Abstract
Plant growth and development are ensured through networks of complex regulatory schemes. Genetic approaches have been invaluable in dissecting these regulatory pathways. This study reports the isolation of a semi-dominant dwarf mutant designated abnormal shoot1-1 dominant (abs1-1D) through an Arabidopsis T-DNA activation tagging mutant screen. It was shown that the overexpression of a novel BAHD family acyltransferase gene, ABS1/At4g15400, was the cause of the dwarf phenotype in abs1-1D. Overexpression of ABS1 led to many phenotypic features reminiscent of brassinosteroid (BR) deficient or signalling mutants, and it was shown that exogenously applied BR could effectively rescue the dwarf phenotype of abs1-1D. Furthermore, genetic analyses indicated that abs1-1D interacted, in different ways, with the BR-deficient mutant det2-1, the constitutive BR response mutant bes1-D and the photomorphogenic mutant phyB-1. Moreover, ABS1 expression was activated by BR treatment or in a bes1-D mutant background. Genome-wide transcriptome profiling of abs1-1D revealed clear reprogramming of metabolic pathways, and it was demonstrated that BR biosynthesis genes were activated in abs1-1D and that the flavonoid biosynthesis pathway was repressed in abs1-1D, as well as in det2-1. This work provides new insights into the possible involvement of BAHD acyltransferase in the regulation of plant growth and development, and indicates a possible role of ABS1 in maintaining BR homeostasis.
Key words: Arabidopsis shoot development, BAHD acyltransferase, brassinosteroid, de-etiolation, dwarfism, genetic interaction
Introduction
The growth and development of higher plants follow stereotypical developmental programmes that are intricately regulated (Steeves and Sussex, 1989). For instance, in the light, plants undergo photomorphogenesis, while in the dark skotomorphogenesis takes place. The failure to properly elaborate these regulatory programmes often leads to phenotypes that have enabled us to probe many facets of these regulations, including the actions of plant photoreceptors and hormones (Koornneef et al., 1980; Chory et al., 1989; Li et al., 1996; Li and Chory, 1997). The need for coordinated hormone actions necessitates perspectives from two opposing fronts. On one hand, plant hormone biosynthesis and signalling are essential for plant development and have long intrigued researchers (Kim and Wang, 2010; Zhao, 2010). However, on the other hand, an equally important strategy is applied by plants to ensure the coordination is through hormone inactivation mechanisms, such as conjugation and/or modification, which have been demonstrated to be important in maintaining the homeostasis of hormone actions (Yamaguchi, 2008).
Genetic screens for mutants with phenotypes indicative of plant hormone deficiency or signalling defects have been one major avenue for elucidating plant hormone biosynthesis and regulatory pathways (Clouse, 1996). One class of mutants that was instrumental in the advancement of plant hormone biology is dwarf mutants (Feldmann et al., 1989). The conspicuous miniature light-grown phenotype of this class of mutants immediately suggests an intimate involvement of the mutated genes in the regulation of plant growth and development. Although many processes have been linked with dwarf phenotypes, the dominant examples are mutants defective in plant hormone biosynthesis and signal transduction, particularly brassinosteroid (BR) and gibberellins (GAs). For example, both BR-deficient and BR-insensitive mutants have been identified, leading to the establishment of DET2 as an important BR biosynthesis gene and BRI1 as the receptor for BR signalling (Li et al., 1996; Li and Chory, 1997). Genetic lesions that can lead to a dwarf phenotype have also been identified in mutants deficient in GAs or defective in GA signalling, and the GA-related dwarf trait has been applied successfully in modern agriculture (Peng et al., 1999).
The majority of dwarf mutants identified so far are genetically recessive, suggesting a positive regulatory role of the underlying genes. In contrast, an increasing number of Arabidopsis mutants are being reported that showed genetically dominant dwarfism. Based on the dominant nature of these mutants, it can be inferred that their gene products typically act as negative regulators of hormone biosynthesis or signalling. Consistent with this genetic nature, factors that are capable of inactivating distinct plant hormones, and thus negatively regulating plant growth, have been documented in Arabidopsis (Neff et al., 1999; Nakamura et al., 2005; Poppenberger et al., 2005; Takahashi et al., 2005; Turk et al., 2005; Varbanova et al., 2007). For instance, the Arabidopsis BAS1 gene was first identified in a genetic screen looking for suppressors of the phytochrome B-defective mutant phyB-4 (Neff et al., 1999). BAS1 encodes a cytochrome P450 (CYP734A1) that probably inactivates BRs through 26-hydroxylation (Neff et al., 1999). Increased expression of a second cytochrome P450 (CYP72C1), which is a close BAS1 homologue known as SHK1/SOB7/CHI2, also leads to a dominant dwarf phenotype (Nakamura et al., 2005; Takahashi et al., 2005; Turk et al., 2005). Another example of an enzyme that can inactivate BR was demonstrated with the overexpression of an Arabidopsis UDP-glycosyltransferase (UGT73C5) (Poppenberger et al., 2005). The overexpression of this enzyme, which probably catalyses the 23-O-glucosylation of brassinolide and castasterone, leads to transgenic plants that are reminiscent of BR-deficient mutants such as det2 (Poppenberger et al., 2005). A similar mode of plant hormone inactivation through modifications has also been observed with GA (Varbanova et al., 2007). The overexpression of Arabidopsis methyltransferase GMAT1 or GMAT2 inactivates active GAs through methylation and leads to conspicuous dwarfism, conferring phenotypes similar to those of recessive GA biosynthesis mutants (Varbanova et al., 2007).
In our previous work, we isolated a series of Arabidopsis mutants that have altered shoot development programmes and we named these mutants abnormal shoot (abs) mutants. One semi-dominant mutant, abs1-1D, that showed dramatic dwarf phenotype in the light was characterized further. We determined that abs1-1D phenotypes were the consequence of the overexpression of ABS1/At4g15400. ABS1 encodes a putative BAHD family of acyltransferases that is expressed in many tissues but with prominent expression in roots. We found that abs1-1D displayed many features of BR-deficient mutants, including dwarfism in the light, a darker green leaf colour, de-etiolation in the dark, and compromised leaf elongation. Moreover, exogenous applied BR could effectively rescue abs1-1D BR-deficient-related phenotypes. Genome-wide abs1-1D transcriptome analysis revealed both expected and previously unknown reprogramming of metabolic pathways at the transcriptional level. These results suggest that ABS1 is capable of regulating plant growth and development, possibly through involvement in BR homeostasis.
Materials and methods
Plant materials and growth conditions
Wild-type, abs1-1D, and the T-DNA insertional line WiscDsLox474E11 (The Arabidopsis Biological Resource Center, OH, USA) are in Columbia ecotype background. The det2-1 (Li et al., 1996), bes1-D (Yin et al., 2002), and phyB-1 (Bo64) (Reed et al., 1993) mutants have been described and are in Columbia, Enkheim-2, and Landsberg erecta backgrounds, respectively. Arabidopsis plants were grown at 22 °C under continuous illumination (~100 μmol m–2 s–1) on commercial soil mix (Pindstrup, Denmark). For plate and liquid culture experiments, seeds were surface sterilized and grown on 0.5× strength Murashige and Skoog (MS) medium supplemented with 1% sucrose and 1% agar (for plates) and grown at 22 °C under continuous illumination (~100 μmol m–2 s–1 for light experiments).
DNA and RNA manipulations
DNA isolation, Southern blot analysis and plasmid rescue were performed as described by Yu et al. (2008). Total cellular RNAs were extracted using Trizol RNA reagent (Life Technologies, CA, USA). Northern blot analyses were performed as described by Yu et al. (2008). For semi-quantitative RT-PCR analysis, first-strand cDNA was synthesized from 1 μg DNase-treated total RNA using a PrimeScript reverse transcription kit (Takara, Japan). The gene-specific primers and RT-PCR conditions used in this study are listed in Supplementary Table S1 (at JXB online). Where possible, we designed RT-PCR primers flanking introns to distinguish genomic DNA and cDNA.
