Abstract
CHR3 (nhr-23, NF1F4), the homologue of Drosophila DHR3 and mammalian ROR/RZR/RevErbA nuclear hormone receptors, is important for proper epidermal development and molting in the nematode Caenorhabditis elegans. Disruption of CHR3 (nhr-23) function leads to developmental changes, including incomplete molting and a short, fat (dumpy) phenotype. Here, we studied the role of CHR3 during larval development by using expression assays and RNA-mediated interference. We show that the levels of expression of CHR3 (nhr-23) cycle during larval development and reduction of CHR3 function during each intermolt period result in defects at all subsequent molts. Assaying candidate gene expression in populations of animals treated with CHR3 (nhr-23) RNA-mediated interference has identified dpy-7 as a potential gene acting downstream of CHR3. These results define CHR3 as a critical regulator of all C. elegans molts and begin to define the molecular pathway for its function.
Keywords: molting, development
Nuclear hormone receptors (NHRs) form a large superfamily of transcription factors that are important for the regulation of cell metabolism, development, and reproduction (1–3). The typical receptor consists of six domains. The most conserved domain is the DNA-binding domain (DBD) and is characteristic for the NHR superfamily. It is formed by two zinc finger motifs that are conserved through evolution (4–6). A second, less conserved domain is the ligand-binding domain, located in the C terminus of the molecule and involved in binding of small hydrophobic molecules, hormones of the steroid class, thyroid hormone, retinoic acid stereoisomers, farnesoids, prostaglandins, and terpenoids (2). The generally accepted classification of NHRs is based on homology in both the DBD and the ligand-binding domain and the specific ligands (if known), which activate the receptors. The majority of NHRs have no known ligands and usually are referred to as orphan receptors.
The NHRs are present in a majority of Metazoan species higher than diploblasts. NHRs apparently evolved from a single gene present in a Metazoan ancestor before the divergence of diploblastic species (4–6). There are about 70 NHRs in vertebrates, 22 in Drosophila (7, 8), about 270 in Caenorhabditis elegans (9, 10), and 1 NHR, a homologue of RXR, in the jellyfish, Tripedalia cystophora (11).
The high number of NHR genes present in the C. elegans genome is surprising, and the function of most of them is not yet known. Of the 270 C. elegans NHRs, only 15 are obvious orthologs of the NHRs of vertebrates or Drosophila; the rest of the C. elegans NHRs are probably nematode-specific. Studies of several of the C. elegans NHRs demonstrated a role for these factors in larval development [daf-12 (12)], neural development [nhr-55 (13) and fax-1 (14)], sex determination [nhr-24/sex-1 (15)], and molting [nhr-23 (CHR3) (16) and nhr-25 (17, 18)].
We previously have cloned the C. elegans orphan NHR CHR3 (19), which is a homologue of Drosophila DHR3 (20), Manduca sexta MHR3 (21), and mammalian ROR/RZR/Rev ErbA (22, 23). For clarity, we refer to it in this paper by using both its common and gene names, CHR3 (nhr-23). The gene is classified as NR1F4 in a unified nomenclature system for the NHR superfamily (24). We previously showed that CHR3 (nhr-23) is expressed in the epidermis of C. elegans throughout development and that it was required for proper molting (16).
Molting is a complex of developmental processes characteristic for a clad Ecdysozoa, which includes arthropods, tardigrades, onychophorans, nematodes, nematomorphs, kinorhynchs, and priapulids (25). At each molt, there is production of the new outer-body cover, the exoskeleton, and the old part is shed. Molting represents critical developmental transitions similar to metamorphosis of amphibians and is accompanied by characteristic behavioral patterns.
In Drosophila, molting is regulated by pulses of a steroid hormone, 20-hydroxyecdysone, and the regulatory cascade involves the ecdysone receptor, EcR (which dimerizes with ultraspiracle receptor, USP, a homologue of mammalian retinoic acid X receptor) and two other NHRs, DHR3 and βFTZ-F1. Drosophila DHR3 is an ecdysone-inducible NHR, which functions during metamorphosis, mainly in the prepupal–pupal transition and is an important regulator of the βFTZ-F1 transcription factor (26–29).
In this study, we show that CHR3 is a critical regulator of each molt of C. elegans development and that disruption of CHR3 function by RNA-mediated interference (RNAi) allows us to screen for genes acting downstream of CHR3. Our data suggest that the dumpy (Dpy) phenotype previously observed by CHR3 inhibition might be due to misregulation of a specific collagen gene. Analysis of a second NHR, whose homolog is important for DHR3 function in Drosophila, suggests divergence of this regulatory cascade through evolution.
Materials and Methods
Strains.
The following strains have been used: the wild-type C. elegans Bristol strain N2, supplied by the Caenorhabitis Genetics Center; strain hlh-8∷gfp-PD 4666[dpy-20(e1282)IV;ayls7(pBH47.70 + pMH86)IV] (30); him-5(e1467) hermaphrodites, which segregate 16% males; EM 599, a transgenic line carrying egl-5∷gfp (kind gift from Scott Emmons, Albert Einstein College of Medicine, Bronx, NY); and atEx32 and atEx35, transgenic lines carrying nhr-25∷gfp (a kind gift from Ann Sluder, University of Georgia, Athens, GA).
