg/ml kanamycin. In brief, bacteria were suspended in 10 mM MgCl2, adjusted to optical density (OD)600 = 0.001 and pressure infiltrated into leaves using a needleless syringe. Leaf discs from four independent plants were combined, ground in 10 mM MgCl2, serial-diluted 1:10 and plated onto King’s B medium containing the appropriate antibiotics. Plates were incubated at 28°C for 2 or 4 d, after which the colonies were counted.
Quantitative PCR and ribonucleic acid gel blot analysis
Total ribonucleic acid (RNA) was isolated using the guanidinium thiocyanate-phenol-chloroform extraction method with subsequent ultracentrifugation (29). Arabidopsis RNA was extracted using LiCl method, and cDNA was synthesized using the SuperScript™ II RNase-Reverse Transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed using the SsoFast EvaGreen Supermix (Bio-Rad) in a CFX96™ Real-Time PCR System (Bio-Rad). The primers used for qPCR are listed in Supplementary Table S2. Expression of CBNAC was detected by RNA gel blot analysis. RNA was separated on 1.5% agarose–formaldehyde gels and transferred to nylon membranes. Membranes were incubated with an (α-32P)dATP-labeled gene-specific probe at 65°C overnight and washed under high stringency conditions as described (29).
Electrophoretic mobility shift assays
For mapping of the CBNAC-binding promoter region of the PR1 gene, DNA probes were generated by PCR amplification with a Klenow fragment polymerase, α-32P-ATP (6000 Ci/mmol; Amersham) and the following primers, for E0, E1, E2, E3, E4, E5, E6, E0-1, E0-2, E0-3, E0-4, E3-1, E3-2, E4-1, E4-2, E4-3, E5-1, E5-2, E6-1 and E6-2 and by end labeling with polynucleotide kinase, γ-32P-ATP and the following primers, for E0-1-1, E0-1-2, E0-4-1, E0-4-2, E3-1-1, E3-1-2, E3-1-3, E4-1-1, E4-1-2, E5-3-1, E5-3-2, E6-1-1 and E6-1-2 (Supplementary Table S2). DNA-binding reactions were conducted at 25°C for 20 min in binding buffer [20 mM HEPES/KOH (pH 7.9), 0.5 mM DTT, 0.1 mM ethylenediaminetetraacetic acid (EDTA)], 50 mM KCl, 15% glycerol, 1 μg poly(dI-dC) and 0.5 μg bacterially produced fusion protein purified with glutathione-Sepharose. 32P-labeled DNA probes (40 000 cpm) were added and incubated with the mixture at 25°C for 30 min. The reactions were separated on an 8% polyacrylamide gel in 0.5× tris-borate-EDTA buffer at 80 V for 3 h. The gel was dried, mounted for autoradiography with intensifying screens and exposed at –70°C.
Chromatin immunoprecipitation assay
Chromatin samples were prepared as described previously (30). Wild-type and 35S:Flag-CBNAC overexpression lines were fixed with 1% formaldehyde for 10 min. The eluted DNA was analyzed by PCR using the following specific primers: E0, E3, E4, E5 and E6 (Supplementary Table S2). The amplified bands were visualized on a 2% agarose gel.
Yeast two-hybrid assay
CBNAC cDNA was digested with EcoRI and XhoI and ligated into the pAS2-1 plasmid (bait vector), which contains the Trp1 selection marker. The SNI1 cDNA was cloned into the pGAD424 plasmid (prey vector), which harbors the Leu2 selection marker. For mapping, the interacting domain deletions of CBNAC were PCR amplified using gene-specific primers and cloned in the pAS2-1 plasmid. Prey and different bait plasmids were co-transformed in the pJ69-4A (31) strain of yeast. Two-hybrid assays were performed as described in CLONTECH’s Yeast Protocols. Positive interactions were verified by the β-galactosidase assay.
Luciferase complementation imaging assay
Luciferase (Luc) complementation imaging (LCI) assay was carried out as described previously (32). CBNAC was fused with the C-terminal fragment of firefly Luc in pCAMBIA NLuc vector (CLuc-CBNAC). SNI1 was fused with the N-terminal fragment of Luc in pCAMBIA NLuc vector (SNI1-NLuc). STG1a-NLuc and CLuc-RAR1 constructs described previously were used as positive interaction controls (32). The constructs were each introduced into A. tumefaciens strain GV3101. Each bacterial strain was grown overnight in LB medium at 30°C, collected by centrifugation, then washed two times with infiltration buffer (10 mM MgCl2, 10 mM MES and 100 μM acetosyringone) and re-suspended in the same buffer. Equal volumes of bacterial suspensions of a CLuc and an NLuc construct were mixed and co-infiltrated into fully expanded leaves of the 3-week-old Nicotiana benthamiana plants using a needleless syringe. After infiltration, plants were placed at 23°C for 48 h. PstDC3000 (OD600 = 0.001 in 10 mM MgCl2) was treated after 24 h as Agro infiltration. The leaves were sprayed with luciferin solution (100 μM luciferin, 0.1% Triton X-100) and kept in the dark for 4 h to quench fluorescence. Luc activity (luminescence) was observed with a low-light cooled CCD imaging apparatus (AndoriXon; Andor).
