Abstract
Iron (Fe) is an essential plant micronutrient, and its deficiency limits plant growth and development on alkaline soils. Under Fe deficiency, plant responses include up-regulation of genes involved in Fe uptake from the soil. However, little is known about shoot responses to Fe deficiency. Using microarrays to probe gene expression in Kas-1 and Tsu-1 ecotypes of Arabidopsis thaliana, and comparison with existing Col-0 data, revealed conserved rosette gene expression responses to Fe deficiency. Fe-regulated genes included known metal homeostasis-related genes, and a number of genes of unknown function. Several genes responded to Fe deficiency in both roots and rosettes. Fe deficiency led to up-regulation of Cu,Zn superoxide dismutase (SOD) genes CSD1 and CSD2, and down-regulation of FeSOD genes FSD1 and FSD2. Eight microRNAs were found to respond to Fe deficiency. Three of these (miR397a, miR398a, and miR398b/c) are known to regulate transcripts of Cu-containing proteins, and were down-regulated by Fe deficiency, suggesting that they could be involved in plant adaptation to Fe limitation. Indeed, Fe deficiency led to accumulation of Cu in rosettes, prior to any detectable decrease in Fe concentration. ccs1 mutants that lack functional Cu,ZnSOD proteins were prone to greater oxidative stress under Fe deficiency, indicating that increased Cu concentration under Fe limitation has an important role in oxidative stress prevention. The present results show that Cu accumulation, microRNA regulation, and associated differential expression of Fe and CuSOD genes are coordinated responses to Fe limitation.
Key words: Copper, iron, microarray, microRNA, miR398, superoxide dismutase
Introduction
Iron (Fe) is a required micronutrient for plants and most other forms of life. The capacity of Fe to accept or donate electrons allows this metal to be a key component of crucial redox enzymes in numerous biochemical processes, including the electron transport chains involved in mitochondrial respiration and photosynthesis (Hansch and Mendel, 2009). Fe is also required for chlorophyll biosynthesis, causing Fe-deficient plants to be chlorotic, and thus less able to absorb light energy (Spiller and Terry, 1980; Terry, 1980, 1983). This can lead to increased production of free radicals (Gill and Tuteja, 2010), which cause oxidative stress if not effectively neutralized. Fe-containing enzymes such as peroxidases, catalase, (Hansch and Mendel, 2009), and Fe-containing superoxide dismutases (FeSODs) help to scavenge reactive oxygen to prevent oxidative damage (Pilon et al., 2011). Some Fe- or Cu-containing enzymes can be functionally interchanged, such as Cu-containing plastocyanin for Fe-containing cytochrome c6 (Raven et al., 1999), and Cu-containing SODs (CuSODs) for FeSOD (Puig et al., 2007; Burkhead et al., 2009).
Several studies have shown that Fe-deficient plants accumulate additional Cu in leaf tissues, including both grasses (Chaignon et al., 2002; Suzuki et al., 2006) and dicots (Welch et al., 1993; Delhaize, 1996; Valdés-López et al., 2010; Waters and Troupe, 2012). The Arabidopsis mutants ysl1ysl3 and opt3, which both exhibit Fe deficiency symptoms, also accumulate excess Cu in shoots (Waters et al., 2006; Stacey et al., 2008; Waters and Grusak, 2008). Some metal uptake genes are regulated by both the Fe and Cu status of Arabidopsis thaliana plants, for example FRO3 and COPT2 were up-regulated by both Fe and Cu deficiency in roots (Wintz et al., 2003; Colangelo and Guerinot, 2004; Mukherjee et al., 2006; Buckhout et al., 2009; Yamasaki et al., 2009; del Pozo et al., 2010; Garcia et al., 2010; Yang et al., 2010), and ZIP2 was up-regulated under Cu deficiency (Wintz et al., 2003) and down-regulated under Fe deficiency (Yang et al., 2010; Stein and Waters, 2011). In rosettes, both the FeSOD gene FSD1 and the CuSOD genes CSD1 and CSD2 are regulated by Cu levels, with high Cu resulting in increased transcript levels of CSD1 and CSD2, and decreased transcript levels of FSD1 (Cohu and Pilon, 2007; Burkhead et al., 2009). The regulation of CSD2 transcripts is not at the transcriptional level (Yamasaki et al., 2007), but rather at the post-transcriptional level by the action of microRNAs (miRNAs) 398a, b, and c, which also regulate CSD1 and CCS1 (Cu Chaperone for SODs) (Sunkar et al., 2006; Beauclair et al., 2010).
