Abstract
CD4+Foxp3+ Treg are essential for immune homeostasis and maintenance of self-tolerance. They are produced in the thymus and also generated de novo in the periphery in a TGFβ dependent manner. Foxp3+ Treg are also required to achieve tolerance to transplanted tissues when induced by co receptor or co stimulation blockade. Using TCR transgenic mice to avoid issues of autoimmune pathology, we show that Foxp3 expression is both necessary and sufficient for tissue tolerance by coreceptor blockade. Moreover, the known need in tolerance induction for TGFβ signalling to T cells can wholly be explained by its role in induction of Foxp3, as such signalling proved dispensable for the suppressive process. We analysed the relative contribution of TGFβ and Foxp3 to the transcriptome of TGFβ-induced Treg and showed that TGFβ elicited a large set of down-regulated signature genes. The number of genes uniquely modulated due to the influence of Foxp3 alone was surprisingly limited. Thus, despite the large genetic influence of TGFβ exposure on iTreg, the crucial Foxp3-influenced signature independent of TGFβ is small. Retroviral mediated conditional nuclear expression of Foxp3 proved sufficient to confer transplant-suppressive potency on CD4+ T cells, and was lost once nuclear Foxp3 expression was extinguished. These data support a dual role for TGFβ and Foxp3 in induced tolerance, where TGFβ stimulates Foxp3 expression, whose sustained expression is then associated with acquisition of tolerance.
INTRODUCTION
Immune homeostasis and maintenance of self-tolerance depends upon constant vigilance by CD4+Foxp3+ Treg. Commitment to the Treg lineage occurs primarily in the thymus (1) (2), but also in the periphery in a TGFß dependent manner (3-6). One of the major goals of modern immunosuppression, be it in autoimmune disease or transplantation, is to harness tolerance mechanisms such as those used by Treg in order to minimise the duration and extent of drug immunosuppression. Short-term co receptor blockade provided the first demonstration that induction of tolerance could be achieved using low-impact intervention in a mature immune system (7). Studied in a transplantation setting, this form of tolerance is totally dependent on the ability of TGFß to signal to T cells (6), and is also associated with de novo induction of antigen specific Foxp3+ iTreg (4, 8). This raises the possibility that the absolute need for TGFß is simply to guarantee conversion of naïve CD4+ T cells to stable Foxp3 expression.
However, TGFß signalling not only induces expression of Foxp3, but many other effector molecules, including CD39, CD73, CTLA4, CD103, neuropilin, perforin and IL-10 (9-12). There are also claims that TGFß is needed for the effector arm of suppression (13) and if so, this too could explain the need for TGFß signalling to T cells.
Despite a large literature on Foxp3+ Treg in self tolerance it is not known whether Foxp3 expression is essential for dominant tolerance induced to foreign antigens, or whether other genetic/expression modalities (e.g. those potentially necessary for Th3, Tr1, iTr35 cells) can operate in its absence. Using a combination of genetically manipulated mouse strains unable to express Foxp3, retroviral constructs facilitating conditional nuclear localisation of Foxp3 and a dominant negative TGFßRII to ablate TGFß signalling in T cells, we have addressed the contributions of TGFß and Foxp3 to the induction and function of iTreg. We show that Foxp3 expression is indispensible for tolerance induction to transplanted tissue, and its continued nuclear expression is necessary for maintenance of tolerance. In contrast, once tolerance is established, prevention of tissue damage does not depend on TGFß signalling to naïve T-cells. This indicates that the major role of TGFß in this form of acquired tolerance largely resides in the induction of Foxp3 expression in naïve CD4+ T cells. Comparative transcriptome analysis of in vitro generated, TGFß-experienced Foxp3+ and Foxp3− CD4+ T cells with TGFß-experienced Foxp3−/− CD4+ T cells not only confirms TGFß signalling in all the populations, but also demonstrates that, although crucial for maintenance of tolerance, the proportion of the transcriptome controlled specifically by Foxp3 in iTreg appears to be relatively limited.
MATERIALS AND METHODS
Experimental animals
CBA/Ca (CBA), C57BL/6J (B6), CBA.Rag1−/−, B6.Rag1−/−, Marilyn.Rag1−/− (Marilyn) (derived from Marilyn.Rag2−/− (14)) , Marilyn.Foxp3hCD2 (8), Marilyn.Foxp3−/− (9), CD4-dnTGFβRII (15), CD4-dnTGFβRII.Rag1−/− and Marilyn.CD4-dnTGFβRII.Rag1−/− mice were bred and maintained under specific pathogen-free conditions at the Sir William Dunn School of Pathology (University of Oxford, Oxford, UK). CD4-dnTGFβRII mice backcrossed to B6 for more than 10 generations were crossed with B6.Rag1−/− mice for two generations to generate CD4-dnTGFβRII.Rag1−/− animals, which were further crossed with Marilyn mice to introduce the Dby-H2Ab specific TCR transgenic element. All experimental procedures received local ethics committee approval and were conducted in accordance with the Home Office Animals (Scientific Procedures) Act of 1986.
Skin grafting
Skin grafting was performed as described previously (16). Maintenance of nuclear Foxp3 in cFoxp3 transduced T cells adoptively transferred into mice was enforced by daily i.p. injections of tamoxifen in sunflower oil (1 mg daily).
Antibodies
Anti-CD4 (L3T4), anti-CD25 (7D4) and anti-CTLA4 (BN13) were purchased from BD Biosciences. Anti-Foxp3 (FJK-16s) anti-CD39 (24DMS1) and anti-CD73 (TY/11.8) were purchased from eBiosciences, CA. Anti-GITR (YGITR-765) was purchased from Serotec, UK.
Cytokines for cell culture and synthetic peptides
Lyophilised recombinant human TGFβ1 was purchased from R&D Systems and reconstituted to 10 μg/ml in 4 mM HCl, containing 1 mg/ml of BSA. Lyophilised peptide HYAb (NAGFNSNRANSSRSS) (14), was re-constituted in PBS and used at a maximum working concentration of 100 nM.
Bone marrow dendritic cell preparation
BMDCs were prepared as described previously (17) . Murine GM-CSF enriched supernatant was harvested from the transfected cell line X63 (provided by D. Gray, Edinburgh) and used at an equivalent of 5 ng/ml.
CD4+ T cell preparation
Splenic CBA or B6 CD4+ T cells were isolated by negative selection using an autoMACs CD4+ T cell isolation kit and AutoMacs separator (Miltenyi Biotec, UK).
In vitro generation of iTreg
Red blood cell depleted splenocytes from Marilyn, Marilyn.Foxp3hCD2, and Marilyn.Foxp3−/− mice were cultured at a 5:1 ratio with mitomycin-treated B6 female BMDCs, in RPMI containing 100 nm of HYAb peptide. For TGFβ conditioned iTreg cultures, recombinant human TGFβ was also added at 2 ng/ml. Foxp3 expression was typically 50-90% for TGFβ conditioned cultures i.e. 50-90% of TGFβ conditioned cultures were Foxp3+ iTregs. For microarray analysis MoFlo sorting was employed to sort Foxp3+ and negative cells from the cultures based on hCD2 expression.
Fluorescence activated cell sorting (FACS)
Cells were washed twice in PBS containing 0.5% w/v BSA, incubated for 10 min at room temperature in PBS with 10 μg/ml of anti-FcR block (2.4G2), and then with primary antibody for 30 min in the dark at 4°C. Following washing and fixation with 2% paraformaldehyde, analysis was performed on a FACSCalibur (Becton Dickinson, NJ) with dual laser excitation (488 nm and 633 nm). Data acquisition was performed with inter-channel compensation using CellQuest version 3.1 software (Becton Dickinson, NJ) and data analysis performed with FlowJo 7.2.4 software (Treestar Inc., Oregon). Foxp3 staining was performed using a commercial kit (eBioscience, CA) according to the manufacturer’s directions.