RESULTS
CBNAC transcripts are induced by pathogen and SA treatment
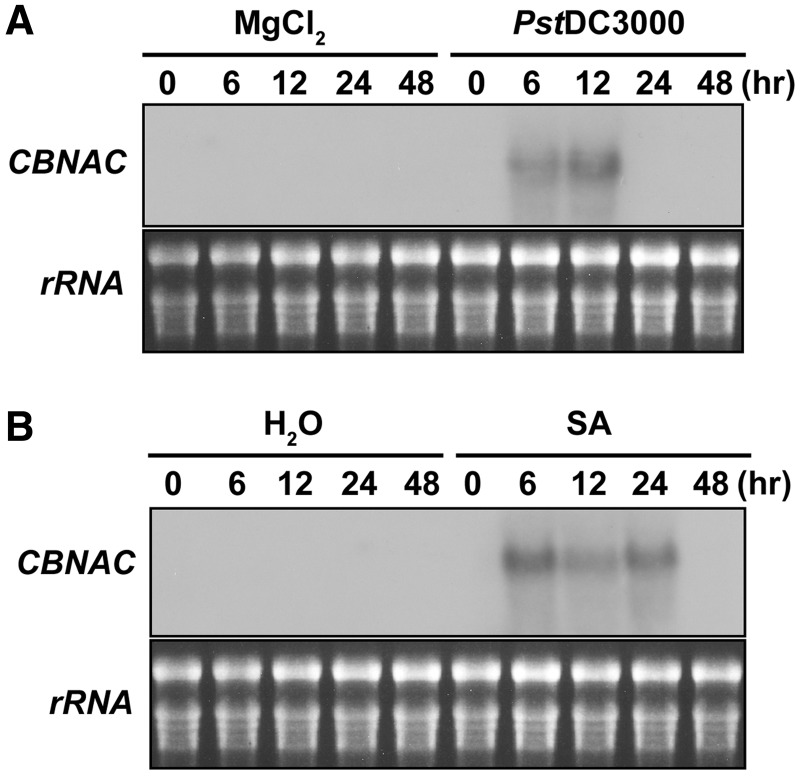
To identify the biological function of CBNAC, we investigated the gene expression of CBNAC in response to environmental stresses. The transcript level of CBNAC was examined after exposure to several biotic and abiotic stresses including bacterial pathogen, SA, jasmonic acid (JA), ABA, drought and NaCl. Interestingly, CBNAC transcript levels were increased in leaves of wild-type Arabidopsis plants after exposure to the virulent bacterial pathogen PstDC3000 or SA (Figure 1). CBNAC transcripts were undetectable in untreated leaves and remained undetectable over a 48-h period in leaves after infiltration with 10 mM MgCl2. In leaves infiltrated with PstDC3000, significant accumulation of CBNAC transcripts was observed at 6–12 h post-inoculation, and it returned to an undetectable level by 24 h (Figure 1A). Expression of CBNAC was also induced by SA, the inducer of systemic acquired resistance (Figure 1B). In SA-treated leaves, CBNAC transcripts reached maximum levels at 6 h, persisted for 24 h and declined to basal levels 48 h after treatment. These results suggested that CBNAC may be involved in SA-mediated pathogen resistance signaling in plants.
Figure 1.
CBNAC expression is induced by pathogen- and SA. (A) Induction of CBNAC gene expression by PstDC3000. Leaves of 4-week-old Arabidopsis plants (Col-0) were infiltrated with a bacterial suspension (OD600 = 0.001 in 10 mM MgCl2). Infitrated leaves were harvested at the indicated times after inoculation. The gel blot analysis of total RNA that was performed with a 32P-labeled CBNAC probe is shown. Ethidium bromide-stained rRNA is shown as loading control. (B) Induction of CBNAC gene expression by SA. Leaves of 4-week-old Arabidopsis plants (Col-0) were treated with 1 mM SA. Leaf collection, RNA isolation and RNA gel blot analysis was performed as in (A).
CBNAC is a negative regulator of the plant defense
The following genetic resources were developed for investigation of CBNAC function in vivo. A Salk line (Salk_065051) carrying a T-DNA insertion in CBNAC (cbnac1) was identified and homozygous F2 progeny derived from this line were used for analyses. Location of the T-DNA insertion at the third exon of the CBNAC gene was confirmed by PCR analyses of genomic DNA (Supplementary Figure S1A). There was no detectable accumulation of CBNAC transcripts in untreated leaves of the wild-type and cbnac1 mutant plants. Complete loss of CBNAC expression was revealed by comparing the transcript abundance in SA-treated leaves of wild-type and cbnac1 plants (Supplementary Figure S1B). Attempts to identify additional independent cbnac mutants were unsuccessful. Thus, a 35S:CBNAC construct was used to transform the cbnac1 mutant, and transgenic lines (cbnac1/CBNAC) exhibiting CBNAC expression levels similar to wild-type plants were chosen for complementation analysis (Supplementary Figure S2A). Transgenic lines constitutively overexpressing CBNAC (35S:Flag-CBNAC) were generated in the wild-type background. Western blot analysis using anti-FLAG antibody revealed several transgenic plants contained elevated levels of CBNAC protein regardless of SA treatment (Supplementary Figure S1C).
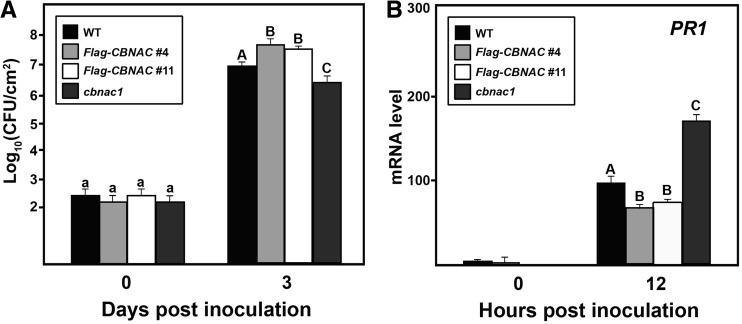
There were no obvious differences in growth or development characteristics of the wild-type, cbnac1 or 35S:Flag-CBNAC plants. The effect of CBNAC expression level on susceptibility to the virulent bacterial pathogen PstDC3000 was then examined. Three days after inoculation, bacterial growth in infiltrated leaves was slightly lower in the null cbnac1 mutant and slightly higher in 35S:Flag-CBNAC overexpression lines compared with the wild type (Figure 2A). There was no significant difference in bacterial growth in leaves of the wild-type plant and a cbnac1/CBNAC complementation line (Supplementary Figure S2B). As loss of CBNAC function is associated with resistance and overexpression of CBNAC is associated with susceptibility in un-induced plants, CBNAC is a negative regulator of basal defense against the bacterial pathogen PstDC3000.
Figure 2.
CBNAC negatively regulates resistance to PstDC3000 and PR1 expression. Leaves of wild type (WT), 35S:Flag-CBNAC and cbnac1 plants were inoculated with a bacterial suspension (OD600 = 0.001 in 10 mM MgCl2). (A) Growth of PstDC3000 in inoculated leaves at 0 and 3 dpi. Mean bacterial densities ± SE were calculated from six to eight replicate plants are shown. Significant differences as calculated by Student’s t test (P < 0.05) are indicated by unique letters. The experiment was repeated at least three times with similar results. (B) qRT-PCR analysis of PR1 expression in inoculated leaves. Values were normalized using the expression level of Tubulin 2 and expressed relative to the expression level in WT at 12 hpi, which is arbitrarily set at 100. Mean relative expression values ± SE from three independent experiments are shown. Data were analyzed by Student’s t test. Different letters indicate statistically significant differences between genotypes (P < 0.05).