Although consequences of Fe deficiency are manifested largely in leaf tissues through effects on photosynthetic capacity and chloroplast development (Spiller and Terry, 1980; Terry, 1980, 1983), most plant Fe deficiency research in recent years has focused on regulation of root responses, uptake mechanisms, and genes involved in increased Fe uptake capacity. This research has greatly increased our understanding of Fe uptake; however, little information is available regarding shoot Fe deficiency responses at the transcriptome level. Likewise, similarities or differences in leaf and root responses to Fe deficiency are largely unknown.
Natural variation in the genomes and phenotypes between accessions or ecotypes within a species has proven to be useful for understanding the relationships between genetic make-up, gene expression, and phenotypic expression of traits. Differences in diverse parental lines and their offspring have long been exploited to understand the underlying genes in quantitative trait locus (QTL) mapping (Doerge, 2002), and more recently expression QTL (eQTL) mapping (Kliebenstein, 2009; Delker and Quint, 2011). These approaches rely on gene expression differences and/or underlying genetic polymorphisms of various types. Likewise, different ecotypes have been shown to have differing constitutive transcriptomes (Kliebenstein et al., 2006; Delker and Quint, 2011), and differing transcriptomes in response to various stimuli or stresses, including salicylic acid (van Leeuwen et al., 2007), auxin (Delker et al., 2010), Pseudomonas syringae type III effector protein (Van Poecke et al., 2007), and drought (Juenger et al., 2010). In previous work, widely diverging transcriptomes were found in Fe-deficient roots of five Arabidopsis ecotypes (Stein and Waters, 2011), and it was proposed that the genes that were commonly expressed in all or most of the ecotypes in response to Fe deficiency were probably the most robust and important for the Fe deficiency response.
Here, gene expression changes in rosettes of Fe-deficient A. thaliana plants of two ecotypes, Kas-1 and Tsu-1, are profiled. These ecotypes were previously shown to respond to Fe deficiency on different time scales, with Kas-1 responding more rapidly than Tsu-1 (Stein and Waters, 2011). Expression changes in miRNAs were also examined in rosettes and roots. Sets of genes that are Fe responsive in rosettes, and those that respond to Fe in both rosettes and roots are presented. A clear link between Fe and Cu was indicated in both the microarray and miRNA results, and follow-up experiments confirmed this Fe–Cu cross-talk. The present results indicate that a major theme in shoot responses to Fe deficiency involves interactions with Cu accumulation that facilitate substitution of Fe-containing enzymes with Cu-containing enzymes.
