Abstract
Activating mutations in leucine-rich repeat kinase 2 (LRRK2) are present in a subset of Parkinson's disease (PD) patients and may represent an attractive therapeutic target. Here, we report that a 2-anilino-4-methylamino-5-chloropyrimidine, HG-10-102-01 (4), is a potent and selective inhibitor of wild-type LRRK2 and the G2019S mutant. Compound 4 substantially inhibits Ser910 and Ser935 phosphorylation of both wild-type LRRK2 and G2019S mutant at a concentration of 0.1–0.3 μM in cells and is the first compound reported to be capable of inhibiting Ser910 and Ser935 phosphorylation in mouse brain following intraperitoneal delivery of doses as low as 50 mg/kg.
Keywords: LRRK2; blood−brain barrier; brain penetrant inhibitor; 2,4-diaminopyrimidine
Parkinson's disease (PD) is the second most common neurodegenerative disease in the world. It affects over 1 million Americans, and more than 60 000 patients are newly diagnosed each year.1,2 Recent genetic studies have revealed an underlying genetic cause in at least 10% of all PD cases,3 which provides new opportunities for the discovery of molecularly targeted therapeutics that may ameliorate neurodegeneration. Among the genes associated with PD, leucine-rich repeat kinase 2 (LRRK2) is unique because a missense mutation, G2019S, is frequently found in both familial and sporadic PD cases.4−9 The G2019S mutation increases kinase activity, which may result in activation of the neuronal death signal pathway, suggesting that small molecule LRRK2 kinase inhibitors may be able to serve as a new class of therapeutics for the treatment of PD.10−13 Transgenic G2019S LRRK2 mice aged to 12–16 months display progressive degeneration of the substantia nigra pars compacta (SNpc) dopaminergic neurons and Parkinson's phenotypes of motor dysfunction, suggesting that this mutation may be functionally relevant to the disease.14
LRRK2 kinase inhibitors are being actively pursued both as “tools” to pharmacologically interrogate normal and pathological LRRK2 biology and as experimental therapeutic agents. For example, LRRK2-IN-1 (1)15 and CZC-25146 (2)16 have been reported as the first-generation “tool” inhibitors that exhibit excellent potency and selectivity for LRRK2. However, none of these compounds are able to efficiently cross the mouse blood–brain barrier (BBB) and inhibit LRRK2 kinase activity, which limits their utility in murine PD models and eventual clinical development.15,16 Here, we report that a lower molecular weight 2,4-diaminopyrimidine, HG-10-102-01 (4), maintains highly potent and selective inhibition of LRRK2 and is, to our knowledge, the first compound reported to be capable of inhibiting LRRK2 phosphorylation in mouse brain.
Several 2,4-diaminopyrimidine-based inhibitors of LRRK2 have been reported including LRRK2-IN-1 (1),15 CZC-25146 (2),16 and TAE684 (3),17 but none of these compounds are capable of effectively inhibiting phosphorylation of Ser910 and Ser935 of LRRK2 in mouse brain at intraperitoneal doses of up to 100 mg/kg. Analysis of predicted docked conformations of these compounds to homology models of LRRK2 suggests that the 4-anilino moiety of each compound occupies quite distinct regions of the adenosine triphosphate (ATP)-binding site. In an attempt to lower the molecular weight and remove possible disfavorable interactions with the protein, we explored compounds where the 4-anilino moiety was removed. We and others18,19 discovered that simplified structures such as 4 maintain the ability to potently inhibit the biochemical activity of wild-type and G2019S mutant LRRK2. Compound 4 exhibited biochemical IC50 values of 20.3 and 3.2 nM against wild-type LRRK2 and LRRK2[G2019S], respectively (Figure 1). The biochemical potency of 4 for inhibition of wild-type LRRK2 and LRRK2[G2019S] is similar to that observed for LRRK2-IN-1 (1); however, 4 maintains inhibition of the A2016T mutation, which induces dramatic resistance to LRRK2-IN-1 (1) (Figure 1). Although both LRRK2-IN-1 (1) and 4 share the aminopyrimidine pharmacophore, a molecular model of 4 docked to a homology model of LRRK2 built based on a previously published crystallographic structure of anaplastic lymphoma kinase (ALK)20 suggests that there is less possibility for steric hindrance with the A2016T mutation (Figure 2a,b).