Analysis of CHR3 (nhr-23) Expression.
A synchronous population of nematodes was prepared by treatment of adult animals with alkaline hypochlorite and hatched overnight. The larvae were placed on 2% agarose plates with the lawn of Escherichia coli OP50. About 10,000 worms were collected from the younger larval stages. For L3, L4, and adult worms, about 5,000 animals were collected for each time point. The plates were incubated at 25°C, and the worms were collected at 2-hr intervals. Three independent collections were done. For one, the worms were collected during L1 stage (hours 0–12). In the second, worms were collected during L2 and L3 stages of development at 2-hr intervals. Finally we collected worms continuously during development. The embryos were collected immediately after hypochlorite treatment of gravid adults.
RNA Preparation.
RNA was prepared essentially as described by Johnstone and Barry (31). The worms were washed several times with water and pelleted by centrifugation for 1 min at 1,000 rpm. All samples were frozen. Each pellet was resuspended in 0.5 ml of buffer containing 0.5% SDS, 5% 2-mercaptoethanol, 10 mM EDTA, 10 mM Tris⋅HCl (pH 7.5), and 0.5 mg/ml proteinase K. The samples were incubated 1 hr at 55°C and extracted with 1 vol of phenol/chloroform (1:1). The RNA was precipitated with ethanol at −80°C overnight, collected by centrifugation, and resuspended in diethyl pyrocarbonate water. The samples were treated with DNase I for 1 hr at 37°C, reextracted with phenol/chloroform, and precipitated. The samples finally were dissolved in 0.1 ml of diethyl pyrocarbonate water.
Reverse Transcription (RT)–PCR.
First-strand cDNA for each developmental stage was prepared from 1 μg of total RNA. The reverse transcriptase (Superscript II; Life Technologies, Gaithersburg, MD) reaction was primed with random hexamers by using conditions recommended by the supplier. The cDNA reaction products were diluted 1:10 in water.
For all genes tested, the diluted cDNA reactions were amplified by using two pairs of the primers. One primer pair was specific for the test gene whereas the other targeted an internal control gene, ama-1, which encodes the large subunit of RNA polymerase II (32). The primers have been designed so that any contaminating genomic DNA products (if present) would include several introns and be larger and less favored than the cDNA products. The number of PCR cycles that gave linear amplification for each gene was determined empirically after testing of each pair of primers individually; 30–33 cycles has been used in most cases. The PCRs started with 94°C for 2 min followed by 30 cycles of 94°C for 30 s, 60°C for 40 s, and 72°C for 1 min and 30 s. Reactions were incubated at 72°C for 3 min after cycling.
Electrophoresis of PCRs was performed on 2% agarose gels. The gels were stained for 30 min with ethidium bromide and destained two times with water. The densitometric analysis of bands was performed by imaging with a Stratagene Eagle Eye digital camera followed by quantitation of bands using ip labs software. The final data are presented as the ratio of the expression of the gene of interest to the expression of internal standard ama-1. Each gel was analyzed separately, and the values were plotted as the ratio of the highest value obtained for CHR3 (nhr-23) in the particular experiment (given as 1) divided by the value at the time point examined.
The following genes were assayed: nhr-23, nhr-25, nhr-41, nhr-85, lrp-1, ldl-1, hint-1, dpy-7, dpy-13, col-12, cp-1, cp-2, and cp-3. The primers ama-1, dpy-7, col-12, and dpy-13 were as described by Johnstone and Barry (31). Other primers were as follows: CHR3 (nhr-23) (5′ end), 5′-GGATGCGAGAGTTCAAATTGCATG and 5′-CTGTGCTGCTGATTGAAAGCTCTG; CHR3 (nhr-23) (ligand-binding domain), 5′-GTTATCAGAGCTTTCAATCAGCAG and 5′-AATTCTTGGCATGACACTTTGGTAC; nhr-25, 5′-CAACAACTGGCCGAATGACAGAAGC and 5′-CACAGCAGTGTATGTGGCTTGTGG; nhr-41, 5′-GATGAGAAGTGAATCAGTAC and 5′-CCATTGCCATTTGATTCGTT; nhr-85, 5′-GAGTGTCGTTGGCACAGGCGATAC and 5′-AACAGCGTATTTGATGTGATGCTTC; lrp-1, 5′-CTTCTTCACAAATCAGCTCAAACTTG and 5′-CCATCAGGACGGATTCTGTAGATAG; ldl-1, 5′-AGTCAGTCACAGTATAGGATATCG and 5′-GCAAAGAGAAGCATCAGCTTCAAG; hint-1, 5′- GCGGCGGCTATCTGCCATCCTGCC and 5′-AGGCGGCGTTCAATTCCTTTGACG; cp-1, 5′-AGACTTCGACTCGGAGGATCTGCC and 5′-CAATCGGATCAGCCCAAACACAATC; cp-2, 5′-GATTCATTCTTCTGGCACTGGTTGC and 5′-CCTTCTTGTAAAGTTGGAAGCTGC; and cp-3, 5′-GATGAAGCTCTTGTGGATATGAGC and 5′-GTCAAGTTGGAAACATCAACATCCG.
Double-Stranded (ds) RNAi.
