Abstract
The 10-subunit RNA exosome is involved in a large number of diverse RNA processing and degradation events in eukaryotes. These reactions are carried out by the single catalytic subunit, Rrp44p/Dis3p, which is composed of three parts that are conserved throughout eukaryotes. The exosome is named for the 3′ to 5′ exoribonuclease activity provided by a large C-terminal region of the Rrp44p subunit that resembles other exoribonucleases. Rrp44p also contains an endoribonuclease domain. Finally, the very N-terminus of Rrp44p contains three Cys residues (CR3 motif) that are conserved in many eukaryotes but have no known function. These three conserved Cys residues cluster with a previously unrecognized conserved His residue in what resembles a metal-ion-binding site. Genetic and biochemical data show that this CR3 motif affects both endo- and exonuclease activity in vivo and both the nuclear and cytoplasmic exosome, as well as the ability of Rrp44p to associate with the other exosome subunits. These data provide the first direct evidence that the exosome-Rrp44p interaction is functionally important and also provides a molecular explanation for the functional defects when the conserved Cys residues are mutated.
INTRODUCTION
The RNA exosome is involved in a wide variety of RNA processing and degradation reactions in both the nucleus and the cytoplasm. First, the nuclear exosome processes a subset of RNAs from longer precursors. For example, it processes a 300-nt 7S precursor into the 160-nt 5.8S rRNA (1). Second, the nuclear exosome completely degrades some RNAs that are byproducts of gene expression, including the 5′-external transcribed spacer that is part of the rRNA precursor (2). Third, the nuclear exosome degrades aberrant RNAs that fail to complete proper processing, including incompletely modified initiator tRNA (3). Fourth, the exosome is involved in one of two general pathways of cytoplasmic mRNA decay (4). Fifth, the cytoplasmic exosome is especially important for degrading aberrant mRNAs, including those that lack a stop codon (nonstop mRNAs) as well as those that are cleaved by a ribozyme (5,6).
Although the exosome has a wide variety of substrates, it acts very specifically on those substrates. For example, the exosome degrades initiator tRNA lacking a single methyl group, but not normal initiator tRNA or other tRNAs that lack a modification (3,7). A second example of the exosome’s specificity is that the exosome degrades both the poly(A) tail and the body of nonstop mRNAs (6), while the poly(A) tail of normal mRNAs can only be removed by dedicated deadenylases and not by the exosome (8). What is not yet known is how the exosome carries out these diverse functions while maintaining specificity.
A series of X-ray crystallography and EM studies have resolved the structural organization of the yeast exosome (9–15). The exosome contains a core of ten proteins that are shared between the nuclear and cytoplasmic exosome. At least in fungi and metazoans, only the Rrp44p subunit (also known as Dis3p) is catalytically active (9,16). Rrp44p was initially identified as an exonuclease with similarity to the RNase II family (1). The similarity to RNase II includes the catalytic RNB domain and three OB-fold RNA-binding domains (CSD1, CSD2 and S1 (17)). We and others have shown that this exonuclease activity is not essential for viability because Rrp44p contains a second domain in its N-terminus (PIN) with endonuclease activity (18–20). Mutations that inactivate each of the nuclease activities individually (hereafter referred to as rrp44–endo− and rrp44–exo−) are viable, but simultaneously mutating both (rrp44–endo−exo−) is lethal, suggesting a functional overlap (18–20). Although the structure containing nine exosome subunits is thought to be conserved in other eukaryotes, whether or not Rrp44p in other eukaryotes stably associates with the other nine subunits is controversial.
The five recognized domains of Rrp44p explain much of the sequence conservation in Rrp44p orthologs. A notable exception is a conserved motif near the N-terminus that has no known function (Figure 1A). This motif has been named CR3 (Cysteine-Rich with three cysteines (21)). In the process of trying to understand how the exosome structure relates to its diverse functions, we initially constructed a mutant Rrp44p in which all three Cys residues were changed to Ser. This rrp44–CR3 allele caused slow growth (19), and severely reduced the exosome’s ability to degrade nonstop mRNAs and ribozyme cleaved mRNAs (22). Interestingly, neither the rrp44–endo− nor the rrp44–exo− mutation caused a similar defect in mRNA decay, suggesting that the rrp44–CR3 mutation somehow affected both catalytic activities. The protein encoded by rrp44–CR3 accumulated to slightly reduced levels (22). However, this is probably not the cause of the observed phenotypes since overexpression of the rrp44–CR3 allele from a high copy plasmid did not restore growth. In addition, haplo-insufficiency of RRP44 did not reduce growth similar to that of rrp44–CR3 (data not shown). All of these data can be explained if the rrp44–CR3 mutation reduces both exo- and endonuclease activities of the exosome, thus causing the slow growth and stabilization of nonstop mRNAs. However, the observation that rrp44–CR3 is viable suggests that it does not completely eliminate catalytic activity.
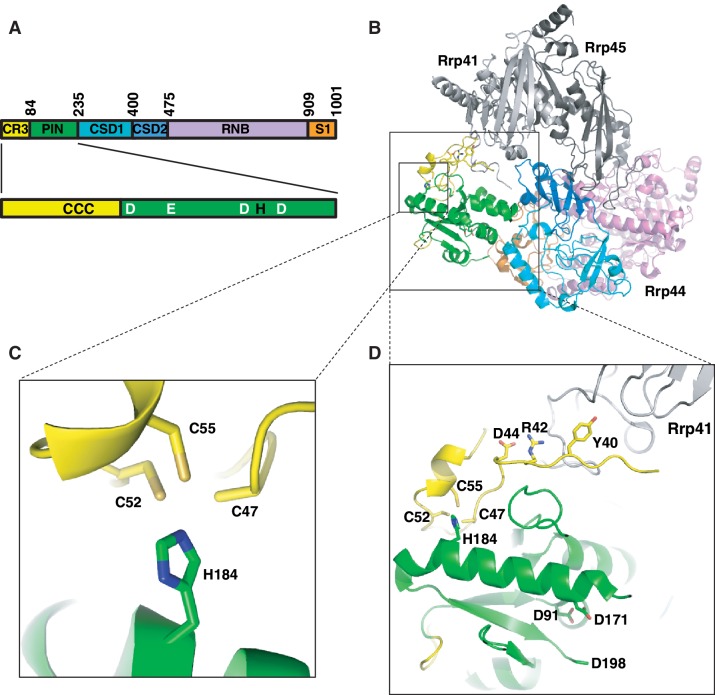
Figure 1.