Figure 1.
LRRK2 inhibitors. (a) Biochemical inhibition of GST-LRRK2 (1326–2527), GST-LRRK2[G2019S] (1326–2527), GST-LRRKT[A2016T] (1326–2527), and GST-LRRK2[G2019S + A2016T] (1326–2527) was assayed using 20 μM Nictide in the presence of 100 μM ATP. Results are averages of triplicate experiments.
Figure 2.

Molecular model of HG-10-102-01 (4) with LRRK2[T2016]. (a) Two hydrogen bonds are predicted between the hinge region A1950 and the aminopyrimidine motif of the inhibitor. One hydrogen bond is predicted between the backbone amide carbonyl of S1954 with the amide carbonyl of the inhibitor. (b) Two potential interactions exist between M1947 and T2016 with the 5-chloro group on the pyrimidine of the inhibitor.
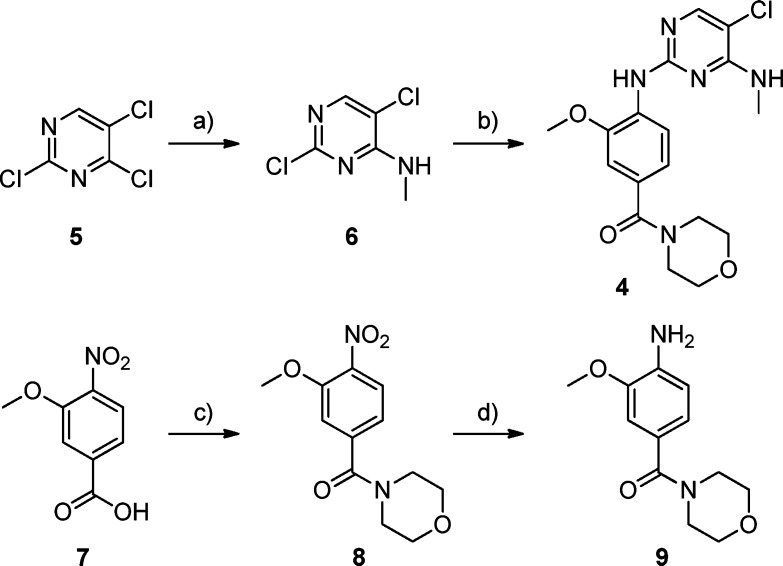
Compound 4 was prepared from commercially available 2,4,5-trichloropyrimidine and 3-methoxy-4-nitrobenzoic acid (Scheme 1). The 3-methoxy-4-nitrobenzoic acid 7 was subjected to chlorination with thionyl chloride followed by reaction with morpholine to form the corresponding amide 8, which was reduced by hydrogenation to yield aniline 9. 2,4,5-Trichloropyrimidine 5 was regioselectively aminated with methylamine to afford to 2,5-dichloro-N-methylpyrimidin-4-amine 6. Compound 6 was aminated with aniline 9 under acidic conditions to furnish the desired compound 4.
Scheme 1.
We next examined the ability of 4 to inhibit LRRK2 in a cellular context in comparison to LRRK2-IN-1 (1). As there are no validated direct phosphorylation substrates of LRRK2, we monitored phosphorylation of Ser910 and Ser935, two residues whose phosphorylation is known to be dependent upon LRRK2 kinase activity21 (Figure 3). Compound 4 induced a dose-dependent inhibition of Ser910 and Ser935 phosphorylation in both wild-type LRRK2 and LRRK2[G2019S] stably transfected into HEK293 cells (Figure 3a). Substantial dephosphorylation of Ser910 and Ser935 was observed at approximately 1 μM concentrations of 4 for wild-type LRRK2 and at a slightly lower dose of 0.3 μM for LRRK2[G2019S] (Figure 3b), which is a similar potency to that observed for LRRK2-IN-1 (1). Consistent with the biochemical results, 4 also induced dephosphorylation of Ser910 and Ser935 at a concentration of 1–3 μM in the drug-resistant LRRK2[A2016T + G2019S] and LRRK2[A2016T] mutants, revealing that the A2016T mutation is not an effective way to induce resistance to 4.
