Abstract
Rex1/Zfp42 is a Yy1-related zinc-finger protein whose expression is frequently used to identify pluripotent stem cells. We show that depletion of Rex1 levels notably affected self-renewal of mouse embryonic stem (ES) cells in clonal assays, in the absence of evident differences in expression of marker genes for pluripotency or differentiation. By contrast, marked differences in expression of several endogenous retroviral elements (ERVs) were evident upon Rex1 depletion. We demonstrate association of REX1 to specific elements in chromatin-immunoprecipitation assays, most strongly to muERV-L and to a lower extent to IAP and musD elements. Rex1 regulates muERV-L expression in vivo, as we show altered levels upon transient gain-and-loss of Rex1 function in pre-implantation embryos. We also find REX1 can associate with the lysine-demethylase LSD1/KDM1A, suggesting they act in concert. Similar to REX1 binding to retrotransposable elements (REs) in ES cells, we also detected binding of the REX1 related proteins YY1 and YY2 to REs, although the binding preferences of the two proteins were slightly different. Altogether, we show that Rex1 regulates ERV expression in mouse ES cells and during pre-implantation development and suggest that Rex1 and its relatives have evolved as regulators of endogenous retroviral transcription.
INTRODUCTION
After undergoing a first differentiation step, the pre-implantation blastocyst is divided in Inner Cell Mass (ICM) that gives rise to the embryo proper and trophectoderm, an external epithelium that contributes to the placenta. Self-renewing stem cells that can be derived from each of these lineages (1) are referred to as embryonic (ES) and trophectoderm (TS) stem cells, respectively. ES cells can be maintained in culture for an apparent unlimited number of cell divisions (self-renewal) and maintain the defining property of pluripotency or the ability to differentiate into cell lineages of all three primary layers of the embryo. Molecular mechanisms that maintain this pluripotent self-renewing state operate at different levels and include (but are not limited to) signalling by leukaemia inhibitory factor (LIF) and BMP4, inhibition of ERK signalling (2), co-operating networks of transcription factors and epigenetic mechanisms (3,4). The Oct4, Sox2 and Nanog transcription factors (5–8) constitute a core transcriptional network to maintain pluripotency through mutual positive regulation (4) and collaborative regulation of target genes. A distinct module whose function is essential for the maintenance of pluripotency and self-renewal consists of Cnot3, Trim28, c-Myc and Zfx (9,10). Trim28 has recently also been shown to participate in the repression of endogenous retroviral elements (ERVs) in mouse ES cells (11).
Rex1 was first discovered as a result of its specific expression in pluripotent F9 embryonal carcinoma (EC) cells (12). Rex1 (for reduced expression-1, also known as Zfp42) was subsequently shown to be expressed in other pluripotent cell types, especially undifferentiated ES cells (13), multipotent adult progenitor cells (14) and amniotic fluid cells (15), in the germ cells of the testis and in the ICM and early trophectoderm derivatives of the mouse embryo (13). Rex1 expression has been positively linked to increased pluripotency in both mES cells (16–18) and human ES and iPS cells (19,20). In contrast, conflicting results have been reported regarding the functional role of Rex-1. Gene silencing by RNA interference results in loss of self-renewal in ES cells (21) and overexpression of Rex1 negatively affects self-renewal (D. Guallar, M. Sánchez and J. Schoorlemmer, unpublished data). However, Rex1 does not have to be provided for efficient reprogramming of differentiated cells towards iPSs (16,17), Rex-1 is dispensable for maintenance of self-renewing pluripotent ES cells (22) and ES cell lines can efficiently be derived from Rex1-deficient blastocyst (23).
Rex1 encodes a protein containing four Cys-His type zinc fingers, which is localized in the nucleus in ES cells (23), and displays significant similarity to the YY1 transcription factor family in the DNA-binding zinc-finger domains (24). Rex1 target genes have been surveyed by gene association and differential expression studies. Target genes identified in ES cells now encompass a circuit of active genes implicated in protein metabolism that coincides partially with Myc targets as opposed to Oct4/Nanog/Sox2 targets (25), binding to (and regulation of) Tsix regulatory elements (26), Ring1B/Rnf2 regulated genes (27) and also imprinted genes during pre-implantation development (28). Interestingly, the absence of Rex1 from an ES cell line has been linked to loss of pluripotency upon prolonged passage and to enhanced expression of retrotransposable elements (RE) (29).
Transposable elements (TEs) are repetitive DNA sequences that are ubiquitous and abundant components of most genomes including mammals and constitute >45% of the human and mouse genome (30,31). They can duplicate and reinsert within genomes, either autonomously or with the assistance of proteins encoded by other (related) elements. As a result, TE profoundly impact genome function and evolution, as transposon-derived promoters also direct expression of alternative transcripts. De-regulated gene expression mediated by the activation of transposon promoters contributes to tumorigenesis and autoimmune disease (32,33).
Most mammalian TEs are REs, which propagate through an RNA intermediate, and 8–10% of those are retrovirus-like long terminal repeat (LTR) elements referred to as ERVs. They constitute a range of similar but clearly distinguishable elements with varying copy numbers, autonomy and expression patterns that together occupy ∼5.4% of the mouse genome (34). The superfamily of ERVs is composed of (but not limited to) muERV-L, IAP, musD, ORR1 and MT families (Supplementary Table SIV), with varying copy numbers ranging from 300 to 200 000 (31,35). Transcription of ERVs in different species is elevated in germ line cells, early embryo and placenta compared with adult or differentiated tissues (36). MaLR and muERV-L family ERV are frequently encountered in chimeric transcripts in the mature oocyte/zygote and two-cell embryos, respectively (37). MuERV-L also displays a transient increased expression during the two-cell stage (38) and normal development to four-cell embryos is compromised when muERV-L levels are attenuated (39). By contrast, IAP and musD/ETn elements are specifically expressed from the blastocyst stage onward (40). As expression of several families of elements is subject to tight regulation during embryonic development and each element was found to act as promoter for subsets of genes at defined stages (37), it has been proposed that MaLR, muERV-L and IAP synchronize stage-specific gene expression.
ERV transcription is limited in most tissues due to methylation-directed silencing. Pre-implantation development is accompanied by general demethylation (41), calling for specific mechanisms to regulate expression of ERV elements that include histone modifications [reviewed in (42)] and RNA interference (43), protection from demethylation (44) or specific repressors including KAP1/Trim28 (11), ESet (45) and LSD1/KDM1A (46). Some of these mechanisms that constrain expression during pre-implantation development are also operative in ES cells (11,45). In contrast to Kap1-deficient ES cells that lose pluripotency in a few generations, Lsd1-deficient ES cells can be stably propagated although muERV-L is severely de-repressed (46).
Expression of Rex1 in the germline (12,47), placenta (28) and during pre-implantation development (13) coincides with high prevalence of gene expression directed by retroviral elements. The presence of Rex1 in tissues displaying high ERV expression prompted us to consider a potential direct role of Rex1 in regulation of REs.
MATERIALS AND METHODS
Immunological reagents and western blot
The α-YY2 and the α-REX1 sera raised in rabbit have been described previously (27). The amino-terminal region of REX1 (amino acids 1–116) that we have used for immunization does not show homology at all to the corresponding region in YY1 (Supplementary Figure S1A) and does not cross-react with YY1 (Supplementary Figure S1B). The rabbit α-REX1 serum was further affinity-purified over REX1Δ-GST protein (ImmunoStep SA, Salamanca, Spain). Monoclonal α-HA (clone HA-7) was obtained as an unpurified ascites fraction (Sigma H9658).
For western blot analysis, cells were scraped with pre-cooled phosphate buffered saline (PBS), pelleted by centrifugation and re-suspended in lysis buffer [50 mM Tris (pH 8.1), 10 mM EDTA, 1% (w/v) sodium dodecyl sulphate (SDS)] containing protease inhibitors (PMSF and Complete; ROCHE). To solubilize REX1, samples were sonicated (Bioruptor®, Diagenode) until homogenous in ice water (0°C). Debris was removed by centrifugation for 15 min at 13 000 rpm at 4°C. Laemmli sample buffer was added to lysates, and samples were separated by SDS–polyacrylamide gel electrophoresis and analysed by standard western blot as described in Supplementary Materials and Methods section.
Co-immunoprecipitation between REX1 and LSD1 was assayed as described in Supplementary Materials and Methods section, using extracts from 293T cells co-transfected with either HA-tagged REX1 or with a mixture of plasmids expressing HA-tagged REX1 and a Flag-tagged LSD1Δ construct (48).