Induction of PR1 is a marker of SA-mediated defense signaling that leads to Arabidopsis resistance to bacterial pathogens (2). PR1 transcript accumulation was not observed in leaves of wild-type, cbnac1, cbnac1/CBNAC or 35S:Flag-CBNAC plants before inoculation with PstDC3000 (Figure 2B and Supplementary Figure S2C). At 12 h post-inoculation when the expression of CBNAC was expected to peak in wild-type plants (Figure 1A), accumulation of PR1 transcript was significantly suppressed in 35S:Flag-CBNAC plants, induced in the cbnac1 mutant and unchanged in the cbnac1/CBNAC complementation line in comparison with wild type (Figure 2B and Supplementary Figure S2C). Thus, the level of CBNAC expression correlates inversely with resistance to pathogen infection and PR1 expression, suggesting that PR1 may be a direct target of CBNAC.
CBNAC binds to the PR1 promoter by a GCTT core element
In cbnac1 knockout plants, the levels of PR1 transcripts were higher compared with those in wild-type plants. To examine whether this effect was attributable to direct binding of the CBNAC to the PR1 promoter, we investigate the direct interaction of CBNAC to the PR promoter by electrophoretic mobility shift assays (EMSA). EMSA using recombinant GST-CBNAC fusion protein and PR1 promoter fragments up to 1041-bp upstream of the PR1 start codon were used to demonstrate that CBNAC protein can bind to the PR1 promoter. Using seven ∼110-bp overlapping fragments of the PR1 promoter sequence (Supplementary Figure S3A), five regions (E0, E3, E4, E5 and E6) were found to bind to CBNAC (Supplementary Figure S3B).
Within the five regions, smaller overlapping fragments (∼52 bp) were used to define the specific target of CBNAC (Supplementary Figure S4A). Six regions (E0-1, E0-4, E3-1, E4-1, E5-3 and E6-1) demonstrated binding to CBNAC (Supplementary Figure S4B). Overlapping oligonucleotides within the E0-1, E0-4, E3-1, E4-1, E5-3 and E6-1 fragments were used to further delineate CBNAC-binding sequences (Supplementary Figure S5A). CBNAC bound six PR1 promoter fragments (Supplementary Figure S5B and Table 1). Of these, E0-1-1 exhibited very strong CBNAC binding. E0-4-2, E3-1-2 and E5-3-1 had moderate affinity. The two remaining elements, E4-1-1 and E6-1-1, bound very weakly. The E0-1-1 and E0-4-2 fragments contain the GCTT core sequence that was previously identified as a CBNAC-binding sequence by the random binding site selection method (26). E3-1-2 has been previously identified as a negative regulatory element on the PR1 promoter (12).
Table 1.
Putative CBNAC-binding cis-acting elements in the PR1 promoter
| cis element | Sequencea | Positionb | Binding affinityc |
|---|---|---|---|
| E0-1-1 | TAATAATGCTTAGTTATAAATTACT | (–) 1209 ∼ (–) 1185 | +++ |
| E0-4-2 | TGTTATTGCTTAGAATCACAGATTC | (–) 994 ∼ (–) 970 | ++ |
| E3-1-2 | CTATTGACTGTTTCTCTACGTCACTATT | (–) 715 ∼ (–) 688 | ++ |
| E4-1-1 | ATACTCATATGCATGAAACACTAAGAAAC | (–) 618 ∼ (–) 590 | + |
| E5-3-1 | ATATACAATGTTTCTTAATAAACTTCATTT | (–) 340 ∼ (–) 311 | ++ |
| E6-1-1 | AAAAAAATATATCAACAATGGCAAAGCT | (–) 288 ∼ (–) 261 | + |
aSequences are indicated from 5′ to 3′. GCTT core sequence is underlined.
bPositions of the cis elements with respect to the translation start site (ATG).
cThe relative binding affinity (+) was determined by densitometry of autoradiograms of DNA-bound CBNAC.
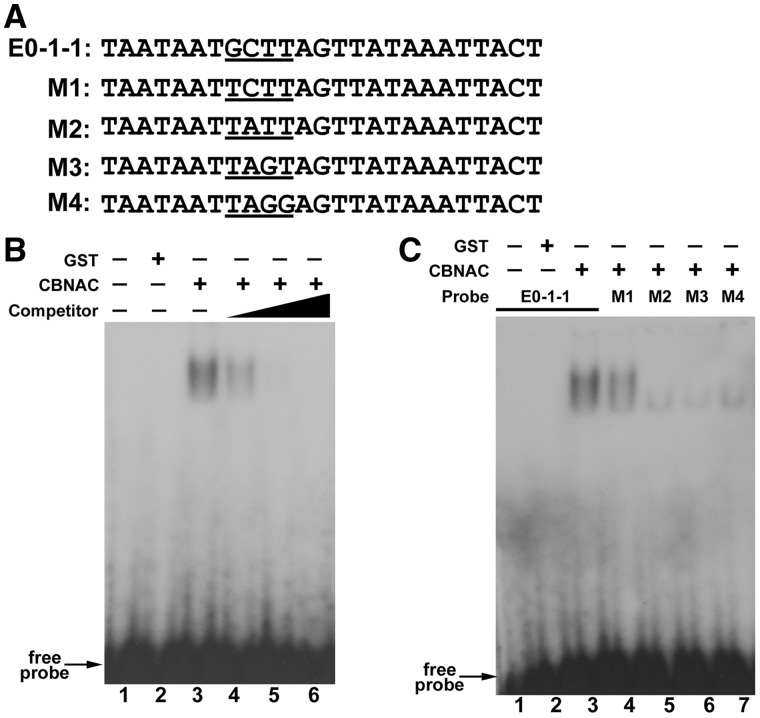
Interactions between CBNAC and the E0-1-1 element were further analyzed. Figure 3A depicts the four mutant E0-1-1 elements (M1–M4) used in the EMSA. As shown in Figure 3B, CBNAC bound strongly and specifically with radiolabeled E0-1-1. A mobility shift was not observed on incubation of the labeled E0-1-1 element with GST alone. Furthermore, addition of unlabeled E0-1-1 element inhibited binding of labeled E0-1-1 to CBNAC in a concentration-dependent manner (Figure 3B). Compared with the interaction of CBNAC with E0-1-1, its binding with M1 was weaker and was almost negligible with M2, M3 and M4 (Figure 3C). These results indicated that the GCTT core sequence of E0-1-1 is critical for CBNAC binding.