MoFlo FACS sorting of cells
Flow cytometry sorting was performed using a MoFlo sorter (Dako Cytomation, Glostrup).
Production of conditional Foxp3 retroviral supernatant
All vectors used except pCL-ECO have previously been described (18). The conditional Foxp3 construct cFoxp3 was made by cloning sequence encoding ERT2, a modified oestrogen binding domain, in-frame with the C-terminus of Foxp3. GFP coding sequence was cloned directly after the fifth codon of Foxp3, in order to produce the fusion protein GFP-Foxp-ERT2. This was then cloned into a Moloney Murine Leukemia Virus (MMLV) backbone, a circular plasmid that also contained a GPI-linked and IRES-driven rat CD8α (rCD8) gene. The empty control vector contained only the GFP and rCD8 segments, but no Foxp3. The packaging vector, pCL-ECO, was purchased from Addgene (Cambridge, MA). The day before transfection, a confluent plate of HEK 293eT cells were split at 1 in 3 and plated in 6-well plates at approximately 40% confluency, with 2 ml per well. Approximately 16 h later the cells were transfected with 2 μg of pCL-ECO packaging plasmid and 2 μg of the retroviral vector cFoxp3 using the CaCl2 method. 6 h after transfection, the medium in the well was replaced with fresh medium. A further 24 h later, the viral supernatant was harvested and filtered through a 0.45 μm filter. Protamine sulphate (600 μg/ml) was added to the viral supernatant.
Retroviral transduction of CD4+ T cells
AutoMACS sorted CD4+ cells were cultured at 2×105 per well in a 96-well plate pre-coated with anti-CD3 (KT3, 4 μg/ml in PBS, 1 h at 37°C). The cells were activated for 24 h and then resuspended in a 1:2 mixture of viral supernatant and complete medium (IMDM containing 10% v/v FCS and 10 μM of β-mercaptoethanol). This mix was supplemented with 5 ng/ml of recombinant mIL-2 (Peprotech) and 5 ng/ml of recombinant mIL-7 (Peprotech, UK). This was followed by centrifugation at 500 g for 2 hr at 32°C. Cells were analysed 40 h later. Typical transduction efficiencies were 30-50%. Following transduction, in most experiments the cells were MoFlo sorted to achieve 99.9% GFP positive cells, as indicated in figure legends. For induction of nuclear localization of Foxp3, transduced cells were cultured in the presence of 50 nM of 4′hydroxytamoxifen (4HT) for at least 24 h.
Confocal microscopy
Confocal microscopy was performed on a Zeiss LSM 510 META laser scanning microscope (Carl Zeiss MicroImaging GmbH, Göttingen) using a x20 objective.
Cytokine Measurement
A 25-component multiplex bead-based assay specific for; Eotaxin, G-CSF, GM-CSF, IFNγ, IL-1α, IL-1ß, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12p40, IL-12p70, IL-13, IL-17, KC, MCP-1, MIP-1α, MIP-1ß, RANTES, TNFα (Bio-Rad Laboratories Inc.), was used according to the manufacturer’s recommended protocol and samples analysed using a Bio-Plex™ System and Bio-Plex Manager Software 4.0 (Bio-Rad Laboratories Inc).
Proliferation and suppression assays
‘Effector’ cells were Marilyn CD4+ cells enriched by negative selection using AutoMACS. ‘Suppressor’ cells were Marilyn CD4+ cells that had been transduced with retroviral vectors for 40 h and then sorted based on CD4 and GFP expression. BMDCs were prepared from female B6 mice. Suppressor cells and BMDCs were incubated with mitomycin C (25 μg/ml) for 30 min at 37°C and washed three times before being added to the assay. Each well of a 96-well plate contained 1×104 BMDCs, 2×104 suppressors and 2.5×104 effectors. HYAb peptide was added as a dilution series (100 nM, 10 nM, 1 nM, 0 nM). Cultures were incubated for 72 h. To measure proliferation, 3H-thymidine (GE Healthcare, UK) was added (0.5 μCi/well) for the final 18 h of culture. 3H-thymidine incorporation was then measured by scintillation counting.
Affymetrix gene expression profiling of total RNA using whole-transcript assay (WTA) and gene/exon 1.0 ST arrays
Cultured splenocytes were used to generate highly purified populations of Marilyn.Foxp3hCD2 activated (HY) and TGFß-induced (HYT) cells sorted as CD4+hCD2+ and CD4+hCD2−, and Marilyn.Foxp3−/− HY and HYT cells sorted as CD4+. Three to five independent biological replicates of the five sorted cell populations above were processed (RNA isolation, labelling and hybridisation to mouse exon 1.0 ST Affymetrix arrays) by Asuragen Inc (Austin, TX). Total RNA was isolated from frozen cell pellets by Asuragen, Inc. according to the company’s standard operating procedure for automated isolation on a KingFisher magnetic particle separator. The procedure included a DNAse treatment step and cleanup prior to elution from the magnetic beads. The purity and quantity of total RNA samples were determined by absorbance readings at 260 and 280 nm using a NanoDrop ND-1000 UV spectrophotometer. The integrity of total RNA was qualified by Agilent Bioanalyzer 2100 microfluidic electrophoresis, using the Nano Assay.
Samples for mRNA profiling studies were processed by Asuragen, Inc. according to the company’s standard operating procedures. Biotin-labeled sense strand cDNA was prepared from 30 μg cRNA generated from either 500 ng or 1 μg total RNA per sample using a modified Affymetrix GeneChip® Whole Transcript (WT) Sense Target Labeling Assay (Affymetrix, Inc). Intermediate cRNA and resulting cDNA yields were quantified by spectrophotometry. Fragmentation and labeling of cDNA was performed using 5 μg for Exon Arrays. Hybridization to arrays was carried out at 45°C for 16 h in an Affymetrix Model 640 hybridization oven. Arrays were washed and stained on an Affymetrix FS450 Fluidics station. The arrays were scanned on an Affymetrix GeneChip Scanner 3000 7G. For every array scanned, .DAT, .CEL, .jpg, and .xml flat files were provided. Data generated from Asuragen for the arrays were further pre-processed in the Affymetrix Expression Console software version 1.2 which was used to normalize CEL files using the quantile normalization method and the data was summarized at both the exon level and gene level by using the Robust Multichip Analysis (RMA) algorithm. All further data analysis was performed in JMP® Genomics software version 5.1 using the gene level log2 data. Low value intensities were filtered out. Differential gene expression analysis was performed on transcript values based on core probe sets. The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE39529 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39529).
Statistical analysis
Statistical significance was determined using one-way ANOVA tests with Dunnett’s multiple comparison post-test or Student’s T test (unpaired, 2-tailed) using GraphPad Prism software, http://www.graphpad.com. In the figures, p values <0.05 are indicated by “*”, p<0.01 by “**” and p<0.001 by “***”, whereas non-significant p values are labeled “ns”.
RESULTS
Foxp3 expression is required for induction of transplantation tolerance mediated by co receptor blockade
It is well established that T cells expressing Foxp3 are critical for maintenance of tolerance to self antigens. The requirement for Foxp3 in the induction of peripheral tolerance to foreign antigens has not, however, been unequivocally demonstrated. It is possible that other cell types such as Th3, Tr1 or iTr35 might emerge and operate in its absence. Transplantation tolerance can be induced in TCR transgenic mice lacking natural Treg (nTreg). This is associated with the development of Foxp3+ iTreg in the periphery. It cannot be induced however, if TGFß signalling to T cells is prevented (6). We asked if alternative routes to tolerance might engage in a situation where Foxp3 induction was prevented by genetic ablation, but where TGFß signalling pathways were left intact. To address this in circumstances where autoimmune pathology could be avoided, we generated Foxp3−/− Marilyn TCR transgenic mice. All of the T cells in normal Marilyn mice are monospecific. They recognise a specific peptide of the male antigen Dby (HYAb) presented in the context of H2-Ab, and naïve T cells do not express Foxp3. Female Marilyn and Marilyn.Foxp3−/− mice were each grafted with male B6.Rag−/− skin under the influence of a short regime of anti-CD4 blocking antibody previously shown to be sufficient to generate tolerance and the induction of peripheral Foxp3+ iTreg in Marilyn mice (8). As expected, the majority of the Foxp3 sufficient mice accepted their male grafts indefinitely, whereas all of the Marilyn.Foxp3−/− mice had rejected their grafts by 60 d (Fig. 1A). Thus the tolerance induced by short term co receptor blockade requires a functional Foxp3 gene, despite an intact TGFß signalling axis.