For RNAi by microinjection and soaking, the dsRNA was prepared by in vitro transcription reaction with T3 and T7 RNA polymerase after linearization of constructs 4133 and 4071 as described (16). For RNAi by soaking, 10 μl of dsRNA at a concentration of about 2 μg/μl was used and the worms were soaked for 12–24 hr.
RNAi by feeding was done essentially as described by Timmons and Fire (33). Feeding construct 4666 consisted of a 550-bp-long CHR3 (nhr-23) cDNA fragment that includes mostly the ligand-binding domain inserted into the vector L4440 (containing two T7 RNA polymerase sites). The construct was transformed into HT115 E. coli and induced with isopropyl-β-d-thiogalactoside.
A visual assay of nhr-25 expression used two transgenic nhr-25∷GFP lines (atEx32 and atEx35), which were kind gifts from Ann Sluder (University of Georgia). dsRNA for nhr-25 RNAi was prepared by using feeding vector L4440 harboring a nhr-25 cDNA fragment from yk342d8 (kindly provided by Y. Kohara, National Institute of Genetics, Mishima, Japan) using EcoRI–XhoI restriction sites. The dsRNA prepared from this construct is directed against both isoforms of nhr-25.
Results
Expression of CHR3 (nhr-23) During Molting Cycles.
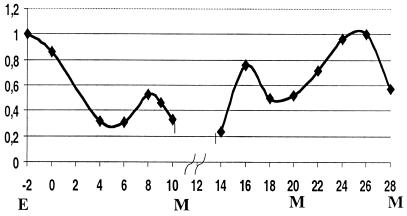
As reported earlier, CHR3 is present in epidermal cells throughout development based on gfp reporter genes, antibody staining (embryos only), and Northern analysis (16, 19). To measure the level of CHR3 (nhr-23) expression during molting cycles, we used synchronized populations of larvae and evaluated the expression of CHR3 (nhr-23) by RT-PCR. As a control, we normalized CHR3 (nhr-23) expression to the ama-1 gene that codes for the large subunit of RNA polymerase II (32). High-level expression of CHR3 (nhr-23) was observed in embryos and persisted in newly hatched larvae, then decreased to a minimum at 4–6 hr after synchronized-hatched L1 larvae were first fed. In the period preceding the first molt, the expression increased for about 2 hr and decreased before the molt (Fig. 1). Similarly, we observed an elevation in CHR3 (nhr-23) expression between successive molts, and minimal values were observed at each molt. CHR3 (nhr-23) transcript is present throughout development, with intermolt peaks reaching two- to five-times-higher levels than the minimal values found at the molts (Fig. 1).
Figure 1.
CHR3 (nhr-23) expression during larval development. Expression of CHR3 (nhr-23) as determined by RT-PCR and normalized against ama-1 expression. Arbitrary units obtained by densitometric analysis of RT-PCR products are plotted against developmental time in hours after synchronized-hatched L1 are plated with food. M indicates time of each molt in the population. The level of the expression in embryos is shown as E, and the level in the starved, synchronized-hatched larvae is shown as time 0. The break in the graph reflects the lack of a 12-hr time point for the combined data sets.
CHR3 Is Required for All Larval Molts.
RNAi is a powerful reverse genetic technique in which dsRNA is used to determine the phenotype resulting from the loss or reduction of gene function (34). It has been shown by Timmons and Fire (33) and Tabara (35) that dsRNA may be introduced to worms by various methods. This includes soaking the animals in a solution containing dsRNA or feeding the worms with bacteria that have been transformed with plasmids coding for the RNA of interest, cloned as an insert flanked on both sides by inducible bacterial T7 promoters. Both soaking and feeding allow the delivery of dsRNA at specific stages of development such that subsequent developmental events can be assayed for defects.
We have shown previously that CHR3 (nhr-23) RNAi performed by injection of dsRNA into the maternal germ line leads to defects in molting and epidermal development among progeny of the injected animal. It was not possible to determine by those experiments whether larval molts that were affected later reflected a requirement for CHR3 (nhr-23) at each specific molt or were merely a secondary consequence of earlier molting defects. To determine whether CHR3 has a role in one, several, or all larval molts, we used soaking and feeding RNAi techniques to deliver the dsRNA at various times during development. Using these additional methods, we were able to affect all four larval molts by CHR3 (nhr-23) RNAi, demonstrating that CHR3 is required for each (Fig. 2). There were differences in the penetrance of molting defects depending on the method, dose, temperature, and exposure time to CHR3 (nhr-23) RNAi (Tables 1 and 2).
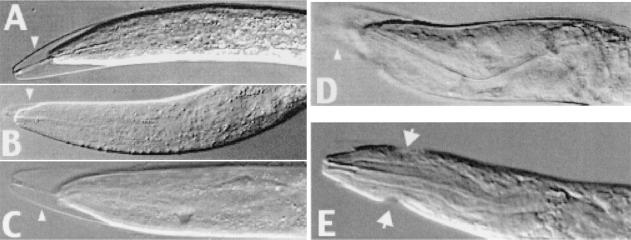
Figure 2.