Materials and methods
Plant materials and growth
Seeds of the A. thaliana ecotypes used in this study, Col-0, Kas-1, and Tsu-1, and the ccs1 mutant SALK_025986c (Chu et al., 2005) were obtained from the Arabidopsis Biological Resource Center (The Ohio State University). Seeds were imbibed in 0.1% agar at 4 °C for 72h. Seeds were planted onto rockwool loosely packed into 1.5ml centrifuge tubes with the bottoms removed. The tubes were floated in foam rafts in containers of nutrient solution, composed of: 0.8mM KNO3, 0.4mM Ca(NO3)2, 0.3mM NH4H2PO4, 0.2mM MgSO4, 50 µM Fe(III)-EDDHA, 25 µM CaCl2, 25 µM H3BO3, 2 µM MnCl2, 2 µM ZnSO4, 0.5 µM CuSO4, 0.5 µM Na2MoO4, and 1mM MES buffer (pH 5.5). Lighting was provided at a photoperiod of 16h of 150 µmol m–2 s–1 4100K fluorescent light (on at 06:00h and off at 22:00h). After 10 d, seedlings and the tubes were transferred to holes in lids of containers containing 0.75 litres of the same nutrient solution with constant aeration for an additional 14 d before plants were transferred to treatments. The +Fe solution contained 50 µM Fe(III)-EDDHA (Sprint 138, Becker-Underwood, Ames, IA, USA). Fe was omitted for the –Fe treatments, and Cu was omitted for the –Cu treatments. All experimental treatments were initiated at 14:00h, 8h before the end of the photoperiod. For microarrays, treatments lasted 24h or 48h. For the miRNA time course, Tsu-1 and Kas-1 samples were collected at time points ranging from 2h to 72h, as indicated in the figures. For miRNA and gene expression under Fe, Cu, and simultaneous Fe and Cu deficiency, Col-0 plants were treated for 72h before sample collection.
Mineral measurements
Roots and rosettes were dried at 60 °C for at least 72h and weighed. Samples were digested with 3ml of concentrated HNO3 (VWR, West Chester, PA, USA, Trace metal grade) at room temperature overnight, then at 100 °C for 1.5h, followed by addition of 2ml of 30% H2O2 (Fisher Scientific, Fair Lawn, NJ, USA) and digestion for 1h at 125 °C, and finally heating the samples to dryness at 150 °C. Dried samples were then resuspended in 5ml of 1% HNO3, and Fe, Zn, and Cu were quantified by inductively coupled plasma mass spectrometry (ICP-MS).
Microarrays and bioinformatics
The rosettes used for microarrays were from the same plants for which root microarrays were performed (Stein and Waters, 2011). Total RNA was isolated from whole rosettes using the RNeasy Plant Kit (Qiagen, Hilden, Germany), and quality was assessed using the Agilent Bioanalyzer 2100 (Agilent). The Affymetrix GeneChip Arabidopsis ATH1 Genome Array was used for microarray analysis, with three biological replicates for each treatment and time point (+Fe, –Fe 24h, –Fe 48h), and each ecotype (Kas-1 and Tsu-1), for a total of 18 arrays. A 5 µg aliquot of DNase I- (Qiagen) treated RNA was used, and all further procedures (hybridization, washing, staining, and scanning) were carried out at the Genomics Core Research Facility of the University of Nebraska according to the manufacturer’s instructions. Probe intensities were imported into R/Bioconductor for analysis, along with probe intensities from Kas-1 and Tsu-1 roots (Stein and Waters, 2011), which were re-analysed for direct comparison. Initial probe intensities were normalized using the robust multichip average algorithm, and differentially expressed genes were identified using the limma package within Bioconductor. Multiple t-testing correction was done using Benjamini and Hochberg’s false discovery rate (FDR). A gene was declared differentially expressed if its corrected P-value was <0.05 in addition to a linear fold change of 2.0. The complete data set is available as GEO Series accession no. GSE39268. The rosette control and Fe deficiency data set from Col-0 (Schuler et al., 2011) was analysed as described above. A union set of Fe deficiency-regulated genes from Col-0 roots was assembled from previously published Affymetrix ATH1 microarray data sets (Colangelo and Guerinot, 2004; Dinneny et al., 2008; Garcia et al., 2010; Long et al., 2010; Yang et al., 2010). Sets of genes were identified using Venny software (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
For miRNA microarrays, miRNA-enriched total RNA was isolated from Kas-1 roots and rosettes 24h after transferring plants to +Fe or –Fe solutions, using the miRNeasy kit (Qiagen). Replicate samples were tested for quality by Agilent Bioanalyzer 2100. Samples labelled and hybridized to miRNA microarrays containing known miRNAs in the Sanger miRbase, Release 14.0, using a dye-swap design by LC Sciences (Houston, TX, USA). Hybridization images were quantified, and data were analysed after subtraction of the background and signal normalization. The ratio of the two sets of detected signals (log2 transformed, normalized) was determined, and P-values were calculated by t-test. Differentially detected signals were those with P ≤ 0.01.