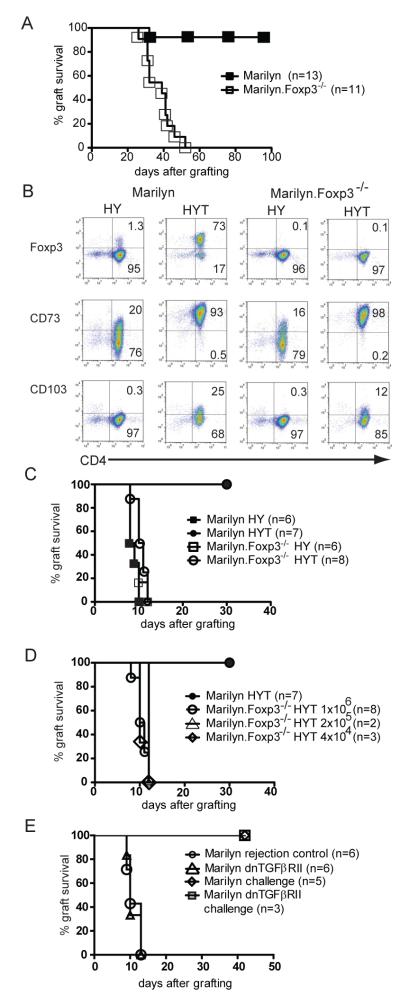
Figure 1. The relative requirements for Foxp3 and TGFß in induced tolerance to foreign antigen.
A. Female Marilyn mice (n=13) were transplanted with male B6.Rag−/− skin and treated with 1 mg of YTS177 (anti-CD4 blocking antibody) at days 0, 3 and 5 after grafting. Identical grafting and treatment was performed using female Marilyn.Foxp3−/− mice (n=11) and the grafts monitored for rejection (log rank p value <0.0001). Data are representative of two separate experiments.
B. Marilyn and Marilyn.Foxp3−/− splenocytes were cultured for 7 d with female B6 BMDC and HYAb peptide in the absence (HY) or presence (HYT) of TGFß. Histopaque-purified cells were co stained with CD4 and either Foxp3, CD73 or CD103 as shown. Numbers represent percentage of cells in the indicated quadrant. Data are representative of two separate experiments.
C. Marilyn or Marilyn.Foxp3−/− splenocytes were stimulated in vitro for 7 d with female B6 BMDCs and HYAb peptide in the absence (HY) or in the presence (HYT) of TGFß. One million Ficoll-purified live cells were injected into female B6.Rag−/− mice and one day later the mice were transplanted with a male B6.Rag−/− skin graft. Rejection was monitored daily. Data are representative of two separate experiments.
D. Using a similar experimental setting, the number of Marilyn.Foxp3−/− HYT splenocytes injected was titrated and is shown compared to the injection of 1×106 Marilyn HYT cells (dark circles).
E. Female Marilyn mice made tolerant to male B6.Rag−/− skin by anti-CD4 antibody treatment (1 mg at days 0, 3 and 5 after grafting) were transplanted with a second male B6.Rag−/− skin graft 95-120 d after the first. At the time of the second graft the mice also received 1×106 AutoMacs purified CD4+ T cells from female Marilyn or female Marilyn.dnTGFßRII mice and rejection was monitored daily. The rejection competency of the transferred Marilyn and Marilyn.dnTGFßRII T cells was validated by demonstrating the capacity of 1×106 cells transferred into naïve female Rag−/− recipients to reject male Rag−/− skin grafts. The x-axis represents days after grafting primary grafts for the rejection controls, and days after second grafting for assessing the requirement for intact TGFß signalling in mediating dominant tolerance.
TGFß driven in vitro generated iTreg require Foxp3 expression in order to become rejection-incompetent
TGFß induces potent anti-inflammatory adenosine production by CD4+ T cells via cell surface expressed CD39 and CD73 ectonucleotidase activity (9). Regulation of these enzymes is independent of Foxp3 expression and it has been suggested may confer suppressive activity in the absence of Foxp3 (19). We asked whether TGFß-conditioned T cells might still be able to reject grafts if they could not express Foxp3, and also whether TGFß was required as a mechanism of suppression. We generated sets of TGFß-conditioned T cells in vitro by stimulating Marilyn or Marilyn.Foxp3−/− splenocytes with BMDCs and cognate peptide in the absence (“HY”) or presence (“HYT”) of TGFß. These cultures typically yielded approximately 70% Foxp3+ cells in the Foxp3 sufficient cells after 7 d (Fig. 1B). CD4+ T cells from both wild-type and Foxp3−/− animals upregulated CD73 and CD103, indicating effective TGFß signalling, consistent with their expression of equivalent surface levels of TGFßRII (Supplementary Fig. 1A). Previous data (Supplementary Fig. 1B) indicated that as few as 2500 Marilyn T cells were sufficient to reject male grafts on adoptive transfer into Rag−/− recipients. One million Marilyn or Marilyn.Foxp3−/− HY or HYT splenocytes were injected into female B6.Rag−/− recipients, followed by grafting with male B6.Rag−/− skin. All seven mice that received TGFß-experienced Foxp3 sufficient splenocytes accepted their grafts, despite co transfer of >30 times more Foxp3− cells than should be necessary for rejection (Fig. 1C). Mice that received either antigen-activated or antigen-activated and TGFß-experienced splenocytes from Foxp3−/− animals rejected their grafts within the same rapid time-frame as that of wild-type antigen-activated, but non TGFß-experienced (Foxp3 negative) splenocytes (Fig. 1C). Even as few as 4×104 HYT Marilyn.Foxp3−/− cells were still able to reject male skin grafts at the same rate as 1×106 cells (Fig. 1D). In short, TGFß conditioning in the absence of Foxp3 expression has little impact on the ability of the T cells to reject grafts, showing the critical role of Foxp3 in extinguishing that function and thus ruling out a major role for alternative TGFß-induced pathways (such as adenosine generation (9)) in iTreg-induced long-term graft acceptance in this setting.
The true role for TGFß in Treg mediated suppression is controversial, with studies arguing for and against its requirement. We have previously shown that TGFß signalling to T cells is essential for the development of dominant tolerance (6). To establish whether in this case TGFß was operating in the inductive phase of tolerance, or at a later stage to sustain dominant tolerance, female Marilyn mice were made tolerant to male B6.Rag−/− skin using anti-CD4 antibody treatment. On re-grafting at 95-120 d after the first graft, the tolerised mice also received 1×106 rejection competent CD4+ T cells from either female Marilyn or female Marilyn.dnTGFßRII mice. In both cases the mice were able to resist rejection by the transferred cells (Fig. 1E), demonstrating that TGFß signalling to T cells was not essential for suppression of rejection once regulatory T cells had been induced.
