Abstract
Galectin-3 is a lectin involved in fibrosis, inflammation and proliferation. Increased circulating levels of galectin-3 have been associated with various diseases, including cancer, immunological disorders, and cardiovascular disease. To enhance our knowledge on galectin-3 biology we performed the first genome-wide association study (GWAS) using the Illumina HumanCytoSNP-12 array imputed with the HapMap 2 CEU panel on plasma galectin-3 levels in 3,776 subjects and follow-up genotyping in an additional 3,516 subjects. We identified 2 genome wide significant loci associated with plasma galectin-3 levels. One locus harbours the LGALS3 gene (rs2274273; P = 2.35×10−188) and the other locus the ABO gene (rs644234; P = 3.65×10−47). The variance explained by the LGALS3 locus was 25.6% and by the ABO locus 3.8% and jointly they explained 29.2%. Rs2274273 lies in high linkage disequilibrium with two non-synonymous SNPs (rs4644; r2 = 1.0, and rs4652; r2 = 0.91) and wet lab follow-up genotyping revealed that both are strongly associated with galectin-3 levels (rs4644; P = 4.97×10−465 and rs4652 P = 1.50×10−421) and were also associated with LGALS3 gene-expression. The origins of our associations should be further validated by means of functional experiments.
Introduction
Galectin-3 (LGALS3) is a lectin and member of the galectin family of carbohydrate binding proteins that have an affinity for beta-galactosides. Galectin-3 plays a role in fibrosis, inflammation, and proliferation [1], [2], [3]. Galectin-3 is secreted into the systemic circulation by unknown mechanisms and is increasingly recognised as a potential biomarker with clinical value. Increased galectin-3 levels have been associated with various diseases, including cancer [4], [5], immunological disorders [6], [7], and cardiovascular traits [8], [9]. Plasma galectin-3 levels are even being considered as a marker of response to cancer treatment [10].
To enhance our knowledge on galectin-3 biology we performed the first genome-wide association study (GWAS) on circulating galectin-3 levels and observed two loci associated with circulating galectin-3 levels. One locus harbours LGALS3 the gene encoding galectin-3 and the other locus harbours the ABO gene which has previously been associated with inflammatory markers, lipids and haematological parameters.
Results
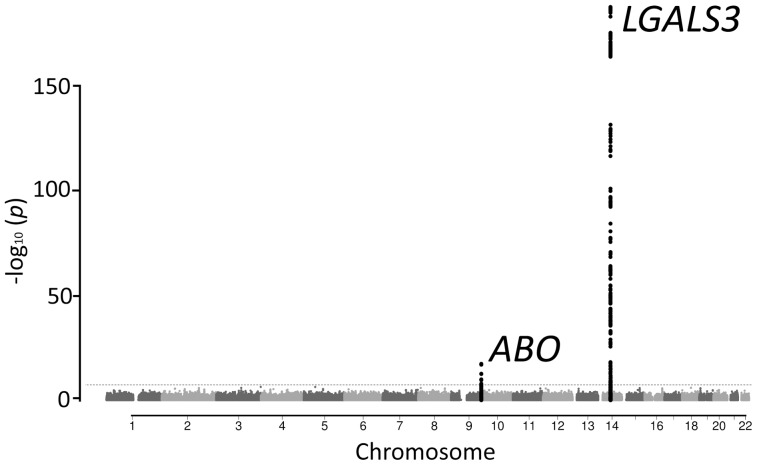
We performed a GWAS analysis of 2,269,099 genotyped or imputed autosomal SNPs (HapMap 2 build 36 CEU panel) in 3,776 subjects of the PREVEND cohort ( Table 1 , Table S1). All included subjects were of European descent. The quantile-quantile plot for association is shown in Figure 1 . There were 2 loci significantly associated with galectin-3 levels (P<5×10−8) and 11 SNPs showing suggestive evidence (P<5×10−6 and P>5×10−8, Figure 2 , Table 2 ). We performed further testing of the lead-SNP of the loci using an additional subset of 3,516 independent subjects derived from the PREVEND cohort ( Table 1 ). Using inverse-variance fixed effect meta-analysis we combined the evidence. None of the suggestive, but both 2 P<5×10−8 loci of the discovery phase, were confirmed in the independent samples ( Table 2 ). One locus harbours the LGALS3 gene and the other locus harbours the ABO gene ( Figure 3 ). The LGALS3 locus accounted for 25.6% of the phenotypic variance. The ABO locus explained 3.8% and together LGALS3 and ABO explained 29.2% of the phenotypic variance. Of note, common genetic variation explained twice the amount of the variation of circulating galectin-3 levels compared to age, age squared (age2), gender, and body mass index combined (11.6%).