Rrp44 contains a conserved CCCH motif. (A) Top: Diagram depicting the five recognized domains in Rrp44 and the CR3 motif. PIN denotes the endonuclease domain, CSD1 and CSD2 denote RNA-binding cold shock domains, RNB denotes the exonuclease domain and S1 denotes an RNA-binding S1 domain. Collectively the CSD1, CSD2, RNB and S1 domains are responsible for exonuclease activity. Bottom: the N-terminus of Rrp44p contains a conserved CCCH motif (black letters) in addition to the catalytic residues of the PIN domain (white letters). (B) Overview of the Rrp44 structure (colored as in panel A) bound to exosome subunits Rrp41 and Rrp45 (PDB ID 2WP8; 13). (C) The three conserved Cys residues and the conserved His residue form a tetrahedral cluster in the crystal structure. Note that the sulfur atom of Cys47 was not modeled. (D) The CR3 motif is physically connected to the exosome-binding site (Y40, R42 and D44) and the endonuclease active site of the PIN domain (D91, D171 and D198). Note that the D198 side chain was not modeled. Structures were rendered using PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.1 Schrödinger, LLC.).
Recently, the x-ray crystal structure of Rrp44p bound to two other exosome subunits was published, providing the first structural insight into the CR3 motif (13). We noted that the CR3 motif is close to a His residue and that these four residues are oriented in a tetrahedral configuration that is reminiscent of a mono-metal-binding site. Here, we present genetic, biochemical and phylogenetic data that offer a coherent explanation of how the CR3 motif alters both the endonuclease and exonuclease activities of Rrp44p in vivo: the direct effect of the CR3 mutation is likely a modest perturbation of the local folding and/or structural stability of Rrp44p, possibly because of the loss of a metal ion. Even though the CR3 motif is distant from both catalytic sites, this folding defect has two important consequences. First, the CR3 motif is close to residues contacting another exosome subunit, and alterations in the CR3 motif reduce copurification of the other exosome subunits. This, in turn, can explain the defect in exonuclease function in vivo. Second, changes in the structure of the CR3 motif appear to cause modest changes in the endonuclease active site, possibly because the two sites are connected by a single α-helix.
MATERIALS AND METHODS
Strains and plasmids
The majority of the experiments used a previously described rrp44 deletion strain with a RRP44, URA3 plasmid (yAV1115; (19)). Various alleles of RRP44 were introduced using the plasmid shuffle as previously described (19). All of these plasmids are low copy plasmids, and Rrp44p is expressed under control of its native promoter and 3′-flanking sequence. The plasmids used are listed in Supplementary Table S1. The dcp1–2 rrp44Δ strain (yAV1143) was previously described (22). To test for synthetic lethality between rrp6Δ and RRP44 alleles, the heterozygous diploid (RRP44/rrp44Δ::KAN; 23) obtained from Open Biosystems was transformed with the wild-type RRP44 plasmid, pAV361 (19). The diploid was then sporulated to generate a haploid rrp44Δ::KAN strain with pAV361. This strain was converted to rrp44Δ::HYG by homologous recombination with pAG32 (24) and selecting for hygromycin resistance. The rrp44Δ::HYG strain with pAV361 was then crossed with the rrp6Δ::KAN strain from the yeast knock out collection (23). The resulting diploid was sporulated resulting in yAV1137 (MATa, leu2-Δ0, ura3-Δ0, his3-Δ1, rrp44Δ::HYG, rrp6Δ::NEO with a RRP44, URA3 plasmid).
Growth assays
Synthetic lethality growth assays were performed essentially as described (25). Briefly, plasmids encoding the rrp44 point mutations were transformed into the rrp44Δ strain, the dcp1–2 rrp44Δ strain or the rrp6Δ rrp44Δ strain containing a URA3 plasmid encoding wild-type Rrp44p. Transformants were grown in SC–URA–LEU overnight at 23°C. Cultures were diluted in the same selection media to an optical density of 0.8 at 600 nm. Next, cells were serially diluted in 96-well plates by a factor of five and spotted onto media containing 5-Fluoroorotic Acid (5-FOA). 5-FOA selects for cells that have lost the URA3 plasmid, and therefore growth indicates that the gene on the LEU2 plasmid is sufficient for viability. Plates were incubated at 23°C, 30°C and 37°C for 2–5 days. The same effects were seen at all temperatures unless otherwise noted in the results section.
The his3-nonstop growth assay was performed essentially as described (6). Briefly, rrp44 deletion strains containing a LEU2 plasmid encoding the rrp44 point mutations were transformed with a URA3 plasmid encoding a his3-nonstop reporter. This reporter contains a 1-bp deletion which removes the stop codon from the HIS3 mRNA. Transformants were grown in SC–URA–LEU overnight at 30°C. Cultures were diluted in the same selection media to an optical density of 0.8 at 600 nm. Next, cells were serially diluted in 96-well plates by a factor of five and spotted onto media lacking histidine, SC–HIS–URA–LEU. The plates were incubated at 30°C for 2–6 days.
Stability of GAL7 mRNA and northern blotting
To determine the half-life of the GAL7 mRNA, duplicate cultures of the dcp1–2 yeast strains were grown in YEP + 2% galactose overnight at 23°C. Cultures were diluted in the same media to an optical density of 0.2 at 600 nm and grown to an optical density of 0.8. Cultures were then shifted to 37°C (a non-permissive temperature for dcp1–2) for 1 h. The medium was replaced with 1/10th volume YEP + 4% glucose. At the time points indicated, 2 ml aliquots were pelleted. RNA was isolated and subjected to formaldehyde agarose gel electrophoresis, followed by northern blot analysis essentially as described (5,26). Northern blots were hybridized with 32P 5′-end-labeled oligonucleotides specific for GAL7 (oAV267; 5′-CTCTTGCTTCTCTGGAGAGATCGTCAGTC) and the RNA subunit of the signal recognition particle, SCR1 (oRP100; 5′-GTCTAGCCGCGAGGAAGG). Signals were detected and quantitated using a STORM PhosphoImager (Amersham), and corrected for loading by quantitating SCR1.