CD4 T cells which fail to convert to Foxp3+ in the presence of TGFß remain able to secrete pro-inflammatory cytokines
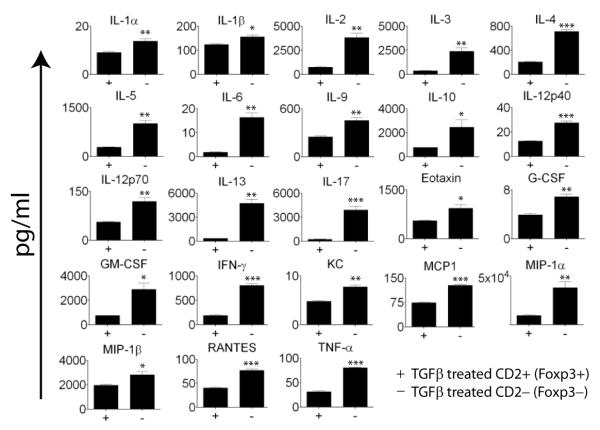
We then asked what had become of CD4 T cells which had not converted to Foxp3 expression by comparing their pattern of cytokine secretion to that of their Foxp3+ counterparts. Marilyn.Foxp3hCD2 CD4 T cells were activated for seven days with dendritic cells and HY peptide in the presence of TGFß. Cells sorted into hCD2 positive and negative populations were then stimulated for 2 days with anti-CD3/anti-CD28 conjugated beads. Culture supernatants were assayed for cytokine content. T cells which had not transcribed Foxp3 in response to TGFß exposure produced 5-20 fold increased amounts of interleukins 2,3,4,5,6,13,17 and IFNγ (Figure 2). Thus in the absence of Foxp3 TGFß treated cells are clearly capable of producing proinflammatory cytokines commensurate with their capacity to reject grafts.
Figure 2. T cells failing to express Foxp3 after exposure to TGFß secrete proinflammatory cytokines.
Marilyn.Foxp3hCD2 splenocytes were cultured in the presence of C57BL/6 female BMDCs and HY peptide in the presence of 10ng/ml TGFß for seven days. TGFß treated cells were then sorted as those which converted to Foxp3 expression, CD4+hCD2+ and those which remained Foxp3 negative, CD4+hCD2−. Sorted cells were stimulated with anti-CD3/anti-CD28 beads for 48 hours prior to assay of cytokines in the cell culture supernatant. Data represent means plus or minus the SEM of triplicate cultures. Statistical significance measured by unpaired t test; *p≤0.05, **p≤0.005, ***p≤0.0005. Results are representative of two separate experiments.
TGFß and Foxp3 induce independent and distinct transcriptional signatures
As TGFß-induced Foxp3 expression is crucial to the continuing function of iTreg in vivo and so closely linked to the biology of induced tolerance, we asked to what extent the gene transcription programme induced by TGFß was related to Foxp3 expression. To answer this question we performed microarray analysis comparing transcriptomes of CD4 T cells from Marilyn.Foxp3hCD2 or Marilyn.Foxp3−/− mice treated (HYT) or untreated (HY) with TGFß in vitro (see materials and methods). In each case CD4+ cells were MoFlo sorted, and in the case of the Marilyn.Foxp3hCD2 derived cells, sorted into Foxp3 expressors and non expressors on the basis of surface expression of hCD2.
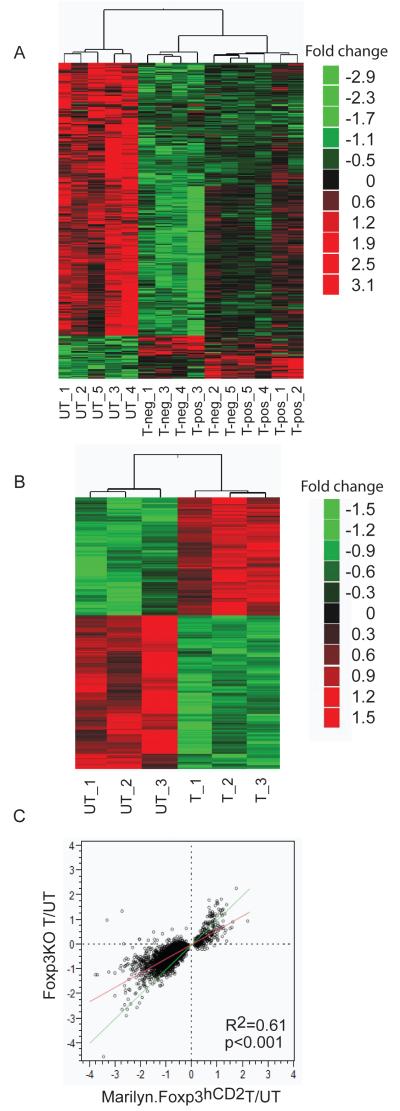
We asked whether the signature of transcripts regulated by TGFß exposure in CD4 T cells was dependent on Foxp3 expression. A global suppression effect was observed upon TGFß treatment in cells of Marilyn.Foxp3hCD2 mice, identified by comparative analysis between TGFß-treated Marilyn.Foxp3hCD2 cells (including both HYT hCD2+ and HYT hCD2− populations) and untreated HY cells (Fig. 3A). 3046 transcripts were found differentially expressed at p<0.01 (Supplementary Table I). A similar analysis performed on Marilyn.Foxp3−/− cells indicated 469 transcripts differentially expressed at p<0.01 (Fig. 3B and Supplementary Table II).
Figure 3. The contribution of Foxp3 to the TGFß induced transcript signature is relatively small.
A. A TGFß transcript signature derived from Marilyn.Foxp3hCD2 mice cultured with HY peptide and dendritic cells and stimulated with TGFß. Data represent TGFß-induced Foxp3+ hCD2+ (T-pos) and cells treated with TGFß in the same cultures which failed to express Foxp3, hCD2− (T-neg) cells, versus cells activated under the same conditions but minus TGFß (UT). Heat map of the differential transcripts derived from t-test between untreated cells and TGFß induced hCD2+ and hCD2−. See Supplementary Table I for gene identities. Five biological replicates for each cell type are shown.
B. A TGFß transcript signature derived from Marilyn.Foxp3−/− mice stimulated under the same conditions as (3A) TGFß untreated (UT) versus treated (T) cells. See Supplementary Table II for gene identities. Three biological replicates for each treatment group are shown.
C. Correlation of TGFß signatures between Marilyn.Foxp3hCD2 and Marilyn.Foxp3−/− mice. The green line indicates an ideal correlation. The red line indicates the actual correlation, R2 value 0.61. Scatter plot of Log2 (fold change) of transcripts in unsorted Marilyn.Foxp3.hCD2 CD4+ T cells treated with or without TGFß (x axis) and Foxp3−/− CD4+ T cells treated with or without TGFß (y axis).
To examine if the presence of Foxp3 wild-type copies could generate additional response to TGFß treatment, the expression profile of the TGFß signature in Marilyn.Foxp3hCD2 mice was examined in Marilyn.Foxp3−/− mice (Fig. 3C). The high correlation (R2=0.61) suggested that there was not a significant additional detectable effect, positive or negative, from the genomic copies of Foxp3. Thus the TGFß signature seems to be independent of Foxp3. These data also show that loss of Foxp3 was not inhibiting TGFß responsiveness of these cells.
