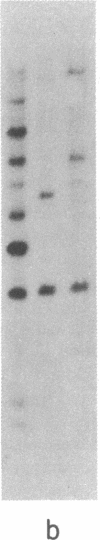
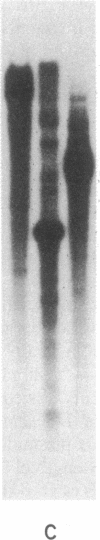
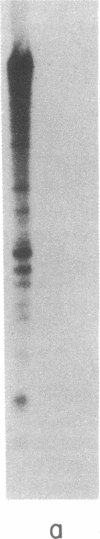
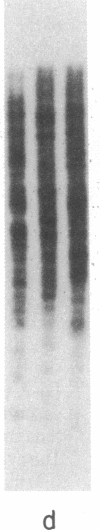
Abstract
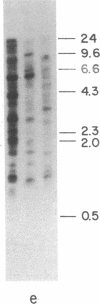
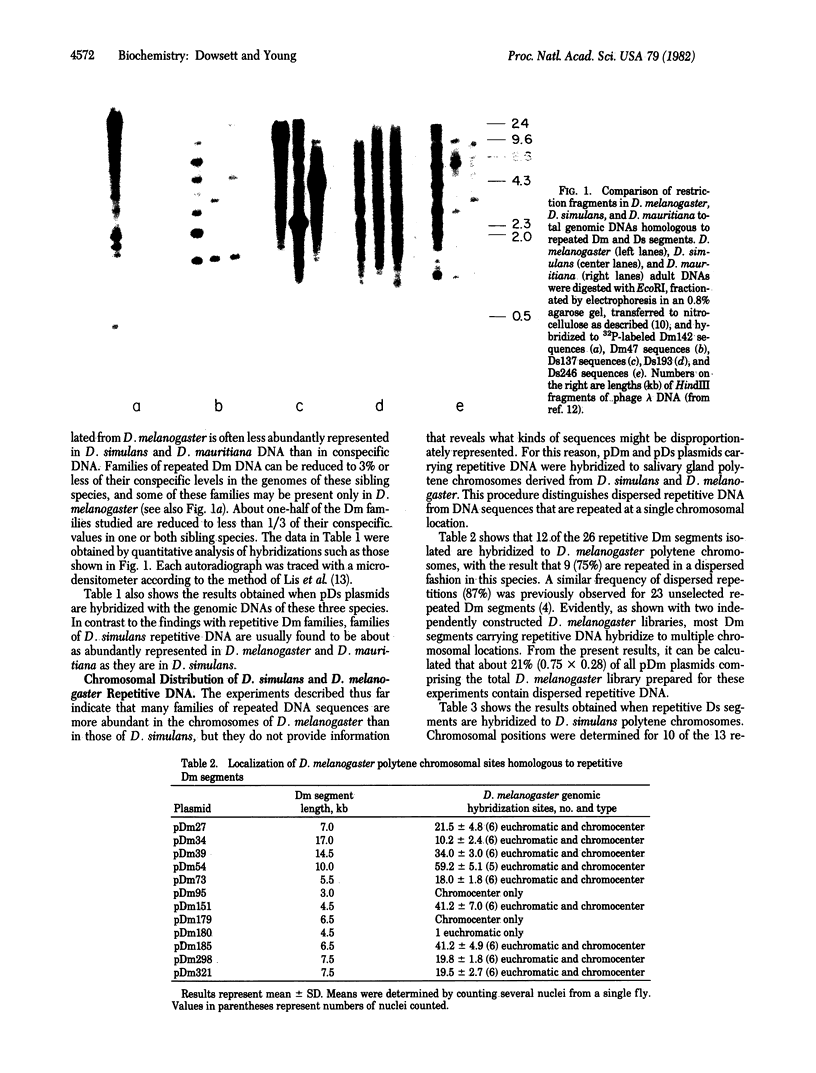
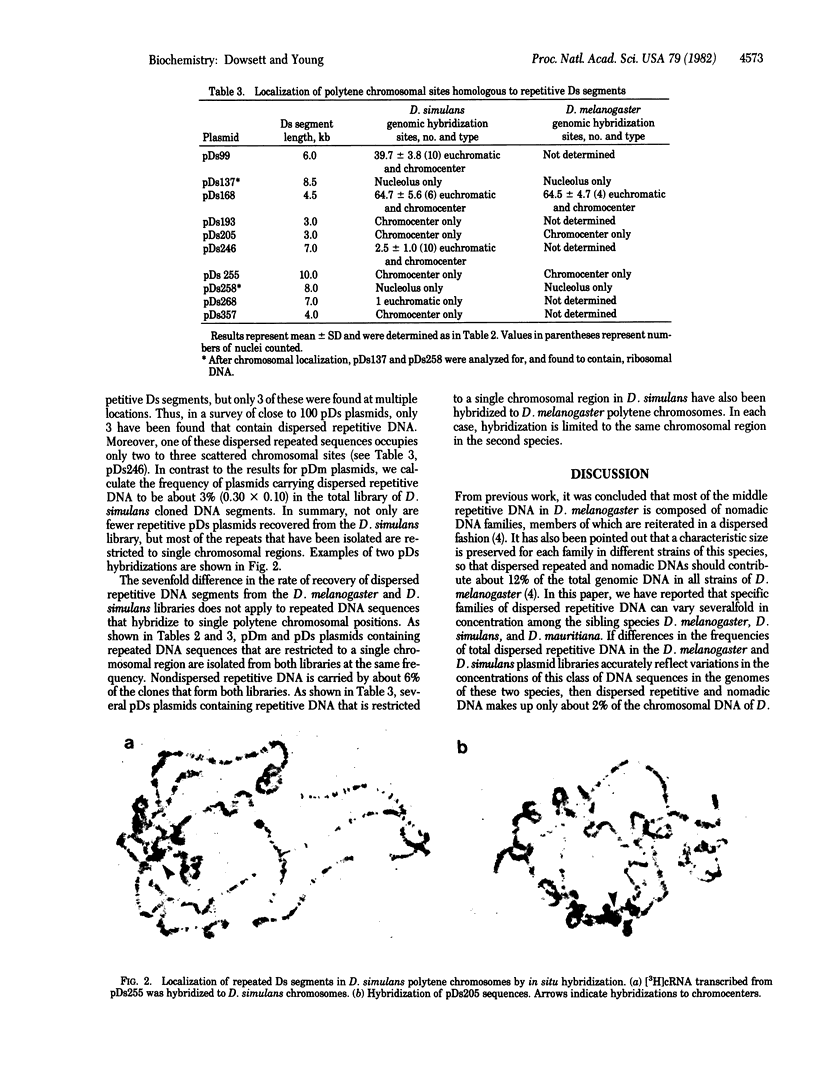
The genomic concentrations of certain middle repetitive DNA sequences vary considerably among closely related species of Drosophila. In fact, the chromosomes of D. melanogaster appear to carry approximately 3 times as much middle repetitive DNA as those of the sibling species D. simulans. Although most of the middle repetitive DNA of D. melanogaster consists of segments of "nomadic" DNA that occupy different dispersed chromosomal locations in different strains of flies, repeated DNA sequences recovered from the D. simulans genome are most often restricted to single chromosomal positions. Apparent differences in the total concentrations of middle repetitive DNA in the two species are most easily attributed to an approximately sevenfold difference in their dispersed repetitive and nomadic DNA contents. These differences may affect the relative mutation rates of these species or contribute to their reproductive isolation or both.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Bregliano J. C., Picard G., Bucheton A., Pelisson A., Lavige J. M., L'Heritier P. Hybrid dysgenesis in Drosophila melanogaster. Science. 1980 Feb 8;207(4431):606–611. doi: 10.1126/science.6766221. [DOI] [PubMed] [Google Scholar]
- David J., Lemeunier F., Tsacas L., Bocquet C. Hybridation d'une nouvelle espèce, Drosophila mauritiana avec D. melanogaster et D. simulans. Ann Genet. 1974 Dec;17(4):235–241. [PubMed] [Google Scholar]
- Davidson E. H., Galau G. A., Angerer R. C., Britten R. J. Comparative aspects of DNA organization in Metazoa. Chromosoma. 1975 Jul 21;51(3):253–259. doi: 10.1007/BF00284818. [DOI] [PubMed] [Google Scholar]
- Endow S. A. On ribosomal gene compensation in Drosophila. Cell. 1980 Nov;22(1 Pt 1):149–155. doi: 10.1016/0092-8674(80)90163-4. [DOI] [PubMed] [Google Scholar]