CHR3 (nhr-23) RNAi-molting defects induced at each larval stage. (A) L2 larva with the characteristic phenotype of partially detached cuticle at the head and the bucal plaque still attached between mouth and cuticle. (B) Larva arrested in L2/L3 molt. (C) Larva arrested in L3/L4. (D) The head of adult hermaphrodite with defects from the L4/A molt. (E) The head of a male arrested in L4/A molt. Arrowheads in A– D indicate incompletely shed cuticle. Arrows in E mark cuticular constrictions from unshed cuticles from previous defective molts.
Table 1.
Developmental arrest and molting defects in CHR3 (nhr-23) RNAi-treated animals by either soaking (Soak) or feeding (Feed) techniques
| Stage and strain of treated animals | Molting/ arrest stage of animals | Soak, % affected | n | Feed, % affected | n |
|---|---|---|---|---|---|
| L4, A N2 | L1/L2 | 2 | 538 | 3 | 463 |
| L2/L3 | 21 | 24 | |||
| L3/L4 | 45 | 65 | |||
| L4/A | 20 | 3 | |||
| L1 N2 | L2/L3 | 10 | 292 | 10 | 320 |
| L3/L4 | 79 | 83 | |||
| L4/A | 7 | 5 | |||
| L1 hlh-8 (nr2061) | L2/L3 | 20 | 231 | 19 | 156 |
| L3/L4 | 77 | 81 | |||
| L4/A | 3 | 0 | |||
| L2 N2 | L3/L4 | 19 | 98 | 3 | 126 |
| L4/A | 62 | 86 | |||
| L2 him-5 (e1467) | L4/A | ND | — | 60 | 132 |
| L3 N2 | L4/A | 50 | 54 | 50 | 46 |
| L3 him-5 (e1467) | L4/A | 50 | 63 | 50 | 48 |
Total number of animals counted in each experiment is given by n.
Table 2.
Developmental arrests and molting defects in CHR3 (nhr-23) RNAi experiments resulting from feeding at 25°C and 15°C
| Stage of treated animals | Molting/arrest stage of animals | 25°C, % affected | n | 15°C, % affected | n |
|---|---|---|---|---|---|
| L4, A | L1/L2 | 0 | 286 | 3 | 275 |
| L2/L3 | 18 | 40 | |||
| L3/L4 | 71 | 55 | |||
| L4/A | 1 | 4 | |||
| L1 | L2/L3 | 15 | 153 | 9 | 202 |
| L3/L4 | 41 | 83 | |||
| L4/A | 23 | 6 | |||
| L2 | L3/L4 | 0 | 79 | ND | — |
| L4/A | 46 | ND | |||
| L3 | L4/A | 0 | 30 | ND | — |
Total number of animals counted in each experiment is given by n.
Soaking late L4 larvae or young adult hermaphrodites results in molting defects among progeny in L2/L3 and later molts; only a small percentage of animals arrest in L1/L2. Soaking the animals at the L1 stage causes the majority of affected animals to have defects in the L3/L4 molt. As expected, soaking the L2 larvae results in a molting defect primarily at the L4/adult (A) transition (Table 1). We also have used a strain with null mutation in the hlh-8 gene (36), which results in a nonlethal constipation defect to see whether slower passage of food through the gut may have an enhanced dsRNA effect. In these animals, the penetrance of RNAi effects was close to 100% for CHR3 (nhr-23). Treating L3 worms led to developmental arrest and a molting defect in L4/A molt. The progeny of these worms also were affected similarly as in experiments with RNAi delivered by microinjections reported earlier (16).
The delivery of dsRNA by feeding animals transformed bacteria proved to be as efficient as the soaking method (Table 1). We tried to influence the dsRNA effect by applying lower or higher worm culture temperature. The cultures maintained at 25°C had lower efficiency of the RNAi effect whereas the lower temperature (15°C) had a more pronounced and earlier effect (Table 2). The ease and efficiency of the feeding method also allowed us to examine entire populations of animals.
A critical feature of CHR3 (nhr-23) loss of function in larvae L2–L4 is the developmental arrest and the subsequent lethality during or shortly after molting. The lethality rate was close to 100% in synchronized cultures treated by continuous feeding of dsRNA-producing bacteria, although not all animals died during the same molt.
CHR3 (nhr-23) Loss of Function Affects Development of Gonad and Associated Epidermal Structures.
About 50% of young larvae exposed to CHR3 (nhr-23) RNAi by soaking or feeding showed defects in gonad development if they survived to adulthood; these defects had not been seen previously by microinjection RNAi. The germ line was often misshapen in these animals, usually folded and constricted. The distal-tip cell position was misplaced as a result of improper migration. These animals often had incompletely shed the L4 cuticle during the L4/A molt, resulting in one or more constrictions of the body at the point at which the shedding cuticle had gotten stuck. In severely affected hermaphrodites, the germ line had an irregular shape and contained small and more numerous nuclei.