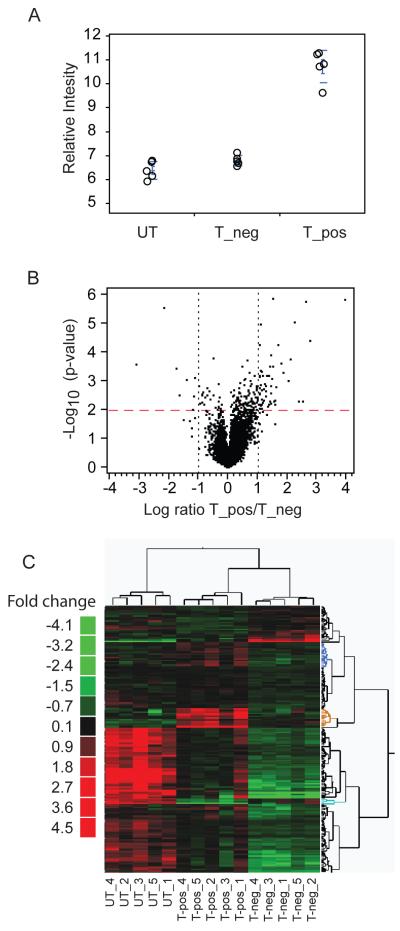
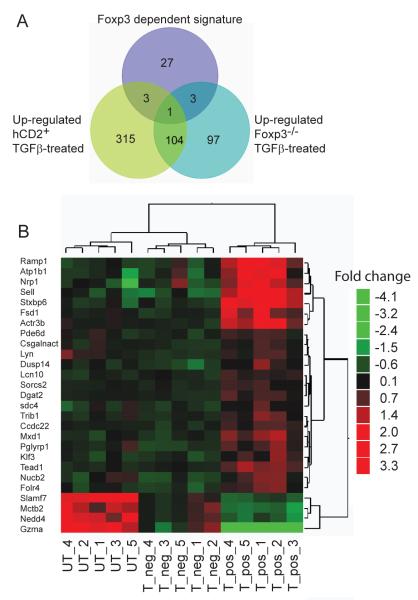
Samples of the TGFß-treated and untreated, sorted hCD2+ and hCD2− groups exhibited the expected relative level of Foxp3 transcript expression (Fig. 4A). To identify a TGFß-induced-Foxp3 effect between the two sorted TGFß-influenced groups (HYT hCD2+ and HYT hCD2−), a comparative analysis was first carried out to identify transcripts differentially expressed between the two populations. 183 transcripts (p<0.01) were found differentially expressed (Fig. 4B, C and Supplementary Table III). Among them, a subset of 34 transcripts was then identified as uniquely associated with the TGFß-induced hCD2+ cells (Fig. 4C, indicated as blue, orange and turquoise). Finally, upon further eliminating general TGFß effects, 27 transcripts were considered to comprise a unique TGFß-induced-Foxp3 signature (Fig. 5A, B).
Figure 4. Identification of a TGFß dependent Foxp3 gene signature.
A. Foxp3 expression by non-TGFß-treated, activated hCD2− CD4+ T cells from Marilyn.Foxp3hCD2 mice (UT), TGFß-treated CD4+ T cells from Marilyn.Foxp3hCD2 mice which failed to express Foxp3, hCD2− (T_neg), and TGFß-treated hCD2+ (T_pos) CD4+ T cells, assessed by microarray. Data from five biological replicates is shown.
B. Volcano plot of differential genes between Foxp3 positive (T_pos) and negative (T_neg) TGFß-treated CD4+ T cells described in (4A).
C. Heat map of statistically differential transcripts derived by t-test between Marilyn.Foxp3hCD2 mice sorted on peptide and dendritic cell-stimulated TGFß-induced hCD2+ (T-pos) and cells treated with TGFß in the same cultures which failed to express Foxp3, hCD2− (T-neg) cells. The same transcripts in activated CD4+ T cells untreated with TGFβ (UT) are shown as a control. See Supplementary Table III for the identities of individual genes. Data from five biological replicates are shown.
Figure 5. The TGFß driven Foxp3 induced gene signature comprises 27 genes.
A. Venn diagram showing the relationship of TGFß signatures of genes in Foxp3 positive and negative populations. CD4+ T cells from Marilyn.Foxp3hCD2 mice, TGFß treated and peptide activated and subsequently FACS sorted into hCD2+ and hCD2− groups, were compared with TGFß and peptide stimulated Foxp3−/− CD4+ T cells. Batch effects were removed and t-tests applied to identify differentially expressed transcripts between the hCD2+ and hCD2− groups. To identify a unique TGFß induced Foxp3 signature (purple circle) the differential transcripts were filtered 1) to include only those with unique expression profiles in hCD2+ cells and 2) to exclude transcripts of Foxp3-independent TGFß signatures from Foxp3−/− cells. Data are from five biological replicate samples.
B. Heat map of statistically significant Foxp3-specific TGFß signature transcripts compared between hCD2+ Foxp3 positive (T_pos) and hCD2− Foxp3 negative (T_neg) TGFß-treated CD4+ T cells and untreated (UT), activated CD4+ T cells, generated as described in (5A). Data are from five biological replicate samples.
Ectopic tamoxifen-mediated nuclear localization of Foxp3 imposes a suppressive phenotype onto naïve T cells
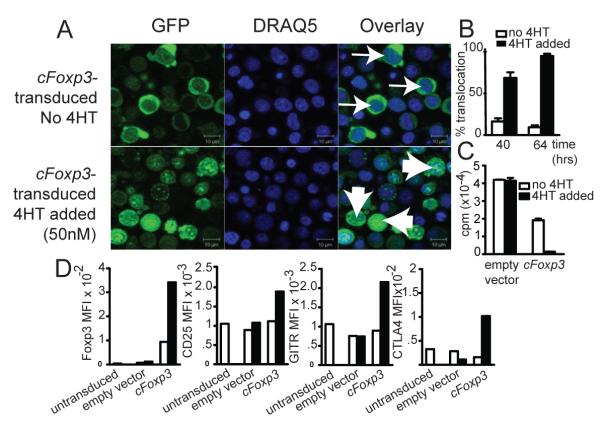
Our demonstration that Foxp3 was indispensible for tolerance induced by TGFß led us to ask whether the need for Foxp3 expression in tolerance was transient, or sustained. Many groups have demonstrated that ectopic expression of Foxp3 leads to suppressive function, and in vitro cre-lox deletion of Foxp3 has been shown to reverse the suppressive properties of Treg. However, it is not known whether iTreg need to express Foxp3 for prolonged periods in order for tolerance to be maintained, nor what effect ‘turning off’ Foxp3 in vivo following tolerance induction would have. For example, Foxp3-influenced T cells may, over time, acquire suppressive features that are Foxp3-independent. We therefore asked two questions, namely, is prolonged Foxp3 expression needed for iTreg to remain rejection incompetent, and is continuous Foxp3 expression required for iTreg to retain their suppressive ability? To address these questions, we used a retroviral construct, cFoxp3, encoding a fusion protein GFP-Foxp-ERT2, whose entry to the nucleus, and therefore Foxp3 activity, requires addition of tamoxifen ((18) and see Materials and Methods). We first established the ability of the construct to translocate to the nucleus following tamoxifen treatment, and to induce attributes consistent with a Treg phenotype, namely, anergy, upregulation of Treg cell surface markers and suppression of pro-inflammatory cytokine production (Fig. 6). We showed that after 64 h, GFP-Foxp3 fusion protein had clearly translocated into the nuclei of over 95% of transduced AutoMacs sorted CD4+ splenocytes, FACS sorted for GFP and treated with 4HT, but remained predominantly cytoplasmic in transduced but non-induced cells (Fig. 6A and B). Tamoxifen exposure rendered the T-cells anergic (Fig. 6C), and was associated with up-regulation of the Treg associated surface markers CTLA4, CD25 and GITR (Fig. 6D). Foxp3 negatively regulates transcription of many cytokines (20, 21), and we found that 4HT-treated transduced T cells also secreted minimal quantities of pro-inflammatory cytokines, such as IL-1, IL-2, IFNγ, IL-4, IL-9 and IL-17, when compared to cells transduced with empty vector or with cFoxp3, but without 4HT treatment (Fig. 7). We term such transduced cells induced to exhibit a regulatory phenotype by using 4HT as ‘conditional Treg’ (cTreg).
Figure 6. cFoxp3 transduced T cells display anergy and upregulate Treg markers in response to 4′HT.
A. Nuclear translocation of GFP-Foxp3-ERT2 64 h from the onset of cFoxp3 transduction, with or without inclusion of 4HT in the culture medium. FACS sorted CD4+CD25− B6 splenic T cells were prepared as per Materials and Methods .Thin arrows point to non-induced cells which show distinct localization of GFP and DRAQ-5 staining, while wide arrows point to 4HT-induced cells which show co localization. Data are representative of three separate experiments.