Figure 3.
CBNAC interacts with the E0-1-1 element of the PR1 promoter. (A) Nucleotide sequence of the native and mutated (M1–M4) E0-1-1 elements used in EMSA. (B) Analysis of binding specificity. EMSA was performed using 32P-labeled native E0-1-1 as probe as above except that GST-CBNAC protein was preincubated with 50- (lane 4), 100- (lane 5) or 200- (lane 6) fold molar excess of cold native E0-1-1 (competitor) before addition of probe. (C) EMSA of CBNAC binding. 32P-labeled native (lanes 1–3) and mutated (lanes 4–7) E0-1-1 probes were incubated with equal amounts of E. coli-expressed GST-CBNAC (lanes 3 to 7) or GST alone (lanes 1 and 2) before electrophoresis.
Chromatin immunoprecipitation (ChIP) experiments were then used to determine whether CBNAC binds the PR1 promoter in vivo. In these experiments, cross-linked chromatin from leaves of wild-type and 35S:Flag-CBNAC plants were incubated with FLAG-specific monoclonal antibodies enriching for CBNAC and PR1 promoter complexes. The five PR1 promoter regions shown to bind CBNAC in vitro (E0, E3, E4, E5 and E6) were then amplified with specific primers. Three independent ChIP experiments were performed, and representative results from a single assay are shown (Supplementary Figure S5C). PCR analyses of input chromatin samples verified that similar quantities of ChIP starting materials were used for each primer pair. As expected, immunoprecipitation reactions lacking anti-FLAG antibody did not result in the recovery of chromatin fragments containing the PR1 promoter (Supplementary Figure S5C, No Ab). Only the E0 (–1209 to –970 bp) region was amplified in immunoprecipitates obtained with anti-FLAG antibody (Supplementary Figure S5C, α-Flag Ab), confirming that CBNAC directly binds to a region of the PR1 promoter that contains the GCTT core sequence.
CBNAC physically interacts with SNI1
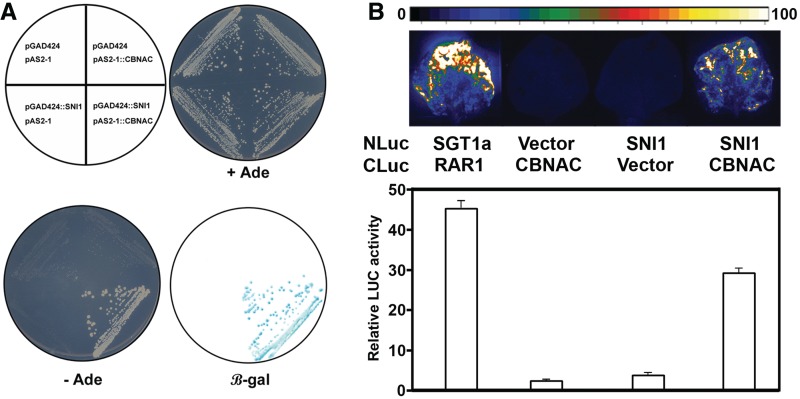
Similar to CBNAC, SNI1 is a negative regulator of PR1 expression. However, unlike CBNAC, SNI1 does not have a DNA-binding domain. Therefore, the possibility of interaction between CBNAC and SNI1 was examined in a yeast two-hybrid assay. The bait construct (pAS2-1::CBNAC) contained full-length CBNAC cDNA fused to the GAL4 DNA-binding domain. The prey construct (pGAD424::SNI1) contained full-length SNI1 cDNA fused to the GAL4 activation domain. As shown in Figure 4A, yeast cells expressing either bait or prey construct alone were not able to grow on selection media. Growth on selective medium and expression of the LacZ reporter gene was observed only if yeast contained both pAS2-1::CBNAC and pGAD424::SNI1, indicating specific interaction between CBNAC and SNI1.
Figure 4.
CBNAC interacts with SNI1. (A) Yeast two-hybrid analysis. Transformants of yeast strain pJ69-4A were grown as indicated (upper left) on minimal medium with (+Ade) or without (–Ade) selection. Adenine prototrophy indicates positive interaction. β-Galactosidase activity in the colonies grown in +Ade medium was determined by filter-lift assay (LacZ). (B) LCI assay for detecting interaction in planta. Tobacco leaves were transformed by Agrobacterium infiltration using a needleless syringe. The indicated NLuc and CLuc construct pairs were used for transformation. Shown are luminescence images (upper panel) and quantitative luminescence measurements (lower panel) depicting luciferase activity in inoculated leaves at 48 hpi.
Interaction of CBNAC with SNI1 in planta was then examined using the LCI assay in N. benthamiana leaves using transient expression (32). The positive control combination of STG1a-NLuc and CLuc-RAR1 resulted in strong Luc activity as previously reported (32) (Figure 4B). The negative control combinations of CLuc-CBNAC/NLuc vector and SNI1-NLuc/CLuc vector did not show Luc activity (Figure 4B). Luc activity was detected with the combination of SNI1-NLuc/CLuc-CBNAC showing that CBNAC interacts with SNI1 in planta.
Additional yeast two-hybrid assays were performed to define the CBNAC domain responsible for interaction with SNI1. A schematic diagram of the CBNAC deletion constructs used for these assays is shown in Supplementary Figure S6A. The NAC domain of CBNAC was not required for interaction with SNI1 (Supplementary Figure S6B). The C-terminal region (301–512 amino acids) of CBNAC interacted with SNI1. The interaction between CBNAC and SNI1 required the CaM-binding domain as well as other sequences.