To see whether CHR3 (nhr-23) loss of function affected males, we treated him-5 animals with CHR3 (nhr-23) RNAi. The him-5 mutation alone results in about 16% of males among the population as opposed to less than 0.5% normally observed in N2 cultures (37). About 50% of him-5 males subjected to CHR3 (nhr-23) RNAi in feeding experiments had male tail defects in addition to molting defects. The main male tail phenotype included problems in fan and sensory ray development, including a failure to retract the tail during morphogenesis (Fig. 3B). About one-half of affected males had vacuoles in the germ line. To see whether the CHR3 (nhr-23) RNAi male tail defects affected cell fates, in addition to morphogenesis, we used a strain carrying the egl-5∷gfp reporter gene that marks the ray cells (38). The ray cells continued to express egl-5∷gfp after CHR3 (nhr-23) RNAi, demonstrating that the fate of these cells, as assayed by expression of this reporter gene, is unaffected and the male tail defects are primarily from problems in morphogenesis.
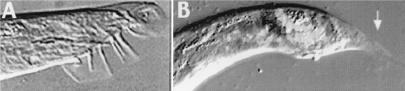
Figure 3.
CHR3 (nhr-23) RNAi male tale defects. (A) A normal male tail fan with the sensory rays and mating spicules. (B) egl-5∷gfp transgenic male treated by CHR3 (nhr-23) RNAi. Note defects in tail retraction (arrow) as well as fan and sensory ray morphogenesis; spicules are formed. The egl-5∷gfp reporter marks neuronal ray cells in the defective male tail (arrow).
Gene Expression in CHR3 (nhr-23) RNAi Cultures.
The high percentage of affected animals resulting from CHR3 (nhr-23) RNAi delivered by feeding in mass culture allowed us to study gene expression by using molecular techniques. Animals were collected after feeding with either CHR3 (nhr-23) dsRNA-producing bacteria or control bacteria, and total RNA subsequently was isolated from the population. The percentage of affected animals was determined at the time of collection and ranged from 98% to 100%. Total RNA was used to make cDNA, and the expression of a variety of genes was assayed by PCR. As a control, all gene-expression levels were normalized to ama-1 that encodes the large subunit of RNA polymerase II (32).
First, we determined how the CHR3 (nhr-23) RNAi influences the expression of CHR3 (nhr-23) itself. We have assayed CHR3 (nhr-23) expression with two sets of primers in RT-PCR experiments. One set of primers partially overlapped the coding region targeted by RNAi. The other primer set amplified the CHR3 coding region located 5′ to the region targeted by RNAi. All experiments were done two to four times, yielding similar results regardless of the region of CHR3 (nhr-23) assayed. CHR3 (nhr-23) expression (relative to ama-1) was reduced, but not eliminated, by CHR3 (nhr-23) RNAi. Reduction of expression continued to decline over time with 25% of control levels observed (Fig. 4). Although some CHR3 (nhr-23) message remained after CHR3 (nhr-23) RNAi, the highly penetrant phenotypic effects observed in the populations suggested that the RNAi feeding method could be used to screen for genes acting downstream of CHR3 (nhr-23).
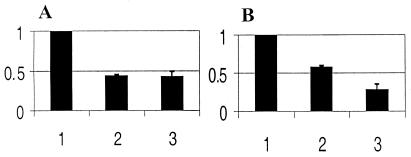
Figure 4.
Expression of CHR3 in animal cultures subjected to CHR3 (nhr-23) RNAi. Animal cultures were fed either bacteria-producing CHR3 (nhr-23) dsRNA or control bacteria. The expression of CHR3 (nhr-23) was determined by RT-PCR and normalized to ama-1 expression. Values were quantified and are plotted as a ratio of values of RNAi-treated to untreated controls. (1) Control experiments arbitrarily set to 100%. (2) CHR3 expression in CHR3 (nhr-23) RNAi-treated populations collected shortly after the L3 molt. (3) CHR3 expression in CHR3 (nhr-23) RNAi-treated populations 3 hr after the L3 molt. The average deviation from duplicates (lanes 2) and triplicates (lanes 3) are indicated. (A) Expression of CHR3 (nhr-23) as determined by RT-PCR, with PCR primers designed to amplify the 5′ region of CHR3 (nhr-23) cDNA (see Materials and Methods). (B) Expression of CHR3 (nhr-23) as determined by RT-PCR primers designed to amplify a fragment of the CHR3 (nhr-23) coding region targeted by RNAi.
CHR3 (nhr-23) RNAi Affects dpy-7 Expression.
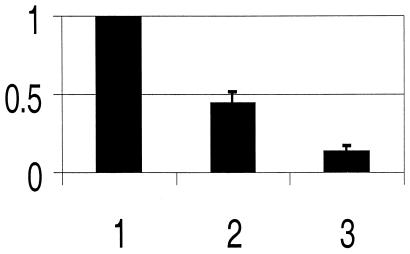
The two most prominent features of the CHR3 (nhr-23) loss of function phenotype are the molting and dumpy phenotypes. Several genes, in addition to CHR3 (nhr-23), have been shown to affect molting or encode candidates for molting factors based on their enzymatic or structural properties. Likewise, numerous genes can be mutated to give a dumpy (Dpy) phenotype, including several collagen genes that are involved in cuticle formation (reviewed in ref. 39). We have studied the expression of 12 genes that are candidates for being involved in the molting and/or Dpy phenotypes observed after CHR3 (nhr-23) RNAi (see Material and Methods). The expression of each gene was measured by RT-PCR in either CHR3 (nhr-23) RNAi-treated or control populations of animals. Only a single candidate gene, dpy-7, showed a dramatic change in expression after CHR3 (nhr-23) RNAi. The levels of dpy-7 expression decreased 5- to 11-fold in treated animals when compared with controls (Fig. 5).