RNase assays on recombinant Rrp44p
Truncated Rrp44p fused to GST was expressed in Escherichia coli, purified, and assayed as previously described (19). Briefly, the Rosetta(DE3) E. coli strain with Rrp44 expression plasmids was grown at 30°C to an optical density of 1.2 at 600 nm. Expression was induced with 0.2 mM IPTG at 18°C overnight. Rrp44 was purified over a GSTtrap FF 1-ml column and a HiTrapTMQ FF column (GE Healthcare). RNase assays used a 5′-end-labeled 30-mer RNA (either A30 or CCCGACACCACCCACUA14), and 20 mM HEPES, pH 7.5, 150 mM NaCl, 3 mM MnCl2, 1 mM DTT.
Rrp44p exosome copurification
The Rrp44p–TAP and Rrp44p–CR3–TAP plasmids were previously described (22). Cultures were grown in 50 ml media to an optical density of 0.8 at 600 nm, pelleted, and resuspended in 250 µl IP50 buffer [50 mM NaCl, 50 mM Tris–HCl pH 7.5, 2 mM MgCl2, 0.1% Triton X100, 0.5 mM β-mercaptoethanol, 0.1 mM PMSF, with complete EDTA-free protease inhibitors (Roche)]. 50 µl glass beads (Sigma) were added to the cell suspension and agitated in a vortex mixer for 7 min at 4°C. Lysates were clarified by centrifugation at 4500g for 7 min at 4°C.
TAP-tagged proteins were purified by adding 10 µl IgG Sepharose 6 Fast Flow beads (Amersham Biosciences) to 200 µl whole-cell lysate. After incubation for 1 h at 4°C, the supernatant was discarded and the beads were washed two times with 200 µl IP50 buffer, and then twice more with 200 µl IP1000 buffer (identical to IP50, but 1 M NaCl). Bound proteins were eluted from beads by heating to 95°C for 3 min in protein sample buffer and analyzed by western blotting using anti-protein A antibodies (Sigma), anti-Rrp4p antibodies (27) and anti-Pgk1p antibodies (Invitrogen).
RESULTS
Three conserved cysteines in Rrp44p cluster with a conserved histidine
In addition to its five recognizable domains, Rrp44p orthologs contain a conserved motif that is found N-terminal to the PIN domain (Figure 1A). We have previously shown that deleting as many as 766 residues from the C-terminus of Rrp44p can be tolerated, but that deleting as little as 84 residues from the N-terminus results in lethality, suggesting that this conserved N-terminal region is important (19). This region was named CR3 (cysteine-rich domain with three cysteines) because it contains a Cx4Cx2C motif that does not resemble any other known domain or motif (21). A 3.0 Å crystal structure of Rrp44p was recently solved, providing the first structural insight into this motif (13). The structure reveals that the three conserved Cys residues are physically adjacent to each other and that H184 is in close proximity to the three conserved Cys residues (Figure 1C).
To examine whether H184 was conserved and co-occurred with the CR3 motif, we performed a multiple sequence alignment of 20 Rrp44p homologs from very diverse eukaryotes using ClustalW. The alignment showed that both the CR3 motif and H184 are conserved in 17 of the 20 included proteins (Supplementary Figure S1A). Strikingly, two of the Rrp44p orthologs (from Ciona intestinalis and Trichomonas vaginalis) lack two or all three of the conserved Cys residues of the CR3 motif and also lack H184. The 20th protein, hDis3L1 (and Dis3L1 from other vertebrates), contains His184 together with a variation on the CR3 motif with five residues separating the second and third Cys instead of just two. Thus, His184 is conserved in 18 Rrp44p homologs that have the CR3 motif, but not in the two homologs that lack the CR3 motif. H184 is part of the PIN endonuclease domain, but as shown in Supplementary Figure S1B, it is not present in PIN domains that are not part of Rrp44p orthologs. Thus, the previously identified CR3 co-occurs and is in close proximity to a conserved histidine. Conserved CCCH motifs in other proteins form binding sites for a metal ion (generally Zn2+). We note, however that the organization in the Rrp44p amino acid sequence and protein fold is distinct from known Zn-binding domains explaining why it previously went unrecognized (See ‘Discussion’ section).
A triple mutation in the CR3 motif affects growth, but single point mutations do not have the same effect
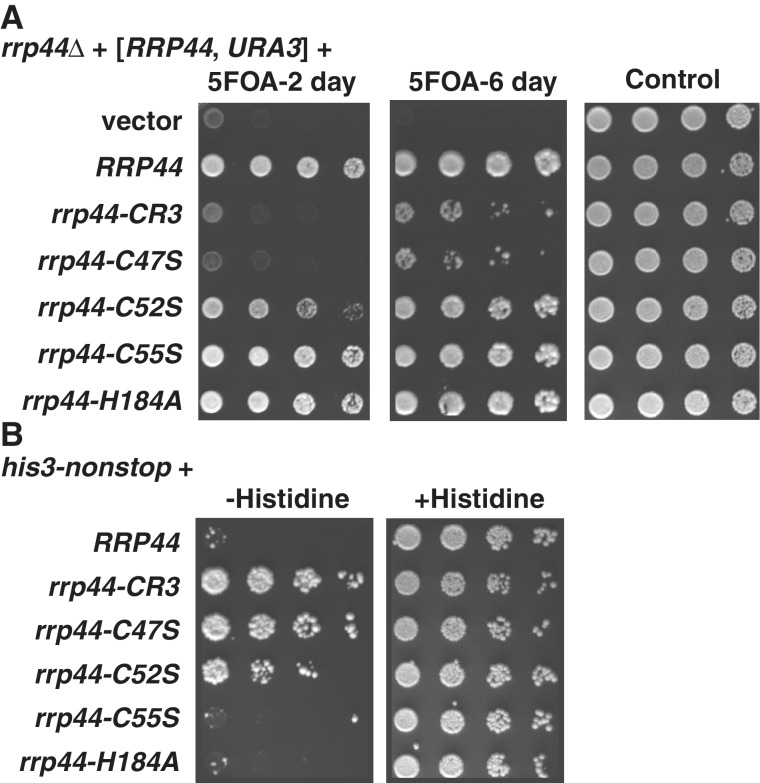
We have previously shown that mutating the three conserved cysteine residues of the CR3 motif to serine (C47S, C52S, C55S; hereafter referred to as rrp44–CR3) resulted in a slow growth phenotype (19). One possibility is that two of the conserved Cys residues form a disulfide bond, which is suggested by the proximity of the modeled sulfur atoms in the Rrp44p crystal structure. If this growth defect was caused by the disruption of a disulfide bond between two of the cysteines, then single point mutations in the two residues forming the bond should have the same effect, whereas the non-interacting Cys would not be expected to affect the disulfide bond, and thus should not affect growth. If, however, the CR3 motif forms a metal-binding site together with H184, any one of the single mutations might decrease affinity for the metal and a triple mutant might further decrease metal affinity. Thus, to distinguish between these two proposed roles of the CR3 motif we created single mutants, C47S, C52S, C55S and H184A, and compared their effect to the rrp44–CR3 triple mutant. For this assay, we used a rrp44Δ strain containing a plasmid with wild-type RRP44 and URA3 genes. This strain was transformed with a second plasmid that contained the mutant RRP44 and LEU2 genes. If this second plasmid can provide functional Rrp44p, then the strain can lose the URA3 plasmid, resulting in growth on plates containing 5-FOA. As shown in Figure 2A, the single mutants showed a variety of growth phenotypes. The C47S mutant grew worse than the other single mutants, with no obvious growth upon short incubation (2 days; Figure 2A left panel). C52S grew slightly worse than wild-type, which was obvious at 2 days. Upon longer incubation (6 days; Figure 2A middle panel) the C47S mutant showed some growth, and the C52S colony size became almost indistinguishable from wild-type. The C55S and H184A single mutants grew as well as the wild-type strain after both short and long incubation.