Table 1. Galectin-3 levels in the PREVEND cohort, indicated for the total population and for the discovery and replication groups.
| Total | Discovery | Follow-up | |
| N = 7,292 | N = 3,776 | N = 3,516 | |
| Galectin-3, median [IQR] | 10.9 [9]–[13] | 10.8 [9]–[13] | 10.9 [9]–[13] |
| Galectin-3 (ng/ml), min-max | 3.4–233 | 3.5–233 | 3.4–200 |
IQR; inter quartile range.
Figure 1. Quantile-quantile plots of observed versus expected p-values for Galectin-3 with and without the GALS3 locus.
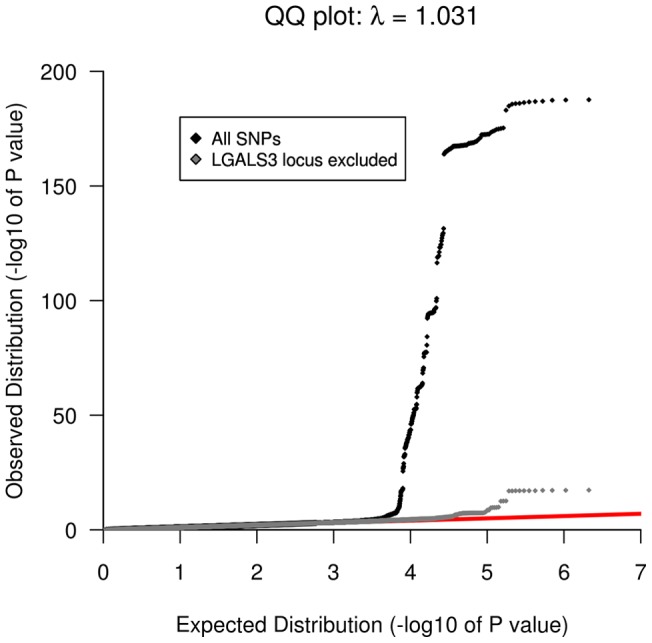
Figure 2. Manhattan plot showing the association of SNPs with circulating galectin-3 levels in a GWAS of 3,776 individuals.
The red dotted line marks the threshold for genome-wide significance (P = 5×10−8). Two loci reached genome-wide significance.
Table 2. Discovery and follow-up genotyping results.
| Discovery | Follow-up | Combined | Annotation | |||||||||
| Chr | SNP | A1/A2 | AF | Effect (se) | P-value | Effect (se) | P-value | Effect (se) | P-value | N | Location | Nearest Gene |
| 2 | rs17775170 | A/G | 0.27 | −4.79E-02 (0.011) | 5.36E-06 | 3.70E-03 (0.008) | 6.218E-01 | −1.38E-02 (0.006) | 2.43E-02 | 7284 | intronic | SLC9A2 |
| 2 | rs2165179 | A/G | 0.33 | −4.85E-02 (0.010) | 1.47E-06 | 7.46E-03 (0.015) | 6.280E-01 | −3.17E-02 (0.008) | 1.77E-04 | 7264 | intronic | SCN3A |
| 2 | rs7586 | T/C | 0.03 | 1.12E-01 (0.025) | 7.79E-06 | 3.75E-03 (0.017) | 8.215E-01 | 3.69E-02 (0.014) | 7.61E-03 | 7278 | 3′-UTR | COL5A2 |
| 3 | rs11919628 | T/G | 0.07 | 6.11E-02 (0.013) | 1.37E-06 | 9.69E-03 (0.012) | 4.218E-01 | 3.43E-02 (0.009) | 8.38E-05 | 7289 | intergenic | FLJ25363, PVRL3-AS1 |
| 4 | rs930956 | C/G | 0.33 | 3.92E-02 (0.009) | 7.84E-06 | 1.34E-02 (0.007) | 5.294E-02 | 2.33E-02 (0.005) | 1.89E-05 | 7277 | intronic | ATP8A1 |
| 5 | rs16902429 | G/T | 0.29 | 3.26E-02 (0.007) | 4.66E-06 | 1.04E-02 (0.007) | 1.227E-01 | 2.09E-02 (0.005) | 1.86E-05 | 7287 | intergenic | COX7C, MIR4280 |
| 6 | rs17658562 | T/C | 0.14 | 5.29E-02 (0.012) | 6.57E-06 | −8.91E-03 (0.010) | 3.651E-01 | 1.67E-02 (0.008) | 2.67E-02 | 7268 | intergenic | MIR548A1, ID4 |