B. Three random microscopy fields were selected per sample for each of the time points, 40 and 60 h from the onset of transduction and cells that showed co localisation of GFP and DRAQ-5 stain were counted as having undergone nuclear translocation of the cFoxp3 encoded fusion protein. Each field examined contained at least 20 cells, with an average of 30 cells counted per field. The error bars represent standard error of the mean, for the three counted fields. Two-way ANOVA was used to compare both time-points and the two conditions (no 4HT versus 4HT added): p<0.01. Data are representative of two separate experiments.
C. Marilyn CD4+ cells were transduced with cFoxp3 or empty vector. 40 h after transduction, cells were MoFlo sorted with respect to expression of CD4 and GFP expression and incubated for a further 66 h with mitomycin C-treated HYAb pulsed female B6 BMDCs , with or without inclusion of 4HT. Cell proliferation was assessed by 3H-thymidine incorporation. The statistical significance of inhibited proliferation by nuclear Foxp3 was calculated using two-way ANOVA with Bonferonni post test: p<0.01. Data are representative of two separate experiments.
D. Conditional Treg stained for CTLA4, CD25 and GITR. FACS sorted CD4+CD25− B6 splenic T cells transduced with cFoxp3 were cultured in the absence or presence of 4HT for 64 h prior to FACS staining. Untransduced cells and cells transduced with empty vector were used as negative controls. Cells were fixed and permeabilised prior to intracellular staining for Foxp3 and CTLA4. Data are represented as mean fluorescence intensities and are representative of two separate experiments. White bars indicate cells with no 4HT added. Black bars indicate cTreg with 50nM 4HT included in the cultures.
Figure 7. Conditional Treg secrete very low levels of proinflammatory cytokines.
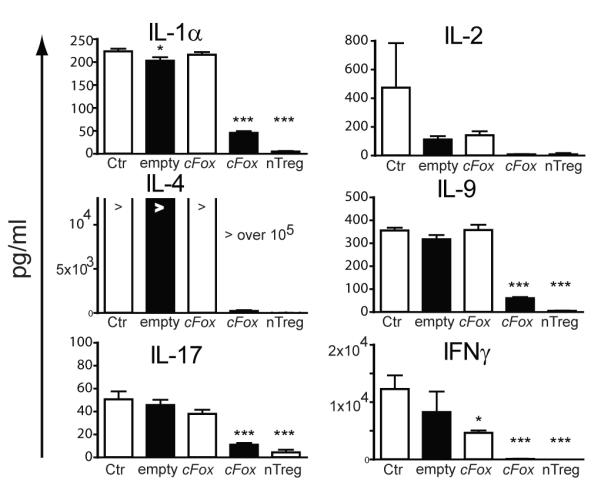
CD4+CD25− cells purified from B6 spleens were transduced with cFoxp3 or empty vector ‘empty’. For comparison, Foxp3+ nTreg were isolated from B6.Foxp3hCD2 mice by MoFlo sorting based on hCD2 expression. Negative control cells ‘Ctr’ were untransfected CD4+CD25− cells. Transduced CD4+ cells were MoFlo sorted based on GFP expression 40 h after the onset of transduction and plated at 2×105 cells/well in fresh medium on anti-CD3 coated 96-well plates with (black bars) or without (white bars) 4HT for a further 48 h. Supernatant was collected and analysed using a custom Bio-Plex Multiplex Cytokine Assay. For statistical analysis, one-way ANOVA with Dunnett’s comparison test was used (all groups compared to untransduced cells). *≤0.05, ***≤0.001. “>” equals more than 105 pg/ml. Data represent technical triplicates plus and minus standard error of the mean. Data are representative of two separate experiments.
Persistent nuclear expression of Foxp3 is required for cTreg to remain rejection incompetent
We next tested whether, in the absence of exogenous addition of TGFß, tamoxifen exposure would suffice to render T cells unable to reject grafts. Naïve Marilyn CD4+ T cells, which do not express Foxp3, were transduced with cFoxp3; half were treated and half untreated with 4HT for 2 d in vitro to induce nuclear localisation of the GFP-Foxp-ERT2 fusion protein. CD4+GFP+ cells (Fig. 8A) were purified by FACS and 2×104 or 1×106 cells transferred into female B6.Rag−/− mice. These mice then each received a male B6.Rag−/− skin graft and half received daily tamoxifen injections, the other half daily injections of vehicle for 28 d post-transplant. All the mice that received 4HT-facilitated cTreg, but without further tamoxifen injections rejected their grafts within 20 d (Fig. 8B). Mice that received either 2×104 or 1×106 transduced cells treated in vitro with 4HT and with further daily injections of tamoxifen, accepted their grafts up to and following cessation of the tamoxifen injections, but rejected their grafts 14-17 d following the last tamoxifen injection. Once the tamoxifen injections ended the speed of rejection was similar to that experienced on transfer of cells with no tamoxifen. Thus persistent expression of nuclear Foxp3 was associated in vivo with T cells maintaining a rejection-impotent phenotype.
Figure 8. Sustained Foxp3 expression is required for maintenance of a suppressive phenotype.
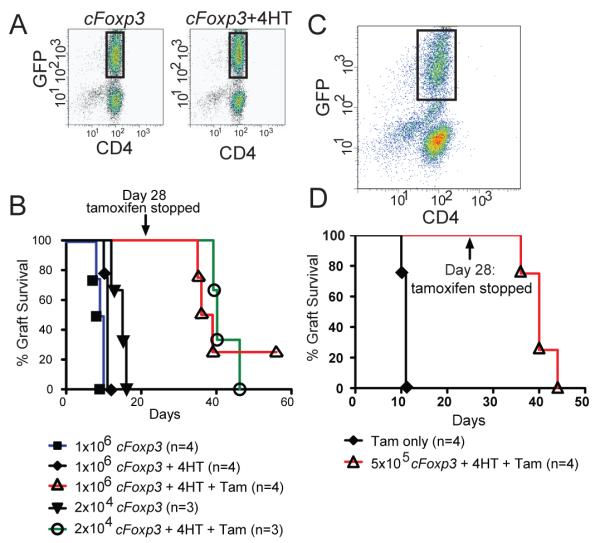
A. Marilyn CD4+ T cells were transduced with cFoxp3 and either treated or not treated with 4HT to induce nuclear localisation of GFP-Foxp3-ERT. After 48 h, transduced cells were sorted based on CD4 and GFP expression (sorted cells indicated by rectangles). Data are representative of three experiments.
B. Splenic CD4+ cells from Marilyn mice were transduced with cFoxp3 and half received in vitro 4HT treatment. 2×104 or 1×106 cells were sorted for expression of GFP and transferred into B6.Rag−/− mice. These mice received daily tamoxifen (1mg i.p.). As controls, some recipients did not receive in vivo tamoxifen treatment, but received vehicle alone. Graft rejection was monitored daily. On day 28, tamoxifen treatment was ceased and graft rejection followed until day 56. Data are representative of two separate experiments.
C. MoFlo FACS profile of sorted GFP+ cTreg used in (4D).
D. 5×105 MoFlo sorted in vitro 4HT-treated cFoxp3-transduced cells were transferred into Marilyn.Foxp3hCD2 mice (n=4) followed the next day by transplantation of a male CBA.Rag−/− graft. Daily tamoxifen (1mg) was administered. Control recipients (n=4) did not receive cells, but were treated with daily tamoxifen. Data are representative of two separate experiments.
Sustained Foxp3 expression is needed to maintain a suppressive phenotype
In order to establish whether sustained expression of Foxp3 in cTreg was needed for their suppressive function, immune competent naive female Marilyn mice, which lack endogenous Treg, were adoptively transferred with 5×105 transduced Marilyn, 4HT-treated, cTreg (Fig. 8C, D). The mice were then grafted with a male CBA.Rag−/− skin graft and injected daily with tamoxifen for 28 d. All Marilyn mice without cTreg cells rejected their grafts within 11 d, as expected. Mice with grafts together with cTreg tolerated their grafts for 40 d, (12 d beyond the last injection of tamoxifen). Thus cTreg can suppress rejection of an allogeneic graft, but this requires sustained exposure to tamoxifen..