Plasmid construction
For the generation of short hairpin RNA (shRNA) vectors, the following fragment were fused into the BamHI-BglII backbone derived from PGKHygro (pHPCAGGS): a doxycyclin-inducible H1 promoter (49), shRNA as BglII-HIII fragment, and a promoterless eGFP derived from pCH-Octi (50). Constructs were verified by DNA sequencing. The shRNAs were designed as described (51), and sequences are as follows (sh1Rex1: 5′-ACGGATACCTAGAGTGCATCA, sh2Rex1: 5′-CACGGAGAGCTCGAAACTAAA, shRNA Gfp: 5′-AAGCGCGATCACATGGTCCTG). Plasmids used for transfections were purified on PureLink™ kits and columns (Invitrogen).
For overexpression, a Rex1/Zfp42 complementary DNA (cDNA) (27) was fused to an IRES-eGFP derived from pIRES2-EGFP (Clontech) and inserted into the chicken βActin promoter-driven expression vector pCAGIP (52). Further details of all plasmids used are available upon request. Plasmids used for injections were linearized and recovered from GeneClean Turbo cartridges (MP Biomedicals) in Tris–EDTA (10/0.1 mM) as described (M. Climent et al., submitted for publication).
Cell culture, differentiation and transfection
293T cells were maintained and transfected with mixtures of plasmids according to the manufacturer’s protocol (Lipofectamine 2000; Invitrogen).
Mouse TS cell line B7 (53) was maintained as described (54) on 1% gelatine-coated tissue culture dishes supplemented with FGF4 (25 ng/ml)(Peprotech) and heparin (1 mg/ml)(Sigma). ES cell line E14T (55) was maintained on gelatine-coated tissue culture dishes in medium supplemented with 10% foetal calf serum, LIF and 2i as described (56). To block HDACs or DNA methylation, ES cells were cultured for 24 h in the presence of 200 nM trichostatin A (TSA; diluted from a 1 mM stock solution) or 3 µM 5-aza-cytidine (diluted from freshly thawed aliquot). RA (Sigma) was stored as a 10 mM stock in dimethyl sulfoxide (DMSO) at −80°C, freshly diluted before use at 0.1 mM in culture medium, and added directly onto the cells to a final concentration of 1 µM. Cells were aggregated in the presence of DMSO as described (57). E14T ES cells were transfected by standard electroporation (5 × 106 cells; 20 µg DNA; 200 V, 960 µF, ∞Ω). ES cells electroporated with plasmids expressing shRNAs were seeded at clonal densities in the presence of LIF and 2i to allow for colony formation as a measure of self-renewal (usually 100 000 cells per six well). After 24 h, transfected cells were subjected to selection (H3274; Sigma at 160 µg/ml) for the Hygro resistance carried on the same plasmid. After selection for 7 days, cells were fixed overnight at 4°C in 80% ethanol, and stained for alkaline phosphatase (AP) using a Sigma kit (85L3R). Alternatively, cells were harvested and processed for expression analysis or chromatin immunoprecipitation (ChIP). AP+ colonies were counted in fields of predetermined size on a Nikon Diaphot Inverted Tissue Culture Microscope. For each condition, 10 fields were counted, colony numbers are represented as mean ± standard deviation (SD).
RT–qPCR analysis
Cells were washed with PBS, scraped and total RNA was extracted using TRIzol® reagent (Invitrogen). After digestion of genomic DNA (RQ1 RNAse-Free DNase; Promega), RNA was extracted with phenol/chloroform, precipitated with ethanol, re-suspended in water and quantified using Nanodrop (Thermo Scientific). cDNA was synthesized from 2 µg of RNA either with Oligo dT or random hexamer primers (ThermoScript® RT–PCR System; Invitrogen) and stored at −20°C until used. cDNA was analysed by quantitative PCR (Platinum® SYBR® Green qPCR SuperMix-UDG; Invitrogen) on an ABI Prism 7000 Real-Time PCR system, reactions were performed in triplicate. Data were processed using the ΔΔCt method (58), using Gapdh as a reference gene. Sequences of all primers used are included in Supplementary Materials and Methods section and Supplementary Table SI. An extensive justification of the localization and origin of primers used for RT–qPCR of TE is provided in Supplementary Materials and Methods section.
siRNA used for microinjection
Stealth siRNA duplexes against mRex1 and control siRNAs (Ref 12935-112) were purchased, annealed according to the manufacturer’s instructions (Invitrogen) and changed to 10 mM Tris/EDTA 0.1 mM as described in the accompanying paper (M. Climent et al., submitted for publication).
Oocyte and embryo collection, culture, microinjection and RNA isolation
All procedures were carried out under Project Licence PI29/08 approved by the in-house Ethic Committee for Animal Experiments from the University of Zaragoza (Spain).
Oocyte and embryo collection, culture and microinjection were performed according to standard procedures (59). A detailed description of microinjection procedures, embryo culture, Rex1 overexpression and gene-expression analysis in embryos will be provided elsewhere (M. Climent et al., submitted for publication). One-cell embryos were microinjected with siRNAs at 100 µM or linearized plasmid at 2 ng/µl and 0,4 mg/ml Dextran-coupled Texas Red (D1829; Invitrogen). In brief, embryos were injected on Day 1, selected for development to at least the two-cell stage on Day 2 and to the five- to eight-cell stage on Day 3. Embryos injected with pCAG-Rex1IRESeGFP were separated on Day 3 in three groups representing low (absent), intermediate and high eGFP expression. Only the groups identified as low and high expressors were used in the experiments.
For the analysis of mouse pre-implantation embryos, an identical number of experimental and control embryos from the same experiment was used (typically 8–10 embryos). Total RNA was reverse-transcribed and two oocyte or embryo equivalents were used as a template for each PCR. PCRs were carried out for 35 cycles of denaturation at 94°C for 30 s, followed by 30 s of annealing, 1 min extension at 72°C; final extension 5 min. Products were separated on agarose gels, visualized using ethidium bromide and photographed on a Gel Doc transiluminator (BioRad). Data were quantified using Quantity One software (BioRad), and expression levels were recalculated using H2afz as a reference gene. Primers used in this study are shown in Supplementary Materials and Methods section and Supplementary Table SI.
ChIP and locus-specific PCR
ChIP assays were carried out as described (27), using per 5 × 106 cells the following sera for immunoprecipitation: Pre-immune serum (75 µg), rabbit anti-REX1 IgG (75 µg), affinity purified rabbit anti-REX1 (25 µg), mouse monoclonal H-10 anti-YY1 (Santa Cruz; 2.5 µg), rabbit anti-YY2 (25 µg). We compared semi-quantitative PCR amplification on amounts of chromatin obtained from the same number of cells after immuno-precipitation using either pre-immune serum or αREX1 serum. PCR products were visualized using ethidium bromide and photographed. Primers used are listed in Supplementary Materials and Methods section and Supplementary Table SII.
Quantitative real-time PCR on ChIPs was performed in 96-well plates using Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen) on an ABI Prism 7000 Real-Time PCR system and processed using the ΔCt method. Amplification was performed in 15 µl reactions using the following parameters: 50°C for 2 min, 95°C for 10 min. followed by 40 cycles of (95°C 15 s, 60°C 60 s). Data presented are derived of triplicate reactions; the results shown are the aggregate of a minimum of three separate ChIPs performed on different days. Data were processed to calculate percentage association as described (www.SABiosciences.com) and are represented as the mean ( ± SD/SEM).
Real-time PCR analysis
Standard curves of all primers (used at 200 nM) were performed to check for efficient amplification (between 90 and 110%). Melting curves were also performed to verify production of single DNA species with each primer pair, except for ERV sequences. Relative levels of expression in each assay were obtained through the standardized ΔΔCt method. We used (i) Gapdh mRNA levels as a reporter in all experiments of Rex1 knockdowns and (ii) the appropriate control cell line (DMSO-treated aggregates) as the reference sample in Figure 2B. Data presented are the mean of three to five independent experiments each performed on different days and using a different preparation of cells. The relative amount of expression between samples was compared using the Tukey’s method (95% confidence interval) following one-way ANOVA (Instat version 3.0, GraphPad Software, San Diego, CA, USA).