Figure 2.
Point mutations in the conserved CCCH motif are inconsistent with a disulfide bond, but are consistent with metal binding. As previously reported, simultaneously mutating the three conserved Cys residues to Ser causes a slow growth phenotype (A) and stabilization of his3-nonstop mRNA (B). The single C47S mutation has a similar, but slightly less severe defect, which can be seen after 6 days on 5-FOA plates in the middle panel. Shorter incubations (e.g. 2 days; left panel) show that the rrp44–C52S allele also has a significant but less severe defect. For panel A a rrp44Δ strain with a low copy plasmid with the wild-type RRP44 and URA3 genes was transformed with a second plasmid with a LEU2 gene and the indicated RRP44 gene (none, wild-type, rrp44–CR3, rrp44–C47S, rrp44–C52S, rrp44–C55S, or rrp44–H184A). Transformants were selected and then serially diluted and spotted on 5FOA containing plates that select for cells that have lost the URA3 plasmid or SC–URA–LEU plates. For Panel B, the colonies that grew on 5FOA were then transformed with a reporter plasmid that contains the his3-nonstop and URA3 genes. Transformants were selected and then serially diluted and spotted on SC–URA–LEU–HIS plates or SC–URA–LEU plates.
We have also shown previously that the rrp44–CR3 mutant has a defect in the rapid degradation of mRNAs that lack a stop codon (22). Because the rrp44–CR3 mutant stabilizes the his3-nonstop mRNA, a rrp44–CR3 his3-nonstop strain produces sufficient histidine to support growth in the absence of added histidine, whereas an isogenic RRP44 his3-nonstop strain fails to grow under these conditions (22). To determine which residues of the CR3 motif are needed for nonstop mRNA decay, we tested whether the C47S, C52S, C55S or H184A single mutants could suppress a his3-nonstop allele. Importantly, the growth results in this assay are counterintuitive: Growth occurs when nonstop mRNA decay is defective, where as functional nonstop mRNA decay limits growth. As shown in Figure 2B, the his3-nonstop defects mirrored the growth defects in Figure 2A. The rrp44–CR3 allele is defective in nonstop mRNA decay, and thus grows. The single C47S mutant shows almost the same defect. C52S showed a less severe defect, and C55S and H184A were similar to wild type.
Results similar to ours, where mutating individual metal-binding residues have different effects, have been reported for various Zn2+-containing proteins (see ‘Discussion’ section). Presumably this indicates that mutating a single Cys residue to Ser reduced the affinity for Zn-ions, but does not completely eliminate Zn2+ binding. Thus, the phenotypes of single amino acid point mutants are consistent with the CCCH motif forming a binding site for a metal ion, with single point mutations decreasing the affinity for the ion and a triple mutation further decreasing this affinity. In contrast, the phenotypes of the single-point mutants appear inconsistent with a disulfide bond.
Genetic interactions indicate that the CR3 motif affects multiple functions of the exosome in vivo
As explained in the Introduction, the known phenotypes of the rrp44–CR3 mutant can best be explained if this mutation decreases both the endo- and exonuclease activities of Rrp44, but does not completely abolish activity. To better understand the connections between the CR3 motif and nuclease activities of the exosome, we tested for genetic interactions between the rrp44–CR3 mutation, and mutations inactivating the catalytic activities of the exosome.
The slow growth phenotype of rrp44–CR3 resembles that of a D551N point mutation that inactivates the exonuclease activity of Rrp44p (hereafter called the rrp44–exo− mutation). One possibility is that the rrp44–CR3 mutation somehow prevents the exonuclease function of Rrp44p, and thereby results in slow growth. This predicts that adding the rrp44–CR3 mutation to the already inactive rrp44–exo− mutation would have no additional effect. Another possibility is that the reason rrp44–CR3 grows slowly is distinct from the reason why rrp44–exo− grows slowly. In this case, combining the two mutations might have an additive effect. To test these possibilities, we combined rrp44–CR3 with rrp44–exo−. Figure 3A shows that an RRP44 plasmid combining the rrp44–CR3 and rrp44–exo− mutations (i.e. C47S, C52S, C55S, D551N) did not support growth. As previously reported, the plasmids encoding either rrp44–CR3 or rrp44–exo− allowed for slow growth compared to wild-type RRP44. We conclude that the rrp44–CR3 mutation is synthetically lethal with the rrp44–exo− mutation. Using the same approach, Figure 3B shows that rrp44–CR3 is also synthetically lethal with a deletion of the exonuclease domain (rrp44–exoΔ). Although rrp44–CR3 and rrp44–exo− have some of the same molecular defects disrupting the exonuclease activity further worsens the growth of rrp44–CR3. We conclude that the rrp44–CR3 mutation does not completely eliminate in vivo exonuclease function, and that the slow growth phenotypes of rrp44–CR3 and rrp44–exo− are distinct. Furthermore, since combing the rrp44–endo− and rrp44–exo− alleles is also synthetic lethal, the rrp44–CR3-exo− synthetic lethality is consistent with rrp44–CR3 affecting endonuclease activity.