| 8 | rs2409784 | C/A | 0.75 | −5.29E-02 (0.011) | 1.45E-06 | 1.10E-03 (0.006) | 8.639E-01 | −1.26E-02 (0.006) | 2.29E-02 | 7225 | intronic | BLK |
| 8 | rs4876386 | C/T | 0.06 | 6.43E-02 (0.014) | 2.51E-06 | −4.79E-04 (0.007) | 9.482E-01 | 1.42E-02 (0.007) | 2.81E-02 | 7277 | intergenic | MED30, EXT1 |
| 9 | rs644234 | G/T | 0.35 | −6.05E-02 (0.007) | 4.52E-18 | −7.55E-02 (0.007) | 2.390E- 30 | −6.84E-02 (0.005) | 3.65E-47 | 7225 | intronic | ABO |
| 11 | rs4351827 | T/C | 0.25 | 4.98E-02 (0.011) | 2.37E-06 | 4.55E-03 (0.008) | 5.655E-01 | 2.10E-02 (0.006) | 9.14E-04 | 7253 | intergenic | CLMP, MIR4493 |
| 13 | rs9512645 | T/C | 0.54 | −5.31E-02 (0.011) | 3.43E-06 | 6.06E-03 (0.007) | 4.185E-01 | −1.18E-02 (0.006) | 5.97E-02 | 7281 | intergenic | RASL11A, GTF3A |
| 14 | rs2274273 | A/G | 0.41 | −1.85E-01 (0.006) | 2.35E-188 | NA | NA | −1.85E-01 (0.006) | 2.35E-188 | 3776 | downstream | DLGAP5 |
| 14 | rs4644 | A/C | 0.41 | −1.85E-01 (0.006) | 2.06E-187 | −1.89E-01 (0.005) | 3.000E-223 | −1.87E-01 (0.004) | 4.97e-465 | 7281 | exonic | LGALS3 |
| 14 | rs4652 | C/A | 0.43 | −1.77E-01 (0.006) | 3.48E-173 | −1.79E-01 (0.006) | 1.160E-202 | −1.78E-01 (0.004) | 1.50e-421 | 7292 | exonic | LGALS3 |
| 15 | rs17223788 | C/T | 0.49 | 5.54E-02 (0.012) | 1.65E-06 | 7.99E-03 (0.006) | 2.144E-01 | 1.93E-02 (0.006) | 5.89E-04 | 7272 | intergenic | LOC283688, ARNT2 |
| 16 | rs8101416 | T/A | 0.17 | 4.60E-02 (0.010) | 1.94E-06 | −8.56E-05 (0.009) | 9.928E-01 | −2.24E-02 (0.007) | 9.52E-04 | 7257 | intergenic | DUS3L, NRTN |
| 19 | rs12443973 | A/C | 0.20 | −4.80E-02 (0.010) | 2.11E-06 | 5.84E-03 (0.008) | 4.405E-01 | −1.35E-02 (0.006) | 2.55E-02 | 7281 | intergenic | MAF, MIR548H4 |
Significant loci (P<5×10−8, bold) and suggestive loci (P<5×10−6) in the discovery phase that were taken forward for follow-up genotyping. In addition, 2 non-synonymous variants (rs4644 and rs4652) were genotyped. AF: Allele Frequency; (se): standard error.
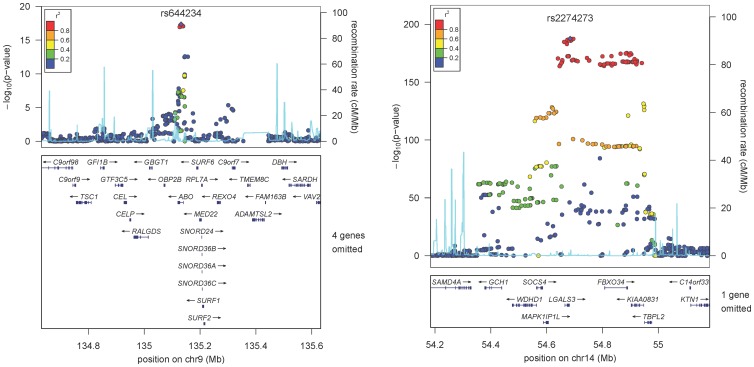
Figure 3. Regional plots at the two significantly associated loci.