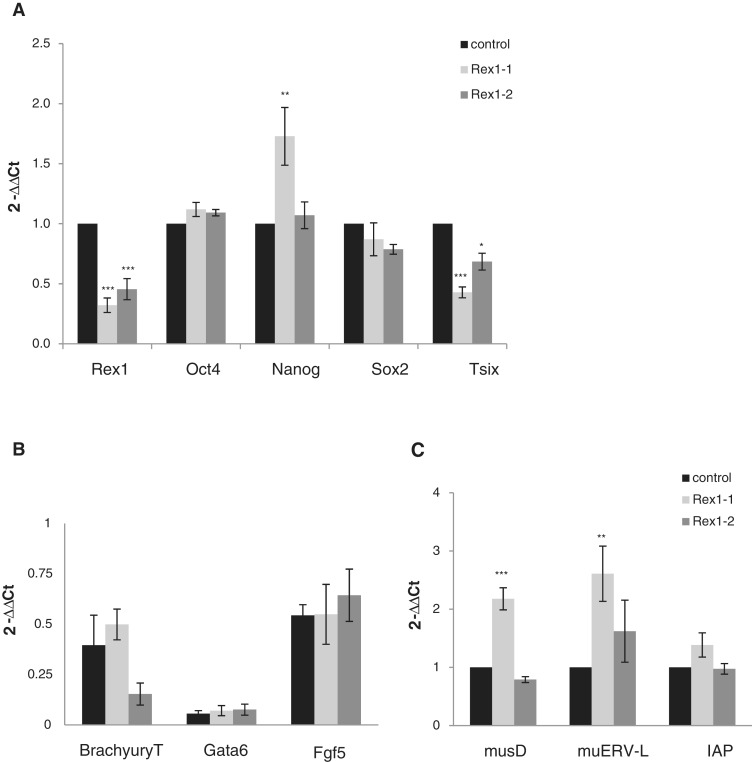
Figure 2.
Altered expression of ERV upon Rex1 depletion in ES cells. Rex1 RNA was depleted using transfection of shRNA-expressing plasmids as indicated in the legend to Figure 1B. After selection for 7 days, changes in gene-expression levels were measured by real-time qPCR: (A) pluripotency-associated genes, (B) differentiation markers, (C) several families of ERVs. Expression levels were normalized to Gapdh as a reference gene and are depicted as fold expression (2-ΔΔCt) relative to wild-type ES cells (that express control shRNA) (A and C) or fold expression (2-ΔΔCt) relative to embryoid bodies (Figure B). Error bars represent the SD over a minimum of three assays. *P < 0.05; **P < 0.01; ***P < 0.001 (ANOVA-Tukey). (A) Changes in expression levels of Rex1, Pou5f1/Oct4, Nanog, Sox2 and Tsix. (B) Changes in expression levels of Gata6, Fgf5 and T/Brachyury. (C) Changes in expression levels of musD, muERV-L and IAP.
Enrichment levels in ChIP assays are expressed as a percentage of immuno-precipitation relative to the input. Essentially, the ΔCt method was used to calculate a ChIP over input ratio, after subtraction of Cts derived from control immunoprecipitations using pre-immune serum. Values obtained were corrected for the appropriate dilution factor of each analysed fraction, and multiplied by 100 to get the percentage of immunoprecipitation (% IP). Alternatively, data are shown as fold enrichment of Rex1 binding for a particular locus compared to control loci as indicated.
RESULTS
Depletion of Rex1 expression causes a loss of self-renewal in mouse ES cells
Although it has been reported that Rex1(Zfp42) null mice appear normal, viable and fertile and normal ES cells can readily be derived from Rex1 null mice or by homologous recombination in vitro (22,23), Rex1 expression is associated with highly pluripotent populations of stem cells (18,20), suggesting an important role for Rex1 in ES cells. We designed shRNAs to attenuate Rex1 expression, which were cloned in appropriate expression vectors. Resulting vectors were tested by co-transfection with HA-Rex1 in 293T cells. Results show that expression of HA-REX1 (Figure 1A, lanes 1 or 4) was abolished by co-transfection of either of the two hairpin constructs (termed Sh1Rex1 and Sh2Rex1). Transfection efficiency was measured using a co-transfected eGFP plasmid and was roughly equal among samples (23–37%).
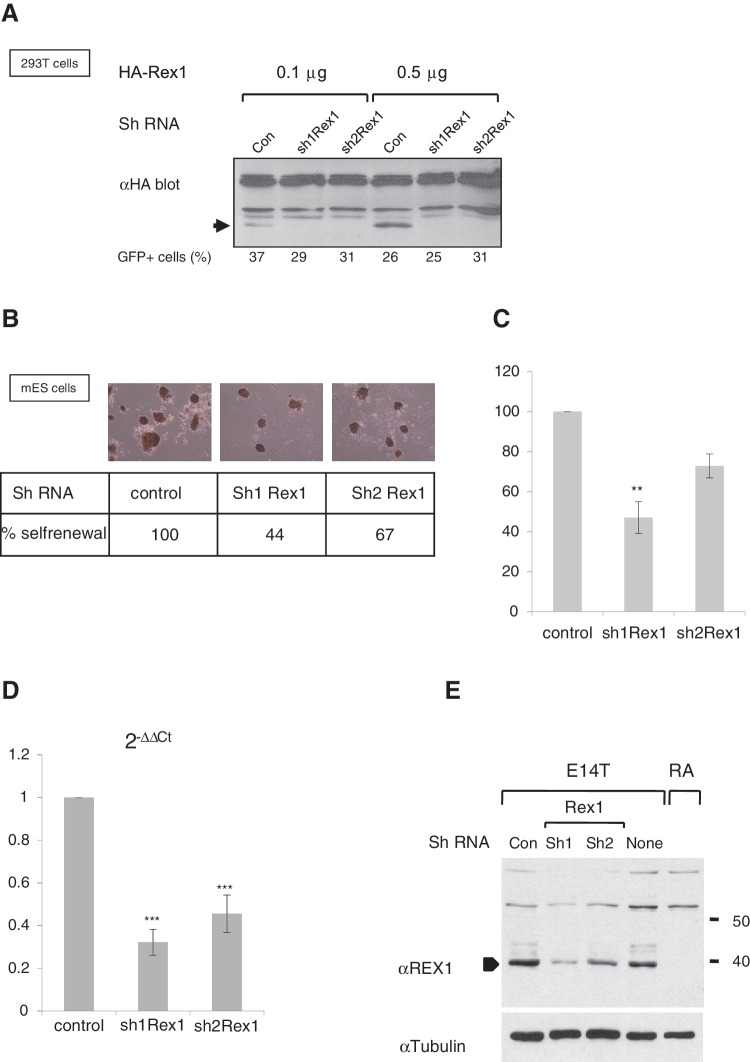
Figure 1.
Attenuation of Rex1 levels causes partial loss of self-renewal in mouse ES cells. (A) Western blot to detect HA-tagged REX1 in 293T cells co-transfected with plasmids that express control shRNAs (con) or two different shRNAs directed against Rex1 (sh1Rex1 and sh2Rex1). The amount of transfected plasmids is indicated above the panel. A band specific for HA-REX1 is indicated with an arrow, as opposed to several non-specific bands. The activity of a co-transfected eGFP expression plasmid served as an indicator of transfection efficiency. The percentage of GFP-positive cells is indicated below each lane. (B) Clonogenic colony formation assay. Equal numbers of E14T mES cells transfected with different shRNA-expressing plasmids tested in (A) were selected for 7 days for plasmid uptake with hygromycin, and resulting colonies were stained for AP. (C) Quantification of colonies obtained in (B). AP+ colonies in a predetermined number of fields were counted; the number of colonies formed by cells expressing control shRNA was set at a 100% and the number of colonies in the presence of shRNAs directed against Rex1 (sh1Rex1 and sh2Rex1) was calculated accordingly. Error bars represent the SD over a minimum of three assays. **P < 0.01 (ANOVA-Tukey). (D) Changes in expression levels of Rex1/Zfp42 after hygromycin-selection for 7 days of cells transfected with plasmids that express shRNAs as described in (B). Expression levels were normalized to Gapdh as a reference gene and are depicted as fraction relative to the control ES cells. Error bars represent the SD over a minimum of three assays. ***P < 0.001 (ANOVA-Tukey). The difference between effects of shRNA1 and shRNA2 is also statistically significant, *P < 0.05 (ANOVA-Tukey). (E) Western blots to detect REX1 in cell lysates of E14T ES cells (E14T), RA-treated ES cells (RA), or cells transfected with shRNAs described in (A). The arrowhead indicates the REX1 protein, which is absent in RA-treated ES cells. Migration of molecular weight standards is indicated to the right. Equivalent loading in each lane is demonstrated by stripping and reprobing the membrane with αTUBULIN antibodies.
We subsequently investigated Rex1 function in ES cells by using stable episomal expression of these shRNAs. ES cells were electroporated and selected for expression of shRNAs, seeded at clonal densities and colony formation was counted as a measure of self-renewal. As expected, no colonies were observed after selection of cells mock-transfected in the absence of plasmids that carry the HygroR gene (data not shown). ES cells that express shRNAs targeting Rex1 (Figure 1B) clearly formed fewer colonies when compared with cells electroporated with control shRNAs, consistently giving rise to 44 and 67% of colonies in control assays for Sh1Rex1 and Sh2Rex1, respectively (Figure 1B and C). All colonies remained undifferentiated as judged by morphology and AP staining (Figure 1B, top panels). The loss of colony formation was not caused by off-target effects, as Sh1Rex1 did not negatively affect colony formation in Rex1 deficient cells (Supplementary Figure S2).
