Abstract
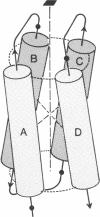
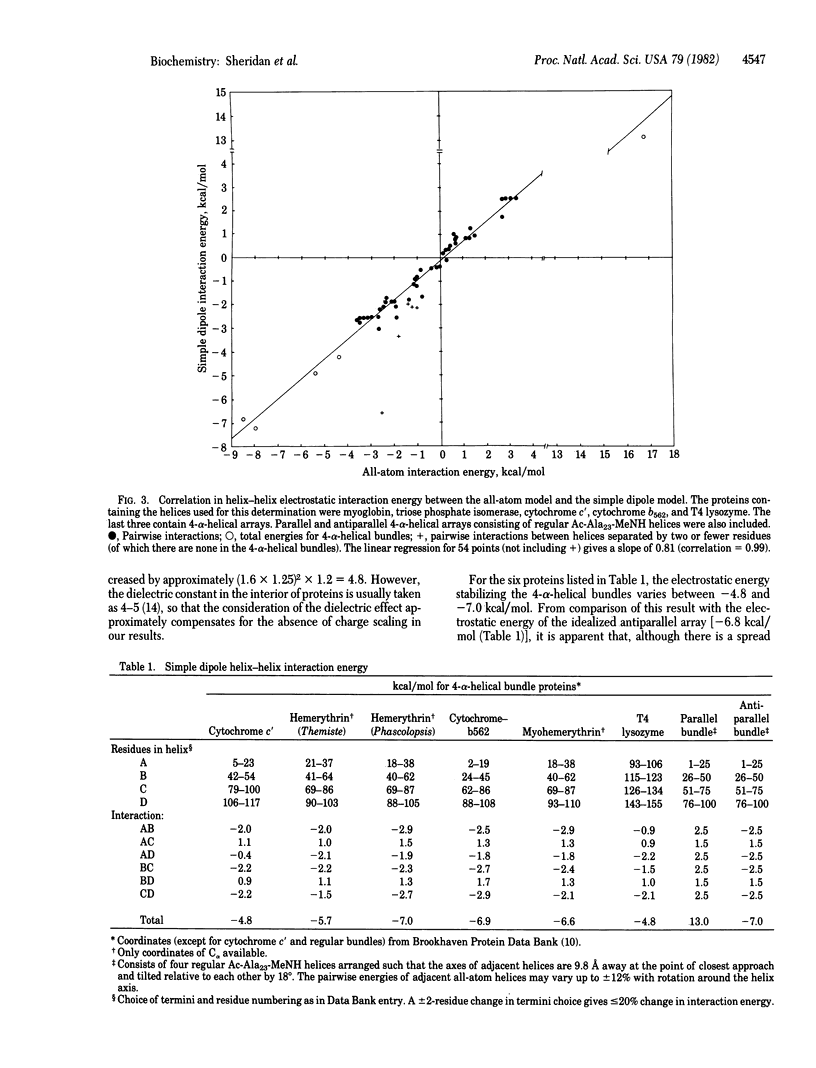
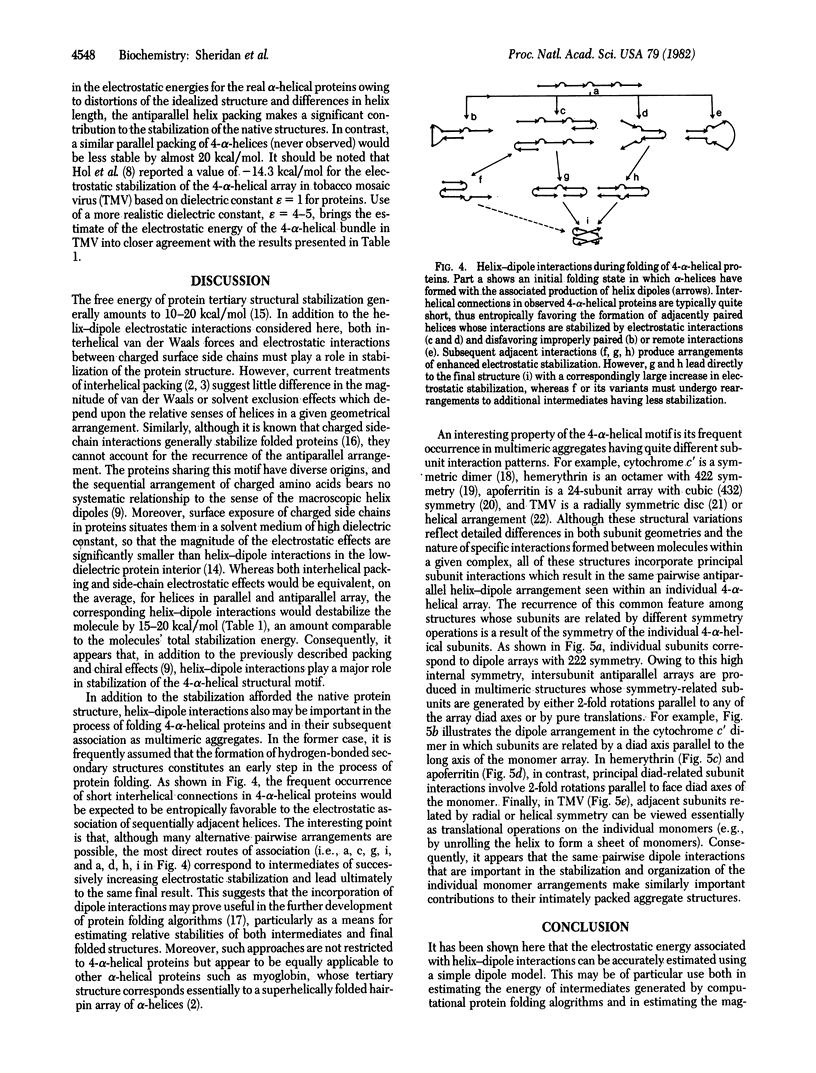
A simple dipole model is developed for estimation of the electrostatic interaction energy between alpha-helices in proteins. This model is used to estimate the electrostatic stabilization in a recurrent protein tertiary structural motif, an array of four closely packed alpha-helices. It is found that, for the proteins examined (cytochrome c', hemerythrin, myohemerythrin, cytochrome b562, and a T4 phage lysozyme domain), their common antiparallel arrangement of adjacent helices confers a stabilization of 5--7 kcal/mol (1 cal = 4.18 J). In contrast, a similarly packed array of parallel helices is relatively destabilized by 20 kcal/mol. These results show that helix-dipole interactions are important in the stabilization of this structural motif. These effects are discussed both in the context of folding pathways for 4-alpha-helical proteins and the stabilization of the higher aggregates.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banyard S. H., Stammers D. K., Harrison P. M. Electron density map of apoferritin at 2.8-A resolution. Nature. 1978 Jan 19;271(5642):282–284. doi: 10.1038/271282a0. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Helix to helix packing in proteins. J Mol Biol. 1981 Jan 5;145(1):215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- Cohen F. E., Richmond T. J., Richards F. M. Protein folding: evaluation of some simple rules for the assembly of helices into tertiary structures with myoglobin as an example. J Mol Biol. 1979 Aug 15;132(3):275–288. doi: 10.1016/0022-2836(79)90260-2. [DOI] [PubMed] [Google Scholar]
- Hayes D. M., Kollman P. A. Electrostatic potentials of proteins. 1. Carboxypeptidase A. J Am Chem Soc. 1976 May 26;98(11):3335–3345. doi: 10.1021/ja00427a048. [DOI] [PubMed] [Google Scholar]
- Hol W. G., Halie L. M., Sander C. Dipoles of the alpha-helix and beta-sheet: their role in protein folding. Nature. 1981 Dec 10;294(5841):532–536. doi: 10.1038/294532a0. [DOI] [PubMed] [Google Scholar]