Figure 5.
Expression of dpy-7 in cultures subjected to CHR3 (nhr-23) RNAi. Animal cultures were fed either bacteria-producing dsRNA to CHR3 (nhr-23) or control bacteria. Expression of dpy-7 was determined by RT-PCR and normalized to ama-1 expression. Results are plotted as in Fig. 4. (1) Expression of dpy-7 in control cultures is arbitrarily set to 100%. (2) Expression of dpy-7 in cultures treated by CHR3 (nhr-23) RNAi collected shortly after the L3 molt. (3) Expression of dpy-7 in cultures treated by CHR3 (nhr-23) RNAi collected 3 hr after the L3 molt. Bars indicate average deviation based on duplicates (2) and triplicates (3).
The nhr-25 gene encodes an orphan nuclear receptor in C. elegans, and loss of function by using microinjection of dsRNA into adult hermaphrodites results in progeny with molting and other defects (17, 18). The Drosophila homologue of nhr-25 is βFTZ-F1. In Drosophila, βFTZ-F1 acts downstream of DHR3 in the ecdysis regulatory cascade (26–29). Surprisingly, we found no dramatic effect in C. elegans on nhr-25 expression in CHR3 (nhr-23) RNAi-treated cultures. As a further test of possible CHR3 and NHR-25 interactions, we used a nhr-25∷gfp reporter (kind gift of Ann Sluder) and monitored gfp expression in control and CHR3 (nhr-23) RNAi-treated animals. No changes in gfp expression were observed in animals treated by CHR3 (nhr-23) RNAi, even in animals with a strong CHR3 (nhr-23) loss of function phenotype. We have retested the effects of nhr-25 RNAi applied to young larvae by using the feeding technique. Although we can induce by feeding many of the phenotypic defects reported previously by microinjection (18, 19), only about 1% of the animals showed signs of impaired molting. The low penetrance of molting defects and lack of effect of CHR3 (nhr-23) RNAi on nhr-25∷gfp expression suggest that nhr-25 is not acting downstream of CHR3 (nhr-23) in the C. elegans molting pathway.
Discussion
CHR3 Regulates Molting in All Four Larval Stages.
This study demonstrates that CHR3 (nhr-23) regulates molting in all larval stages of C. elegans development. This regulation may be a consequence of the cyclical pattern of CHR3 (nhr-23) expression that we observe. In larval stages L1–L3, there are maximum values of CHR3 (nhr-23) expression detected 2–4 hr before each molt and minimal values in the time of molting. In larval stage L4, the CHR3 expression is also high in the middle of molting cycle and decreases immediately after this point but starts to increase toward the in L4/A molt. Consistent with this expression pattern, we previously detected strong expression of CHR3 the germ line of adult hermaphrodites (16). We do not know how the CHR3 (nhr-23) expression pattern is regulated. Ecdysone and EcR are responsible for metamorphosis regulation in Drosophila; neither of these has been detected in C. elegans. However, it is possible that the cyclical pattern of CHR3 (nhr-23) expression is regulated by fluctuating levels of a ligand that binds CHR3, leading to autoregulation similar to that seen for the thyroid hormone receptor during metamorphosis of amphibians (40). Alternatively, a fluctuating ligand could act through another NHR that, in turn, regulates CHR3 (nhr-23) expression.
Molting is a developmental change during which the cuticle is exchanged, the animal enlarges, and the animal undergoes developmental transitions (25, 41). In many respects, this process is similar to metamorphosis of insects. The homologue of CHR3 in Drosophila, DHR3, is a critical factor in the regulatory cascade of metamorphosis, where it acts, in part, to reset the molecular cascade triggered by pulses of the hormone 20-hydroxyecdysone (42–45). The regulation of all molts in C. elegans by CHR3 thus is strikingly similar to the regulation of metamorphosis in Drosophila by DHR3. In Drosophila, the βFTZ-F1 orphan NHR acts downstream of DHR3 (26–29, 46). Using three different assays, we find no evidence that the C. elegans homolog of βFTZ-F1, NHR-25, is regulated by CHR3. In addition, the lack of a highly penetrant molting defect in nhr-25 RNAi-treated animals makes it unlikely that nhr-25 acts immediately upstream of CHR3 (nhr-23) in the molting process.
Although CHR3 inhibition affects the L1/L2 molt, the lethal effect is rare and a majority of animals are able to proceed to later stages. Contrary to that, published data showed a strong, lethal effect of nhr-25 RNAi (delivered by microinjection) at the L1/L2 molt (17, 18). It is possible that in an early stage, nhr-25 regulates development of steroidogenic structures, which may be important for CHR3 function. Nevertheless, the regulation of a hypothetical steroidogenic signal by nhr-25 is not likely to be responsible for regulation of the cycling in molts, because the disruption of nhr-25 function in molts L2–L4 doesn't interfere with molting cycles but has a strong, developmental effect on germ-line and related structures. It would appear, then, that although both CHR3 and DHR3 trigger related events in C. elegans and Drosophila, respectively, at least one (and likely many more) of the downstream targets has diverged through evolution.
dpy-7 Is Affected by CHR3 Loss of Function.