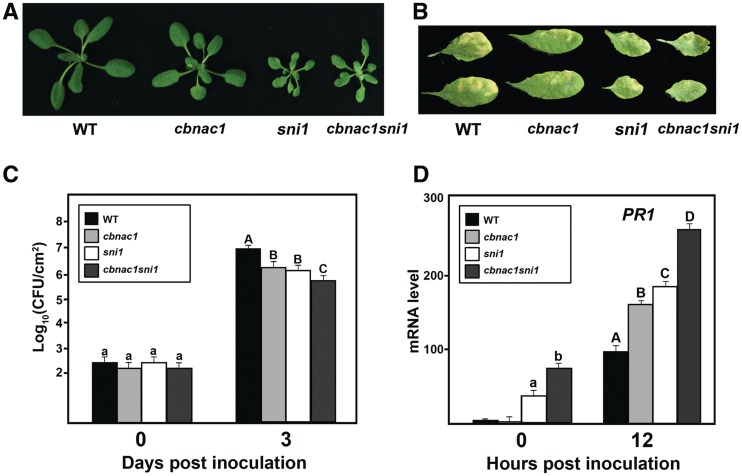
CBNAC and SNI1 function synergistically as negative regulators of disease resistance
A cbnac1 sni1 double mutant was generated through genetic crossing and analyzed for potential interactions between CBNAC and SNI1. Morphological phenotypes of the cbnac1 sni1 mutant resembled those of the sni1 mutant rather than the cbnac1 mutant. Both sni1 and cbnac1 sni1 plants had smaller rosettes compared with the wild type (Figure 5A). Similar to the sni1 mutant, cbnac1 sni1 plants exhibited pleiotropic phenotypes, including decreased leaf size and altered leaf texture (Figure 5A and B). In contrast, the cbnac1 plants showed no differences in growth, development or morphology in comparison with the wild type (Figure 5A and B). As shown in Figure 5C, the cbnac1 and sni1 mutants had similar levels of basal resistance to PstDC3000 (Figure 5C). Bacterial counts in inoculated leaves of both lines were slightly lower than in leaves of the wild type 3 days post-inoculation. Disease symptoms were also less severe in the cbnac1 and sni1 mutants than in wild-type leaves (Figure 5B). The cbnac1 sni1 double mutant was markedly more resistant to PstDC3000 than either the cbnac1 or sni1 single mutants (Figure 5C). Bacterial counts in inoculated leaves of the cbnac1 sni1 mutant were 10-fold lower than in leaves of the wild type at 3 days post-inoculation. This marked reduction in bacterial growth in inoculated leaves of the cbnac1 sni1 mutant compared with the cbnac1 or sni1 mutants was accompanied by the substantially reduced disease symptom development (Figure 5B).
Figure 5.
Altered responses of the cbnac1 sni1 double mutant to PstDC3000. (A) Morphology of 5-week-old wild-type (WT), cbnac1, sni1 and cbnac1 sni1 plants grown on MS agar plates. (B–D) Disease resistance responses in leaves inoculated with bacterial suspension as in Figure 3. Disease symptoms in inoculated leaves at 5 dpi are depicted (B). Bacterial growth in inoculated leaves at 0 and 3 dpi are compared (C). Mean bacterial densities ± SE were calculated from six to eight replicate plants. Significant differences as calculated by Student’s t test (P < 0.05) are indicated by unique letters. The experiment was repeated at least three times with similar results. PR1 expression was monitored in inoculated leaves by qRT-PCR (D). Values were normalized using the expression level of Tubulin 2 and expressed relative to the expression level in WT at 12hpi, which is arbitrarily set at 100. Mean relative expression values ±SE values from three independent experiments are shown. Data were analyzed by Student’s t test. Different letters indicate statistically significant differences between genotypes (P < 0.05).
To investigate the enhanced basal resistance of the double mutant, PR1 expression was analyzed following bacterial pathogen infection. The level of PR1 mRNA was comparable in uninoculated leaves of wild type and cbnac1 plants (Figure 5D). SNI1 has been reported to repress basal PR1 expression (18). Accordingly, the level of PR1 mRNA was higher in uninoculated leaves of the sni1 mutant than in wild type. At 12 h post-inoculation, PR1 transcript abundance was elevated in the cbnac1 and sni1 mutants compared with wild type, as expected if they function as transcriptional repressors. The PR1 levels in uninoculated and inoculated leaves of the cbnac1 sni1 double mutant were significantly higher than in leaves of cbnac1 or sni1 single mutants under the same condition, suggesting a synergistic effect of the combined cbnac1 and sni1 mutations on PR1 expression (Figure 5D). Together, these results show that CBNAC and SNI1 function synergistically as negative regulators of basal resistance to the virulent bacterial pathogen PstDC3000 due, in part, to their overlapping activities as negative regulators of PR1 expression.