Figure 3.
Compound HG-10-102-01 (4) inhibits LRRK2 in cells. HEK293 cells stably expressing (a) wild-type GFP-LRRK2, (b) GFP-LRRK2[G2019S], (c) GFP-LRRK2[A2016T], and (d) GFP-LRRK2[G2019S + A2016T] were treated with DMSO or increasing concentrations of 4 for 90 min (1 μM of LRRK2-IN-1 was used as a control). Cell lysates were subjected to immunoblotting for detection of LRRK2 phosphorylated at Ser910 and Ser935 and for total LRRK2.
We next examined the effect of 4 on endogenously expressed LRRK2 in human lymphoblastoid cells derived from a control and Parkinson's patient homozygous for the LRRK2[G2019S] mutation (Figure 4a). We found that increasing doses of 4 led to similar dephosphorylation of endogenous LRRK2 at Ser910 and Ser935, as was observed in HEK293 cells stably expressing wild-type LRRK2 or LRRK2[G2019S] (compare Figure 3a to Figure 4a). Moreover, endogenous LRRK2 was also more sensitive to 4 than LRRK2-IN-1 (1), which is consistent with the trend that we observed in HEK293 cells. We also found that 4 induced similar dose-dependent Ser910 and Ser935 dephosphorylation of endogenous LRRK2 in mouse Swiss 3T3 cells and mouse embryonic fibroblast cells (Figure 4b,c).
Figure 4.
Compound 4 inhibits endogenously expressed LRRK2. (a) Endogenous LRRK2 from EBV immortalized human lymphoblastoid cells from a control subject (LRRK2+/+), a PD patient homozygous for the LRRK2[G2019S] mutation. After treatment of the cells with DMSO or the indicated concentration of 4 [or LRRK2-IN-1 (1)] for 90 min, cell lysates were subjected to immunoblot analysis with the indicated antibody for western analysis. Immunoblots were performed in duplicate, and the results were representative of at least two independent experiments. (b) As in a, except mouse Swiss 3T3 cells were used. (c) As in a, except mouse embryonic fibroblast cells were used.
The mouse pharmacokinetic profile of 4 demonstrated good oral bioavailability (%F = 67), a short half-life of 0.13 h, and low plasma exposure [502 h*ng/mL, area under the concentration–time curve (AUC)last] [complete pharmacokinetics (PK) parameters are in the Supporting Information]. The short half-life is likely the consequence of rapid first-pass metabolism as incubation with mouse liver microsomes also revealed a short T1/2 of 13 min.22 We next investigated the pharmacodynamic properties of 4 by monitoring inhibition of LRRK2 Ser910/Ser935 phosphorylation in kidney, spleen, and brain following intraperitoneal delivery of 100 mg/kg of 4. We observed near complete dephosphorylation of Ser910 and Ser935 of LRRK2 in all tissues including brain at this dose. We then repeated the study at lower doses of 50, 30, and 10 mg/kg, where we observed near complete inhibition in all tissues at 50 mg/kg but only partial inhibition in brain at the 30 and 10 mg/kg doses. These results indicate that 4 is a promising chemotype for achieving dephosphorylation of Ser910 and Ser935 in the brain.
Figure 5.
Pharmacodynamic analysis for HG-10-102-01 (4). Pharmacodynamic study of HG-10-102-01 (4) from brain, spleen, and kidney following intraperitoneal administration at the indicated doses. Tissues were collected, and endogenous LRRK2 was resolved by SDS-PAGE and blotted with a phospho-specific antibody directed against Ser910, Ser935, and total LRRK2 (the quantitative analysis is included in the Supporting Information).