The CHR3 (nhr-23) loss of function phenotype includes a dumpy phenotype that is particularly evident in older worms that escape early larval molting defects and lethality. Because several of the nearly 150 collagen genes in C. elegans are associated with a Dpy phenotype when mutated (47–49), we included several collagen genes in our limited survey of gene expression in CHR3 (nhr-23) RNAi-treated populations. One of these, dpy-7, has an expression pattern that correlates with CHR3 (nhr-23) (31). Moreover, dpy-7 is expressed in some epidermal cells at most developmental stages and its expression cycles (31, 49). Interestingly, the peak of dpy-7 expression is in the middle of each molt just as we have found for CHR3 (nhr-23). The drop in dpy-7 expression in CHR3 (nhr-23) RNAi animals and their associated Dpy phenotype makes it tempting to speculate that dpy-7 is a target of CHR3. An examination of cis-acting regulatory sequences required for dpy-7 transgene expression reveals several possible CHR3-binding sites. However, we have yet to determine any in vivo CHR3-binding site and can discriminate between sites based only on limited in vitro binding data (16). Experiments aimed to address potential CHR3 binding to dpy-7 gene sequences are ongoing. It is worth noting that the regulation of dpy-7 by CHR3 may be indirect, and further work is necessary to discriminate between these possibilities.
CHR3 (nhr-23) Message Is Not Eliminated by RNAi.
Surprisingly, CHR3 (nhr-23) message was never completely eliminated by RNAi, even though almost all animals in the treated culture had severe phenotypic defects. This demonstrates that reduction of CHR3 (nhr-23) function is sufficient to cause a phenotype and that the processes it regulates are dose-sensitive. The wide application of RNAi to affect gene function in C. elegans and other organisms makes this observation more generally of interest. Although near-complete loss of endogenous message after RNAi has been shown for a maternal gene product (50), message levels usually are not assayed after RNAi. We have tested message levels directly and shown that the persistent CHR3 (nhr-23) message after RNAi is not due to preferential stability of coding regions not targeted by dsRNA. One possibility for the persistent message is that CHR3 (nhr-23) is expressed in a subset of cells that are not responsive to dsRNA-mediated message disruption; mature neurons have been shown to be less amenable to RNAi (51). Alternatively, RNAi might routinely result in the reduction, but not complete loss, of targeted messages and the level of message depletion may be dependent on a variety of conditions. This might explain the variable degree of penetrance and phenotype observed from experiment to experiment with particular targeted genes.
Conclusion.
Our data support the existence of the clad Ecdysozoa (25) by finding that the related genes CHR3 and DHR3 are in the same regulatory pathway of molting with a critical role in the transmission and timing of the cycling signal. However, these pathways are not conserved absolutely because CHR3 appears not to act through the gene nhr-25, thereby distinguishing the C. elegans pathway from that found in Drosophila. Thus, although there is conservation of genes involved in regulation of molting in Ecdysozoa, their position in the regulatory cascade may differ.
Acknowledgments
We thank Andy Fire and Lisa Timmons for providing us with the vector and host for RNAi, Ann Sluder and Scott Emmons for strains and information, Yuji Kohara for clone yk342d8, and Jack Robbins for critically reading the manuscript. The work of M.K. was supported partially by Grants 304-99-0682, NC 5261-3, and 11110000-3 from the Grant Agency of the Czech Republic, Ministry of Health of the Czech Republic, and Ministry of Education of the Czech Republic, respectively.
Abbreviations
- NHR
nuclear hormone receptor
- RNAi
RNA-mediated interference
- EcR
ecdysone receptor
- RT
reverse transcription
- dsRNA
double-stranded RNA
References
- 1.Thummel C S. Cell. 1995;83:871–877. doi: 10.1016/0092-8674(95)90203-1. [DOI] [PubMed] [Google Scholar]
- 2.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 4.Amero S A, Kretsinger R H, Moncrief N D, Yamamoto K R, Pearson W R. Mol Endocrinol. 1992;6:3–7. doi: 10.1210/mend.6.1.1738368. [DOI] [PubMed] [Google Scholar]
- 5.Laudet V, Hanni C, Coll J, Catzeflis F, Stehelin D. EMBO J. 1992;11:1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laudet V. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- 7.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 8.Celniker S E. Curr Opin Genet Dev. 2000;10:612–616. doi: 10.1016/s0959-437x(00)00140-4. [DOI] [PubMed] [Google Scholar]
- 9.Chervitz S A, Aravind L, Sherlock G, Ball C A, Koonin E V, Dwight S S, Harris M A, Dolinski K, Mohr S, Smith T, et al. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sluder A E, Mathews S W, Hough D, Yin V P, Maina C V. Genome Res. 1999;9:103–120. [PubMed] [Google Scholar]