DISCUSSION
We had set out to test the extent to which induction of Foxp3 expression explained the need for TGFß signalling to T cells in eliciting tolerance to transplanted tissues (6). We made use of TCR transgenic mice on a Rag−/− background. Untreated, these mice have no Foxp3+CD4+ T cells, so that any Foxp3+ cells emerging from treatment are de facto iTreg cells. Furthermore, the use of TCR transgenic Foxp3−/− mice sidesteps any potentially confounding issues of autoimmune pathology. We used Foxp3-reporter mice on the Marilyn background, Marilyn.Foxp3−/− mice and microarray analysis to demonstrate that although the number of genes affected by TGFß exposure is large, the set of genes in iTreg whose expression is correlated specifically with Foxp3 expression is surprisingly low. Finally, ectopic inducible nuclear Foxp3 expression was used to demonstrate that continuous expression of Foxp3 on a per cell basis was required for maintenance of induced tolerance.
We present five novel findings. First, the potential to express Foxp3 is essential for this form of induced peripheral tolerance. Second, a TGFß ’experience’ in the absence of Foxp3 is insufficient for tolerance despite inducing a significant TGFß. gene signature. Third, TGFß signalling to T cells, although essential for induction of tolerance, is not essential for suppression by iTreg. Fourth, sustained functional expression of Foxp3 in vivo is necessary to ensure prolonged graft survival. Fifth, iTreg have independent TGFß and Foxp3 gene signatures, the latter consisting of a relatively small number of genes.
The literature describes many different CD4+ T cell subsets exhibiting regulatory properties. These include TGFß secreting Th3 cells (22, 23), IL-10 secreting Tr1 cells (24) and IL-35 secreting iT(R)35 cells (25). We have previously shown that maintenance of tolerated allogeneic skin grafts required constant suppression by Foxp3+ cells (8) . Here we show for the first time that Foxp3 expression is essential for the induction of tolerance. Any other potentially suppressive mechanisms such as Tr1 cells did not emerge as effective substitutes for Foxp3+ T cells.
In Marilyn mice, tolerance by co receptor blockade relies on the TGFß dependent de novo differentiation of iTreg (6). We propose therefore, that failure to induce tolerance in the Marilyn.Foxp3−/− mice was due to failure to generate Foxp3-expressing iTreg cells. Consistent with that, antigen-activated Marilyn.Foxp3−/− CD4+ T cells cultured in the presence of TGFß (9) remained responsive to antigen in vitro, and could still reject grafts in vivo, suggesting that any other TGFß-induced mechanisms were insufficient to both calm the cells, and to convert them to potent regulatory function.
It is still unclear whether iTreg play a major role in the control of immune homeostasis. Numerous studies have shown that Foxp3+ iTreg can arise in a number of experimental settings. These include exposure of T cells to cognate antigen in the absence of microbial or endogenous ‘danger’ signals as in the case of mucosally (26), as well as parenterally delivered antigens (27). It has however been difficult to separate the need for nTreg away from that for iTreg in experimental models. Whatever emerges as the normal physiological role for iTreg, it is clear that they play a crucial role in therapeutic tolerance in TCR transgenic mice, as demonstrated here. This coupled with the absolute need for TGFß signalling to T cells for transplantation tolerance in conventional mice suggests that iTreg are also very relevant in that context.
Foxp3 has been shown to have binding regions on approximately 700 genes in nTreg, as well as forming a supramolecular complex with other transcription factors and chromatin modifiers (21). Persistent Foxp3 expression is likely needed to maintain the transcriptional changes and epigenetic alterations of target genes which it controls. We performed extensive transcript analysis using exon tiling microarrays and demonstrated that T cells have approximately 3000 transcripts significantly altered in expression by TGFß. However subtractive comparisons of the transcriptome of TGFß-experienced T cells which did not convert to Foxp3 expression (HYT CD4+hCD2− cells from Marilyn.Foxp3hCD2) or TGFß-experienced Foxp3−/− CD4+ T cells, showed a very limited signature of 27 genes influenced solely by Foxp3 in iTreg. Thus although Foxp3 can bind hundreds of genes in thymically derived Treg, resulting in up and down gene regulation, in our populations of iTreg the number of genes significantly altered in expression by Foxp3 was much more limited. Foxp3 expression is controlled in part by variable DNA methylation at the Treg Specific Demethylated Region (TSDR) (28, 29). The transcriptional and epigenetic modifications initiated by Foxp3 may not be sufficient to render the cell regulatory in the absence of continued Foxp3 expression. Cre-lox deletion of the Foxp3 gene in mature Treg was found to result in reversal of their regulatory characteristics and acquisition of ability to produce pro-inflammatory cytokines (30).
We have shown that sustained nuclear Foxp3 expression is required to maintain a cellular program compatible with regulation. For a regulatory cell to suppress, one desirable function would be that it limits its own production of proinflammatory cytokines. We observed such behaviour for IL-1, IL-2, IL-4, IL-9, IL-17 and IFNγ, all of which were reduced to low levels following tamoxifen induced nuclear localisation of GFP-Foxp3-ERT2 in cFoxp3-transduced Marilyn CD4+ T cells. The requirement for sustained suppression to maintain tolerance has already been argued where tolerance was lost when Foxp3 expressing cells were deleted (8, 31). We have shown that simple withdrawal of tamoxifen in this system is sufficient to revert cTreg to an effector phenotype. Tamoxifen in an equivalent setting in human T cells has been shown to induce nuclear localisation as well as increase the stability of the fusion protein (32). Withdrawal of tamoxifen resulted in reversal of the inhibition of IFNγ production (32). Thus lowering the expression and changing the sub-cellular localisation of Foxp3 from nucleus to cytoplasm is a potent switch to mediate loss of suppressive activity.
Two recent reports have shown that N-terminal addition of GFP to Foxp3 can alter the interaction of Foxp3 with HIF1α, Eos, HDAC7 and TIP60, whereas interaction with NFAT is unaltered (33, 34). This led to a decrease in proteasomal degradation of Foxp3 and subtle alterations of the genes inhibited by Foxp3. The result of these alterations was increased suppression of Th17 responses resulting in increased susceptibility to diabetes on the NOD background, consistent with previous reports showing Th17 cells to be protective in this model. We have shown that Th17 cells mediate damage in allogeneic skin graft rejection (35), thus the potency of our conditional Treg which have an N-terminal GFP tagged Foxp3 may be explained in part by this phenomenon.
These studies reinforce and extend our earlier findings that Foxp3+ cells are critical for clinically relevant modes of tolerance induction. The role of TGFß can be adequately accounted for by the induction of Foxp3 gene expression, and a narrow set of Foxp3 influenced genes, and no other feature of TGFß signalling. The implications of these findings are that clinical efforts to harness induced Foxp3+ cells for tolerance will need to ensure the stable expression of Foxp3 as its role in Treg is not a transient one, but one requiring persistent expression.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Dr Nigel Rust for expert FACS sorting, the PSB staff for excellent animal husbandry and Dr. Amin Moghaddam for expert assistance with cytokine assays.
Abbreviations used in this manuscript
- Treg
regulatory T cell
- iTreg
induced Treg
- B6
C57BL/6J
- Marilyn
Marilyn.Rag1−/−
- HYAb
peptide of male antigen Dby (NAGFNSNRANSSRSS)
- BMDCs
bone marrow derived dendritic cells
- 4HT
4′ hydroxytamoxifen
- cTreg
conditional Treg
Footnotes
Supported by a programme grant from the Medical Research Council and the European Framework 7 Betacell programme .