To further explore the ability of 4 to inhibit LRRK2 in vivo, we used a chemical proteomics approach, KiNativ, to monitor kinase inhibition in tissues from the inhibitor-treated animals. Brains and spleens from animals treated with 4 at 3, 10, 30, 50, and 100 mg/kg and LRRK2-IN-1 (1) at 100 mg/kg were evaluated and compared to vehicle-treated animal brains. Briefly, the brains were lysed in a 5× volume of buffer (no detergent) and labeled with an ADP-acylphosphate probe. Following sample workup and trypsin digestion, kinase labeling by the acylphosphate probe was quantitatively determined by mass spectrometry. Data were collected for >150 protein and lipid kinases (Supporting Information). Of the kinases profiled, only LRRK2 showed a clear and significant dose-related increase in engagement with 4. LRRK2 inhibition was ∼40% at 30 mg/kg and ∼70% at 50 and 100 mg/kg in the brain. Greater LRRK2 target engagement was observed in the spleen: ∼40% at 3 mg/kg, ∼80–90% at 10 mg/kg, and greater than 90% in the 30–100 mg/kg treated animals. In contrast, no LRRK2 inhibition was observed in brains of mice treated with 100 mg/kg LRRK2-IN-1 (1), consistent with the inability of the compound to induce dephosphosphorylation of the Ser910 and Ser935 sites of LRRK2 in the brain. Considering these inhibition values correspond to a 5× dilution of material relative to the intact brain, these engagement values correlate very well with the observed inhibition of Ser910 and Ser935 phosphorylation of LRRK2 by 4. No other kinases showed >50% inhibition at any dose of 4, and only JNK1/2/3 and TBK1 showed potential dose-related inhibition (∼35–40% at 50 and 100 mg/kg), suggesting that compound 4 is a highly selective inhibitor of LRRK2.
The kinase selectivity of 4 was further assessed using standard radioactivity-based enzymatic assays against a panel of 138 kinases (Dundee profiling).23 At a concentration of 10 μM, compound 4 only inhibited the kinase activities of mixed-lineage kinase 1 (MLK1) and MAP kinase-interacting serine/threonine-protein kinase 2 (MNK2) to greater than 80% of the dimethyl sulfoxide (DMSO) control (complete results are presented in the Supporting Information). Dose–response analysis revealed inhibition of MLK1 with an IC50 = 2.1 μM and MNK2 with an IC50 = 0.6 μM. KinomeScan analysis against a near comprehensive panel of 451 kinases at a concentration of 1 μM resulted in no interactions detected with kinases other than LRRK2[G2019S] with the exception of one mutant form of c-Kit (L576P) demonstrating the outstanding selectivity of this inhibitor (complete profiling results are provided in the Supporting Information).24 These results suggest that 4 is a highly selective LRRK2 inhibitor; however, further profiling of additional kinases and other ATP-dependent enzymes is still warranted.
In summary, we have discovered that 4 is a potent biochemical and cellular inhibitor of LRRK2 kinase activity. Detailed characterization of 4 using LRRK2-IN-1 as a bench mark revealed that 4 significantly inhibited phosphorylation of wild-type LRRK2 and LRRK2[G2019S] mutant at Ser910 and Ser935 at 0.3–1.0 μM in cell culture, which is approximately the same potency as LRRK2-IN-1 (1). Compound 4 is relatively insensitive to the A2016T mutation, which suggests that this mutant will not be useful to validate whether the pharmacological effects of the compound are LRRK2-dependent. Compound 4 exhibits excellent kinase selectivity as assessed by recombinant kinases (138), KinomeScan (451 kinases), and KiNativ profiling. Compound 4 can inhibit phosphorylation of Ser910 and Ser935 of LRRK2 in brain and peripheral tissues following intraperitoneal doses of 50 mg/kg. Further optimization of this chemotype especially in regards to in vivo half-life will be reported in due course.
Acknowledgments
We thank the staff at the National Centre for Protein Kinase Profiling (www.kinase-screen.mrc.ac.uk) for undertaking Dundee kinase specificity screening, SAI Advantium for performing pharmacokinetic studies, Faycal Hentati (Institut National de Neurologie, Tunis, Tunisia) and GlaxoSmithKline Pharmaceuticals R&D for providing EBV immortalized human lymphoblastoid cells, and the antibody purification teams [Division of Signal Transduction Therapy (DSTT), University of Dundee] coordinated by Hilary McLauchlan and James Hastie for generation of antibodies. Microsome stability studies were performed by Michael Cameron (TSRI Florida), and we thank Sara Buhrlage for critical reading of the manuscript.