Over-represented gene ontologies in sets of differentially expressed genes from Kas-1 and Tsu-1 were identified using BiNGO, a Cytoscape plugin. Ontologies were generated for the up- and down-regulated differentially expressed genes independently for each genotype. The GOslim-generic GO annotation was used, with an FDR-corrected P-value threshold of 0.05. The figures were generated using the Cytoscape Hierarchical layout and then adjusted for spacing without changing the graph structure.
Real-time RT-PCR
Total RNA was extracted from roots and rosettes using the Plant Total RNA kit (IBI Scientific, Peosta, IA, USA). A 1 µg aliquot of DNase-treated RNA was used for cDNA synthesis, using the High Capacity cDNA Reverse Transcription kit (ABI, Foster City, CA, USA). cDNA corresponding to 50ng of total RNA was used in a 15 µl real-time PCR performed in a MyIQ (BioRad, Hercules, CA, USA) thermal cycler using SYBR GreenER qPCR SuperMix (Invitrogen Technology, Carlsbad, CA, USA). Reactions were performed and data were analysed according to Stein and Waters (2011). Primer sequences are given in Supplementary Table S1 available at JXB online.
MiRNAs from roots and rosettes were extracted using the miRNeasy® Kit (Qiagen), and 5 µg of miRNA-enriched DNase-treated RNA samples were used for cDNA synthesis using the High Capacity cDNA Reverse Transcription kit (ABI), using the stem–loop priming strategy (Chen et al., 2005). cDNA corresponding to 250ng of miRNA-enriched total RNA was used for 15 µl real-time PCRs. The relative gene expression was assessed using the equation Y=2–ΔCt, where ΔCt = Ct–Fe–Ct+Fe. The forward primers used for the real-time PCR analysis are given in Supplementary Table S1 at JXB online, using the universal miRNA reverse primer (5’-GTGCAGGGTCCGAGGT-3’).
Oxidative stress measurements
Plants (Col-0 and ccs1) were cultivated as described above, and 72h after withdrawal of Fe (–Fe), Cu (–Cu), or both simultaneously (–Fe–Cu), were sprayed with 20 µM methyl viologen in 0.01% Tween-20. After 48h, rosettes were sampled and the lipid peroxidation estimated through the quantification of thiobarbituric acid-reactive substances (TBARS), determined according to Hodges et al. (1999).
Results
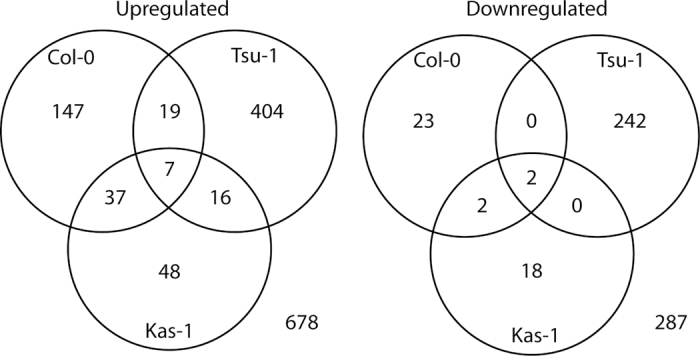
A total of 130 genes exhibited altered expression (≥2.0 fold) under Fe deficiency in Kas-1 rosettes, while Tsu-1 had a much higher number of Fe-regulated genes, 690 in total (Supplementary Fig. S1 at JXB online). Most of these genes were differentially expressed transiently at the 24h time point in Tsu-1. The numbers of up-regulated genes exceeded the number of down-regulated genes. Comparing the Fe regulon between ecotypes, there were largely dissimilar sets of genes with altered expression in Fe-deficient rosettes (Fig. 1), as was observed for Fe deficiency-regulated gene expression in roots (Stein and Waters, 2011). In addition to Kas-1 and Tsu-1, rosette Fe deficiency-regulated genes from Col-0 (Wintz et al., 2003; Schuler et al., 2011) were included in this analysis. For up-regulated genes, the majority (599 of 678; 88%) were observed in only one of the three ecotypes. For down-regulated genes, 98.6% of all differentially regulated genes were observed in rosettes of only one ecotype. The different Fe-regulated transcriptomes between ecotypes were associated with differences in over-represented GO-slim categories (Supplementary Fig. S2). Over-represented categories for up-regulated genes in Tsu-1 rosettes were found for all three primary GO categories. Up-regulated genes in Kas-1 fell into only two of the primary categories. Up-regulated Col-0 genes contained over-represented genes in all three primary GO categories, but were largely different from those of Tsu-1 or Kas-1. For down-regulated genes in Tsu-1 rosettes, all three primary categories contained over-represented classes. This is in contrast to down-regulated genes in Kas-1, where no GO-slim categories were over-represented. Only one category was over-represented for Col-0 down-regulated genes.