Generation of transgenic lines
To generate an ABS1 overexpression construct, a genomic DNA fragment encompassing the full-length ABS1/At4g15400 sequence was amplified with Pfu Turbo DNA polymerase (Stratagene, CA, USA) using primers 15400F and 15400R. The amplified fragment was cloned into pBluescript KS+ vector and sequenced before subcloning into the binary vector pBI111L (Yu et al., 2004). The resulting construct was transformed into Agrobacterium tumefaciens by electroporation, and the floral dip method was used to transform wild-type or phyB-1 mutant plants (Clough and Bent, 1998). T1 transgenic lines were screened on solid 0.5× MS medium containing 50mg l–1 of kanamycin. The phenotypes of the transgenic lines were examined in both T1 and T2 generations.
To generate an ABS1 promoter–β-glucuronidase (GUS) fusion construct, a genomic DNA fragment of 1149bp upstream of the start codon of the ABS1 gene was amplified using primers 15400PF and 15400PR and cloned into the XbaI and BamHI sites of pCB308 (Xiang et al., 1999). The resulting construct was designated P ABS1 ::uidA and introduced to wild-type plants. T1 transgenic plants were screened for Basta resistance. GUS activity was examined in T2 plants from independent lines. Histochemical GUS staining was performed following the procedure of Jefferson (1987).
Protoplast transient expression assay and confocal microscopy
The control green fluorescent protein (GFP) vector pTF486 containing the open reading frame of enhanced GFP (eGFP) driven by the cauliflower mosaic virus (CaMV) 35S promoter was designated P35S::GFP (Yu et al., 2008). To generate a C-terminal GFP-tagged ABS1 fusion protein, the open reading frame of ABS1 was amplified using primers 15400F and 15400GFPR and subcloned into pTF486, and the resulting construct was designated P35S::ABS1–GFP. Arabidopsis leaf protoplast preparation and transient expression of GFP constructs were performed as described by Yoo et al. (2007). For nuclei visualization, protoplasts were resuspended in 0.16M CaCl2 (pH 7.4). Hoechst 33342 (Sigma, St Louis, MO, USA) was added to a final concentration of 20 μg ml–1 to stain the nuclei for 15min in the dark (Meadows and Potrykus, 1981). GFP, Hoechst 33342, and chlorophyll fluorescence signals were monitored using a Nikon A1 confocal microscope.
BR treatment
Synthetic BR epi-brassinolide (epiBL; Sigma) was used in BR treatment experiments. For testing the response of Arabidopsis seedlings to exogenous BR, plants were germinated and grown on 0.5× MS plates without epiBL for 6 days and then transferred to 0.5× MS plates with various concentrations of epiBL for 6 days prior to photographing. To analyse the effect of BR on hypocotyl elongation, plants were germinated and grown on vertically placed 0.5× MS plates with various concentrations of epiBL in the dark or in the light for 1 week in the growth room before photographing and hypocotyl length measurements. A one-sided t-test was used to compare the mean hypocotyl length of abs1-1D or det2-1 with that of the wild type under the same treatment conditions. For BR treatment in liquid medium, seedlings were grown on solid 0.5× MS medium for 6 days and then transferred into 0.5× MS liquid medium. After 5 days of liquid culture, seedlings were treated with 0.2 μM or 0.5 μM epiBL for 4h and then collected and flash frozen in liquid N2 for RNA extraction.
Light microscopy
The middle section of leaf petioles from the first pair of true leaves were hand cut and fixed in 4% (v/v) glutaraldehyde in 0.1mM sodium phosphate buffer (pH 6.8) for 12h at 4 °C. After fixation, samples were dehydrated in a dilution series of acetone and embedded in SPI-PON 812 resin (Structure Probe, West Chester, PA, USA). Semi-thin sections were stained with 1% (v/v) toluidine blue O and observed under an Olympus BX51 light microscope equipped with a DP70 digital camera.
Chlorophyll concentration measurement
Chlorophyll concentration was determined on a fresh-weight basis. In brief, 2-week-old seedlings were harvested, weighed, and finely ground in liquid N2. Total chlorophyll was extracted with 95% ethanol, and chlorophyll concentrations were calculated according to the method of Lichtenthaler (1987). Differences in chlorophyll concentration between the wild-type and mutants were evaluated using a P value generated by a one-sided t-test.
Microarray analysis
Total RNAs were prepared from the aerial parts of 3-week-old wild-type and abs1-1D plants. Two biological replicates were used for microarray experiments with Affymetrix ATH1 GeneChip. RNA purification, probe labelling, hybridization, gene chip scanning, and microarray data normalization using Affymetrix MicroArray Suite (MAS) 5.0 software were performed at Shanghai Biotechnology Co., Shanghai, China. A single technical replicate was performed for each biological sample. Genes with a minimum twofold change in expression were considered to be differentially regulated.
Results
Isolation of abs1-1D
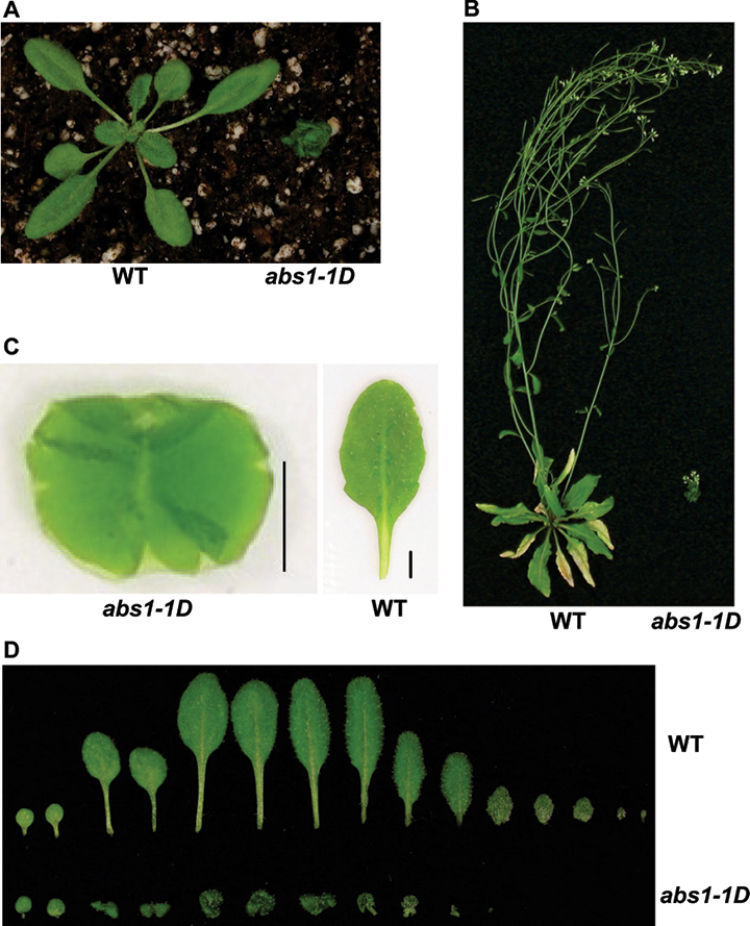
In our efforts to identify activation tagged gain-of-function yellow variegated (var2) suppressors, we recovered numerous mutants with altered shoot development (Yu et al., 2008). We named these mutants abnormal shoot (abs) mutants, and the abs1-1D (D denotes dominant) mutant was investigated further. Because all of our mutants were in a var2-5 background, we first crossed out the var2-5 background and obtained the abs1-1D mutation in a wild-type background. Genetic analysis indicated that abs1-1D behaved in a semi-dominant manner (Supplementary Fig. S1 at JXB online). The mutant phenotype was stable during our experiments. Compared with the wild type, the most striking feature of the abs1-1D homozygous mutant was its conspicuous dwarf phenotype at the 3-week-old light-grown stage (Fig. 1A). This became even more dramatic at late developmental stages (Fig. 1B). Closer examination of individual leaves from abs1-1D plants revealed that, unlike wild-type leaves, the leaves of abs1-1D plants showed abnormal curvature and could not be flattened without overlapping between two solid surfaces (Fig. 1C). At 3 weeks old, abs1-1D plants developed fewer leaves with significantly shortened petioles compared with those of the wild type (Fig. 1D). We also observed that abs1-1D leaves appeared to be darker green under our growth conditions (Fig. 1A).