We did not observe complete elimination of Rex1 mRNA levels, but a clear reduction to 33% or 45 % of wild-type levels for Sh1Rex1 or Sh2Rex1-expressing ES cells, respectively (Figure 1D). More important, REX1 protein levels in E14T ES cells (Figure 1E, lane None) were severely reduced in cultures overexpressing either hairpin RNA (Figure 1E, lanes marked ShRex1), as opposed to cultures expressing control ShRNA (Lane Con). Together, these data show that reduced expression of Rex1 was coincident with diminished self-renewal of ES cells.
Gene expression in Rex1-depleted ES cells
To analyse gene expression in the Rex1-depleted, AP+ colonies, we assayed mRNA for expression of known pluripotency markers or (Polycomb-regulated) differentiation markers by RT–qPCR. Confirming a previous report (26), Tsix RNA was down-regulated to a level proportional to the reduction in Rex1 levels (Figure 2A). We measured expression of several pluripotency markers and detected similar or identical levels, independent of the level of Rex1 for Oct4/Pou5f1 and Sox2 mRNAs (Figure 2A). A slight increase was observed in the levels of Nanog mRNA upon expression of Sh1Rex1. This difference was not detected upon expression of Sh2Rex1, pointing towards a weak dose-dependent effect of Rex1 on Nanog expression. No up-regulation was observed for several differentiation markers previously shown to bind REX1 in ChIP assays: Fgf5, Gata6, T (Brachyury) (Figure 2B). We detected small differences in expression levels of T upon Rex1 depletion (Figure 2B), which is consistent with published data showing reciprocal expression of Rex1 and T (23). We conclude that Rex1-depleted ES cells do not exhibit major differences in expression of pluripotency markers and differentiation markers.
RE and ERV are prominently expressed during pre-implantation development in the mouse (37,38). As Rex1 is expressed throughout pre-implantation development (M. Climent et al., submitted for publication) we envisaged a potential function for Rex1 in regulation of ERV. Therefore, we tested de-regulation of ERV in Rex1-depleted cells as compared to ES cells with wild-type Rex1 levels. We initially focused on several autonomous LTR retrotransposons, which are highly expressed in either the two-cell (muERV-L) or blastocyst stage (IAP, musD) mouse pre-implantation embryo.
Using primers that amplify regions conserved among several copies of REs, we detected a clear difference for several families in expression level in Rex1 depleted cells as opposed to control cells (Figure 2C). Expression of both muERV-L and musD was up-regulated ∼2- to 3-fold, as opposed to IAP, whose overall expression level was not affected. We conclude that expression of ERVs is altered upon attenuation of Rex1 levels in mouse ES cells.
REX1 binds ERVs of different families to a varying degree in mES cells
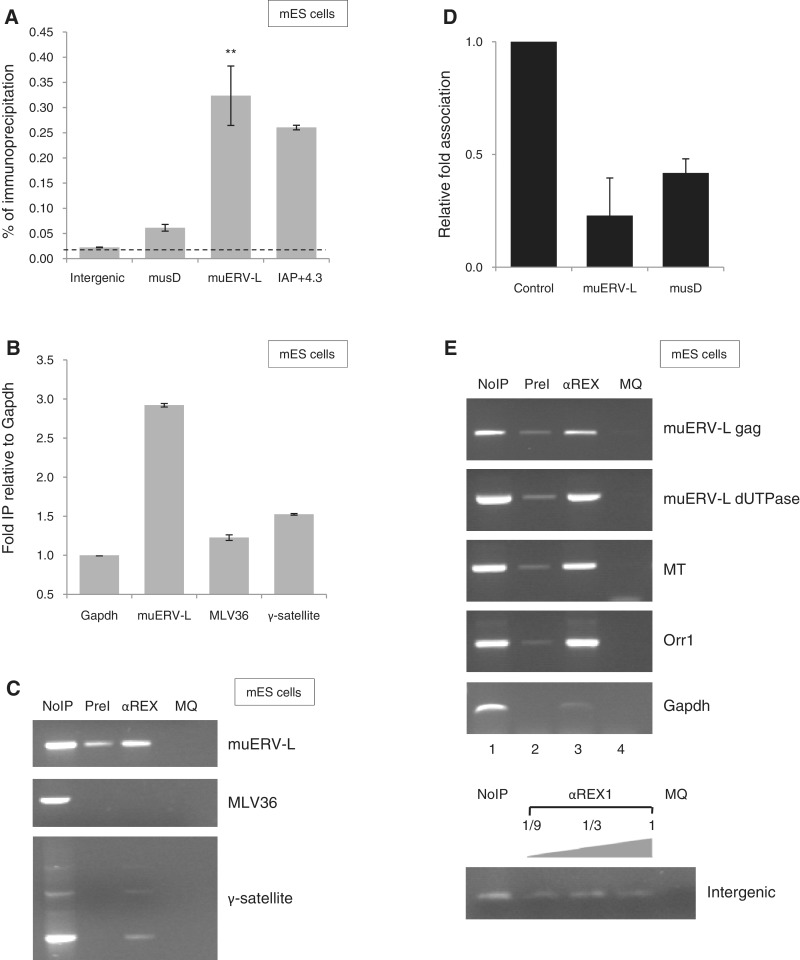
We subsequently carried out ChIP assays in combination with locus-specific primers to interrogate REX1 association to a subset of ERV in ES cells. Enrichment was measured by quantitative real-time PCR and association was calculated as percentage association relative to control chromatin (Figure 3A). We started out testing binding to muERV-L elements, previously demonstrated to be highly expressed in the blastocyst (38). After subtraction of background values and normalization against an intergenic fragment on chromosome 6 (60), we calculated a reproducible 8- to 15-fold enrichment of this locus in chromatin immunoprecipitated using αREX1 in ES cells (Figure 3A). To a lesser extent, reproducible enrichment was also detected for MusD and IAP elements (Figure 3A). Similar results were obtained for muERV-L and musD in experiments using the Gapdh promoter or an intron in the β-actin gene as controls (Supplementary Figure S3). Enrichment for muERV-L was also observed when association was compared with MLV36 or γ-satellite multi-copy sequences, either in qPCR (Figure 3B) or semi-quantitative PCR (Figure 3C). Weaker, reproducible enrichment was also detected for an IAP-related element IAP-RLTR10 (Supplementary Figure S3).
Figure 3.
REX1 binds RE/ERV elements in ES cells. (A) REX1 association to RE in ES cells. Binding was assessed in ChIP assays using αREX1 serum followed by quantification of precipitated DNA using real-time qPCR amplification. The figure shows analysis of a non-binding reference site on chromosome 6 as a control (intergenic), and the ERV elements indicated. Association was assessed to the Gag gene in muERV-L (Supplementary Materials and Methods section). Association is represented as percentage bound (relative to purified chromatin extract from the same lysate). Error bars indicate SEM; *P < 0.05. (B) REX1 binding does not extend to other repeated elements. ChIP assays as in (A) except that binding to several sequences present in the genome as multiple copies (MLV36, γ-satellite) is represented as fold binding relative to a non-binding reference gene Gapdh. Error bars indicate SEM. (C) REX1 binding does not extend to other repeated elements. Binding of REX1 in ES cells to several sequences present in the genome as multiple copies (MLV36, γ-satellite) was assessed by PCR analysis after ChIP by Pre-immune serum (PreI), or αREX1 serum (αREX1). The figure shows repeat-specific PCR amplification, using primers specific for the elements indicated on the right. PCRs without input DNA served as a negative control (MQ). PCRs using a fraction of purified chromatin extract from the same lysate are shown as positive controls on the left (No IP). (D) Loss of REX1 binding to selected ERV elements in RA-treated ES cells. Levels of REX1 binding were determined by ChIP as in A using either pluripotent or RA-treated ES cells and site-specific qPCR. The relative amount of immunoprecipitated DNA is shown as the ratio of αREX1 immunoprecipitations in RA-treated versus non-treated cells, normalized to non-bound control genes (Intergenic or Gapdh). Error bars indicate SD. (E) REX1 association to muERV-L and MaLR. Binding of REX1 in ES cells was assessed by PCR analysis after ChIP by Pre-immune serum (PreI), or αREX1 serum (αREX). The figure shows gene-specific PCR amplification, using primers specific for the ERV indicated on the right. PCRs without input DNA served as a negative control (MQ). PCRs using a fraction of purified chromatin extract from the same lysate are shown as positive controls on the left (NoIP).