SNI1 enhances the DNA-binding activity of the CBNAC
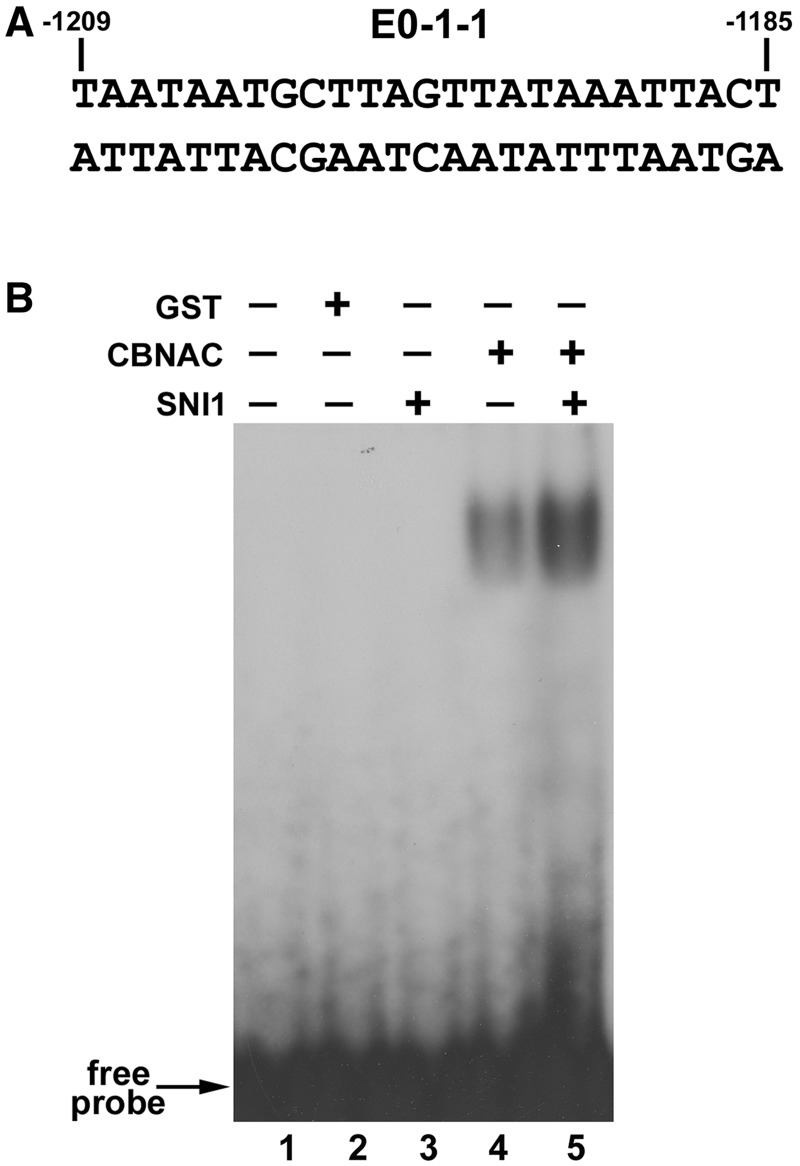
Although SNI1 functions as a transcriptional repressor of PR1 expression, it lacks a DNA-binding domain (23). Therefore, the possibility that SNI1 could influence the DNA-binding activity of CBNAC toward the PR1 promoter cis-element (E0-1-1) was investigated because SNI1 interacts with CBNAC.
The PR1 promoter element (E0-1-1), previously identified by EMSA, contains the preferred GCTT core binding sequence for CBNAC (Figure 6A). Incubation of SNI1 with the E0-1-1 probe did not result in a retarded band in an EMSA (Figure 6B, lane 3) indicating that SNI1 does not directly bind to this element. Addition of SNI1 to the CBNAC-binding mixture increased CBNAC binding to the E0-1-1 element without producing a supershift band, indicating that the DNA–protein complex contained CBNAC but not SNI1 (Figure 6B, lanes 5 and 4). These results suggested that SNI1 enhances the binding of CBNAC to the E0-1-1 element.
Figure 6.
SNI1 enhances binding of CBNAC to the E0-1-1 element. (A) Nucleotide sequence of the PR1 promoter indicating E0-1-1 element. The numbers indicate the position of the element relative to the PR1 translation start site. (B) EMSA analysis of the effect of SNI1. EMSA was performed using 32P-labeled E0-1-1 element (lanes 1 to 5), without (lane 1) or with the addition GST (lane 2; negative control), CBNAC (lanes 4 and 5) and SNI1 (lane 5). Equal amounts of CBNAC were used in the two lanes.
DISCUSSION
CBNAC acts as a negative regulator of plant defense responses
Discrepancies exist in several studies that had reported on the biochemical properties of CBNAC/NTL9 (26,33). First, CBNAC/NTL9 had been reported to contain a C-terminal transmembrane domain and that CBNAC/NTL9 was localized in the plasma membrane (33). However, the predicted C-terminal transmembrane domain of CBNAC/NTL9 was identified as a CaM-binding domain (26). Furthermore, the GFP-tagged CBNAC/NTL9 protein was considered to be dominantly localized in nuclei (26). Finally, on a different level of function, CBNAC/NTL9 was reported to be involved in regulating signaling during leaf senescence (33). The biological function of CBNAC/NTL9 was based on the phenotypes of transgenic plants overexpressing the C-terminal truncated form of CBNAC/NTL9, but the cbnac1/ntl9-1 mutant and transgenic plants overexpressing the full-length CBNAC/NTL9 did not exhibit any phenotypes. It seems likely that the biological function of CBNAC/NTL9 could be misinterpreted from the phenotypes reported for transgenic plants overexpressing CBNAC/NTL9 devoid of its regulatory domain. In essence, the physiological function of CBNAC/NTL9 has not been elucidated to sufficient detail.
CBNAC is a CaM-binding transcriptional repressor that interacts with DNA through a GCTT core sequence flanked on both sides by frequently repeating sequences (26). This study demonstrates CBNAC binds to the promoter of PR1, a marker gene for SA-induced defense responses and also inhibits pathogen-induced increase in PR1 expression. CBNAC alone, and in synergism with SNI1, negatively regulates basal resistance to PstDC3000. As CaM regulates the repressor function of CBNAC (26), these results imply that CBNAC is a point of crosstalk between calcium- and SA-mediated defense signaling.
Linker-scanning (LS) mutagenesis of the PR1 promoter revealed at least three cis-elements (LS5, LS7 and LS10) displaying positive or negative characters (34). LS5 and LS7 both contain the binding sequence (TGACG) for TGA transcription factors and TGA2 independently binds the LS5 and LS7 promoter elements in vitro (10). The LS5 element seems to contribute to the negative regulation of PR1 expression both in the absence and presence of SA, whereas LS7 is required for SA-mediated PR1 induction (34). The GCTT cis-element for CBNAC binding to the PR1 promoter represents a novel PR1 regulatory module. Interestingly, CBNAC is recruited to the PR1 promoter in both non-treated and SA-treated tissues (Supplementary Figures S5C and S7). SA-independent binding of CBNAC to the PR1 promoter suggests that the DNA-binding activity of CBNAC is not altered by SA.
Both PstDC3000 and SA induced CBNAC quickly in a transient way (Figure 1). The induction of the negative regulator during the early stages of infection might serve as a mechanism to prevent gratuitous or even harmful overactivation of pathogen-induced defense mechanisms when the population of the pathogen remains at relatively low levels (35). As pathogen growth increases, enhanced defense mechanisms would be necessary, and this could be achieved at least partially by suppressed expression and inactivation of negative regulators such as CBNAC and WRKY factors.