- Engels W. R., Preston C. R. Identifying P factors in Drosophila by means of chromosome breakage hotspots. Cell. 1981 Nov;26(3 Pt 1):421–428. doi: 10.1016/0092-8674(81)90211-7. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Kidwell M. G., Kidwell J. F., Sved J. A. Hybrid Dysgenesis in DROSOPHILA MELANOGASTER: A Syndrome of Aberrant Traits Including Mutation, Sterility and Male Recombination. Genetics. 1977 Aug;86(4):813–833. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R., Marzluf G. A. Characterization of the sequence complexity and organization of the Neurospora crassa genome. Biochemistry. 1979 Aug 21;18(17):3705–3713. doi: 10.1021/bi00584a011. [DOI] [PubMed] [Google Scholar]
- Lemeunier F., Ashburner M. A. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc R Soc Lond B Biol Sci. 1976 May 18;193(1112):275–294. doi: 10.1098/rspb.1976.0046. [DOI] [PubMed] [Google Scholar]
- Lis J. T., Prestidge L., Hogness D. S. A novel arrangement of tandemly repeated genes at a major heat shock site in D. melanogaster. Cell. 1978 Aug;14(4):901–919. doi: 10.1016/0092-8674(78)90345-8. [DOI] [PubMed] [Google Scholar]
- Marx J. L. Genes that control development. Science. 1981 Sep 25;213(4515):1485–1488. doi: 10.1126/science.6792705. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Kramer R. A., Davis R. W. Cloning of the yeast ribosomal DNA repeat unit in SstI and HindIII lambda vectors using genetic and physical size selections. J Mol Biol. 1978 Aug 15;123(3):371–386. doi: 10.1016/0022-2836(78)90085-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Tartof K. D. Evolution of transcribed and spacer sequences in the ribosomal RNA genes of Drosophila. Cell. 1979 Jul;17(3):607–614. doi: 10.1016/0092-8674(79)90268-x. [DOI] [PubMed] [Google Scholar]
- Timberlake W. E. Low repetitive DNA content in Aspergillus nidulans. Science. 1978 Dec 1;202(4371):973–975. doi: 10.1126/science.362530. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Young M. W. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. W., Schwartz H. E. Nomadic gene families in Drosophila. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):629–640. doi: 10.1101/sqb.1981.045.01.081. [DOI] [PubMed] [Google Scholar]