Fig. 1.
Phenotype of the Arabidopsis abs1-1D mutant. (A, B) Phenotypes of representative 3-week-old (A) and 7-week-old (B) wild-type (WT) and abs1-1D plants. (C) An individual rosette leaf of abs1-1D and wild-type plants. Leaves were flattened between glass slides before photographing; overlapping leaf areas are darker green. Bar, 0.25cm. (D) Comparison of rosette leaves from 3-week-old wild-type and abs1-1D plants. Leaves were arranged from left to right in the order of their initiation.
Cloning of ABS1
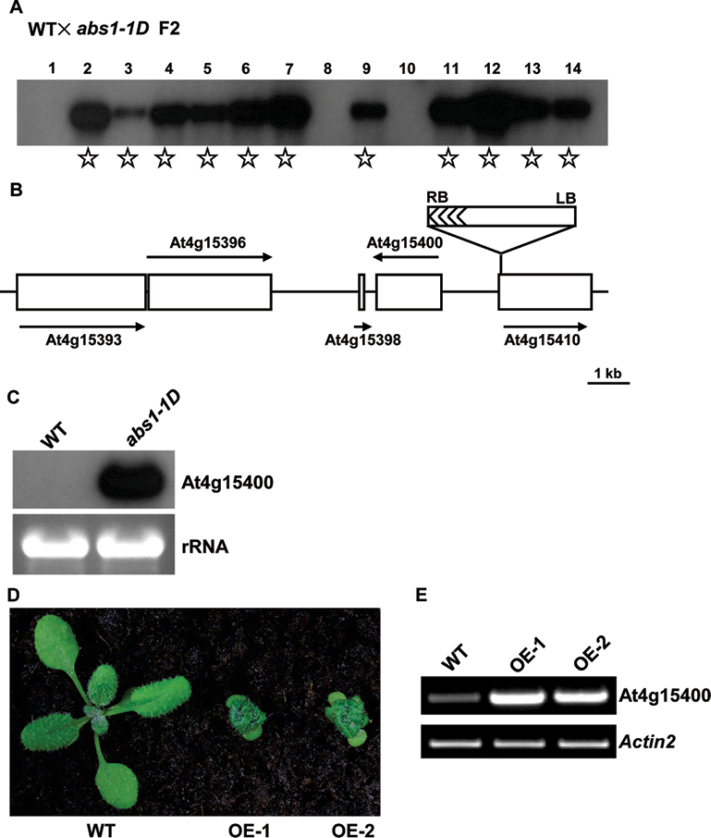
We next tested whether the abs1-1D dwarf phenotype was linked with activation tagging T-DNA insertions. Southern blotting revealed the presence of a single T-DNA insertion in abs1-1D that co-segregated with the mutant phenotype, suggesting that abs1-1D was tagged with a T-DNA (Fig. 2A). Applying a standard plasmid rescue procedure, we recovered a plasmid that harboured both plant and T-DNA vector sequences. Subsequent sequencing revealed that the T-DNA was inserted in Arabidopsis chromosome 4 between At4g15400 and At4g15410 (Fig. 2B). As At4g15400 is the gene immediately adjacent to the right border region, where the 35S enhancer sequences are located, we examined the expression of At4g15400, and northern blotting indicated that the expression level of At4g15400 was greatly enhanced in abs1-1D (Fig. 2C).
Fig. 2.
Cloning of the ABS1 gene. (A) Co-segregation analysis of abs1-1D. Fourteen F2 plants derived from a cross between wild type and abs1-1D were randomly selected to score phenotype. Genomic DNAs from these plants were digested with BamHI. A Southern blot was probed with a 32P-labelled BAR gene sequence (Yu et al., 2008). Samples from plants showing the abs1-1D phenotype are indicated with stars. (B) Schematic representation of the T-DNA insertion site in abs1-1D. The positions of genes in the vicinity of the T-DNA are shown as open boxes. Arrows represent the orientation of the open reading frames of these genes. (C) Accumulation of At4g15400 transcripts in wild-type and abs1-1D plants. Equal amounts of total RNA (5 μg) extracted from 2-week-old seedlings were separated on a formaldehyde gel and transferred to a nylon membrane. The RNA blot was hybridized with 32P-labelled full-length At4g15400 cDNA. The ethidium bromide-stained RNA gel shown below served as a loading control. (D) Phenotypes of representative 2-week-old wild-type and two independent transgenic lines overexpressing At4g15400 (OE-1, and OE-2). (E) Semi-quantitative RT-PCR analysis of At4g15400 transcript accumulation in the plants shown in (D). All primers and RT-PCR conditions used in this study are listed in Supplementary Table S1. Actin2 expression was used as a control.
To evaluate whether overexpression of At4g15400 was the cause of the abs1-1D dwarf phenotype, we recapitulated the abs1-1D phenotype with independent CaMV 35S promoter-driven At4g15400 overexpressing transgenic lines (Fig. 2D). Multiple transgenic lines showed the characteristic dwarf phenotype of abs1-1D (Fig. 2D) and the phenotypes were associated with elevated At4g15400 expression revealed by semi-quantitative RT-PCR (Fig. 2E). RT-PCR also revealed that At4g15400 was expressed in shoot tissues (Fig. 2E). These data indicated that overexpression of At4g15400 was the cause of the abs1-1D dwarf phenotype and that ABS1 is At4g15400.
To investigate the impact caused by a loss-of-function mutation of ABS1, we sought a T-DNA insertional line of ABS1 from The Arabidopsis Biological Resource Center and obtained one such mutant, WiscDsLox474E11 (Supplementary Fig. S2 at JXB online). The T-DNA insertion site in WiscDsLox474E11 is near the 3’ region of the ABS1 coding sequence, 49bp upstream of the stop codon (Supplementary Fig. S2A). Full-length ABS1 transcripts were not detectable in plants that were homozygous for the T-DNA insertion (Supplementary Fig. S2B, C). WiscDsLox474E11 homozygous mutants were indistinguishable from the wild type when grown under our conditions (Supplementary Fig. S2D). However, dark-grown WiscDsLox474E11 seedlings showed modest yet significant enhanced elongation of the hypocotyl compared with that of the wild type (Supplementary Fig. S2E, F). WiscDsLox474E11 was recently identified as brassinosteroid inactivator1-1 (bia1-1) in another study (Roh et al., 2012), and our findings are in line with their results.
ABS1 encodes a putative BAHD family of acyltransferases
BlastP analysis using ABS1/At4g15400 amino acid sequences revealed that it is a putative member of the Arabidopsis BAHD family of acyltransferases (D’Auria, 2006; Yu et al., 2009). BAHD was named based on the first letters of four canonical enzymes in this family: benzylalcohol O-acetyltransferase, BEAT; anthocyanin O-hydroxycinnamoyltransferase, AHCT; anthranilate N-hydroxycinnamoyl/benzoyltransferase, HCBT; and deacetylvindoline 4-O-acetyltransferase, DAT (D’Auria, 2006; Yu et al., 2009). In Arabidopsis, there are ~60 putative BAHD acyltransferases that are grouped phylogenetically into five distinctive clades (D’Auria, 2006; Yu et al., 2009). ABS1/At4g15400 was assigned to clade IV, which was previously designated clade III in a different naming system (D’Auria, 2006; Yu et al., 2009). The closest homologues of ABS1/At4g15400 in Arabidopsis are At5g47980 and At5g23970, which share 51.7 and 42.1% amino acid identity with ABS1, respectively (Supplementary Fig. S3 at JXB online).
BAHD family acyltransferases share two major features. First is the HXXXD motif, where the histidine residue is probably the catalytic centre (D’Auria, 2006; Yu et al., 2009). In ABS1/At4g15400, the HXXXD motif corresponds to HKICD and the universally conserved histidine residue is His151 in ABS1/At4g15400 (Supplementary Fig. S3). At the second conserved motif of BAHD, ABS1 has a DFGSG arrangement (Supplementary Fig. S3), rather than the more prevalent DFGWG, and this seems to agree with data suggesting that the DFGWG domain can tolerate a certain degree of variability in BAHD family acyltransferases (Yu et al., 2009).