Fig. 1.
Three-way Venn diagrams of expression of Fe-regulated genes in rosettes. Numbers represent counts of up-regulated or down-regulated genes in Col-0, Tsu-1, and Kas-1 ecotypes under control or Fe deficiency conditions.
Genes that were up-regulated in both Kas-1 and Tsu-1 are presented in Table 1. Seven genes were up-regulated in Kas-1, Tsu-1, and Col-0 (Table 1), including the ferric-chelate reductase FRO3, the E3 ligase BTS, and the oligopeptide transporter OPT3. A larger number of genes were up-regulated only in two of the three ecotypes, 72 in total (Fig. 2). Several metal-related genes were found in this data set (Table 1), such as the transcription factors bHLH039, bHLH101, and PYE, the nicotianamine (NA) transporter ZIF1, and the nicotianamine synthase gene NAS4. Only two genes were down-regulated in all three ecotypes (Fig. 1, Table 2); the FeSOD FSD1, and the ferritin gene FER4, while two additional genes were down-regulated in Col-0 and Kas-1; the ferritin gene FER1 and the nicotianamine synthase gene NAS3.
Table 1.
Genes up-regulated under Fe deficiency in both Tsu-1 and Kas-1 rosettes Data for Col-0 are included for genes that were differentially expressed under Fe deficiency. Numbers represent fold change under –Fe relative to +Fe.
| Locus | Tsu-1 24 h | Tsu-1 48 h | Kas-1 24 h | Kas-1 48 h | Col-0 5 da | Col-0 8 db | |
|---|---|---|---|---|---|---|---|
| At1g23020 | 18.4 | 10.3 | 1.9 | 2.9 | 3.3 | 6.9 | FRO3; ferric-chelate reductase |
| At3g18290 | 3.5 | 3.3 | 2.7 | 4.8 | BTS; putative E3 ligase protein | ||