Our assay is specific for REX1, as binding was abolished in ES cells with attenuated levels of Rex1 (data not shown). Also, we compared binding between undifferentiated ES cells and their RA-treated counterparts, the latter having lost REX1 expression [Figure 1E; (27)]. Results from ChIP assays (Figure 3D) demonstrate Rex1 association to muERV-L and musD elements in ES cells (Figure 3A), as opposed to RA-treated cells (Figure 3D).
Efficient binding to muERV-L was confirmed by amplification using different primer sets (Supplementary Materials and Methods section), of sequences in the 5′-Gag gene or in the 3′-dUTPase gene (Figure 3E). We subsequently tested a potential association of REX1 to ORR1 and MT elements of the MaLR family (Supplementary Table SIV). While efficient amplification was observed using control templates for each of the loci analysed (Figure 3E, lanes NoIP), hardly any amplification was observed for the majority of markers after immunoprecipitation with pre-immune serum (lane PreI). By contrast, we observed reproducible association of REX1 to ORR1 and MT elements, as opposed to several intergenic regions used as controls (data not shown), an intergenic fragment in chromosome 6 (Figure 3E, bottom panel) and the Gapdh promoter, which were not amplified above background (Figure 3E).
In conclusion, REX1 associates with a subset of genomic repeats derived from LTR retrotransposons, including muERV-L and IAP elements in mouse ES cells.
YY1 and YY2 associate with RE
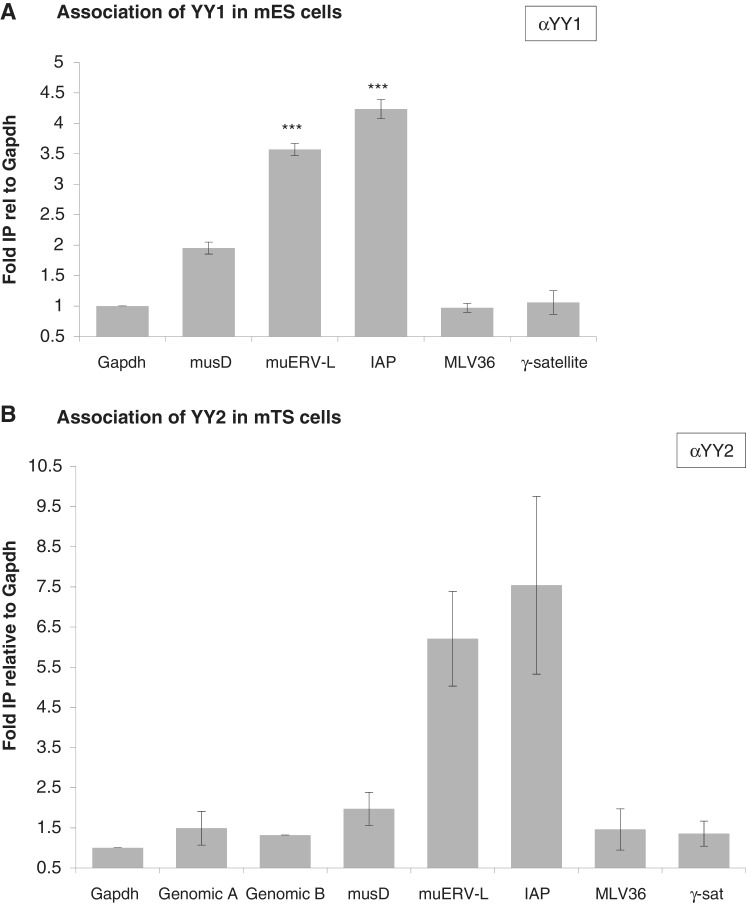
Although Rex1 is expressed in both ES and TS cells in culture (27), levels of REX1 association to the same ERVs were lower in TS cells as opposed to ES cells (data not shown), in line with reduced expression of Rex1 in TS cells (27). As REX1 shares high homology in the DNA-binding zinc fingers with YY1 and YY2 (24), we also tested their binding to ERVs. Hardly any enrichment was observed for several control markers, i.e. a genomic region in chromosome 8 (Genomic A/B), the Gapdh promoter or different multicopy controls (MLV36, y-satellites) in both cases (Figure 4). After normalization against a control promoter, we calculated reproducible enrichment of several ERV elements. In chromatin immunoprecipitated using αYY1 (Figure 4A), we demonstrate a 3-, 5- to 4- and 5-fold enrichment of both IAP and muERV-L elements, and a weaker 2-fold enrichment of musD elements. We interrogated YY2 association in TS cells, considering its high expression in this cell type (data not shown). In chromatin immunoprecipitated using αYY2 (Figure 4B), we detected a 6- to 8-fold enrichment of both IAP and muERV-L elements, and a weaker but significant enrichment of musD elements. In contrast to REX1 in ES cells, association of both YY1 and YY2 to IAP was slightly stronger when compared with muERV-L, indicating that the binding specificities are not the same as the one observed for REX1.
Figure 4.
YY1 and YY2 association to ERV elements. Association of YY1 in ES cells (A) and YY2 in TS cells (B) to potential target genes was assessed by locus-specific qPCR analysis after ChIP using αYY1 or αYY2, respectively. The Gapdh promoter, non-binding control elements in chromosome 8 and several sequences present in the genome as multiple copies (MLV36, γ-satellite) are included as negative controls. Association was assayed for musD, IAP and muERV-L. The amount of immunoprecipitated DNA as a percentage of input DNA was recalculated as fold association normalized to the Gapdh promoter. Error bars represent SEM over a minimum of three assays.
REX1 associates with LSD1
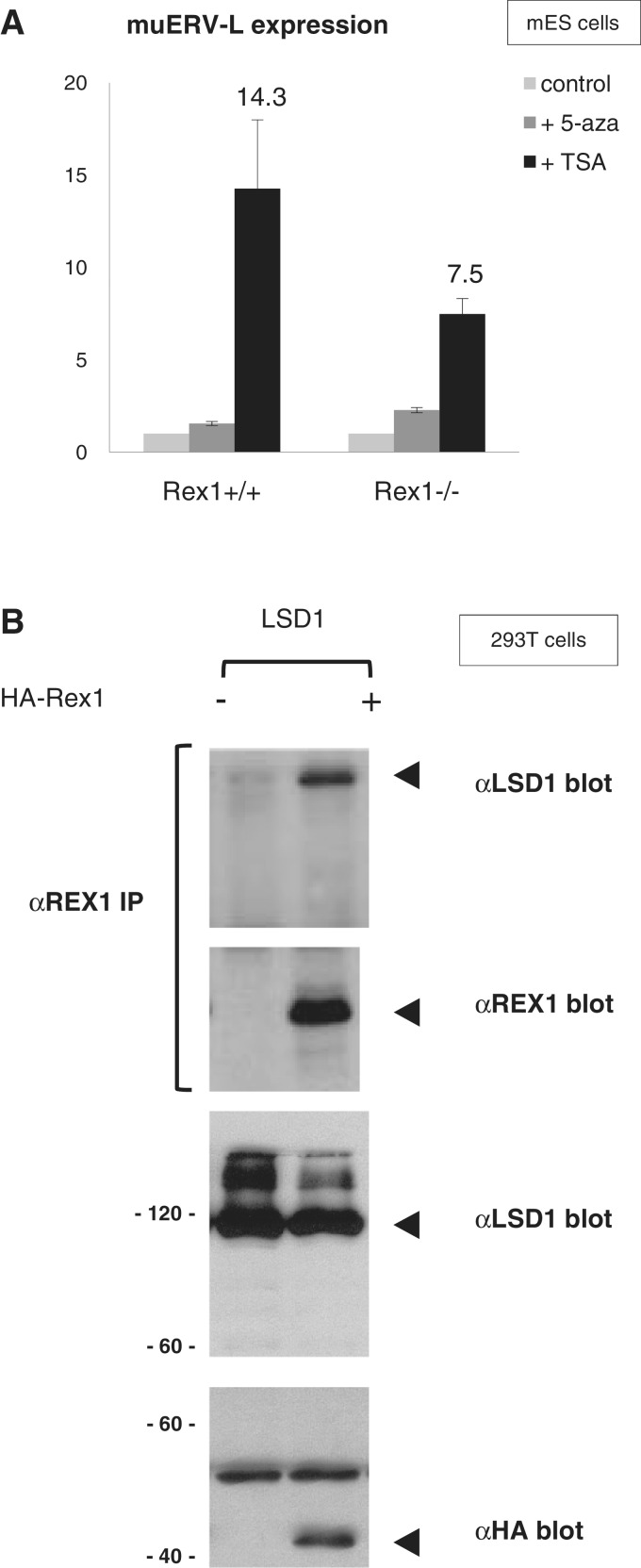
Transcriptional repression by YY1 is mediated by interaction with HDACs (61). To provide more mechanistic insight into the Rex1-dependent repression of retroviral elements, we wished to address a potential interplay between HDACs and Rex1. We therefore tested the effect of the HDAC inhibitor TSA on regulation of muERV-L expression. As reported before (46), TSA treatment caused a significant increase in muERV-L mRNA levels in wild-type and Rex1-deficient ES cells (Figure 5A). Surprisingly however, the increase caused by TSA was attenuated in Rex1-deficient ES cells, suggesting HDACs function in concert with Rex1 (Figure 5A). This effect of Rex1 was specific, as Rex1 deficiency did not influence TSA-mediated induction of Dub1 (data not shown). The fold induction by TSA in Rex1+/+ versus Rex1−/− cells was 1.90 and 1.17 for muERV-L and Dub1, respectively. In agreement with recent reports (46), we did not observe differences in levels of muERV-L expression in the presence of the DNA demethylating agent 5-azacytidine (5Aza) (Figure 5A). The resistance to 5Aza was not affected by Rex1 deficiency, in line with DNA methylation-independent pathways being responsible for repression.