Figure 3.
Synthetic lethality of rrp44–CR3 with mutations in the exosome. Simultaneously mutating the three conserved Cys residues of Rrp44p to Ser is synthetically lethal with point mutations disrupting the Rrp44p exonuclease (A), with deletion of the Rrp44p exonuclease domains (B) or with deletion of the Rrp6p exonuclease (D), but not point mutations disrupting the Rrp44p endonuclease (C) For panels A, B and C synthetic lethality was tested using a plasmid shuffle assay as in Figure 2. Failure to grow on 5-FOA plates in panels A to C indicates that the tested RRP44 allele is inviable. For panel D, for the top row and bottom rows an rrp6Δ, rrp44Δ double mutant with a wild-type RRP44, URA3 plasmid was transformed with either a wild-type RRP44, LEU2 plasmid (top row), or a rrp44–CR3, LEU2 plasmid (bottom row). For the middle row an rrp44Δ mutant with a wild-type RRP44, URA3 plasmid was transformed with a rrp44–CR3, LEU2 plasmid. All three double transformants were then serially diluted and spotted on 5FOA containing plates that were then incubated at the indicated temperatures.
Next, to test the genetic interaction between the CR3 motif and the endonuclease domain of Rrp44p, we combined the rrp44–CR3 allele with rrp44–endo− mutant. Figure 3C shows that this combination has the same growth phenotype as the rrp44–CR3 single mutant. This is also consistent with the rrp44–CR3 mutation not completely eliminating exonuclease function, since elimination of exonuclease function (e.g. rrp44–exo−) is synthetic lethal with rrp44–endo−.
We next tested for genetic interactions with rrp6Δ. Rrp6p is an exoribonuclease that associates with the nuclear form of the exosome, and thus provides it with a third catalytic domain. Figure 3D shows that the rrp6Δ rrp44–CR3 double mutant grows significantly slower than either single mutant at 23°C and fails to grow at 37°C, indicating that rrp6Δ has a synthetic growth defect with rrp44–CR3.
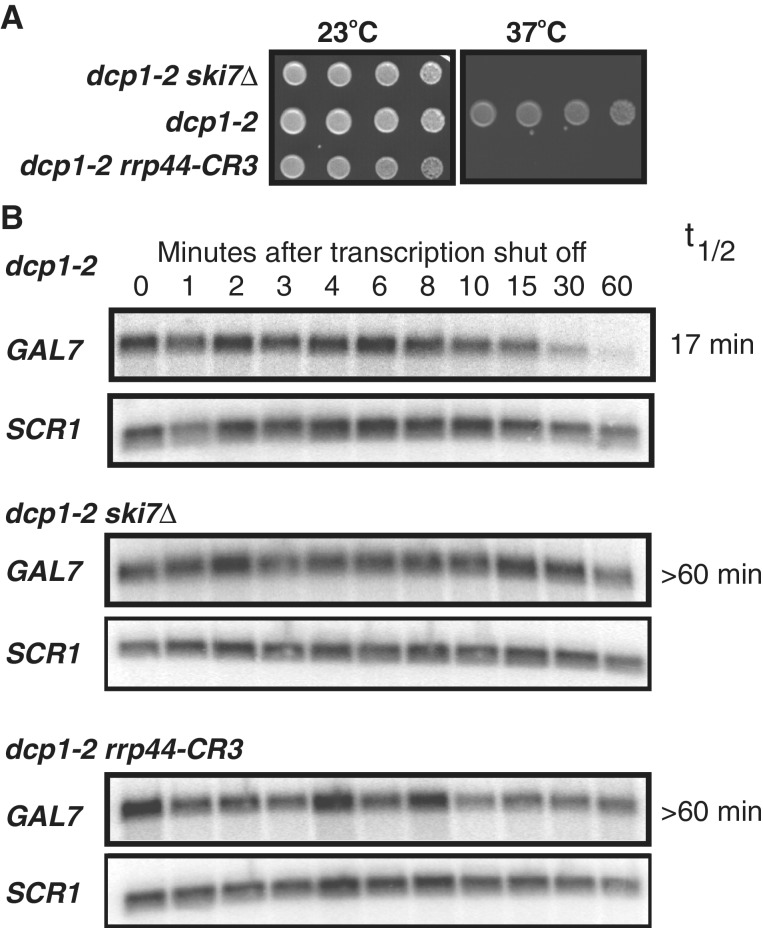
Since the nuclear exosome is essential, but the cytoplasmic exosome is not, the above growth phenotypes reflect effects of the mutations on nuclear exosome function. To extend our observations to the cytoplasmic exosome, we tested for effects on mRNA decay. Normal mRNAs are mainly degraded through a decapping pathway. In the absence of decapping activity, normal mRNAs are degraded by the exonuclease activity of the exosome (4). Thus, mutations that inactivate decapping are synthetically lethal with mutations that inactivate the exonuclease activity of the exosome (22). To test whether the rrp44–CR3 allele reduces exosome-mediated decay of normal mRNAs, we initially looked for genetic interactions with dcp1–2. dcp1–2 is a temperature sensitive mutation in a subunit of the decapping enzyme. Since decapping is not required for viability, this strain is viable at all temperatures, but requires exosome-mediated mRNA decay at the restrictive temperature. Figure 4A shows that, as expected, the dcp1–2 rrp44–CR3 double mutant is viable at the permissive temperature (23°C), but inviable at the restrictive temperature for dcp1–2 (37°C). Therefore, the rrp44–CR3 mutation likely disrupts exosome-mediated decay of normal mRNAs.
Figure 4.