| At4g16370 | 2.4 | 3.1 | 2.2 | OPT3; oligopeptide transporter | |||
| At3g27060 | 2.2 | 2.1 | 2.6 | 14.8 | TSO2; ribonucleutide reductase small subunit | ||
| At1g33960 | 3.2 | 2.4 | 12.2 | AIG1; AVRRPT2-induced gene | |||
| At1g47400 | 6.5 | 5.1 | 5.7 | 11.9 | 42.2 | Unknown protein | |
| At5g05250 | 5.8 | 4.5 | 2.7 | 5.6 | 7.5 | Unknown protein | |
| At3g56980 | 10.0 | 2.7 | bHLH039; transcription factor | ||||
| At5g04150 | 8.8 | 2.6 | bHLH101; transcription factor | ||||
| At3g47640 | 2.1 | 2.2 | 2.3 | PYE; bHLH transcription factor | |||
| At5g13740 | 2.3 | 2.4 | 2.1 | ZIF1; zinc-induced facilitator 1 | |||
| At1g56430 | 3.2 | 2.4 | NAS4; nicotianamine synthase | ||||
| At5g53450 | 4.3 | 3.6 | 3.6 | ORG1 (OBP3-RESPONSIVE GENE 1); kinase | |||
| At2g26400 | 2.8 | 2.4 | 2.8 | ARD3; acireductone dioxygenase | |||
| At5g59320 | 5.4 | 5.0 | 2.6 | LTP3; lipid transfer protein | |||
| At1g19250 | 2.6 | 2.5 | 2.5 | FMO1; flavin-dependent monooxygenase | |||
| At5g44420 | 2.3 | 2.4 | LCR77; ethylene- and jasmonate-responsive defensin | ||||
| At2g24850 | 3.4 | 3.3 | 2.3 | TAT3; tyrosine aminotransferase | |||
| At5g24660 | 2.5 | 2.1 | LSU2; response to low sulphur | ||||
| At2g21650 | 2.2 | 2.2 | RSM1; MYB family transcription factor | ||||
| At3g02480 | 2.2 | 2.3 | Late embryogenesis abundant protein (LEA) family protein | ||||
| At3g56360 | 2.1 | 2.1 | Unknown protein | ||||
| At5g67370 | 3.7 | 3.0 | 3.3 | Unknown protein |
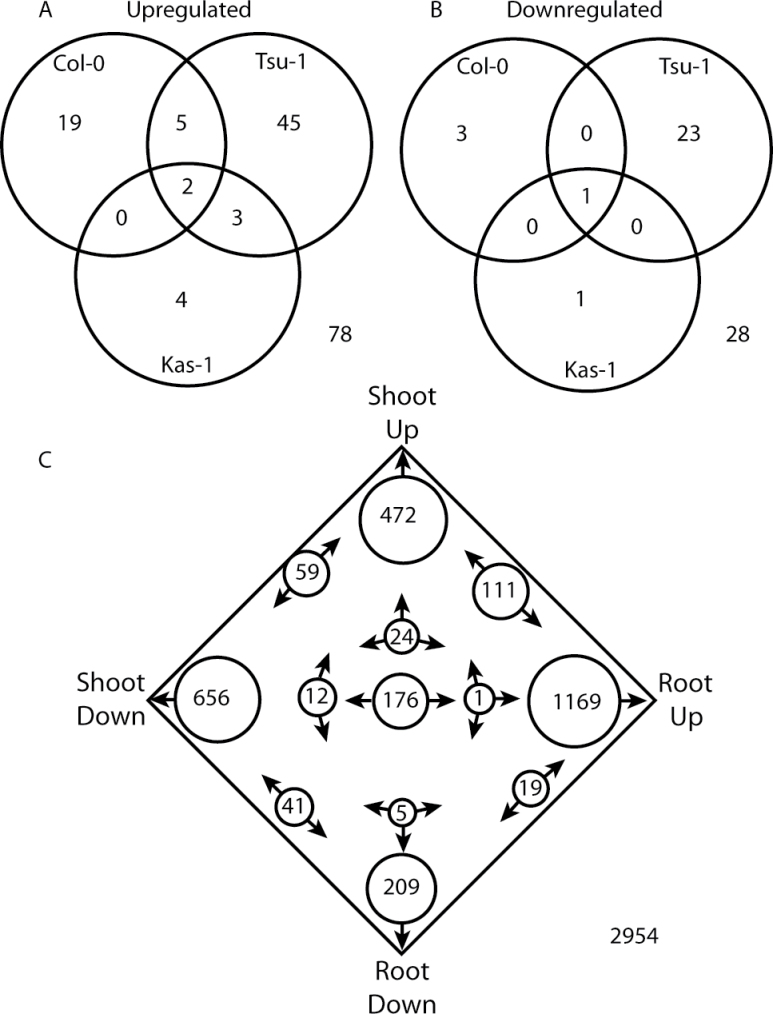
Fig. 2.