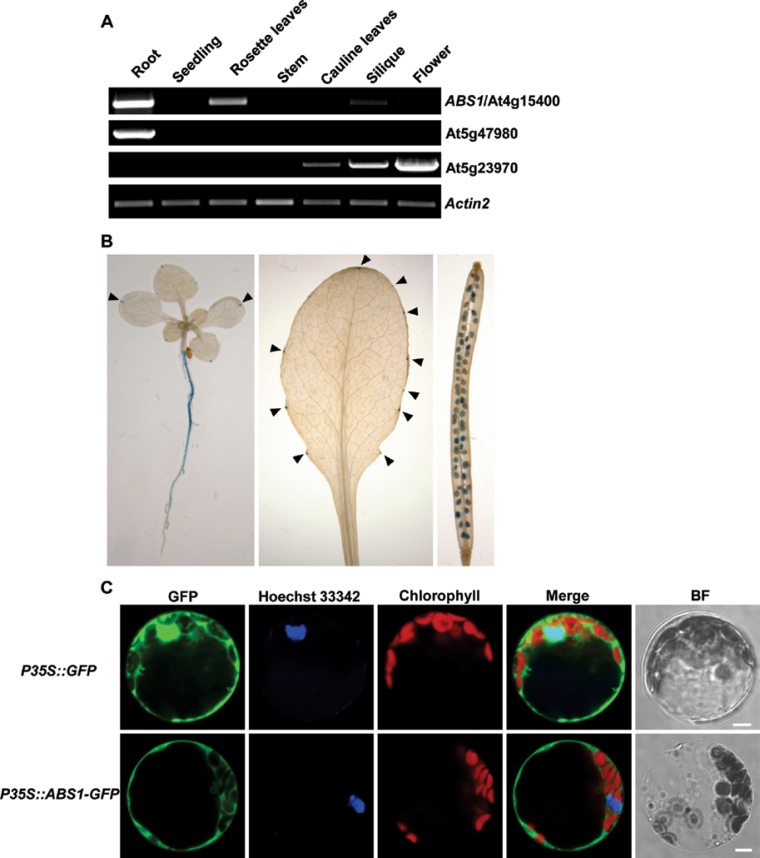
We next examined the tissue expression pattern of ABS1/At4g15400 and its two closest homologues: At5g47980 and At5g23970 (Yu et al., 2009). Semi-quantitative RT-PCR showed distinct expression patterns for these three genes. ABS1/At4g15400 was predominantly expressed in Arabidopsis root tissues, with relatively low expressions also detected in seedling, rosette leaves, silique, and flower tissues (Fig. 3A). For At5g47980, we detected major expression in the root tissue but not in other tissues examined (Fig. 3A). Overexpression of At5g47980 also led to transgenic plants with reduced sizes; however, the phenotypes of transgenic lines we recovered were not as dramatic as ABS1/At4g15400 overexpression lines (Supplementary Fig. S4). Interestingly, At5g23970 showed an expression pattern associated with floral tissues, with expression in cauline leaves, silique, and the highest level in the flower (Fig. 3A).
Fig. 3.
Expression analysis of ABS1. (A) Semi-quantitative RT-PCR analysis of the expression of ABS1 and its two close homologues in the indicated tissues from wild-type plants. (B) Histochemical GUS staining of wild-type plants expressing the ABS1 promoter–GUS fusion construct. Shown are a 10-day-old seedling, fully expanded rosette leaf and silique tissues. Arrowheads indicate GUS staining in the cotyledons and rosette leaf. (C) Cellular localization of the ABS1 protein. Protoplasts prepared from 4-week-old wild-type Arabidopsis leaves were transformed with P35S::GFP or P35S::ABS1-GFP. Hoechst 33342 was used to stain the nucleus. The GFP, Hoechst 33342, and chlorophyll autofluorescence signals were monitored by confocal microscopy. Bright field (BF) images served as controls for protoplast integrity. Representative images of a single protoplast are shown. Bar, 5 μm).
We next examined the tissue expression profile of ABS1 through promoter–GUS reporter analysis. Transgenic lines harbouring GUS driven transcriptionally by a ~1.1kb ABS1 endogenous promoter showed strong expression in root tissues (Fig. 3B). In leaves, ABS1 expression was distributed as distinct foci at the edge of cotyledons and rosette leaves, reminiscent of the distribution of hydathodes along the leaf edge (Fig. 3B; Figueroa-Balderas et al., 2006). ABS1 expression was also observed in developing seeds in the silique (Fig. 3B). Promoter–GUS tissue expression results were in line with the RT-PCR results (Fig. 3A, 3B).
Arabidopsis BAHD family members have been predicted to be targeted into various subcellular compartments including the cytosol, nucleus, chloroplast, and mitochondria (Yu et al., 2009). To test the subcellular localization of ABS1, we carried out transient expression analysis of ABS1–GFP fusion protein in Arabidopsis leaf protoplasts. A control vector containing only the GFP coding sequence under the control of the CaMV 35S promoter (P35S::GFP) expressed GFP in the cytosol and nucleus, consistent with previous findings (Fig. 3C; Vert et al., 2002). Transient expression of P35S::ABS1–GFP showed green fluorescence distribution mostly in the cytosol, suggesting that ABS1 is probably a cytosolic protein (Fig. 3C).
The abs1-1D mutant shows a typical BR-deficient phenotype
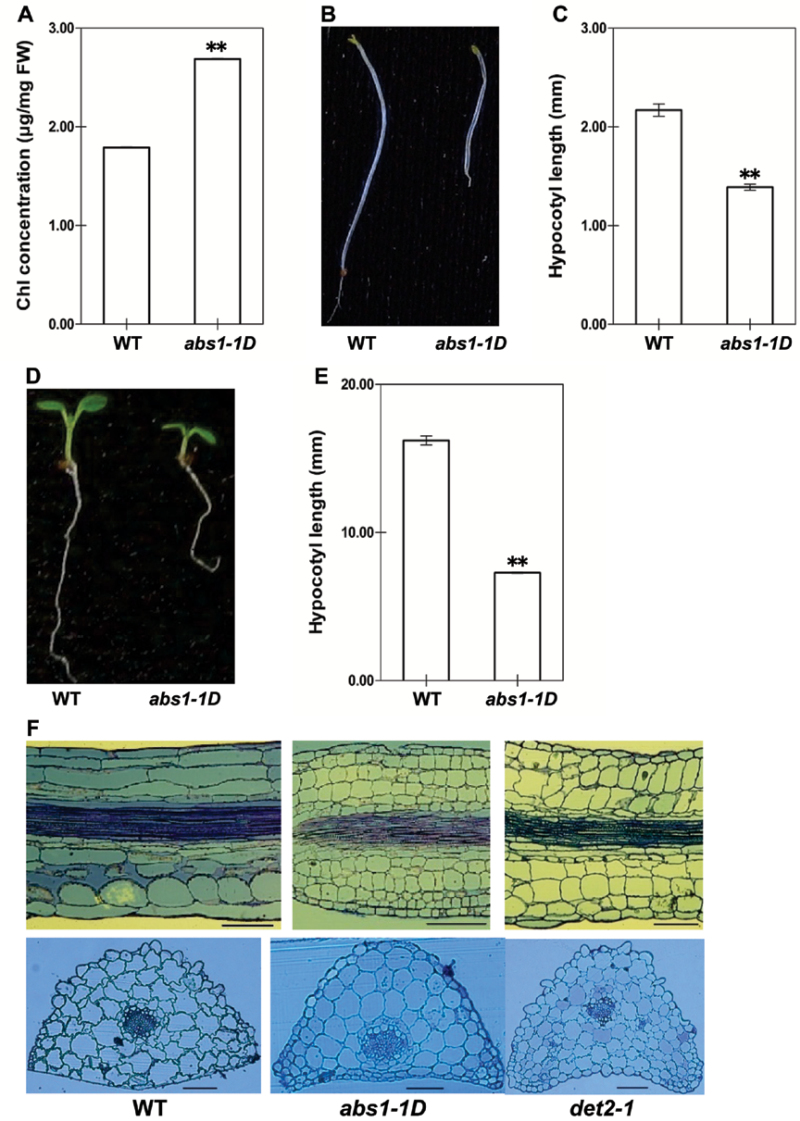
The dwarf phenotype of abs1-1D closely resembles the canonical BR-deficient or -signalling mutants, such as det2 (Li et al., 1996). BR-deficient or -signalling mutants have a number of stereotypical phenotypes including dwarfism, a darker green leaf colour, and de-etiolation in the dark (Fujioka and Yokota, 2003). During our routine growing of abs1-1D, we observed casually that abs1-1D had a darker green appearance, and quantitative analysis of the chlorophyll concentration confirmed that abs1-1D had a significantly higher accumulation of chlorophyll when compared with wild-type plants (Fig. 4A). Chlorophyll accumulation in ABS1 overexpressing plants was also higher than that of wild-type plants (Supplementary Fig. S5A). We next examined the dark-growth features of abs1-1D. After 1 week of dark growth, abs1-1D seedlings had significantly shortened hypocotyls when compared with those of wild-type plants (Fig. 4B, 4E). We also examined the hypocotyl lengths of seedlings grown in the light (Fig. 4C, 4D). Light-grown abs1-1D plants displayed shortened hypocotyls (Fig. 4C, 4D). A shorter hypocotyl phenotype was also observed in ABS1 overexpressing plants (Supplementary Fig. S5B, C). Abnormal hypocotyl elongation of abs1-1D in both the light and dark prompted us to examine the state of cell elongation in abs1-1D. det2-1, a known BR deficient mutant defective in a steroid 5-α-reductase, was included as a control (Li et al., 1996). For both abs1-1D and det2-1, the longitudinal sections of the leaf petioles showed clear abnormal elongation along the leaf axis, with the lengths of cells greatly reduced when compared with those of wild-type plants (Fig. 4F). However, cross-sections of abs1-1D and det2-1 showed that the sizes of cells along the axis perpendicular to the leaf petiole were comparable in the two mutants when compared with those of the wild type (Fig. 4F). Taken together, these data indicated that the phenotypes of the abs1-1D mutant, including dwarfism, a darker green leaf colour, de-etiolation, and cell elongation defects, mimics BR-deficient or -signalling mutants.