Simultaneously mutating the three conserved Cys residues of Rrp44p to Ser stabilizes cellular mRNAs. (A) The rrp44–CR3 mutation is synthetically lethal with a temperature-sensitive mutation in the decapping enzyme, dcp1–2. The experiment was performed as described for Figures 2 and 3. (B) The rrp44–CR3 mutation reduces the rate of exosome-mediated degradation of the GAL7 mRNA. duplicate cultures of the indicated strains were grown in media containing galactose, and then incubated for 1 h at 37°C to inactivate the decapping enzyme. The media was then replaced with media containing glucose to shut off transcription of the GAL7 gene and RNA was isolated at the indicated time points and analyzed by northern blotting. The RNA subunit of the signal recognition particle (encoded by the SCR1 gene) was used as a loading control. RNA levels were quantitated using a STORM PhosphoImager and half-lives determined. Both half-life measurements for the dcp1–2 strain were 17 min. In the dcp1–2 ski7Δ and dcp1–2 rrp44–CR3 strains the half-lives were more than 60 min.
The dcp1–2 rrp44–CR3 double mutant also allowed us to directly measure the effect of rrp44–CR3 on the degradation of an endogenous mRNA, GAL7. For this experiment we grew dcp1–2 strains at the permissive temperature in the presence of galactose to mid-log phase. The cultures were then incubated at the restrictive temperature for 1 h to inactivate the decapping enzyme, followed by the addition of glucose to inhibit further transcription of the GAL7 gene. Figure 4B shows that in a dcp1–2 single mutant, the endogenous GAL7 mRNA decays with a half-life of about 17 min. In contrast, in the dcp1–2 rrp44–CR3 double mutant, the GAL7 mRNA half-life is greater than 60 min, which is similar to the dcp1–2 ski7Δ control strain. Both the dcp1–2 synthetic lethality and GAL7 mRNA half-lives suggest that the rrp44–CR3 mutation interferes with exosome-mediated degradation of normal cellular mRNAs.
Overall, these genetic interactions are consistent with the hypothesis that the rrp44–CR3 mutation reduces, but does not eliminate, the exonuclease activity in vivo. Furthermore, the synthetic interactions with rrp6Δ and dcp1–2 suggest that the rrp44–CR3 allele affects both cytoplasmic and nuclear exosome function.
Mutation of the CR3 motif affects the endoribonuclease activity of Rrp44p in vitro
While the genetic analysis above indicates that the CCCH motif is important in vivo, it can not address whether these amino acids play a direct role or an indirect role in Rrp44p nuclease activity. In fact, since the CCCH motif is very well separated from the exonuclease domain, we consider it unlikely that these amino acids have a direct role in exonuclease activity. At first glance, the CCCH motif is also far (>25 Å) away from the endonuclease active site, however it is important to note that H184 of the CCCH motif and D171 of the endonuclease active site are part of the same α-helical structure (Figure 1D and Supplementary Figure S1A). Because D171 is a key residue in the endonuclease active site (18–20), we considered the possibility that perturbation to the CCCH site may have an indirect effect on the orientation or flexibility of this α-helix, and thereby affect endonuclease activity. To test this, we purified three truncated Rrp44 proteins as glutathione S-transferase (GST) fusions from E. coli. As we and others have previously shown, a truncated Rrp44 protein that contains the PIN domain and CR3 motif (residues 1–235), has endonuclease activity in vitro (18–20). The second recombinant protein purified is the same, except that the three conserved Cys residues were mutated to Ser. This CR3 mutant enzyme was less active in in vitro assays containing the same amount of enzyme of similar purity (Figure 5). Specifically, Figure 5 shows that the wild-type enzyme degraded most of the substrate in the first 5 min, with all of the substrate gone after 15 min. In comparison, more substrate was left in the reaction with the same amount of the enzyme with all three Cys residues mutated after 5 or 15 min. The third recombinant protein we purified contains only the part of Rrp44p that is similar to other PIN domains (residues 84–235) and lacks the CR3 motif. This PIN domain only protein also exhibited less nuclease activity in vitro (Figure 5). Quantitation of the experiment shown in Figure 5 indicated that the wild-type enzyme was approximately twice as active as either mutant version. Although individual batches of purified protein can have variations in the amount of activity, the differences between the three forms of Rrp44p were reproducible with independent repeat purifications.
Figure 5.
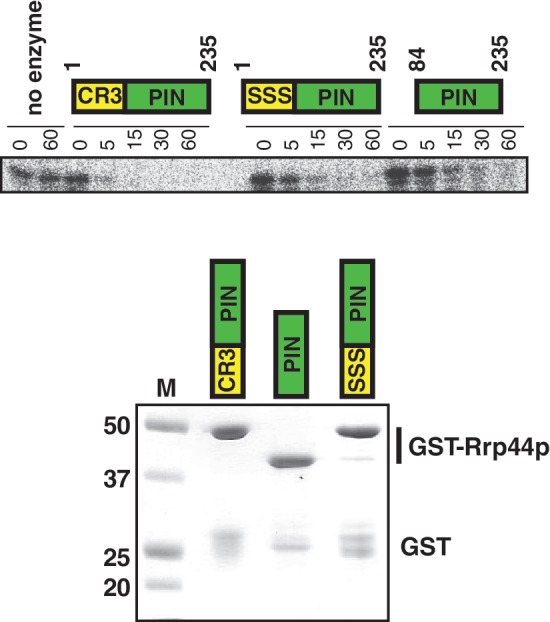
Mutating the CCCH motif affects the in vitro endonuclease activity of Rrp44p. Recombinant truncated Rrp44 proteins that either contain both an intact CR3 motif and PIN domain (i.e. residues 1–235), a similar protein with the Cys residues in the CR3 motif mutated to Ser, or a protein containing only the PIN domain (i.e. residues 84–235) were used in in vitro RNase assays with an A30 substrate as described in materials and methods. Similar results were obtained with a CCCGACACCACCCACUA14 substrate. The bottom panel show a Coomassie stained gel of the purified recombinant proteins.