Glossary
Abbreviations
- LRRK2
leucine-rich repeat kinase 2
- PD
Parkinson's disease
- ATP
adenosine triphosphate
- ALK
anaplastic lymphoma kinase
- THF
tetrahydrofuran
- rt
room temperature
- TFA
trifluoroacetic acid
- 2-BuOH
2-butanol
- DIEA
N,N-diisopropylethylamine
- PK
pharmacokinetics
- F
bioavailability
- AUC
area under the concentration–time curve
- CL
clearance
- Clast
last measured concentration
- Cmax
maximum concentration observed
- T1/2
elimination half-life
- Vss
volume in steady state
- MLK1
mixed-lineage kinase 1
- MNK2
MAP kinase-interacting serine/threonine-protein kinase 2
- DMSO
dimethyl sulfoxide
Supporting Information Available
Experimental procedures and compounds characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
# School of Life Sciences, Xiamen University, Fujian 361005, People's Republic of China.
Author Contributions
∇ These authors contributed equally to this work.
This work was supported by NIH Grant P41 GM079575-03 (N.S.G.), the Medical Research Council (D.R.A.), the Michael J. Fox foundation for Parkinson's disease research (N.S.G. and D.R.A.), and the pharmaceutical companies supporting the DSTT (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KgaA, and Pfizer) (D.R.A.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Gandhi P. N.; Chen S. G.; Wilson-Delfosse A. L. Leucine-rich repeat kinase 2 (LRRK2): A key player in the pathogenesis of Parkinson's disease. J. Neurosci. Res. 2009, 87, 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey E. R.; Constantinescu R.; Thompson J. P.; Biglan K. M.; Holloway R. G.; Kieburtz K.; Marshall F. J.; Ravina B. M.; Schifitto G.; Siderowf A.; Tanner C. M. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology 2007, 68, 384–386. [DOI] [PubMed] [Google Scholar]
- Daniëls V.; Baekelandt V.; Taymans J. M. On the Road to Leucine-Rich Repeat Kinase 2 Signalling: Evidence from Cellular and in vivo Studies. Neurosignals 2011, 19, 1–15. [DOI] [PubMed] [Google Scholar]
- Healy D. G.; Falchi M.; O'Sullivan S. S.; Bonifati V.; Durr A.; Bressman S.; Brice A.; Aasly J.; Zabetian C. P.; Goldwurm S.; Ferreira J. J.; Tolosa E.; Kay D. M.; Klein C.; Williams D. R.; Marras C.; Lang A. E.; Wszolek Z. K.; Berciano J.; Schapira A. H. V.; Lynch T.; Bhatia K. P.; Gasser T.; Lees A. J.; Wood N. W. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: A case-control study. Lancet Neurol. 2008, 7, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dächsel J. C.; Farrer M. J. LRRK2 and Parkinson Disease. Arch. Neurol. 2010, 67, 542–547. [DOI] [PubMed] [Google Scholar]
- Lee B. D.; Dawson V. L.; Dawson T. M. Leucine-rich repeat kinase 2 (LRRK2) as a potential therapeutic target in Parkinson's disease. Trends Pharmacol. Sci. 2012, 337365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Hamamichi S.; Lee B. D.; Yang D.; Ray A.; Caldwell G. A.; Caldwell K. A.; Dawson T. M.; Smith W. W.; Dawson V. L. Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson’s disease models. Hum. Mol. Genet. 2011, 20, 3933–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. D.; Shin J. H.; VanKampen J.; Petrucelli L.; West A. B.; Ko H. S.; Lee Y. I.; Maguire-Zeiss K. A.; Bowers W. J.; Federoff H. J.; Dawson V. L.; Dawson T. M. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat. Med. 2010, 16, 998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. W.; Pei Z.; Jiang H.; Dawson V. L.; Dawson T. M.; Ross C. A. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 2006, 9, 1231–1233. [DOI] [PubMed] [Google Scholar]