Pathogen-induced expression of negative defense regulators could be explained by their involvement in the antagonistic crosstalk of distinct signaling pathways against various types of microbial pathogens. SA-mediated signaling activates defense mechanisms effective against biotrophic pathogens but can suppress ET/JA-mediated signaling in defense against necrotrophic pathogens (3). For example, overexpression of the transcription factor WRKY33 enhances susceptibility to the biotrophic pathogen PstDC3000 and increases resistance to necrotrophic fungal pathogens (36). cbnac1 plants showed enhanced resistance to biotrophic pathogen PstDC3000 but no difference to necrotrophic fungal pathogens. Consistently, the transcription levels of JA-responsive marker genes, PDF1.2 and LOX2, were similar in cbnac1 mutant and wild-type plants (Supplementary Figure S8). These results suggest that CBNAC is not involved in the antagonistic crosstalk of defense signaling pathways against biotrophic and necrotrophic pathogens. It is possible that CBNAC is involved in crosstalk between SA- and NPR1-mediated defense signaling and as yet unidentified environmental stress signaling.
CBNAC acts synergistically with SNI1 as a transcriptional repressor of the PR1 gene
SNI1, a transcriptional repressor of PR gene expression, has no apparent DNA-binding domain and is predicted to bind to DNA indirectly by interactions with DNA-binding proteins (23). CBNAC has been identified as one DNA-binding protein that interacts with SNI1 (Figure 5). SNI1 enhances the DNA-binding activity of CBNAC to a PR1 promoter element (Figure 6B). There are several reported instances where an interaction partner enhances or inhibits DNA-binding activity of a transcription factor. The human viral protein TAX stimulates the DNA binding of bZIP proteins (37), whereas Arabidopsis NPR1 stimulates the DNA binding of TGA factors to the LS7 element (10). The tobacco ankyrin repeat protein ANK1 inhibits the binding of the bZIP factor BZI-1 to its cognate promoter element in EMSA without altering the complex mobility (38). Similarly, SNI1 enhanced the binding of CBNAC to its cognate promoter element in EMSA without altering the complex mobility (Figure 6B). There are two possible explanations. First, SNI1 may have been released from the complex by a conformational change resulting from CBNAC binding to DNA. Second, other interacting proteins may be required for a stable interaction between CBNAC and SNI1.
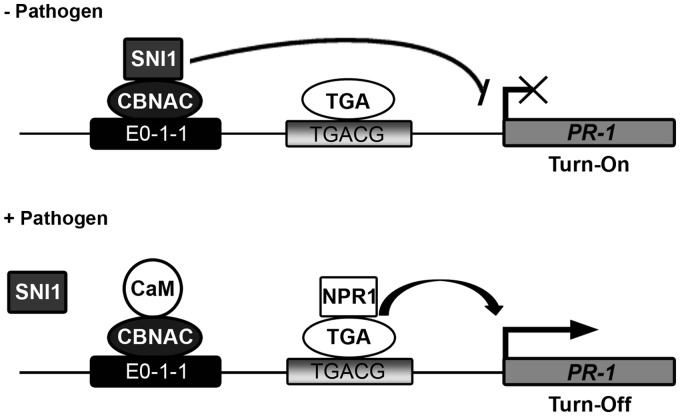
Model for PR1 regulation
Based on the information generated in this and previous studies, the following model is proposed (Figure 7). The PR1 promoter contains both positive TGA-binding (TGACG) and negative CBNAC-binding (E0-1-1) elements. Genetic analyses showed that CBNAC interacts synergistically with SNI1 as a transcriptional repressor of PR1 gene expression and basal resistance to PstDC3000 (Figure 5C and D). Unlike cbnac1 plants, sni1 and cbnac1 sni1 plants exhibit constitutive PR gene expression and other pleiotropic phenotypes (Figure 5A). Therefore, in absence of pathogen induction, SNI1 is proposed to function as the major negative regulator of PR gene expression whose activity is governed, in part, by its interaction with the transcriptional repressor CBNAC at the E0-1-1 element. The constitutive binding of transcriptional activators (such as TGA1, TGA3 and TGA6) to the positive element in a non-induced state is still a possibility. However, this binding would not lead to gene expression in the presence of CBNAC and SNI1.
Figure 7.
Model for the regulation of PR1 by the CBNAC-SNI1 complex. In non-induced conditions (–Pathogen), because SNI1 does not contain a known DNA-binding domain, we postulate that SNI1 binds to CBNAC and is thereby recruited the E0-1-1 element of PR1 promoter. SNI1 enhances the DNA-binding activity of CBNAC and somehow this enhances repression of PR1 by SNI1. In the presence of inducer (+Pathogen), PR1 gene expression is induced by the translocation of a large amount of active NPR1 to the nucleus and its interaction with TGA transcription factors. The SNI1/CBNAC protein complex can be removed by NPR1, CaM or other unknown mechanisms.
On pathogen infection, transcription of PR1 is activated and repression is terminated. Transcriptional activation of PR1 by NPR1 is well characterized and is mediated by its association with the positive TGA transcription factors. However, mechanisms by which repression of PR1 expression are terminated following pathogen infection are poorly understood. On the basis of the data presented herein, three possibilities can be considered. First, CBNAC and SNI1 could be degraded by a pathogen signal, but there is no strong evidence in support of this hypothesis. Second, CBNAC and SNI1 could be removed by the binding of other regulator proteins or by covalent modifications because the interaction between CBNAC and SNI1 is reduced or removed by pathogen treatment (Supplementary Figure S9). CaM could also be involved in the termination of transcriptional repression through the PR1 promoter E0-1-1 element because CaM interacts with CBNAC (26), and the CaM-binding domain of CBNAC is required for interaction with SNI1 (Supplementary Figure S6). This scenario is rendered likely because influx of Ca2+ followed by the activation of Ca2+/CaM signaling processes represents early and essential events in the response to pathogen infection (39,40). Several CaM-binding proteins have already been shown to be involved in plant defense responses (41,42). Third, CBNAC and SNI1 repression is removed by NPR1. It has been proposed that the role of NPR1 in a wild-type plant is to inactivate SNI1-mediated transcriptional repression of PR genes (18). As such, PR gene expression is restored and is inducible in the sni1 npr1-1 double mutant. However, a physical interaction between SNI1 and NPR1 has never been demonstrated so it is unclear whether NPR1 regulates the SNI1 repression of PR genes. Further molecular and genetic studies are required to precisely characterize the regulatory mechanisms that converge on the PR1 gene, resulting in the precise control of PR1 expression in response to environmental stimuli.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–9 and Supplementary Tables 1 and 2.