Horizontal axis indicates chromosomal location and P-values are indicated in the left y-axis. Each plot shows approximately ± 500 kb around each lead SNP and has known gene transcripts annotated at the bottom. The SNPs are colored according to their degree of linkage disequilibrium (r 2) with the lead SNP which is highlighted with a purple diamond and displayed by rs number and significance level achieved in the discovery analysis.
Putative causal genetic variants
The lead SNP (rs2274273) of the LGALS3 locus lies in high LD with two non-synonymous variants (rs4644; r2 = 1.0 and rs4652; r2 = 0.91). As these variants were not present on our platform and not well imputed we wet-lab genotyped these variants and confirmed their association ( Table 2 ). We next considered potential confounding by the specific galectin-3 assay used and noticed the epitopes of the antibodies used are directed against the region harbouring the non-synonymous variant ( Figure 4 ). Therefore, this variant might affect the affinity of the antibody and not represent a true difference in circulating galectin-3 levels. We did not find variants in high LD (r2>0.8) associated with the lead SNP (rs644234) of the ABO locus. Next, we searched for eQTLs in 1,469 samples from peripheral blood for which gene expression levels were obtained using illumine HT12V3 and illumine H8v2 platforms [11]. rs2274273, rs4644, and rs4652 were all associated with LGALS3 gene expression levels ( Table 3 ) and rs2274273 and rs4644 were also the strongest SNP associated with that particular LGALS3 probe. Finally, to gain further insights we queried the catalogue of published genome wide association studies [12] for our loci and observed no previous associations for the LGALS3 locus but many previous genome wide associations findings have been reported for the ABO locus. Previous SNP associations in or near ABO are in high linkage disequilibrium with our lead SNP and include associations with inflammatory markers, lipids and haematological parameters as well as diseases such as cancer and coronary heart disease (Table S2).
Figure 4. Amino acid sequence (41 to 100) of Galectin-3.
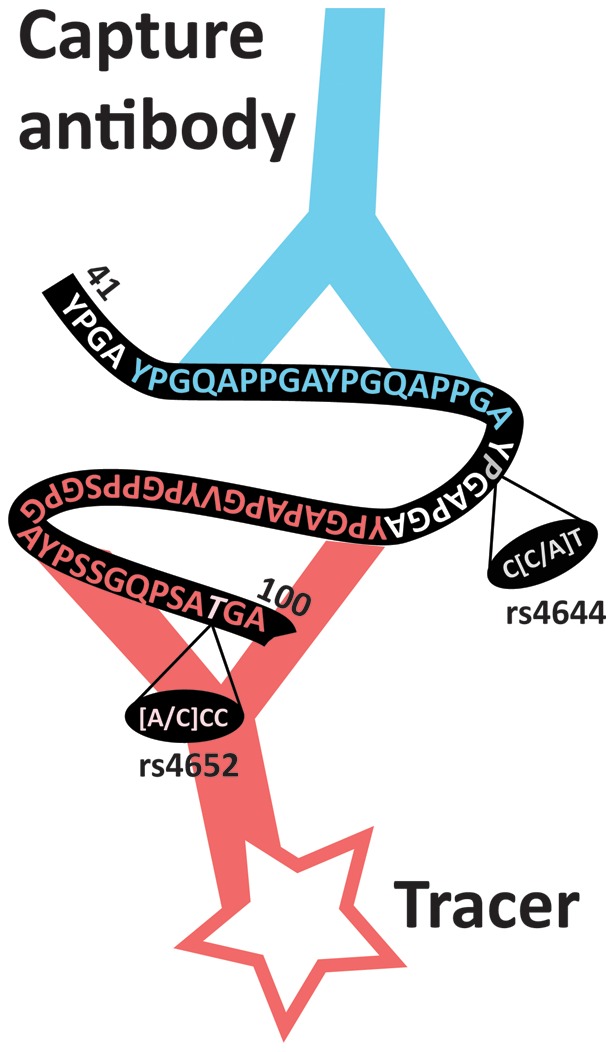
The two antibodies used in the Galectin-3 assay recognize epitopes within the N-terminus of the protein (white-colored amino acids). The capture antibody of the galectin-3 assay binds to amino acids number 45 to 62 and the tracer antibody binds to amino acids number 70 to 100. Indicated are the non-synonymous SNPs rs4644 and rs4652.