Mutation of the CR3 motif affects the interaction between Rrp44p and the core exosome
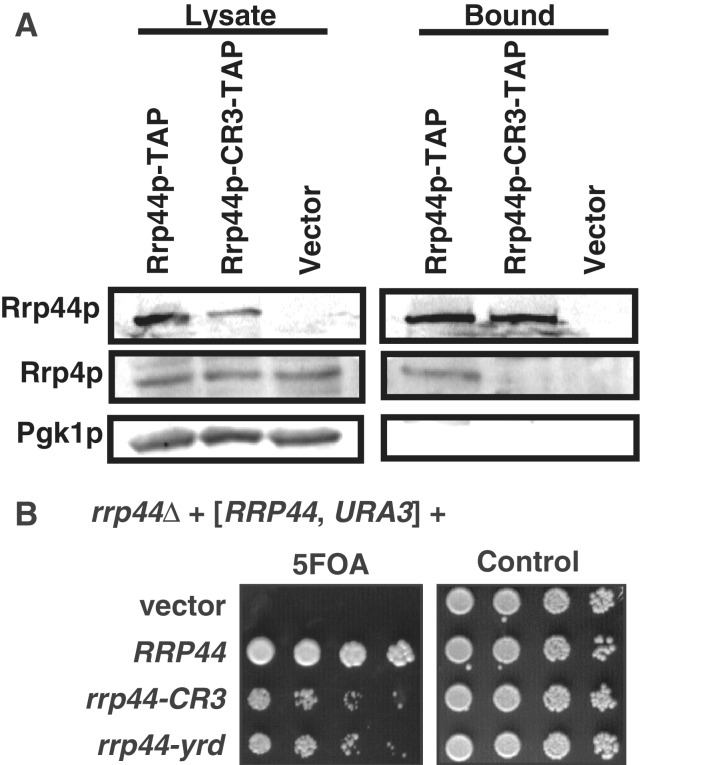
We noted that the CCCH motif is close to three residues that interact with the core exosome. Specifically, in the crystal structure of a trimer of Rrp41p, Rrp45p and Rrp44p, the conserved Rrp44p Y40, R42 and D44 residues directly interact with Rrp41p (Figure 1D; (13)) and only two residues separate this YxRxD motif from the Cx4Cx2C motif (Supplementary Figure S1A). This led us to hypothesize that mutating the conserved Cys residues to Ser might disorder this part of the protein and reduce the interaction with the core exosome. To test this idea, TAP-tagged wild-type Rrp44p and Rrp44p–CR3 proteins were purified using IgG sepharose beads, and exosome interaction was determined using an anti-Rrp4p antibody. Figure 6A shows that the exosome subunit Rrp4p copurifies with the wild-type Rrp44p, but no Rrp4p could be detected in the Rrp44p–CR3–TAP purification. Thus, even though we suspect that the three conserved Cys residues do not directly contact other exosome subunits, mutating them reduces the ability of Rrp44p to interact with the rest of the exosome, possibly by altering the conformation of Y40, R42 and/or D44.
Figure 6.
Simultaneously mutating the three conserved Cys residues to Ser reduces interaction with the exosome. (A) TAP-tagged Rrp44 containing either a wild-type or mutated CR3 motif were purified from yeast. The lysate (left) and bound (right) fractions were analyzed by western blot with antibodies for the TAP tag (top), the exosome subunit Rrp4p (middle) or the control protein Pgk1p (bottom). (B) The rrp44–yrd allele that contains Y40A, R42A and D44A mutations has a slow growth phenotype that resembles that of rrp44–CR3. The experiment was performed using the plasmid shuffle as described in Figure 2.
If the effects of the rrp44–CR3 mutation are due to an alteration of the interaction of Y40, R42 and/or D44 with Rrp41p, then mutating these residues should have a similar effect. We tested this by creating three single point mutations in these residues (rrp44–Y40A, rrp44–R42A and rrp44–D44A) as well as a triple-point mutant (rrp44–yrd) combining these mutations. The single-point mutants did not have an obvious growth defect (data not shown), but the triple rrp44–yrd mutant had a slow growth phenotype that closely resembled the growth phenotype of the rrp44–CR3 allele (Figure 6B), consistent with the idea that mutating the CCCH motif affects the conformation of Y40, R42 and/or D44, which in turn affects exosome interaction.
DISCUSSION
We have previously shown that mutating the CR3 motif leads to a slow growth phenotype, a slightly reduced expression level of Rrp44p, and a stabilization of nonstop mRNAs (19,22). Although these results hinted at the importance of the conserved motif, it was not clear how changing these Cys residues to Ser affected Rrp44p function. The crystal structure (13) provided important insight as the published structural model indicated that the cysteines are clustered together. The structure further revealed that, although in the primary sequence His184 is far from the CR3 motif, the histidine is in fact positioned in a tetrahedral arrangement with the cysteine residues. While the proximity of Cys residues raises the potential for disulfide bond formation, disulfide bonds are rare in the reduced environment of the cytoplasm and nucleus. Furthermore, mutating individual Cys residues to Ser should completely disrupt such a disulfide bond, but such mutations did not mimic the rrp44–CR3 phenotype. This strongly argues against the formation of a disulfide bond in the CR3 motif. Instead the structural clustering of CR3 with His184 suggested a metal ion-binding site, since four Cys and/or His residues form Zn2+ ion-binding sites in a wide variety of proteins. In such metal-binding sites, mutations of individual residues to serine can have a variety of effects. For example, in the Rpc10p subunit of RNA polymerase III, mutating the first Zn2+-binding Cys residue to Ser is lethal, but changing either the third or fourth Cys (or both) to Ser has no obvious effect on growth (28). Consistent with the idea that a single Cys to Ser change does not always completely eliminate Zn2+ binding, is the natural presence of Ser in some Zn2+-binding sites (29). The suggestion that the CCCH motif forms a metal-binding site is further reinforced by the observation that His184 is conserved in most Rrp44p orthologs, but is lacking in two orthologs that also lack the CR3 motif. In addition, although H184 is part of the PIN domain, it is not conserved in PIN domains that are not part of Rrp44p homologs. Based on all of the available genetic, biochemical, structural and phylogenetic data we suggest that binding a monoatomic metal ion to this CCCH motif stabilizes folding of this part of Rrp44p. Interestingly, Lebreton et al. (18) have previously shown that the endonuclease activity of Rrp44p in vitro is stimulated by addition of either Mn2+ or Zn2+ ions. One possibility is that either metal ion can bind to the active site. However, an alternative explanation is that one or both of these ions bind to the CCCH motif, and the active site may contain a metal ion that copurifies with the recombinant protein. So far we have not been able to clearly distinguish between these and other possibilities using in vitro assays. Moreover, although the CCCH motif most closely resembles a Zn2+-binding site, binding of some other metal ion is also possible, further complicating interpretation of such in vitro experiments.