Fig. 4.
BR-deficient related morphological phenotypes in abs1-1D. (A) Chlorophyll content of 2-week-old wild-type and abs1-1D plants. Measurements were calibrated on a fresh weight (FW) basis. Error bars represent standard error of three independent pooled whole seedling samples (**, P <0.01). (B, D) Comparison of hypocotyl length of 1-week-old wild-type and abs1-1D plants grown in the dark (B) and in the light (D). (C, E) Statistical analysis of hypocotyl length measurements of plants grown on the same plates as shown in (D) and (B), respectively. Each bar represents the mean hypocotyl length of 12 plants. Error bars represent ±standard error (**, P<0.01). (F) Longitudinal sections (upper panels) and cross-sections (lower panels) of petioles of the first two true leaves from wild-type, abs1-1D and det2-1 plants. Bars, 50 μm.
Exogenous BR rescues abs1-1D
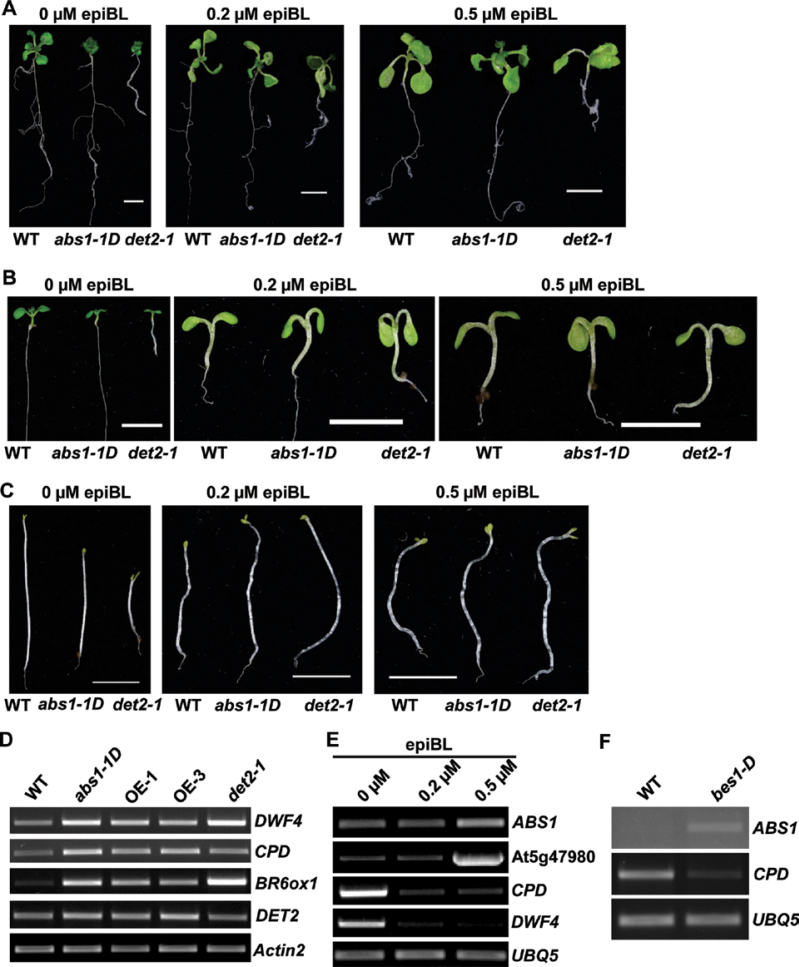
This overall resemblance of abs1-1D to BR-deficient or -signalling mutants prompted us to test whether exogenously applied BR could rescue the abs1-1D phenotypes, a common procedure for distinguishing BR-deficient from BR-signalling mutants (Clouse, 1996). We tested the responses of abs1-1D and det2-1 to the applications of various concentrations of synthetic BR, epiBL. Fig. 5A shows that, when transferred to control medium, both abs1-1D and det2-1 maintained the characteristic dwarf phenotype. However, when seedlings were transferred to 0.2 μM epiBL medium, both abs1-1D and det2 showed elongation of leaf petioles, a clear sign of the rescue of the dwarf phenotype (Fig. 5A). The rescue effect could also be observed with 0.5 μM epiBL treatment (Fig. 5A).
Fig. 5.
BR-deficient-related molecular characteristics in abs1-1D. (A) Representative phenotypes of wild-type, abs1-1D, and det2-1 plants after transplanted to epiBL-supplemented plates. Plants were germinated on 0.5× MS plates and grown for 6 days prior to transplanting to plates supplemented with 0, 0.2, or 0.5 μM epiBL. Photographs were taken 6 days after transplantation. Bars, 0.5cm. (B, C) Hypocotyl elongation of 1-week-old wild-type, abs1-1D, and det2-1 plants germinated on 0.5× MS plates containing 0, 0.2, or 0.5 μM epiBL in the light (B) and in the dark (C). Bars, 0.5cm. (D) Semi-quantitative RT-PCR analysis of expression of the indicated BR biosynthesis genes in wild-type, abs1-1D, two ABS1 overexpression lines (OE-1 and OE-3) and det2-1 plants. (E) Semi-quantitative RT-PCR analysis of expression of the indicated genes in response to exogenously applied epiBL in wild-type plants. (F) Semi-quantitative RT-PCR analysis of expression of the indicated genes in wild-type and bes1-D plants. Actin2 or UBQ5 expression was used as a control.
We next tested the responses of other aspects of abs1-1D phenotypes to epiBL. The hypocotyl lengths of light-grown abs1-1D and det2-1 seedlings were significantly shorter than those of the wild type (Fig. 5B; statistical analysis in Supplementary Fig. S6A). However, when germinated on MS medium containing various concentrations of epiBL, both abs1-1D and det2-1 showed enhanced growth of hypocotyls, and at 0.2 μM epiBL, the hypocotyl lengths of abs1-1D and det2-1 were comparable with those of the wild type, which were also enhanced to a lesser degree (Fig. 5B; statistical analysis in Supplementary Fig. S6A). The de-etiolation phenotypes of abs1-1D and det2-1 in the dark were also rescued by exogenously applied epiBL (Fig. 5C; statistical analysis in Supplementary Fig. S6B). Overall, these data indicated that exogenously applied BR can rescue the dwarf and hypocotyl defects of abs1-1D, as well as those of det2-1, suggesting that abs1-1D phenotypes may be associated with BR deficiency.