Table 3. Relationship between identified SNPs with expression of cis-genes in peripheral blood in 1,469 samples.
| Chr | SNP | Position | Gene | Probe | P-Value | FDR |
| 14 | rs4644 | 54673724 | LGALS3 | 5420377 | 2.42×10−7 | 0 |
| 14 | rs4652 | 54674789 | LGALS3 | 5420377 | 3.10×10−6 | 0 |
| 14 | rs2274273 | 54673724 | LGALS3 | 3450685 | 4.45×10−6 | 0 |
cis-genes: genes within ±1 MB around the lead SNP. FDR: False discovery rate.
Relevance of LGALS3 variant for prognostic value of plasma galectin-3 levels
To study the relevance of the rs2274273 and rs4644 (r2 = 1) variant in the LGALS3 locus for the prognostic value of the galectin-3 assay on mortality in the general population we repeated our earlier reported analyses [13] with and without rs2274273 as a covariate in the model. Knowledge of the genotype did not appear to change the prognostic value of plasma galectin-3 levels ( Table 4 ).
Table 4. Effect of LGALS3 genotype on association of plasma galectin-3 levels on mortality.
| All cause | Cardiovascular | Cancer | ||||||||||
| Model | HR without rs2274273 (95% CI) | P-value | HR with rs2274273 (95% CI) | P-value | HR without rs2274273 (95% CI) | P-value | HR with rs2274273 (95% CI) | P-value | HR without rs2274273 (95% CI) | P-value | HR with rs2274273 (95% CI) | P-value |
| Unadjusted | 1.46 (1.37–1.56) | 0 | 1.52 (1.43–1.62) | 0 | 1.56 (1.39–1.75) | 0 | 1.60 (1.44–1.78) | 0 | 1.41 (1.28–1.56) | 0 | 1.47 (1.34–1.62) | 0 |
| Age and gender adjusted | 1.12 (1.03–1.21) | 0.007 | 1.20 (1.09–1.31) | 0 | 1.16 (1.01–1.35) | 0.044 | 1.27 (1.07–1.49) | 0.005 | 1.10 (0.97–1.23) | 0.128 | 1.16 (1.02–1.33) | 0.027 |
| Multivariable classical risk factor adjusted* | 1.09 (1.01–1.19) | 0.036 | 1.17 (1.06–1.28) | 0.001 | 1.10 (0.94–1.28) | 0.228 | 1.22 (1.03–1.45) | 0.02 | 1.08 (0.96–1.22) | 0.222 | 1.13 (0.99–1.30) | 0.067 |
HR indicates hazard ratio per SD change in plasma galectin-3 level. * the multivariable model included age, sex, hypertension, hypercholesterolemia, diabetes, and smoking.
Discussion
We report the first genetic association study on galectin-3 levels and identified 2 genome-wide significant loci; one including the galectin-3 encoding gene (LGALS3) and the other gene being ABO.
Galectin-3 is a member of the galectin family that comprises of lectins with affinity for beta-galactosidases containing carbohydrates. The galectin gene family is evolutionarily ancient and can be found in vertebrates, invertebrates, and even in protists suggesting an important role in biology [14]. All galectins have a carbohydrate-recognition domain (CRD) consisting of many conserved sequence elements and each galectin has an individual carbohydrate-binding preference [15]. Galectin-3 is an unique galectin as it contains a non-lectin N-terminal region which is connected to the CRD. Galectin-3 is therefore referred to as a chimera-like galectin [1]. Galectin-3 does not contain a signal sequence and is primarily localised within the cytoplasm. It can be externalized by a mechanism independent of the endoplasmic reticulum (ER)-Golgi complex [16], [17]. Galectin-3 has high affinity for lactose and N-acetyllactosamine but can also interact with a wide array of other carbohydrates, membrane and extracellular matrix proteins [18]. Upon ligand binding, galectin-3 (and its ligands) forms cross-links, is involved in strengthening cell-cell interactions, and is associated with stiffening of the extracellular matrix and fibrogenesis. Galectin-3 has been shown to play a role in inflammatory diseases, cancer and heart failure [3], [15], [19], [20]. Little is known about the regulation of galectin-3. The galectin-3 promoter contains several responsive elements, including Sp-1, AP-1 and cAMP responsive elements [21].