Figure 5.
Association of REX1 and LSD1 proteins. (A) Expression levels of muERV-L. Rex1+/+ or Rex1−/− ES cells were treated with TSA or 5Aza for 24 h. Levels of muERV-L expression were determined by qRT–PCR as explained in the legend to Figure 2C. Results are depicted as fold expression in each cell line after normalization to Gapdh. The fold change was then plotted relative to untreated cells, with error bars representing SD. The fold muERV-L induction by TSA in Rex1+/+ versus Rex1−/−cells was 1.90. (B) Human kidney 293T cells were co-transfected with a plasmid expressing HA-tagged-intact REX1 proteins as indicated, in combination with a plasmid expressing LSD1/Kdm1a. Proteins were immunoprecipitated with αREX1 antibodies and analysed by western blot (top panels) together with proteins in total extracts (bottom panels) with monoclonal αHA antibody, αREX1 antibodies or αLSD1 antibodies as indicated on the right. Bands corresponding to HA-REX1 or LSD1 are marked on the right, protein molecular weight markers are indicated to the left in the bottom panels.
Methylation-independent silencing of muERV-L retroviral elements in ES cells is dependent on the lysine demethylase LSD1 (46). Other LSD1 target genes are also de-regulated in Rex1-deficient ES cells (22,46). As these data suggest co-operation between REX1 and LSD1, we probed for direct interactions. To do so, we introduced plasmids that drive expression of either HA-tagged REX1, together with plasmids that express cDNAs encoding LSD1 protein (48) in tissue culture cells. Interacting proteins were identified by western blot after immunoprecipitation. Using αREX1 serum, we detected LSD1 co-immunoprecipitated with HA-REX1 (Figure 5B, lanes marked LSD1). Detection of the interaction was dependent on the use of αREX1 serum as opposed to Pre-immune serum (Supplementary Figure S5B), and on the presence of REX1 proteins (Figure 5B). Furthermore, in the reverse experiment, we easily detected HA-REX associated with immunoprecipitated Flag-tagged LSD1 (Supplementary Figure S5C). These results indicate that REX1 is capable of association with LSD1 protein. We propose that REX1 may function in regulation of muERV-L expression in concert with LSD1 and associated HDACs.
Rex1 represses expression of ERV elements in gain- and loss-of-function mouse embryos
As we had observed an increase in expression of LTR retrotransposons in Rex1-depleted ES cells when compared with wild-type, we measured directly whether de-repression of retroelements was associated with mobilization (Supplementary Figure S4). As Rex1 participates in Polycomb regulation and on the other hand Rnf2 deletion was reported to severely affect retro-transposition of MLV36, a mouse leukaemia virus (ERV Class I) (62), we also tested mobilization of this retrovirus in Rex1-depleted ES cells.
We did not observe increases in the copy number of IAP, musD, Etn, muERV-L elements or of the MLV provirus in ES cells with attenuated levels of Rex1 as opposed to control cells (Supplementary Figure S4). We infer that although musD and muERV-L elements were overexpressed when Rex1 was depleted (Figure 2), they could not integrate into the genome at increased levels.
MuERV-L is expressed during pre-implantation development and displays a transient increased expression during the two-cell stage (38). To prove a potential role for Rex1 in control of muERV-L expression, we generated Rex1 loss-of-function embryos by micro-injection of zygotes with siRNAs directed against Rex1. Embryos were further cultured in vitro, and gene expression was assayed by RT–PCR at different stages using expression of H2afz as a reference gene, comparing embryos injected with control siRNAs or anti-Rex1 siRNAs (see ‘Materials and Methods’ section). As opposed to H2afz, expression of Rex1 was clearly affected by the siRNAs (Figure 6A, lanes 3 and 5). Rex1 mRNA levels were moderately attenuated in both two-cell embryos and morulas. Under these conditions, muERV-L levels were not altered in two-cell embryos (Figure 6A, lane 3). By contrast, we observed a significant difference in morula, as Rex1 attenuation resulted in ectopic expression of muERV-L.
Figure 6.
Rex1 gain-and-loss of function in mouse embryos. (A) Effect of attenuation of Rex1 levels on muERV-L expression. One-cell embryos were microinjected with control siRNAs (C) or Rex1-specific siRNAs (R), cultured to either the two-cell or the morula stage and analysed by semi-quantitative RT–PCR. cDNA from equivalent amounts of embryos was amplified with primers specific for the genes indicated; H2afz was used as a control for input. The top panel shows an ethidium bromide stained gel of amplification products. No I refers to no cDNA input. RNA extracted from E14T ES cells was processed alongside as a positive control. Bottom panel: quantification of Rex1 and muERV-L levels (grey and black bars for C and R, respectively). Data are represented as the average fold induction in Rex1 depleted morulas (black bars) using H2afz as a reference gene (two independent experiments). (B) Top panel: muERV-L and IAP mRNA expression in wild-type or Rex1 overexpressing mouse blastocysts. One-cell embryos were microinjected with plasmids that direct expression of Rex1-IRES-eGFP. Embryos were separated in groups according to eGFP expression levels at the eight-cell stage, and 4 days after injection RNA expression was analysed by semiquantitative RT–PCR as described in Figure 6(A). The following templates were used for amplification: E14T ES cells (lane 1), embryos with low or neglectable Rex1 overexpression levels (lane 2), embryos with high Rex1 overexpression (lane 3) or MQ water as a negative control (lane 4). Bottom panel: quantification of Rex1, Pou5f1/Oct4 and muERV-L levels, represented as fold repression in Rex1 overexpressing embryos using H2afz as a reference gene.
To test the effects of Rex1 gain-of-function, we also injected zygotes with expression vectors that overexpress Rex1 or eGFP as a control (See ‘Materials and Methods’ section). After continued development in vitro, we selected embryos with either high or low expression of GFP (from an attached IRES-eGFP in the case of Rex1) and assayed differences in gene expression between Rex1 and eGFP overexpressing embryos by RT–PCR. No differences were observed between wild-type, eGFP overexpressing embryos and embryos that express absent or low levels of the injected Rex1–IRES–eGFP plasmid (data not shown). Therefore, we compared embryos that express either neglectable or high levels of eGFP, after injection with Rex1-IRES-eGFP plasmid. Results are presented in Figure 6B. We easily observed expression in blastocysts of H2afz (control), IAP and muERV-L as described (38). Overexpression of Rex1 in these embryos (Figure 6B, lane 3) suppressed expression of both IAP and muERV-L elements. This reduction was not caused by a general effect on transcription, as expression of Oct4/Pou5f1 was not affected by Rex1 overexpression.
We conclude that attenuation of Rex1 levels interferes with repression of ERVs, while overexpression attenuates normal expression levels. These results demonstrate that Rex1 controls expression of ERV, particularly of the muERV-L family, during pre-implantation mouse development. These data indicate that Rex1 has a crucial role in controlling ERVs both during early embryonic development and in ES cells.
DISCUSSION
Rex1 regulates expression of ERVs
We show that attenuation of Rex1 levels de-regulates expression of ERV elements both in mouse ES cells and embryos. As REX1 also specifically binds these elements in mouse ES cells, we provide evidence for regulation by Rex1 of ERVs, especially muERV-L elements. To a lesser extent, binding was also observed to musD, IAP elements. It should be taken into account that the primers used were designed against conserved regions and therefore amplify several copies of each family. We suggest that this property may underestimate changes in both gene expression and association of REX1, as non-regulated copies may also be assayed. In mouse ES cells and in the presence of attenuated but detectable levels of Rex1 (Figure 1), regulation did not involve retro-transposition itself (Supplementary Figure S4). We cannot rule out the possibility that during pre-implantation development, or in ES cell truly deficient in Rex1, retro-transposition is affected by Rex1.