One hallmark feature of BR-deficient mutants is the feedback upregulation of BR biosynthesis genes (Choe et al., 2001; Tanaka et al., 2005; Song et al., 2009). We thus tested the expression of four classic BR biosynthesis genes (DWF4, CPD, BR6ox1, and DET2) in abs1-1D, two ABS1 overexpressing transgenic lines (OE-1 and OE-3), and det2-1 with semi-quantitative RT-PCR (Fig. 5D). Three of the four genes examined (DWF4, CPD, and BR6ox1) showed clearly elevated expression in abs1-1D, OE-1, OE-3, and det2-1 plants when compared with the wild type (Fig. 5D), while expression of DET2 appeared to be only slightly higher in the mutants (Fig. 5D). These data indicated that transcripts of some BR biosynthesis genes accumulate at higher levels in abs1-1D.
We next looked at the expression of ABS1 and its homologue At5g47980 in response to exogenously applied epiBL. When treated with epiBL, expression of the BR biosynthesis genes CPD and DWF4 was repressed as a result of the negative-feedback regulation (Fig. 5E). On the other hand, we observed that, when treated with 0.5 μM epiBL, both ABS1 and At5g47980 were induced, while the expression of both genes at 0.2 μM epiBL treatment was not significantly altered (Fig. 5E). Furthermore, the expression of ABS1 in the bes1-D mutant, in which BR signalling is constitutively active, was also elevated, while the BR biosynthesis gene CPD was repressed (Fig. 5F; Yin et al., 2002). The induction of ABS1 in bes1-D is in agreement with a recent report of upregulated genes in a bes1-D background through microarray analysis (Yu et al., 2011). Our results indicated that ABS1 is induced by activated BR signalling, either by applying BR or by activating BR-signalling components.
Genetic interactions between abs1-1D, det2-1, bes1-D, and phyB-1
If abs1-1D is defective in BR accumulation, we should be able to test its genetic interactions with known mutants that are related to BR biosynthesis or signalling. Using this concept, we surveyed the genetic interactions between abs1-1D and mutants including det2-1, bes1-D, and phyB-1.
We first tested the genetic interaction between abs1-1D and det2-1. Under our growth conditions, det2-1 displayed a weaker dwarf phenotype when compared with abs1-1D (Fig. 6A; Li et al., 1996). We obtained abs1-1D det2-1 double mutants (Supplementary Fig. S6C), which showed extreme dwarfism and were sterile (Fig. 6A). Genetic interaction between abs1-1D and det2-1 could also be observed with dark-grown seedlings (Fig. 6B). Compared with the de-etiolation phenotypes of either parent, abs1-1D or det2-1, the double mutant showed a more dramatic de-etiolation phenotype (Fig. 6B). The synergistic interaction between abs1-1D and det2-1 suggested that ABS1 and DET2 may be functionally related.
Fig. 6.
Genetic interactions between abs1-1D and various mutants with altered photomorphogenic growth. (A) Phenotypes of representative 3-week-old wild-type, abs1-1D, det2-1 and abs1-1D det2-1 double mutant plants. (B) Comparison of hypocotyl elongation of 1-week-old wild-type, abs1-1D, det2-1, and abs1-1D det2-1 double mutant plants grown in the dark. (C) Phenotypes of representative 2-week-old wild-type, abs1-1D/+ heterozygote, abs1-1D homozygote, bes1-D/+ heterozygote, bes1-D/+ abs1-1D/+ double heterozygote and bes1-D homozygote plants. (D) Comparison of hypocotyl elongation of 1-week-old Arabidopsis Landsberg erecta ecotype (Ler), phyB-1 mutant, two independent transgenic lines overexpressing ABS1 in a phyB-1 mutant background, and Arabidopsis Columbia ecotype (Col) grown in the light.
The semi-dominant BR constitutive response mutant bes1-D was originally isolated in a genetic screen for suppressors of bri1-119, which is a weak mutant allele of the BR receptor gene BRI1 (Li and Chory, 1997; Yin et al., 2002). BES1 encodes a transcription factor that is a positive BR-signalling component downstream of BRI1 (Yin et al., 2002). bes1-D can suppress many features of bri1-119, including the dwarfism and de-etiolation phenotypes (Yin et al., 2002). When compared with a heterozygous abs1-1D/+ plant, the double heterozygote bes1-D/+ abs1-1D/+ plant was much enlarged in size, suggesting that bes1-D is a robust abs1-1D genetic suppressor (Fig. 6C).
Compromised BR biosynthesis can also rescue the elongated hypocotyl phenotype of phyB-1 mutant, as demonstrated by the phyB-1 det2-1 double mutant (Chory, 1992). To test the effect of ABS1 overexpression on phyB-1 (Landsberg erecta ecotype background), we generated transgenic lines overexpressing ABS1 in a phyB-1 mutant background. Multiple transgenic plants showed phenotypes that resembled those of abs1-1D (Fig. 6D). The hypocotyls of transgenic plants were significantly shortened when compared with phyB-1, suggesting that the overproduction of ABS1 can suppress phyB-1, reminiscent of what has been observed with the interaction between det2 and phyB-1 and of phyB-1 genetic suppressors such as bas1-D (Chory, 1992; Neff et al., 1999).
Global gene expression profile of abs1-1D
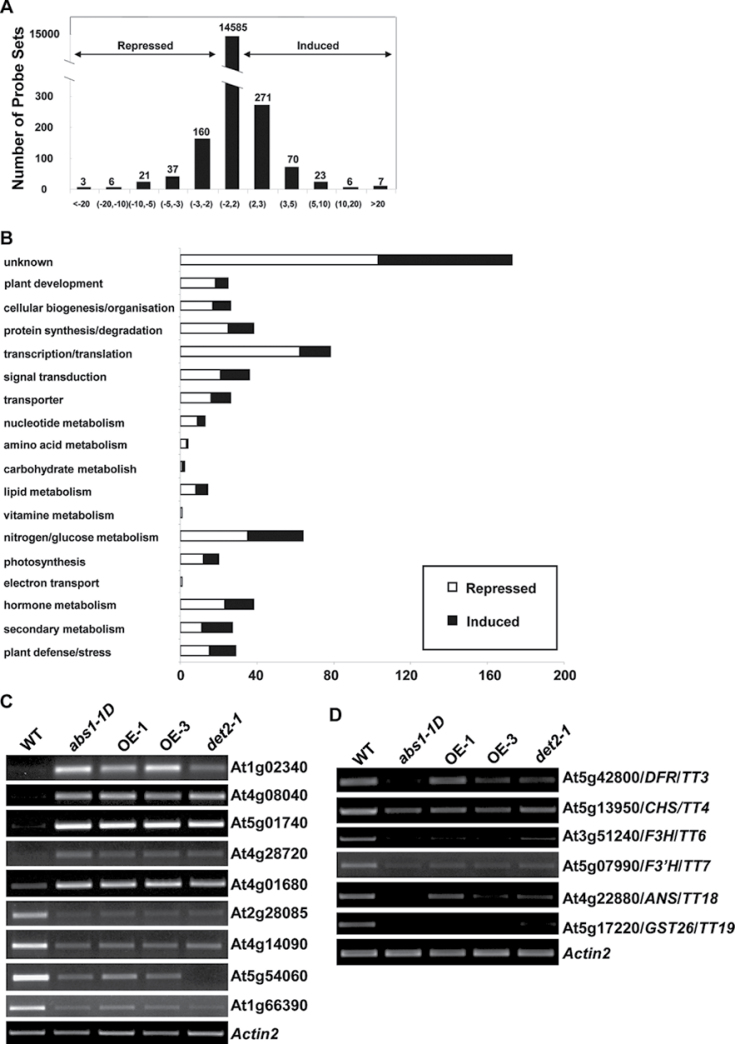
To examine the global impact of the overexpression of ABS1 on plant development, we obtained Affymetrix ATH1 gene chip data of both wild-type and abs1-1D mutant plants. Despite the dramatic developmental phenotypes of abs1-1D, we only detected a relatively small impact on the transcriptome, with 383 genes upregulated and 238 genes downregulated, using a two-fold cut-off level (Fig. 7A; Supplementary Table S2). The gene ontology of the genes with up- or downregulated expression appeared to cover many different cellular processes, as with gene chip data from other mutants with developmental phenotypes (Fig. 7B). The expression levels of the major up- and downregulated genes in the microarray were examined using semi-quantitative RT-PCR (Fig. 7C). Similar expression pattern changes of these genes were observed in abs1-1D plants and two ABS1 overexpression lines compared with wild-type plants, confirming the reliability of the microarray data (Fig. 7C).