We now report the first 2 genome wide associations with circulating galectin-3 levels. The strongest locus is within the LGALS3 gene. The lead SNP (rs2274273) is in full LD with two non-synonymous SNPs (rs4644 and rs4652) which were confirmed by follow-up genotyping. Both rs2274273 and rs4644 affected LGALS3 gene-expression providing a potential explanation for the observed effect. In the current study we also tested whether knowledge of the lead variant in the LGALS3 gene might obscure the association of plasma galectin-3 levels with outcome but it did not alter our previously published associations further supporting a true effect of these variant on galectin-3 [13]. However, some note of caution is warranted. Associations of coding SNPs (e.g. rs4644 and rs4652) that structurally change the properties of its encoded protein can give rise to false positive associations when that protein is also the phenotype under investigation. The non-synonymous SNPs identified in our study also lies within or near the epitopes of the antibodies used for the galectin-3 assay ( Figure 4 ). These antibodies might have different affinities for the amino acid change and therefore this association could also be artifactual. Interference of antibody based assays with epitopes directed against regions harbouring non-synonymous variants are not novel and have previously been reported for the NPPA-NPPB locus when ANP levels were measured [22]. Although gene-expression analyses and association with outcome are suggesting a true effect, additional work will be required to define the precise mechanisms of our reported association at the LGALS3 locus.
Our second genome wide locus is the ABO locus. The ABO locus is becoming an increasingly complex and pleiotropic locus. Variants in ABO, and in high LD with our lead SNP (rs644234), have been associated by genome wide association studies with various blood measured traits and diseases. This includes several inflammatory markers, lipids, hematological parameters, cancer, inflammatory diseases, and cardiovascular diseases (Table S2). Interestingly, galectin-3 levels also are associated with many of these conditions. Galectin-3 can indeed bind to polysaccharides of the ABO epitopes and even more strongly to the A- or B-histo-blood group epitopes versus the O group [23]. However, this does not explain how the ABO gene variant affects circulating galectin-3 levels.
In summary, we performed a GWAS on plasma galectin-3 levels and identified two genome wide significant loci, one including the LGALS3 gene and the other the ABO gene. The origins of these associations should be further validated by means of functional experiments.
Materials and Methods
Study population
We studied subjects included in the PREVEND cohort. The PREVEND cohort has been described in detail elsewhere [13], [24], [25]. In brief, 8,592 subjects were enrolled in the PREVEND cohort in 1997–1998. Subjects were asked to refrain from eating and drinking prior to their visit (fasting) in the outpatient clinic (between 8:00 a. um and 1:00 pm) and blood samples were drawn and stored at −80C. The PREVEND study was approved by the local medical Ethical Committee, and is conducted in accordance with the guidelines of the Declaration of Helsinki. All subjects provided written informed consent.
Galectin-3 Measurements
For 7,968 subjects plasma was available to measure plasma galectin-3 levels [13]. The galectin-3 assay is an enzyme-linked immunosorbent assay (BG Medicine, Inc., Waltham, USA). This assay quantitatively measures the concentration of human galectin-3 levels in EDTA plasma. This assay has high sensitivity (lower limit of detection 1.13 ng/mL) and exhibits no cross reactivity with collagens or other members of the galectin family [26]. Commonly used medication like ACE-inhibitors, beta blockers, spironolactone, furosemide, acetylsalicylic acid, warfarin, coumarines, and digoxin have no interference with the assay [26]. All samples were assayed in duplicate. Two standard controls were included in all runs: a lower control (expected value: 13.0–23.1 ng/mL) and a higher control (expected value: 48.9–81.5 ng/mL). The average lower control results were 16.65±1.13 (coefficient of variance: 6.8%), and the average higher control results were 68.17±3.20 (coefficient of variance: 4.7%).
Genotyping, Quality control & Imputation
Genotyping in 4,016 of the total number of participants in PREVEND was carried out using Illumina HumanCytoSNP-12 arrays. SNPs were called using Illumina Genome Studio software. Forty-seven subjects were excluded from analyses because call rates were <0.95. Another 65 subjects were excluded because they were closely related as judged based on Identity-By-Descent estimation using PLINK v1.07. Population structure was assessed using PCA based on 16,842 independent SNPs. Based on this analysis, an additional 2 samples were excluded that diverged from the mean with at least 3 standard deviations (Z-score >3) for the first 5 PCAs. Another 35 subjects were excluded based on sex inconsistencies. We excluded samples with a genetic similarity >0.1. Of 87 subjects no phenotype was available because of missing plasma samples for assessment of Galectin-3. As a consequence 3,776 (1,927 males, 1,849 females) were available for GWAS analysis. SNPs were excluded with a minor allele frequency of <0.01, call rate <0.95, or deviation from Hardy Weinberg equilibrium (P<1×10−5). Genome wide genotype imputation was performed using Beagle v. 3.3.1 [27], 232,571 genotyped SNPs were imputed up to 2,269,099 autosomal SNPs with NCBI build 36 of Phase II HapMap CEU data (release 22) as reference panel. Replication genotyping was performed by KBiosciences (KBiosciences, Herts, UK) utilizing the SNPline system in an additional 3,516 independent subjects of the PREVEND study.