- Hol W. G., van Duijnen P. T., Berendsen H. J. The alpha-helix dipole and the properties of proteins. Nature. 1978 Jun 8;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Matthew J. B., Hanania G. I., Gurd F. R. Electrostatic effects in hemoglobin: hydrogen ion equilibria in human deoxy- and oxyhemoglobin A. Biochemistry. 1979 May 15;18(10):1919–1928. doi: 10.1021/bi00577a011. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Stability of proteins: small globular proteins. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Richards F. M. Packing of alpha-helices: geometrical constraints and contact areas. J Mol Biol. 1978 Mar 15;119(4):537–555. doi: 10.1016/0022-2836(78)90201-2. [DOI] [PubMed] [Google Scholar]
- Sheridan R. P., Allen L. C. The electrostatic potential of the alpha helix (electrostatic potential/alpha-helix/secondary structure/helix dipole). Biophys Chem. 1980 Apr;11(2):133–136. doi: 10.1016/0301-4622(80)80015-9. [DOI] [PubMed] [Google Scholar]
- Stenkamp R. E., Sieker L. C., Jensen L. H., McQueen J. E., Jr Structure of methemerythrin at 2.8-Angstrom resolution: computer graphics fit of an averaged electron density map. Biochemistry. 1978 Jun 27;17(13):2499–2504. doi: 10.1021/bi00606a007. [DOI] [PubMed] [Google Scholar]
- Stubbs G., Warren S., Holmes K. Structure of RNA and RNA binding site in tobacco mosaic virus from 4-A map calculated from X-ray fibre diagrams. Nature. 1977 May 19;267(5608):216–221. doi: 10.1038/267216a0. [DOI] [PubMed] [Google Scholar]
- Wada A., Nakamura H. Nature of the charge distribution in proteins. Nature. 1981 Oct 29;293(5835):757–758. doi: 10.1038/293757a0. [DOI] [PubMed] [Google Scholar]
- Wada A. The alpha-helix as an electric macro-dipole. Adv Biophys. 1976:1–63. [PubMed] [Google Scholar]
- Weber P. C., Howard A., Xuong N. H., Salemme F. R. Crystallographic structure of Rhodospirillum molischianum ferricytochrome c' at 2.5 A resolution. J Mol Biol. 1981 Dec 5;153(2):399–424. doi: 10.1016/0022-2836(81)90286-2. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Salemme F. R. Structural and functional diversity in 4-alpha-helical proteins. Nature. 1980 Sep 4;287(5777):82–84. doi: 10.1038/287082a0. [DOI] [PubMed] [Google Scholar]