Fig. 7.
Global transcriptome profiling of abs1-1D. (A) Distribution of genes with altered expression in abs1-1D compared with the wild type. (B) Functional classification of genes that are significantly repressed or induced (by at least twofold) in abs1-1D versus wild-type plants. (C) Semi-quantitative RT-PCR analysis of expression of the major up- and downregulated genes shown in microarray data in wild type, abs1-1D, two ABS1 overexpression lines (OE-1 and OE-3), and det2-1. (D) Semi-quantitative RT-PCR analysis of the expression of indicated flavonoid biosynthesis genes from the same samples as in (C). Actin2 expression was used as a control.
Upon closer examination of the microarray data, two major features of the abs1-1D transcriptome stood out. Firstly, many genes that are involved in the BR biosynthesis pathway were upregulated in abs1-1D, including DWF4, CPD, and BR6ox1, and this is consistent with our earlier data showing that several BR biosynthesis genes were upregulated in abs1-1D (Figs 5D, 7B; Supplementary Table S2). Secondly, many genes related to the flavonoid biosynthesis pathway were downregulated in abs1-1D (Fig. 7B; Supplementary Table S2). Semi-quantitative RT-PCR analysis verified the downregulation of a representative set of such genes, including many TRANSPARENT TESTA (TT) genes (Fig. 7D). A similar expression level change in all genes tested was also observed in det2-1 plants, suggesting that the phenotypic similarity of abs1-1D and det2-1 is associated with some similar alterations in their transcriptomes (Fig. 7C, 7D).
Discussion
In this work, we have reported the isolation of a semi-dominant Arabidopsis mutant, abs1-1D, which displayed a dramatic dwarf stature when grown in the light. We subsequently established that overexpression of a BAHD family of acyltransferases, ABS1/At4g15400, was the cause of the dwarf phenotype. The BAHD family of acyltransferases typically catalyses the acylation of a diverse array of substrates, with acyl-CoA thioesters serving as the acyl group donor and the majority of substrates being plant secondary metabolites including flavonoid pathway intermediates (D’Auria, 2006). Currently, we do not know the exact substrate or the nature of the acyl group that ABS1 might be able to exert enzymatic activity towards. However, amino acid sequence alignment with known BAHD family members shows the presence of conserved motifs of BAHD acyltransferases in ABS1, particularly the universally conserved histidine residue that has been shown to be the catalytic centre (Ma et al., 2005). With our genetic data, it is obvious that ABS1 is capable of regulating plant growth and development. To our knowledge, this is the first case where the overexpression of a BAHD family acyltransferase was shown to induce such dramatic dwarf phenotypes in the light.
Although defects in many regulatory pathways can lead to dwarf mutant phenotypes, three lines of evidences suggest that abs1-1D is intimately associated with BR-deficient phenotypes. Firstly, abs1-1D mutants demonstrate many phenotypes of BR-deficient or -signalling mutants such as det2 and bri1. These features include extreme dwarfism when grown in the light, de-etiolation in the dark, higher accumulation of chlorophyll in the leaves, and aberrant cell elongation in leaf petioles. One distinction between hormone-deficient and -signalling mutants is whether exogenously applied plant hormone can rescue the mutant phenotype. We found that exogenously applied BR could rescue the abs1-1D mutant phenotypes, and this certainly indicates that the BR-signalling pathway is normal in abs1-1D. Although we did not measure the endogenous level of BR, our data suggested that the dwarf phenotype caused by the overexpression of ABS1 is probably associated with the lack of active endogenous BR.
Secondly, genetic evidence further strengthens the intimate relationship between ABS1 and plant growth, and developmental programmes regulated by BR homeostasis and signalling. Double mutants of abs1-1D and det2-1 are extreme dwarf and sterile, suggesting that ABS1 and DET2 may contribute to the same genetic pathway. In addition, when the abs1-1D allele was introduced into a bes1-D mutant background, in which the BR-signalling pathway is constitutively active, the dwarf phenotype conferred by the abs1-1D allele was effectively suppressed. This suggested that the constitutively active BR-signalling pathway in bes1-D clearly suppresses the abs1-1D dwarf phenotype. Moreover, overexpressing ABS1 in a phyB-1 mutant background reversed the long-hypocotyl phenotype of the phyB-1 mutant. In essence, abs1-1D can be seen as a dominant genetic suppressor of the phyB-1 mutant, similar to what has been observed in other phyB-1 suppressor genes, BAS1 and other SUPPRESSOR OF PHYB-4 (SOB) genes (Neff et al., 1999; Turk et al., 2005). Interestingly, BAS1 encodes a cytochrome P450 that inactivates BRs, representing one of the few examples of BR-inactivating mechanisms (Neff et al., 1999; Poppenberger et al., 2005).
Lastly, we also established through semi-quantitative RT-PCR and gene chip analysis that key BR biosynthesis genes are upregulated in abs1-1D, reminiscent of the feed-back upregulation of these genes in BR-deficient mutants such as det2-1 (Choe et al., 2001; Tanaka et al., 2005; Song et al., 2009). Both ABS1/At4g15400 and its close homologue At5g47980 were also induced by exogenous BR treatment, providing further evidence for the linkage between BAHD acyltransferases and BR actions. There are at least two possibilities. One is that both genes are downstream targets that are involved in the response of the plant cell to BR. It is known that some BAHD family members may be involved in cell-wall biosynthesis, and many BR-responsive genes are involved in cellular processes such as cell-wall biosynthesis (Goda et al., 2002; Müssig et al., 2002; Hoffmann et al., 2004; Besseau et al., 2007; Gou et al., 2009). Another scenario is that the induction of these genes may represent a mechanism for BR inactivation, much like the effect of GA treatment on the expression of GA-inactivating oxidases (Thomas et al., 1999). The dominant nature of abs1-1D and its phenotype do show that it is probably a negative regulator of plant growth and development, lending more support to the latter hypothesis. However, in either case, these genes are linked with BR regulation. We found that ABS1 expression was elevated in the bes1-D mutant, consistent with what was observed previously with bes1-D genome-wide transcriptome analysis (Yu et al., 2011). This seems to suggest that BES1 positively regulates ABS1, directly or indirectly. However, two recent reports profiling direct target genes for BZR1 or BES1 did not identify ABS1 as a direct target of either BZR1 or BES1 (Sun et al., 2010; Yu et al., 2011). This seems to suggest that there may be additional regulatory components between BZR1 and/or BES1 and ABS1.
While this work was under review, At4g15400 was identified as BIA1 (BRASSINOSTEROID INACTIVATOR1) in an independent study and was shown to be involved in BR homeostasis (Roh et al., 2012). Results from this new report, particularly the reduced levels of active BRs in At4g15400 overexpression plants, and our genetic data, certainly argue a role for At4g15400/BIA1/ABS1 in regulating BR homeostasis.