We have previously shown that either the endo- or exonuclease activity of Rrp44p is sufficient for rapid nonstop mRNA degradation, but that the rrp44–CR3 mutation results in stabilization of nonstop mRNAs. This suggests that the rrp44–CR3 mutation reduces both nuclease activities. However, since rrp44–CR3 is viable and Rrp44p nuclease activity is required, the rrp44–CR3 mutation cannot completely eliminate both functions. Here we test this genetically. If rrp44–CR3 completely eliminates the exonuclease function, combining it with another mutation that eliminates the exonuclease activity should not have an additional effect. Instead we show that rrp44–CR3 is synthetically lethal with rrp44–exo−. Thus, although rrp44–CR3 and rrp44–exo− have some of the same molecular defects, we conclude that rrp44–CR3 does not completely eliminate exonuclease function in vivo. Furthermore, since eliminating both endo- and exonuclease activities is lethal, the synthetic lethality of rrp44–CR3 with rrp44–exo− is consistent with a reduced endonuclease activity. Further synthetic lethality of rrp44–CR3 with dcp1–2 and with rrp6Δ suggests that both the cytoplasmic and nuclear exosomes are affected, since dcp1–2 synthetic lethality reflects a role in cytoplasmic mRNA decay, and Rrp6p is exclusively nuclear. Therefore we conclude that the CR3 motif affects many facets of Rrp44p function.
Since the CCCH site is far removed from both catalytic active sites of Rrp44p, it is likely not directly involved in either catalytic activity. In many Zn2+-containing proteins, the metal ion stabilizes the overall fold of the protein. The available evidence suggests that the overall structure of Rrp44p can fold in the absence of a metal ion bound to the CCCH motif (13,18–20), but we suggest that binding of a metal ion to the CCCH site has a modest effect on folding and/or flexibility of this part of the protein. This then indirectly affects endo- and exonuclease activities, possibly because the CCCH motif is well-connected to two other important parts of Rrp44p (Figure 1D). First, His184 is part of an α-helix that also contains the endonuclease active site residue D171. Thus, perturbation of the CCCH motif may slightly alter the position or flexibility of this α-helix, and thereby affect endonuclease activity. Second, we show that the rrp44–CR3 mutation weakens the interaction between Rrp44p and the remainder of the exosome. Mutations in or depletion of the other nine exosome subunits all cause defects in RNA processing that are similar to those of rrp44–exo−. In contrast, the rrp44–endo− allele does not affect growth or any RNA decay or processing reaction, and therefore an endonuclease defect cannot explain the phenotypes of rrp44–CR3. Thus, we suggest that the reduced in vivo exonuclease activity is a result of the reduced exosome interaction. Although the YxRxD motif is conserved and in the crystal structure interacts with Rrp41p, the importance of these residues had not previously been tested. Our data indicate that this motif indeed is a functionally important contact between the core exosome and Rrp44p. Strikingly, of the single Cys mutations, C47S would be expected to have the largest effect on the YxRxD motif which may explain why it had the largest effect on exosome function. Interestingly, in many cases Rrp44p orthologs fail to copurify with the exosome (30–33) even though exosome-interacting residues, such as the YxRxD motif, are conserved (Supplementary Figure S1A and data not shown). Since the YxRxD motif is conserved in Rrp44p orthologs and hDis3L1 orthologs, our results suggest that all of these proteins interact with the core exosome, even though such interaction has proven difficult to demonstrate. Our results further suggest that in some cases the failure to copurify may be because copurification was tested in buffers that included the Zn2+ chelator EDTA. Strikingly, two papers reported that hDIS3 does not copurify with the exosome using EDTA containing buffers, while a third paper reports hDIS3-exosome copurification using EDTA-free buffers (32–34).
Overall we can now offer a coherent model for the importance of the CR3 motif: The primary effect of mutating the CR3 motif is probably modest local misfolding and/or structural instability of Rrp44p, possibly due to loss of a metal ion. Because the CR3 motif is structurally connected to both the endonuclease active state and to exosome-interacting residues, this effect on local structure has surprisingly far-reaching effects on exosome function. Furthermore, it opens the possibility that the activity of Rrp44p may be regulated by similar structural alterations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1 and Supplementary Table 1.
FUNDING
The National Institutes of Health (NIH) [GM069900 and GM099790 to A.v.H.]; the National Science Foundation [MCB1020739 to A.v.H.]; an EMBO short term fellowship [to D.S.]; grants from Fundação para a Ciência e Tecnologia [FCT; PEst-OE/EQB/LA0004/2011 to C.M.A.]; Ph.D. fellowship from FCT (to F.P.R.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Antibodies against Rrp4p were kindly provided by David Tollervey. The authors thank members of the van Hoof and Arraiano labs and Lisa Berreau for fruitful discussions.
REFERENCES
- 1.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3'–>5' exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 2.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3' end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs Anderson JS, Parker R. The 3' to 5' degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3' to 5' exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meaux S, van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 7.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 10.Lorentzen E, Conti E. Structural basis of 3' end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol. Cell. 2005;20:473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 12.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 13.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 14.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3' end processing. Proc. Natl Acad. Sci. USA. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 17.Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 18.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 19.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairrao F, Arraiano C, Newbury S. Drosophila gene tazman, an orthologue of the yeast exosome component Rrp44p/Dis3, is differentially expressed during development. Dev. Dy./ 2005;232:733–737. doi: 10.1002/dvdy.20269. [DOI] [PubMed] [Google Scholar]
- 22.Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc. Natl Acad. Sci. USA. 2011;108:2366–2371. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 25.van Hoof A, Staples RR, Baker RE, Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3'-to-5' degradation of mRNA. Mol. Cell Biol. 2000;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caponigro G, Muhlrad D, Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3' processing of stable RNAs. Mol. Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubbi L, Labarre-Mariotte S, Chedin S, Thuriaux P. Functional characterization of ABC10alpha, an essential polypeptide shared by all three forms of eukaryotic DNA-dependent RNA polymerases. J. Biol. Chem. 1999;274:31485–31492. doi: 10.1074/jbc.274.44.31485. [DOI] [PubMed] [Google Scholar]
- 29.Karlin S, Zhu ZY. Classification of mononuclear zinc metal sites in protein structures. Proc. Natl Acad. Sci. USA. 1997;94:14231–14236. doi: 10.1073/pnas.94.26.14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 31.Estevez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 34.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.