FUNDING
World Class University Program [R32-10148] and Basic Science Research Program [2010-0010607] of the National Research Foundation (NRF) funded by the Ministry of Education, Science and Technology, and a grant from the Next Generation BioGreen 21 Program (in part); Rural Development Administration, Republic of Korea [#PJ008173]; scholarships from the BK21 program of the Ministry of Education, Science and Technology (to H.S.K., H.J.H., K.E.K., S.B. and J.A.); KRCF Research Fellowship for Young Scientists program (to H.S.K., in part). Funding for open access charge: Basic Science Research Program [2010-0010607].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Xinnian Dong for critical reading of the manuscript and providing sni1 seed, Dr. Hans J. Bohnert for helpful discussions and critique of the manuscript and the Laboratory of JianMing Zhou (National Institute of Biological Sciences, China) for providing the CAMBIA1300-NLuc/CLuc vector. They also thank the Salk Institute Genomic Analysis Laboratory for seeds and for providing T-DNA insertion flanking sequence information.
REFERENCES
- 1.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 2.Durrant WE, Dong X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 3.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 4.Cao H, Bowling SA, Gordon S, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe. Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 7.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 8.Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Després C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 2000;13:191–202. doi: 10.1094/MPMI.2000.13.2.191. [DOI] [PubMed] [Google Scholar]
- 12.Pontier D, Miao ZH, Lam E. Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J. 2001;27:529–538. doi: 10.1046/j.1365-313x.2001.01086.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Tessaro MJ, Lassner M, Li X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell. 2003;15:2647–2653. doi: 10.1105/tpc.014894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt MD, Delaney TP, Dietrich RA, Weyman KB, Dangl JL, Ryals JA. Salicylate-independent lesion formation in Arabidopsis lsd mutants. Mol. Plant Microbe Interact. 1997;10:531–536. doi: 10.1094/MPMI.1997.10.5.531. [DOI] [PubMed] [Google Scholar]
- 16.Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE. Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 2001;26:409–420. doi: 10.1046/j.1365-313x.2001.2641041.x. [DOI] [PubMed] [Google Scholar]
- 17.Maleck K, Neuenschwander U, Cade RM, Dietrich RA, Dangl JL, Ryals JA. Isolation and characterization of broad-spectrum disease-resistant Arabidopsis mutants. Genetics. 2002;160:1661–1671. doi: 10.1093/genetics/160.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Zhang Y, Clarke JD, Li Y, Dong X. Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell. 1999;98:329–339. doi: 10.1016/s0092-8674(00)81962-5. [DOI] [PubMed] [Google Scholar]
- 19.Weigel RR, Pfitzner UM, Gatz C. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell. 2005;17:1279–1291. doi: 10.1105/tpc.104.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Journot-Catalino N, Somssich IE, Roby D, Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. 2006;18:3289–3302. doi: 10.1105/tpc.106.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim KC, Fan B, Chen Z. Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 2006;142:1180–1192. doi: 10.1104/pp.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durrant WE, Wang S, Dong X. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. USA. 2007;104:4223–4227. doi: 10.1073/pnas.0609357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosher RA, Durrant WE, Wang D, Song J, Dong X. A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell. 2006;18:1750–1765. doi: 10.1105/tpc.105.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy AS, Ali GS, Celesnik H, Day IS. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23:2010–2032. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy AS, Poovaiah BW. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature. 2009;457:1154–1158. doi: 10.1038/nature07612. [DOI] [PubMed] [Google Scholar]
- 26.Kim HS, Park BO, Yoo JH, Jung MS, Lee SM, Han HJ, Kim KE, Kim SH, Lim CO, Yun DJ, et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 2007;282:36292–36302. doi: 10.1074/jbc.M705217200. [DOI] [PubMed] [Google Scholar]
- 27.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 28.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 29.Kim HS, Jung MS, Lee SM, Kim KE, Byun H, Choi MS, Park HC, Cho MJ, Chung WS. An S-locus receptor-like kinase plays a role as a negative regulator in plant defense responses. Biochem. Biophys. Res. Commun. 2009;381:424–428. doi: 10.1016/j.bbrc.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 30.Johnson C, Boden E, Arias J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell. 2003;15:1846–1858. doi: 10.1105/tpc.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon HK, Kim SG, Kim SY, Park CM. Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol Cells. 2008;25:438–445. [PubMed] [Google Scholar]
- 34.Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim KC, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Z, Qamar SA, Chen Z, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 37.Wagner S, Green M. HTLV TAX protein stimulation of DNA binding of bZIP proteins by enhancing dimerization. Science. 1993;262:395–399. doi: 10.1126/science.8211160. [DOI] [PubMed] [Google Scholar]
- 38.Kuhlmann M, Horvay K, Strathmann A, Heinekamp T, Fischer U, Böttner S, Dröge-Laser W. The alpha-helical D1 domain of the tobacco bZIP transcription factor BZI-1 interacts with the ankyrin-repeat protein ANK1 and is important for BZI-1 function, both in auxin signaling and pathogen response. J. Biol. Chem. 2003;278:8786–8794. doi: 10.1074/jbc.M210292200. [DOI] [PubMed] [Google Scholar]
- 39.Xu H, Heath MC. Role of calcium in signal transduction during the hypersensitive response caused by basidiospore-derived infection of the cowpea rust fungus. Plant Cell. 1998;10:585–597. doi: 10.1105/tpc.10.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurusu T, Yagala T, Miyao A, Hirochika H, Kuchitsu K. Identification of a putative voltage-gated Ca2+ channel as a key regulator of elicitor-induced hypersensitive cell death and mitogen-activated protein kinase activation in rice. Plant J. 2005;42:798–809. doi: 10.1111/j.1365-313X.2005.02415.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature. 2002;416:447–451. doi: 10.1038/416447a. [DOI] [PubMed] [Google Scholar]
- 42.Kang CH, Jung WY, Kang YH, Kim JY, Kim DG, Jeong JC, Baek DW, Jin JB, Lee JY, Kim MO, et al. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 2006;13:84–95. doi: 10.1038/sj.cdd.4401712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.