Genome-wide transcriptome profiling of abs1-1D revealed clear reprogramming of many metabolic and regulatory genes in abs1-1D. The most prominent changes were upregulation of the BR biosynthesis pathway and downregulation of the flavonoid biosynthesis pathway. Intriguingly, the downregulation of flavonoid biosynthesis genes could also be observed in the BR-deficient mutant det2-1, establishing a link between flavonoid biosynthesis and BR actions. Although members of the BAHD family of acyltransferases have been shown previously to be involved in the flavonoid biosynthesis pathway, there are few reports regarding the link between BR homeostasis and the flavonoid pathway. In a genetic screen for dominant bri1 enhancers, a mutant ben1-1D was isolated (Yuan et al., 2007). Interestingly, BEN1 encodes a dihydroflavonol 4-reductase-like protein, an enzyme presumably involved in the flavonoid pathway (Yuan et al., 2007). Although the precise mechanism is still unclear regarding how BEN1 is involved in BR homeostasis or how the overexpression of ABS1 is associated with the flavonoid pathway, there are certainly increasing data suggesting the existence of a link between BR and the flavonoid pathway.
Supplementary data
Supplementary Fig. S1. Phenotypes of 3-week-old wild-type, abs1-1D homozygote, and abs1-1D/+ heterozygote plants.
Supplementary Fig. S2. Identification of a loss-of-function allele of ABS1.
Supplementary Fig. S3. Multiple alignments of amino acid sequences of ABS1, At5g47980, At5g23970, and four canonical BAHD enzymes.
Supplementary Fig. S4. Phenotypes of At5g47980 overexpression lines.
Supplementary Fig. S5. Phenotypic analysis of ABS1 overexpression lines.
Supplementary Fig. S6. Hypocotyl elongation measurements of wild-type, abs1-1D, and det2-1 following epiBL treatment in the light and dark, and genotyping of the abs1-1D det2-1 double mutant.
Supplementary Table S1. Primers and PCR conditions used in this study.
Supplementary Table S2. List of genes that showed significant expression changes (at least twofold) in abs1-1D compared with wild-type plants.
Acknowledgements
We thank Dr Yanhai Yin (Iowa State University) for kindly providing the det2-1 and bes1-D seeds. This work was supported by the National Natural Science Foundation of China (31100864 to X.L.) and Chinese Ministry of Education Program for New Century Excellent Talents in University (NCET-09-0657 to F.Y.).
Glossary
Abbreviations:
- BR
brassinosteroid
- CaMV
cauliflower mosaic virus
- epiBL
epi-brassinolide
- GA
gibberellin
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- MS
Murashige and Skoog
References
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M. 2007. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth The Plant Cell 19 148––162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. 2001. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis The Plant Journal 26 573––582 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. 1989. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light Cell 58 991––999 [DOI] [PubMed] [Google Scholar]
- Chory J. 1992. A genetic model for light-regulated seedling development in Arabidopsis Development 115 337––354 [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana The Plant Journal 16 735––743 [DOI] [PubMed] [Google Scholar]
- Clouse SD. 1996. Molecular genetic studies confirm the role of brassinosteroids in plant growth and development The Plant Journal 10 1––8 [DOI] [PubMed] [Google Scholar]
- D’Auria J. 2006. Acyltransferases in plants: a good time to be BAHD Current Opinion in Plant Biology 9 331––340 [DOI] [PubMed] [Google Scholar]
- Feldmann KA, Marks MD, Christianson ML, Quatrano RS. 1989. A dwarf mutant of Arabidopsis generated by T-DNA insertion mutagenesis Science 243 1351––1354 [DOI] [PubMed] [Google Scholar]
- Figueroa-Balderas RE, García-Ponce B, Rocha-Sosa M. 2006. Hormonal and stress induction of the gene encoding common bean acetyl-coenzyme A carboxylase Plant Physiology 142 609––619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T. 2003. Biosynthesis and metabolism of brassinosteroids Annual Review of Plant Biology 4 137––164 [DOI] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. 2002. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis Plant Physiology 130 1319––1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J Y, Yu XH, Liu CJ. 2009. A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis Proceedings of the National Academy of Sciences, USA 106 18855––18860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M. 2004. Silencing of hydroxycinnamoyl-coenzyme a shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis The Plant Cell 16 1446––1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. 1987. Assaying chimeric genes in plants: the GUS gene fusion system Plant Molecular Biology Reporter 5 387––405 [Google Scholar]
- Kim TW, Wang ZY. 2010. Brassinosteroid signal transduction from receptor kinases to transcription factors Annual Review of Plant Biology 61 681––704 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. 1980. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh Zeitschrift Fur Pflanzenphysiologie 100 147––160 [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0 Bioinformatics 23 2947––2948 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. 1997. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction Cell 90 929––938 [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. 1996. A role for brassinosteroids in light-dependent development of Arabidopsis Science 272 398––401 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987). Chlorophylls, carotenoids: pigments of photosynthetic biomembranes Methods in Enzymology 148 350––382 [Google Scholar]
- Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J. 2005. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily Journal of Biological Chemistry 280 13576––13583 [DOI] [PubMed] [Google Scholar]
- Meadows MG, Potrykus I. 1981. Hoechst 33258 as a vital stain for plant cell protoplasts Plant Cell Reports 1 77––79 [DOI] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T. 2002. Brassinosteroid-regulated gene expression Plant Physiology 129 1241––1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Satoh T, Tanaka SI, Mochizuki N, Yokota T, Nagatani A. 2005. Activation of the cytochrome P450 gene, CYP72C1, reduced the levels of active brassinosteroids in vivo Journal of Experimental Botany 56 833––840 [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, et al. 1999. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis Proceedings of the National Academy of Sciences of the USA 96 15316––15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, et al. 1999. ‘Green revolution’ genes encode mutant gibberellin response modulators Nature 400 256––261 [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Fujioka S, Soeno K, et al. 2005. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids Proceedings of the National Academy of Sciences, USA 102 15253––15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. 1993. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development The Plant Cell 5 147––157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh H, Jeong CW, Fujioka S, Kim YK, Lee S, Ahn JH, Choi YD, Lee JS. 2012. Genetic evidence for the reduction of brassinosteroid levels by a BAHD acyltransferase-like protein in Arabidopsis Plant Physiology10.1104/pp.112.197202 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhou XY, Li L, Xue LJ, Yang X, Xue HW. 2009. Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis Molecular Plant 2 755––772 [DOI] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in plant development. New York: Cambridge University Press; 1989. [Google Scholar]
- Sun Y, Fan XY, Cao DM, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis Developmental Cell 19 765––777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Nakazawa M, Shibata K, Yokota T, Ishikawa A, Suzuki K, Kawashima M, Ichikawa T, Shimada H, Matsui M. 2005. shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels The Plant Journal 42 13––33 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S. 2005. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism Plant Physiology 138 1117––1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. 1999. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation Proceedings of the National Academy of Sciences, USA 96 4698––4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk E M, Fujioka S, Seto H, et al. 2005. BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms The Plant Journal 42 23––34 [DOI] [PubMed] [Google Scholar]
- Varbanova M, Yamaguchi S, Yang Y, McKelvey K, Hanada A, Borochov R, Yu F, Jikumaru Y, Ross J, Cortes D, et al. 2007. Methylation of gebberellins by Arabidopsis GAMT1 and GAMT2 The Plant Cell 19 32––45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. 2002. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth The Plant Cell 14 1223––1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. 1999. A mini binary vector series for plant transformation Plant Molecular Biology 40 711––717 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. 2008. Gibberellin metabolism and its regulation Annual Review of Plant Biology 59 225––251 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. 2002. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation Cell 109 181––191 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis Nature Protocols 2 1565––1572 [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S. 2008. Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis The Plant Cell 20 1786––1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR. 2004. The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes The Plant Journal 37 864––876 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, et al. 2011. A brassinosteroid transcriptional network revealed by genome-wide identification of BES1 target genes in Arabidopsis thaliana The Plant Journal 65 634––646 [DOI] [PubMed] [Google Scholar]
- Yu XH, Gou JY, Liu CJ. 2009. BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: bioinformatics and gene expression Plant Molecular Biology 70 421––442 [DOI] [PubMed] [Google Scholar]
- Yuan T, Fujioka S, Takatsuto S, Matsumoto S, Gou X, He K, Russell SD, Li J. 2007. BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana The Plant Journal 51 220––233 [DOI] [PubMed] [Google Scholar]
- Zhao Y. 2010. Auxin biosynthesis and its role in plant development Annual Review of Plant Biology 61 49––64 [DOI] [PMC free article] [PubMed] [Google Scholar]