Gene-expression analyses
We investigated whether each of the associated variants had an effect on gene expression levels by mapping cis-expression quantitative trait loci (cis-eQTL) in 1,469 samples from peripheral blood, for which gene expression level measurements were obtained using Illumina HT12v3 and Illumina H8v2 platforms [11]. Since the genotypes were imputed using the CEU population of HapMap 2 release 24 as reference, eQTL effects were tested using the imputation dosage values. Effects for SNPs (MAF >5%, HWE >0.001) were considered cis-eQTLs when the distance between the SNP and the midpoint position of the probe was smaller than 1 MB. As multiple testing correction, we controlled the false discovery rate (FDR) at 0.05, by comparing observed p-values to the null distribution obtained from permuting the expression phenotype labels relative to genotype labels 100 times. We also determined the top eQTL SNP for each given probe and tested whether the GWAS SNP had an independent effect on the associated gene expression probe after removing the effect of the top eQTL SNP.
Statistical analysis
Galectin-3 was non-normally distributed and was log transformed before regression analyses. We calculated residuals of galectin-3 levels after adjustment for age, age2, and gender. GWAS analyses were performed on residuals using an additive genetic model in PLINK (v 1.07) [28]. The most significant (P<5×10−8) SNPs (lead SNP) at each locus was taken forward for further testing. The explained variance of the significant associations was analysed using the directly genotyped variants from the replication stage. Fixed-effect meta-analysis was performed using the variance weighting method of the METAL software package to calculate the overall p-value. The Cox proportional-hazards model was used to calculate the hazard ratio and 95% confidence intervals (CI) of galectin-3. Based on our previous work, sequential models were fitted without and with the SNP of interest [13]. The first model including no covariates (unadjusted) and the second model adjusted for age and gender and the third model adjusted for: age, gender, previous myocardial infarction, previous stroke, hypertension, hypercholesterolemia and diabetes. The assumptions underlying the proportional hazards model were tested and found valid. Analyses were performed using STATA version 11.0 for Windows software (StataCorp LP, College Station, TX, USA).
Supporting Information
SNPs significantly associated with galectin-3 levels at the Discovery stage.
(DOC)
Previous reported traits with genome wide associations at the ABO locus.
(DOC)
Funding Statement
PREVEND genetics is supported by the Dutch Kidney Foundation (Grant E033), the National Institutes of Health (grant LM010098), The Netherlands organisation for health research and development (NWO VENI grant 916.761.70 & ZonMw grant 90.700.441), and the Dutch Inter University Cardiology Institute Netherlands (ICIN). R.A. de Boer is supported by the Netherlands Heart Foundation (grant 2007T046) and the Innovational Research Incentives Scheme program of the Netherlands Organization for Scientific Research (NWO VENI grant 916.10.117). BG Medicine Inc. provided an unrestricted research grant to the Department of Cardiology of the University Medical Centre Groningen. N. Verweij is supported by the Netherlands Heart Foundation (grant NHS2010B280). I. Mateo Leach is supported by the Netherlands Heart Foundation (grant NHS2008B065) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yang RY, Rabinovich GA, Liu FT (2008) Galectins: structure, function and therapeutic potential. Expert Rev Mol Med 10: e17. [DOI] [PubMed] [Google Scholar]
- 2. Dumic J, Dabelic S, Flogel M (2006) Galectin-3: an open-ended story. Biochim Biophys Acta 1760: 616–635. [DOI] [PubMed] [Google Scholar]
- 3. de Boer RA, Voors AA, Muntendam P, van Gilst WH, van Veldhuisen DJ (2009) Galectin-3: a novel mediator of heart failure development and progression. Eur J Heart Fail 11: 811–817. [DOI] [PubMed] [Google Scholar]
- 4. Nangia-Makker P, Balan V, Raz A (2012) Galectin-3 binding and metastasis. Methods Mol Biol 878: 251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Califice S, Castronovo V, Van Den Brule F (2004) Galectin-3 and cancer (Review). Int J Oncol 25: 983–992. [PubMed] [Google Scholar]
- 6. Dhirapong A, Lleo A, Leung P, Gershwin ME, Liu FT (2009) The immunological potential of galectin-1 and -3. Autoimmun Rev 8: 360–363. [DOI] [PubMed] [Google Scholar]