Venn diagrams of genes regulated in both rosettes and roots of Arabidopsis in response to Fe deficiency. Numbers represent the counts of genes (A) up-regulated or (B) down-regulated in both tissues specifically in Col-0, Tsu-1, and Kas-1 ecotypes. (C) Four-way Venn diagram of genes that were Fe regulated in any of the Kas-1, Tsu-1, or Col-0 ecotypes.
Table 2.
Genes that were down-regulated in rosettes under Fe deficiency in multiple ecotypes
| Locus | Tsu-1 24 h | Tsu-1 48 h | Kas-1 24 h | Kas-1 48 h | Col-0 5 da | Col-0 8 db | |
|---|---|---|---|---|---|---|---|
| At4g25100 | -9.1 | –34.1 | –51.5 | –144.8 | FSD1; iron superoxide dismutase | ||
| At2g40300 | -2.2 | –2.8 | –3.6 | –4.6 | FER4; ferritin 4 | ||
| At5g01600 | –2.8 | –4.3 | –8.9 | FER1; ferritin 1 | |||
| At1g09240 | –2.1 | –6.3 | NAS3; nicotianamine synthase | ||||
Within each ecotype, Fe-regulated gene expression in rosettes was compared with that of roots (from Stein and Waters, 2011; Table 3). Considering genes that were up-regulated or down-regulated in both roots and rosettes within a given ecotype, a sum total of 78 genes were up-regulated in both tissues across the three ecotypes (Fig. 2). However, only two of these (FRO3 and At1g47400) were up-regulated in all three ecotypes, and eight were Fe up-regulated in two of the three ecotypes (Fig. 2A, Table 3). A total of 28 genes were down-regulated by Fe deficiency in both roots and rosettes within the specific ecotypes, but only one (FSD1) was observed in all three ecotypes (Fig. 2B, Table 3). Union sets of all rosette Fe-regulated genes and all root Fe-regulated genes were also made, regardless of ecotype source, and these root and shoot sets were then compared. In this comparison, 111 genes were up-regulated in both roots and rosettes, while 41 were down-regulated in both roots and rosettes (Fig. 2C). There were also 188 genes (6.4%) that had opposite patterns of Fe regulation in rosettes and roots (up-regulated in one tissue and down-regulated in the other). Two genes that were up-regulated in roots and down-regulated in rosettes are IREG3 (roots 2.9-fold, Tsu-1 48h; rosettes –2.7-fold, Tsu-1 24h) and the copper transporter COPT2 (roots 2.6-fold, Tsu-1 48h; rosettes –2.4-fold, Kas-1 24h). Another 108 genes (3.7%) had contradictory expression patterns (e.g. found to be both up-regulated or down-regulated in different time points or ecotypes). The majority of Fe-regulated genes (2584, 87%) were expressed in a tissue-specific manner. Many of the genes that were Fe regulated similarly in both roots and leaves are known metal-related genes (e.g. involved in metal transport or homeostasis) (Table 3) that were differentially expressed under Fe deficiency in rosettes in multiple ecotypes (Table 1). Some additional genes that were expressed similarly in both tissues include the metal transporter NRAMP4, the copper chaperone CCH, and the metal–NA transporter YSL2.
Table 3.