- Greggio E.; Cookson M. R. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN Neuro 2009, 11e00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Cookson M. R. Role of LRRK2 kinase dysfunction in Parkinson disease. Expert Rev. Mol. Med. 2011, 13, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonet D.; Daher J. P.; Lin B. M.; Stafa K.; Kim J.; Banerjee R.; Westerlund M.; Pletnikova O.; Glauser L.; Yang L.; Liu Y.; Swing D. A.; Beal M. F.; Troncoso J. C.; McCaffery J. M.; Jenkins N. A.; Copeland N. G.; Galter D.; Thomas B.; Lee M. K.; Dawson T. M.; Dawson V. L.; Moore D. J. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLoS One 2011, 64e18568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusonchet J.; Kochubey O.; Stafa K.; Young S. M. Jr.; Zufferey R.; Moore D. J.; Schneider B. L.; Aebischer P. A rat model of progressive nigral neurodegeneration induced by the Parkinson’s disease-associated G2019S mutation in LRRK2. J. Neurosci. 2011, 31, 907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y.; Weng Y. H.; Chien K. Y.; Lin K. J.; Yeh T. H.; Cheng Y. P.; Lu C. S.; Wang H. L. (G2019S) LRRK2 activates MKK4-JNK pathway and causes degeneration of SN dopaminergic neurons in a transgenic mouse model of PD. Cell Death Differ. 2012, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.; Dzamko N.; Prescott A.; Davies P.; Liu Q.; Yang Q.; Lee J.-D.; Patricelli M. P.; Nomanbhoy T. K.; Alessi D. R.; Gray N. S. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat. Chem. Biol. 2011, 7, 203–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden N.; Perrin J.; Ren Z.; Lee B. D.; Zinn N.; Dawson V. L.; Tam D.; Bova M.; Lang M.; Drewes G.; Bantscheff M.; Bard F.; Dawson T. M.; Hopf C. Chemoproteomics-Based Design of Potent LRRK2-Selective Lead Compounds That Attenuate Parkinson Disease-Related Toxicity in Human Neurons. ACS Chem. Biol. 2011, 6, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Deng X.; Choi H. G.; Alessi D. R.; Gray N. S. Characterization of TAE684 as a potent LRRK2 kinase inhibitor. Bioorg. Med. Chem. Lett. 2012, 22, 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Glenn C.; Burdick D. J.; Chambers M.; Chan B. K.; Chen H.; Estrada A.; Gunzner J. L.; Shore D.; Sweeney Z. K.; Wang S.; Zhao G.. Aminopyrimidine derivatives as LRRK2 modulators and their preparation and use for the treatment of Parkinson's disease. WO2011151360A1, 2011.
- Chen H.; Chan B. K.; Drummond J.; Estrada A. A.; Gunzner-Toste J.; Liu X.; Liu Y.; Moffat J.; Shore D.; Sweeney Z. K.; Tran T.; Wang S.; Zhao G.; Zhu H.; Burdick D. J. Discovery of Selective LRRK2 Inhibitors Guided by Computational Analysis and Molecular Modeling. J. Med. Chem. 2012, 55, 5536–5545. [DOI] [PubMed] [Google Scholar]
- Bossi R. T.; Saccardo M. B.; Ardini E.; Menichincheri M.; Rusconi L.; Magnaghi P.; Orsini P.; Avanzi N.; Borgia A. L.; Nesi M.; Bandiera T.; Fogliatto G.; Bertrand J. A. Crystal Structures of Anaplastic Lymphoma Kinase in Complex with ATP Competitive Inhibitors. Biochemistry 2010, 49, 6813–6825. [DOI] [PubMed] [Google Scholar]
- These images were produced using a free version of Pymol software.
- Madoux F.; Li X.; Chase P.; Zastrow G.; Cameron M. D.; Conkright J. J.; Griffin P. R.; Thacher S.; Hodder P. Potent, Selective and Cell Penetrant Inhibitors of SF-1 by Functional Ultra-High-Throughput Screening. Mol. Pharmacol. 2008, 73, 1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J.; Plater L.; Elliott M.; Shpiro N.; Hastie C. J.; McLauchlan H.; Klevernic I.; Arthur J. S.; Alessi D. R.; Cohen P. The selectivity of protein kinase inhibitors: A further update. Biochem. J. 2007, 408, 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. I.; Hunt J. P.; Herrgard S.; Ciceri P.; Wodicka L. M.; Pallares G.; Hocker M.; Treiber D. K.; Zarrinkar P. P. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1046–1051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.