We demonstrate an important role for Rex1 in control of ERV, as Rex1 depletion in pre-implantation embryos provoked up-regulation of muERV-L, while the gain-of-function experiments caused down-regulation of several ERVs, including muERV-L (Figure 6). The de-regulation of muERV-L (and IAP) ERVs represents the earliest phenotypic abnormality described in Rex1 loss of function embryos.
During pre-implantation development, REs and particularly ERVs are prominently expressed in a precise temporal pattern (37–40), while general demethylation (41) removes the major repressing mechanisms active in adult tissues (63). Control of ERV expression during pre-implantation development involves protection of methylation patterns imposed by Dppa3/Stella (44) and the expression of ERV-specific siRNAs (43).
Recently, KDM1A/LSD1 has been discovered as a methylation-independent epigenetic regulator of ERV and ERV LTR-linked genes (46). Reminiscent of Rex1, Lsd1-deficient ES cells could be derived and deficiency caused de-regulation of pre-implantation-specific gene expression, in turn interfering with lineage determination upon differentiation. In addition, genome-wide analysis of binding sites in ES cells has revealed preferential binding of REX1 to RE-linked genes expressed during pre-implantation development (Guallar,D., García-Tuñon,I., Climent,M., Muniesa,P. and Schoorlemmer,J. manuscript in preparation). LSD1 is associated with HDACs (46,64) and HDAC-mediated repression of muERV-L retroviruses in ES cells is affected by Rex1 depletion (Figure 5A). Although we show that REX1 and LSD1 can interact (Figure 5B), the interrelationship of REX1 with either of these mechanisms in the control of ERV expression has not been molecularly defined yet. We speculate that similar to or even in concert with LSD1 and/or associated HDACs, REX1 may play a general role in control of transcription from ERV and ERV-derived elements during mouse pre-implantation development.
Rex1 and self-renewal/pluripotency in ES cells
Although it has been reported that Rex1 null mice appear normal, viable and fertile and that normal ES cells can readily be derived either from Rex1 null mice or by homologous recombination in vitro (22,23), an important role for Rex1 in ES cells is suggested by the high coincidence of Rex1 expression and enhanced pluripotency (18,20). We observe that the attenuation of Rex1 levels negatively affects self-renewal in mouse ES cells (Figure 1), in contrast to results reported earlier (22,23). The discrepancy may be due to functional redundancy with Yy1 and Yy2 proteins. Alternatively, the diminished self-renewal ability caused by depletion of Rex1 levels we demonstrate may only be revealed in the clonogenic assays we use. Surprisingly, we found no evidence that expression of muERV-L is elevated in Rex1-deficient ES cells (data not shown), and TSA treatment relieved repression of muERV-L in Rex1-deficient cells (Figure 5A). Therefore, in addition to repression through a Rex1/HDAC-dependent mechanism, a parallel pathway is operative to repress muERV-L in mouse ES cells. As repression of muERV-L is not notably influenced by demethylating agents, even in the absence of Rex1, we suggest that in ES cells neither mechanism is dependent on DNA methylation. We propose that a potential player in the second mechanisms might be RYBP (65).
As we fail to detect major differences in ES cell-specific gene expression upon Rex1 depletion, we suggest that, rather than directing pluripotency- and self-renewal-related gene expression, Rex1 may contribute to pluripotency and pre-implantation development via mechanisms that affect or read chromatin structure around ERVs. Such a role is also exemplified by the regulation of imprinted genes by Rex1 (28) at stages preceding the generation of pluripotent cells and by the epigenetic role of Rex1 in de-regulation of differentiation (22), possibly through association of REX1 to Polycomb group proteins (27).
ERVs and retroviral LTRs have been co-opted by cellular genes as promoter elements. In this way, ERVs contribute to ∼25% of POU5F1/OCT4 and NANOG-binding sites, both in human and mouse ES cells (66). ERVs, and especially ERV-K (IAP, musD, ETn), are over-represented among binding sites for NANOG, SOX2 and OCT4 (67). We speculate that Rex1 interaction with the principal pluripotency network (10) is centered at RE/ERVs and/or LTR-linked genes. Indirectly, Rex1 may influence posterior differentiation, similar to endodermal differentiation resulting from KDM1A/LSD1 deficiency (46).
A function for YY1-family members in control of REs/ERVs?
We show that REX1 is significantly enriched at genomic loci encoding members of the ERV2 and ERV3 superfamilies of LTR-containing ERVs) in mouse ES cells (Figure 3 and Supplementary Table SIV). Specifically, we observed binding of REX1 to muERV-L, ORR1 and MT elements, and to a lesser extent to IAP and musD elements. We do not know at present to which extent REX1 associates with a wider subset of ERV, but conclude that REX1 associates with at least a distinct subset of ERVs. Searches for REX1-binding sites within IAP/MusD/muERV-L sequences using position weight matrices revealed the presence of several potential binding sites, suggesting that regulation likely results from direct association of REX1 to DNA (B. Moreiras, data not shown). More extended analysis will be required to determine whether these associations are (super)family-dependent or are determined by the sequence and/or epigenetic environment of individual elements. Alternatively, the association of REX1 may be tissue-specific, as relative affinity for IAP and muERV-L elements is changed in placental tissue (M. Climent, R. Pérez-Palacios, S. Climent, P. Muniesa and J. Schoorlemmer, unpublished data).
REX1 has been described to activate Tsix expression through binding a cis-acting element discovered as DxPas34 (68,69). Not surprisingly, DxPas34 shares homology to the ERV3/ERV-L family of ERVs (69,70). Homology to this element is conserved in and restricted to human, mus musculus, rodents and eutherian mammals in general. These homologies support an origin of DxPas34 as an ERV-originated element and are compatible with a generic function for Rex1 in control of transcription from cis-elements that originate from ERVs. It has been suggested that ERV-L inserted more than 70 million years ago in the Tsix locus and was adopted as a regulatory module for Xi by early eutherians (69). Interestingly, Rex1 was duplicated from Yy1 by retro-transposition. This event also happened exclusively in eutherian mammals about 60–100 million years ago (24). We propose that similar to co-option of ERV as a regulatory element for Xi, Rex1 was co-recruited as a regulator of such elements.
All three Yy1 family members are present during pre-implantation development [(13,71), R. Pérez and J. Schoorlemmer, unpublished data] and in the germ line (13,72). REX1 shares extensive homology in the DNA-binding zinc fingers with YY1 and YY2, which prompted us to test association of YY1 and YY2 to ERVs bound by REX1 (Figure 4). We demonstrate appreciable binding of both proteins to muERV-L and IAP elements, hinting at a shared function of family members. As specificity was slightly altered compared to REX1, we hypothesize that each protein may regulate a specific subset of REs. De novo binding sites generated by transposons have contributed significantly to species-specific gene regulatory networks during pre-implantation development (67). As ERVs are less active in human as opposed to mouse (31) and certainly less prominent in the region corresponding to DXPas34 (69,70), it will be of interest to survey the RE specificity of YY1-family members in human.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables I–IV, Supplementary Figures 1–5, Supplementary Materials and Methods, and Supplementary References [73–76].
FUNDING
FIS/CarlosIII, Ministry of Health [PI07119]; the Ministerio de Educación y Ciencia [BFU2004-00467]; PAMER, Aragon Health Sciences Institute, Spain; Dpto de CTU, Gobierno de Aragón [PI110/09] the Government of Aragon/European Social Funds [B77] B77; PhD fellowships from the DGA [to D.G. and R.P-P.]; CIBER OBN [to P.M.]. Funding for open access charge: Aragon Health Sciences Institute [PIPAMER1003].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank A. Benítez for additional tissue culture, M. Strunk (I+CS) for quantification, I. García-Tuñón for expert advice, M. Sanz (I+CS) for embryo manipulation and B. Contreras Moreira for binding site analysis. The authors acknowledge A. de Diego (Transgenic Core Facility, I+CS) and D. García-Domingo (Cell Culture Core Facility, I+CS) for technical assistance with zygote injections and tissue culture, respectively. Jon Schoorlemmer wishes to express his gratitude towards the Departamento de Anatomía, Embriología y Genética Animal, Facultad de Veterinaria, Universidad de Zaragoza for its hospitality. The authors thank N. Brockdorf, A. Smith, H. Niwa, R. Velkey, M. Gassmann, I. Chambers, MJ. Barrero and A. Adamo for generously providing cells and expression vectors.