- 7. Henderson NC, Sethi T (2009) The regulation of inflammation by galectin-3. Immunol Rev 230: 160–171. [DOI] [PubMed] [Google Scholar]
- 8. Weigert J, Neumeier M, Wanninger J, Bauer S, Farkas S, et al. (2010) Serum galectin-3 is elevated in obesity and negatively correlates with glycosylated hemoglobin in type 2 diabetes. J Clin Endocrinol Metab 95: 1404–1411. [DOI] [PubMed] [Google Scholar]
- 9. de Boer RA, Lok DJ, Jaarsma T, van der Meer P, Voors AA, et al. (2011) Predictive value of plasma galectin-3 levels in heart failure with reduced and preserved ejection fraction. Ann Med 43: 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saussez S, Lorfevre F, Lequeux T, Laurent G, Chantrain G, et al. (2008) The determination of the levels of circulating galectin-1 and -3 in HNSCC patients could be used to monitor tumor progression and/or responses to therapy. Oral Oncol 44: 86–93. [DOI] [PubMed] [Google Scholar]
- 11. Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, et al. (2011) Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 7: e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 106: 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Boer RA, van Veldhuisen DJ, Gansevoort RT, Muller Kobold AC, van Gilst WH, et al. (2012) The fibrosis marker galectin-3 and outcome in the general population. J Intern Med 272: 55–64. [DOI] [PubMed] [Google Scholar]
- 14. Cooper DN (2002) Galectinomics: finding themes in complexity. Biochim Biophys Acta 1572: 209–231. [DOI] [PubMed] [Google Scholar]
- 15. de Boer RA, Yu L, van Veldhuisen DJ (2010) Galectin-3 in cardiac remodeling and heart failure. Curr Heart Fail Rep 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elola MT, Wolfenstein-Todel C, Troncoso MF, Vasta GR, Rabinovich GA (2007) Galectins: matricellular glycan-binding proteins linking cell adhesion, migration, and survival. Cell Mol Life Sci 64: 1679–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mehul B, Hughes RC (1997) Plasma membrane targetting, vesicular budding and release of galectin 3 from the cytoplasm of mammalian cells during secretion. J Cell Sci 110 (Pt 10): 1169–1178. [DOI] [PubMed] [Google Scholar]
- 18. Krzeslak A, Lipinska A (2004) Galectin-3 as a multifunctional protein. Cell Mol Biol Lett 9: 305–328. [PubMed] [Google Scholar]
- 19. Rabinovich GA, Liu FT, Hirashima M, Anderson A (2007) An emerging role for galectins in tuning the immune response: lessons from experimental models of inflammatory disease, autoimmunity and cancer. Scand J Immunol 66: 143–158. [DOI] [PubMed] [Google Scholar]
- 20. Newlaczyl AU, Yu LG (2011) Galectin-3–a jack-of-all-trades in cancer. Cancer Lett 313: 123–128. [DOI] [PubMed] [Google Scholar]
- 21. Kadrofske MM, Openo KP, Wang JL (1998) The human LGALS3 (galectin-3) gene: determination of the gene structure and functional characterization of the promoter. Arch Biochem Biophys 349: 7–20. [DOI] [PubMed] [Google Scholar]
- 22. Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, et al. (2009) Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet 41: 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feizi T, Solomon JC, Yuen CT, Jeng KC, Frigeri LG, et al. (1994) The adhesive specificity of the soluble human lectin, IgE-binding protein, toward lipid-linked oligosaccharides. Presence of the blood group A, B, B-like, and H monosaccharides confers a binding activity to tetrasaccharide (lacto-N-tetraose and lacto-N-neotetraose) backbones. Biochemistry 33: 6342–6349. [DOI] [PubMed] [Google Scholar]
- 24. Pinto-Sietsma SJ, Janssen WM, Hillege HL, Navis G, De Zeeuw D, et al. (2000) Urinary albumin excretion is associated with renal functional abnormalities in a nondiabetic population. J Am Soc Nephrol 11: 1882–1888. [DOI] [PubMed] [Google Scholar]
- 25. Boger CA, Chen MH, Tin A, Olden M, Kottgen A, et al. (2011) CUBN is a gene locus for albuminuria. J Am Soc Nephrol 22: 555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christenson RH, Duh SH, Wu AH, Smith A, Abel G, et al. (2010) Multi-center determination of galectin-3 assay performance characteristics: Anatomy of a novel assay for use in heart failure. Clin Biochem 43: 683–690. [DOI] [PubMed] [Google Scholar]
- 27. Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNPs significantly associated with galectin-3 levels at the Discovery stage.
(DOC)
Previous reported traits with genome wide associations at the ABO locus.
(DOC)