- 11.Kostrouch Z, Kostrouchova M, Love W, Jannini E, Piatigorsky J, Rall J E. Proc Natl Acad Sci USA. 1998;95:13442–13447. doi: 10.1073/pnas.95.23.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antebi A, Culotti J G, Hedgecock E M. Development (Cambridge, UK) 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- 13.Zhou H M, Walthall W W. J Neurosci. 1998;18:10438–10444. doi: 10.1523/JNEUROSCI.18-24-10438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Much J W, Slade D J, Klampert K, Garriga G, Wightman B. Development (Cambridge, UK) 2000;127:703–712. doi: 10.1242/dev.127.4.703. [DOI] [PubMed] [Google Scholar]
- 15.Carmi I, Kopczynski J B, Meyer B J. Nature (London) 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- 16.Kostrouchova M, Krause M, Kostrouch Z, Rall J E. Development (Cambridge, UK) 1998;125:1617–1626. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- 17.Gissendanner C R, Sluder A E. Dev Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- 18.Asahina M, Ishihara T, Jindra M, Kohara Y, Katsura I, Hirose S. Genes Cells. 2000;5:711–723. doi: 10.1046/j.1365-2443.2000.00361.x. [DOI] [PubMed] [Google Scholar]
- 19.Kostrouch Z, Kostrouchova M, Rall J E. Proc Natl Acad Sci USA. 1995;92:156–159. doi: 10.1073/pnas.92.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koelle M R, Segraves W A, Hogness D S. Proc Natl Acad Sci USA. 1992;89:6167–6171. doi: 10.1073/pnas.89.13.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palli S R, Hiruma K, Riddiford L M. Dev Biol. 1992;150:306–318. doi: 10.1016/0012-1606(92)90244-b. [DOI] [PubMed] [Google Scholar]
- 22.Carlberg C, Hooft van Huijsduijnen R, Staple J K, DeLamarter J F, Becker-Andre M. Mol Endocrinol. 1994;8:757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- 23.Forman B M, Chen J, Blumberg B, Kliewer S A, Henshaw R, Ong E S, Evans R M. Mol Endocrinol. 1994;8:1253–1261. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- 24.Nuclear Receptors Nomenclature Committee. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 25.Aguinaldo A M, Turbeville J M, Linford L S, Rivera M C, Garey J R, Raff R A, Lake J A. Nature (London) 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 26.Kageyama Y, Masuda S, Hirose S, Ueda H. Genes Cells. 1997;2:559–569. doi: 10.1046/j.1365-2443.1997.1460344.x. [DOI] [PubMed] [Google Scholar]
- 27.Lam G, Hall B L, Bender M, Thummel C S. Dev Biol. 1999;212:204–216. doi: 10.1006/dbio.1999.9343. [DOI] [PubMed] [Google Scholar]
- 28.Lam G T, Jiang C, Thummel C S. Development (Cambridge, UK) 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- 29.White K P, Hurban P, Watanabe T, Hogness D S. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- 30.Harfe B D, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A. Genes Dev. 1998;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnstone I L, Barry J D. EMBO J. 1996;15:3633–3639. [PMC free article] [PubMed] [Google Scholar]
- 32.Rogalski T M, Riddle D L. Genetics. 1988;118:61–74. doi: 10.1093/genetics/118.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timmons L, Fire A. Nature (London) 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 34.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 35.Tabara H, Grishok A, Mello C C. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- 36.Corsi A K, Kostas S A, Fire A, Krause M. Development (Cambridge, UK) 2000;127:2041–2051. doi: 10.1242/dev.127.10.2041. [DOI] [PubMed] [Google Scholar]
- 37.Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 243–279. [Google Scholar]
- 38.Ferreira H B, Zhang Y, Zhao C, Emmons S W. Dev Biol. 1999;207:215–228. doi: 10.1006/dbio.1998.9124. [DOI] [PubMed] [Google Scholar]
- 39.Johnstone I L. Trends Genet. 2000;16:21–27. doi: 10.1016/s0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- 40.Tata J R. Insect Biochem Mol Biol. 2000;30:645–651. doi: 10.1016/s0965-1748(00)00035-7. [DOI] [PubMed] [Google Scholar]
- 41.Ambros V. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 42.Thummel C S. BioEssays. 1997;19:669–672. doi: 10.1002/bies.950190806. [DOI] [PubMed] [Google Scholar]
- 43.Lan Q, Hiruma K, Hu X, Jindra M, Riddiford L M. Mol Cell Biol. 1999;19:4897–4906. doi: 10.1128/mcb.19.7.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S J, Park J G, Lee C C. Mol Cells. 1999;9:61–66. [PubMed] [Google Scholar]
- 45.Kozlova T, Thummel C S. Trends Endocrinol Metab. 2000;11:276–280. doi: 10.1016/s1043-2760(00)00282-4. [DOI] [PubMed] [Google Scholar]
- 46.Carney G E, Wade A A, Sapra R, Goldstein E S, Bender M. Proc Natl Acad Sci USA. 1997;94:12024–12029. doi: 10.1073/pnas.94.22.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Mende N, Bird D M, Albert P S, Riddle D L. Cell. 1988;55:567–576. doi: 10.1016/0092-8674(88)90215-2. [DOI] [PubMed] [Google Scholar]
- 48.Johnstone I L, Shafi Y, Barry J D. EMBO J. 1992;11:3857–3863. doi: 10.1002/j.1460-2075.1992.tb05478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilleard J S, Barry J D, Johnstone I L. Mol Cell Biol. 1997;17:2301–2311. doi: 10.1128/mcb.17.4.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montgomery M K, Xu S, Fire A. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fire A. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]