REFERENCES
- 1.Smith A. The battlefield of pluripotency. Cell. 2005;123:757–760. doi: 10.1016/j.cell.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Surface L, Thornton S, Boyer L. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Boyer L, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr. Opin. Genet. Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 6.Avilion A, Nicolis S, Pevny L, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 8.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 9.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 11.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 12.Hosler B, LaRosa G, Grippo J, Gudas L. Expression of REX-1, a gene containing zinc finger motifs, is rapidly reduced by retinoic acid in F9 teratocarcinoma cells. Mol. Cell. Biol. 1989;9:5623–5629. doi: 10.1128/mcb.9.12.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers M, Hosler B, Gudas L. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Y, Jahagirdar B, Reinhardt R, Schwartz R, Keene C, Ortiz-Gonzalez X, Reyes M, Lenvik T, Lund T, Blackstad M, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 15.Karlmark K, Freilinger A, Marton E, Rosner M, Lubec G, Hengstschläger M. Activation of ectopic Oct-4 and Rex-1 promoters in human amniotic fluid cells. Int. J. Mol. Med. 2005;16:987–992. [PubMed] [Google Scholar]
- 16.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 19.Brivanlou A, Gage F, Jaenisch R, Jessell T, Melton D, Rossant J. Stem cells. Setting standards for human embryonic stem cells. Science. 2003;300:913–916. doi: 10.1126/science.1082940. [DOI] [PubMed] [Google Scholar]
- 20.Chan E, Ratanasirintrawoot S, Park I, Manos P, Loh Y, Huo H, Miller J, Hartung O, Rho J, Ince T, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Gao W, Yang H, Zhang B, Zhu Z, Xue Y. Screening for genes essential for mouse embryonic stem cell self-renewal using a subtractive RNA interference library. Stem Cells. 2006;24:2661–2668. doi: 10.1634/stemcells.2006-0017. [DOI] [PubMed] [Google Scholar]
- 22.Scotland K, Chen S, Sylvester R, Gudas L. Analysis of Rex1 (zfp42) function in embryonic stem cell differentiation. Dev. Dyn. 2009;238:1863–1877. doi: 10.1002/dvdy.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masui S, Ohtsuka S, Yagi R, Takahashi K, Ko M, Niwa H. Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells. BMC Dev. Biol. 2008;8:45. doi: 10.1186/1471-213X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Faulk C, Kim J. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007;35:3442–3452. doi: 10.1093/nar/gkm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Chu J, Shen X, Wang J, Orkin S. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarro P, Oldfield A, Legoupi J, Festuccia N, Dubois A, Attia M, Schoorlemmer J, Rougeulle C, Chambers I, Avner P. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468:457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Tuñon I, Guallar D, Alonso-Martin S, Benito AA, Benítez-Lázaro A, Pérez-Palacios R, Muniesa P, Climent M, Sánchez M, Vidal M, et al. Association of Rex-1 to target genes supports its interaction with Polycomb function. Stem Cell Res. 2011;7:1–16. doi: 10.1016/j.scr.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Kim JD, Kim H, Ekram MB, Yu S, Faulk C, Kim J. Rex1/Zfp42 as an epigenetic regulator for genomic imprinting. Hum. Mol. Genet. 2011;20:1353–1362. doi: 10.1093/hmg/ddr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez MA, Pericuesta E, Fernandez-Gonzalez R, Moreira P, Pintado B, Gutierrez-Adan A. Transcriptional and post-transcriptional regulation of retrotransposons IAP and MuERV-L affect pluripotency of mice ES cells. Reprod. Biol. Endocrinol. 2006;4:55. doi: 10.1186/1477-7827-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin-Drubin ME, Munger K. Viruses associated with human cancer. Biochim. Biophys. Acta. 2008;1782:127–150. doi: 10.1016/j.bbadis.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perl A. Role of endogenous retroviruses in autoimmune diseases. Rheum. Dis. Clin. North Am. 2003;29:123–143, vii. doi: 10.1016/s0889-857x(02)00098-4. [DOI] [PubMed] [Google Scholar]
- 34.Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 36.Taruscio D, Mantovani A. Factors regulating endogenous retroviral sequences in human and mouse. Cytogenet. Genome Res. 2004;105:351–362. doi: 10.1159/000078208. [DOI] [PubMed] [Google Scholar]
- 37.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 2004;269:276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 39.Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- 40.Brûlet P, Condamine H, Jacob F. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc. Natl Acad. Sci. USA. 1985;82:2054–2058. doi: 10.1073/pnas.82.7.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 42.Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell. Mol. Life Sci. 2008;65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 2010;38:5141–5151. doi: 10.1093/nar/gkq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 45.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 46.Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kristensen DM, Nielsen JE, Skakkebaek NE, Graem N, Jacobsen GK, Rajpert-De Meyts E, Leffers H. Presumed pluripotency markers UTF-1 and REX-1 are expressed in human adult testes and germ cell neoplasms. Hum. Reprod. 2008;23:775–782. doi: 10.1093/humrep/den010. [DOI] [PubMed] [Google Scholar]
- 48.Adamo A, Sesé B, Boue S, Castaño J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 49.van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velkey JM, O'Shea KS. Oct4 RNA interference induces trophectoderm differentiation in mouse embryonic stem cells. Genesis. 2003;37:18–24. doi: 10.1002/gene.10218. [DOI] [PubMed] [Google Scholar]
- 51.Santoyo J, Vaquerizas JM, Dopazo J. Highly specific and accurate selection of siRNAs for high-throughput functional assays. Bioinformatics. 2005;21:1376–1382. doi: 10.1093/bioinformatics/bti196. [DOI] [PubMed] [Google Scholar]
- 52.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mak W, Baxter J, Silva J, Newall A, Otte A, Brockdorff N. Mitotically stable association of polycomb group proteins eed and enx1 with the inactive x chromosome in trophoblast stem cells. Curr. Biol. 2002;12:1016–1020. doi: 10.1016/s0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka S, Kunath T, Hadjantonakis A, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 55.Aubert J, Dunstan H, Chambers I, Smith A. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat. Biotechnol. 2002;20:1240–1245. doi: 10.1038/nbt763. [DOI] [PubMed] [Google Scholar]
- 56.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wobus AM, Kleppisch T, Maltsev V, Hescheler J. Cardiomyocyte-like cells differentiated in vitro from embryonic carcinoma cells P19 are characterized by functional expression of adrenoceptors and Ca2+ channels. In Vitro Cell Dev. Biol. Anim. 1994;30A:425–434. doi: 10.1007/BF02631310. [DOI] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 60.Boyer L, Plath K, Zeitlinger J, Brambrink T, Medeiros L, Lee T, Levine S, Wernig M, Tajonar A, Ray M, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 61.Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl Acad. Sci. USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 64.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hisada K, Sánchez C, Endo TA, Endoh M, Román-Trufero M, Sharif J, Koseki H, Vidal M. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol. Cell. Biol. 2012;32:1139–1149. doi: 10.1128/MCB.06441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 67.Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010;20:804–815. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vigneau S, Augui S, Navarro P, Avner P, Clerc P. An essential role for the DXPas34 tandem repeat and Tsix transcription in the counting process of X chromosome inactivation. Proc. Natl Acad. Sci. USA. 2006;103:7390–7395. doi: 10.1073/pnas.0602381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cohen DE, Davidow LS, Erwin JA, Xu N, Warshawsky D, Lee JT. The DXPas34 repeat regulates random and imprinted X inactivation. Dev. Cell. 2007;12:57–71. doi: 10.1016/j.devcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 70.Malakhova AA, Piatkova MS, Elisafenko EA, Shevchenko AI, Kel' A, Zakiian SM. [Comparative analysis of the DXPas34 regulatory region in rodents] Genetika. 2010;46:1401–1404. [PubMed] [Google Scholar]
- 71.Donohoe M, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo C, Lu X, Stubbs L, Kim J. Rapid evolution of a recently retroposed transcription factor YY2 in mammalian genomes. Genomics. 2006;87:348–355. doi: 10.1016/j.ygeno.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 73.Changolkar LN, Singh G, Pehrson JR. macroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol. Cell. Biol. 2008;28:2059–2065. doi: 10.1128/MCB.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gassmann M, Donoho G, Berg P. Maintenance of an extrachromosomal plasmid vector in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 1995;92:1292–1296. doi: 10.1073/pnas.92.5.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baust C, Gagnier L, Baillie GJ, Harris MJ, Juriloff DM, Mager DL. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J. Virol. 2003;77:11448–11458. doi: 10.1128/JVI.77.21.11448-11458.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.