REFERENCES
- 1.Klein L, Jovanovic K. Regulatory T cell lineage commitment in the thymus. Semin Immunol. 2011;23:401–409. doi: 10.1016/j.smim.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Lio CW, Hsieh CS. Becoming self-aware: the thymic education of regulatory T cells. Curr Opin Immunol. 2011;23:213–219. doi: 10.1016/j.coi.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25-naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobbold SP, Castejon R, Adams E, Zelenika D, Graca L, Humm S, Waldmann H. Induction of foxP3+ regulatory T cells in the periphery of T cell receptor transgenic mice tolerized to transplants. J Immunol. 2004;172:6003–6010. doi: 10.4049/jimmunol.172.10.6003. [DOI] [PubMed] [Google Scholar]
- 5.Kretschmer K, Apostolou I, Hawiger D, Khazaie K, Nussenzweig MC, von Boehmer H. Inducing and expanding regulatory T cell populations by foreign antigen. Nature immunology. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 6.Daley SR, Ma J, Adams E, Cobbold SP, Waldmann H. A key role for TGF-beta signaling to T cells in the long-term acceptance of allografts. Journal of immunology. 2007;179:3648–3654. doi: 10.4049/jimmunol.179.6.3648. [DOI] [PubMed] [Google Scholar]
- 7.Benjamin RJ, Waldmann H. Induction of tolerance by monoclonal antibody therapy. Nature. 1986;320:449–451. doi: 10.1038/320449a0. [DOI] [PubMed] [Google Scholar]
- 8.Kendal AR, Chen Y, Regateiro FS, Ma J, Adams E, Cobbold SP, Hori S, Waldmann H. Sustained suppression by Foxp3+ regulatory T cells is vital for infectious transplantation tolerance. The Journal of experimental medicine. 2011;208:2043–2053. doi: 10.1084/jem.20110767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regateiro FS, Howie D, Nolan KF, Agorogiannis EI, Greaves DR, Cobbold SP, Waldmann H. Generation of anti-inflammatory adenosine byleukocytes is regulated by TGF-beta. European journal of immunology. 2011;41:2955–2965. doi: 10.1002/eji.201141512. [DOI] [PubMed] [Google Scholar]
- 10.Cobbold SP, Graca L, Lin CY, Adams E, Waldmann H. Regulatory T cells in the induction and maintenance of peripheral transplantation tolerance. Transpl Int. 2003;16:66–75. doi: 10.1007/s00147-003-0545-y. [DOI] [PubMed] [Google Scholar]
- 11.Rao PE, Petrone AL, Ponath PD. Differentiation and expansion of T cells with regulatory function from human peripheral lymphocytes by stimulation in the presence of TGF-{beta} J Immunol. 2005;174:1446–1455. doi: 10.4049/jimmunol.174.3.1446. [DOI] [PubMed] [Google Scholar]
- 12.Haribhai D, Lin W, Edwards B, Ziegelbauer J, Salzman NH, Carlson MR, Li SH, Simpson PM, Chatila TA, Williams CB. A central role for induced regulatory T cells in tolerance induction in experimental colitis. Journal of immunology. 2009;182:3461–3468. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson J, Tran DQ, Pesu M, Davidson TS, Ramsey H, O’Shea JJ, Shevach EM. CD4+ FoxP3+ regulatory T cells confer infectious tolerance in a TGF-beta-dependent manner. The Journal of experimental medicine. 2008;205:1975–1981. doi: 10.1084/jem.20080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lantz O, Grandjean I, Matzinger P, Di Santo JP. Gamma chain required for naive CD4+ T cell survival but not for antigen proliferation. Nature immunology. 2000;1:54–58. doi: 10.1038/76917. [DOI] [PubMed] [Google Scholar]
- 15.Gorelik L, Flavell RA. Abrogation of TGFbeta signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12:171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 16.Qin SX, Wise M, Cobbold SP, Leong L, Kong YC, Parnes JR, Waldmann H. Induction of tolerance in peripheral T cells with monoclonal antibodies. Eur J Immunol. 1990;20:2737–2745. doi: 10.1002/eji.1830201231. [DOI] [PubMed] [Google Scholar]
- 17.Nolan KF, Strong V, Soler D, Fairchild PJ, Cobbold SP, Croxton R, Gonzalo JA, Rubio A, Wells M, Waldmann H. IL-10-conditioned dendritic cells, decommissioned for recruitment of adaptive immunity, elicit innate inflammatory gene products in response to danger signals. J Immunol. 2004;172:2201–2209. doi: 10.4049/jimmunol.172.4.2201. [DOI] [PubMed] [Google Scholar]
- 18.Andersen KG, Butcher T, Betz AG. Specific immunosuppression with inducible Foxp3-transduced polyclonal T cells. PLoS biology. 2008;6:e276. doi: 10.1371/journal.pbio.0060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annual review of immunology. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 22.Carrier Y, Yuan J, Kuchroo VK, Weiner HL. Th3 cells in peripheral tolerance. I. Induction of Foxp3-positive regulatory T cells by Th3 cells derived from TGF-beta T cell-transgenic mice. J Immunol. 2007;178:179–185. doi: 10.4049/jimmunol.178.1.179. [DOI] [PubMed] [Google Scholar]
- 23.Zheng SG, Gray JD, Ohtsuka K, Yamagiwa S, Horwitz DA. Generation ex vivo of TGF-beta-producing regulatory T cells from CD4+CD25-precursors. J Immunol. 2002;169:4183–4189. doi: 10.4049/jimmunol.169.8.4183. [DOI] [PubMed] [Google Scholar]
- 24.Levings MK, Bacchetta R, Schulz U, Roncarolo MG. The role of IL-10 and TGF-beta in the differentiation and effector function of T regulatory cells. Int Arch Allergy Immunol. 2002;129:263–276. doi: 10.1159/000067596. [DOI] [PubMed] [Google Scholar]
- 25.Collison LW, Chaturvedi V, Henderson AL, Giacomin PR, Guy C, Bankoti J, Finkelstein D, Forbes K, Workman CJ, Brown SA, Rehg JE, Jones ML, Ni HT, Artis D, Turk MJ, Vignali DA. IL-35-mediated induction of a potent regulatory T cell population. Nature immunology. 2010;11:1093–1101. doi: 10.1038/ni.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Verginis P, McLaughlin KA, Wucherpfennig KW, von Boehmer H, Apostolou I. Induction of antigen-specific regulatory T cells in wild-type mice: visualization and targets of suppression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3479–3484. doi: 10.1073/pnas.0800149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, Reid SP, Levy DE, Bromberg JS. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 31.Lahl K, Loddenkemper C, Drouin C, Freyer J, Arnason J, Eberl G, Hamann A, Wagner H, Huehn J, Sparwasser T. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. The Journal of experimental medicine. 2007;204:57–63. doi: 10.1084/jem.20061852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allan SE, Song-Zhao GX, Abraham T, McMurchy AN, Levings MK. Inducible reprogramming of human T cells into Treg cells by a conditionally active form of FOXP3. European journal of immunology. 2008;38:3282–3289. doi: 10.1002/eji.200838373. [DOI] [PubMed] [Google Scholar]
- 33.Bettini ML, Pan F, Bettini M, Finkelstein D, Rehg JE, Floess S, Bell BD, Ziegler SF, Huehn J, Pardoll DM, Vignali DA. Loss of epigenetic modification driven by the foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36:717–730. doi: 10.1016/j.immuni.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darce J, Rudra D, Li L, Nishio J, Cipolletta D, Rudensky AY, Mathis D, Benoist C. An N-terminal mutation of the foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity. 2012;36:731–741. doi: 10.1016/j.immuni.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agorogiannis EI, Regateiro FS, Howie D, Waldmann H, Cobbold SP. Th17 cells induce a distinct graft rejection response that does not require IL-17A. Am J Transplant. 2012;12:835–845. doi: 10.1111/j.1600-6143.2011.03971.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.