Genes up- or downregulated in both roots and rosettes of multiple ecotypes
| Roots | Shoots | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tsu-1 24 h | Tsu-1 48 h | Kas-1 24 h | Kas-1 48 h | Col-0 (Garcia) | Col-o (Yang) | Col-0 (Long) | Col-0 (Colangelo) | Tsu-1 24 h | Tsu-1 48 h | Kas-1 24 h | Kas-1 48 h | Col-0 (Schuler) | ID | ||||
| Up-regulated in all ecotypes | |||||||||||||||||
| At1g47400 | 3.5 | 7.2 | 6.2 | 13.6 | 5.2 | 6.5 | 5.1 | 5.7 | 11.9 | 42.2 | Unknown protein | ||||||
| At1g23020 | 2.9 | 4.7 | 2.6 | 7.8 | 4.8 | 18.4 | 10.3 | 1.9 | 2.9 | 6.9 | FRO3; ferric-chelate reductase | ||||||
| Up-regulated in Col-0 and Tsu-1 | |||||||||||||||||
| At2g18690 | 2.4 | 2.5 | 5.7 | Unknown protein | |||||||||||||
| At2g43570 | 2.0 | 14.9 | 2.8 | 8.0 | Endochitinase isologue | ||||||||||||
| At3g18290 | 3.0 | 2.7 | 2.2 | 3.5 | 3.3 | 2.7 | 4.8 | BTS; putative E3 ligase protein | |||||||||
| At5g05250 | 3.3 | 6.6 | 6.0 | 3.0 | 5.8 | 4.5 | 2.7 | 5.6 | 7.5 | Unknown protein | |||||||
| At5g42830 | 2.6 | 2.5 | 2.7 | HXXXD-type acyl-transferase family protein | |||||||||||||
| Up-regulated in Kas-1 and Tsu-1 | |||||||||||||||||
| At1g19250 | 2.6 | 2.6 | 2.5 | 2.5 | FMO1; flavin-containing monooxygenase | ||||||||||||
| At3g56980 | 5.6 | 10.5 | 2.8 | 24.5 | 9.5 | 10.0 | 2.7 | BHLH039; DNA binding/transcription factor | |||||||||
| At5g53450 | 3.3 | 2.1 | 5.3 | 4.4 | 4.3 | 3.6 | 3.6 | ORG1 (OBP3-responsive gene 1); protein kinase | |||||||||
| Down-regulated in all ecotypes | |||||||||||||||||
| At4g25100 | –14.0 | –27.8 | –5.3 | –9.1 | –34.1 | –51.5 | –144.8 | FSD1; Fe superoxide dismutase | |||||||||
| Metal related genes up- or down-regulated in both roots of any ecotype and rosettes of any ecotype | |||||||||||||||||
| At1g56430 | 3.5 | 4.8 | 2.9 | 3.2 | 2.4 | NAS4; nicotianamine synthase | |||||||||||
| At3g47640 | 2.6 | 1.5 | 2.1 | 2.2 | 2.3 | PYE; bHLH transcription factor | |||||||||||
| At4g16370 | 2.6 | 6.4 | 7.3 | 5.2 | 2.4 | 3.1 | OPT3; oligopeptide transporter | ||||||||||
| At5g04150 | 11.3 | 13.9 | 4.1 | 8.8 | 2.6 | bHLH101; DNA binding/transcription factor | |||||||||||
| At5g13740 | 2.5 | 2.6 | 2.5 | 2.3 | 2.4 | 2.1 | ZIF1; nicotianamine transporter | ||||||||||
| At5g43450 | 100.2 | 5.8 | 2.7 | 1-Aminocyclopropane-1-carboxylate oxidase | |||||||||||||
| At5g47220 | 2.1 | 2.0 | ERF2 (Ethylene Responsive Element binding Factor 2); transcription factor | ||||||||||||||
| At5g47910 | 3.0 | 4.2 | 2.9 | 3.8 | 2.4 | RBOHD; NAD(P)H oxidase | |||||||||||
| At5g67330 | 3.7 | 2.2 | 2.0 | 3.4 | 3.2 | ATNRAMP4; metal ion transmembrane transporter | |||||||||||
| At2g40300 | –3.4 | –1.5 | –2.2 | –2.8 | –4.6 | ATFER4; ferritin | |||||||||||
| At3g56240 | -2.9 | –1.7 | –2.0 | CCH; copper chaperone | |||||||||||||
| At5g01600 | –3.8 | –6.7 | –2.5 | –2.8 | –8.9 | FER1; ferritin | |||||||||||
| At5g24380 | –4.1 | –2.1 | YSL2 (Yellow Stripe-Like 2); oligopeptide transporter | ||||||||||||||
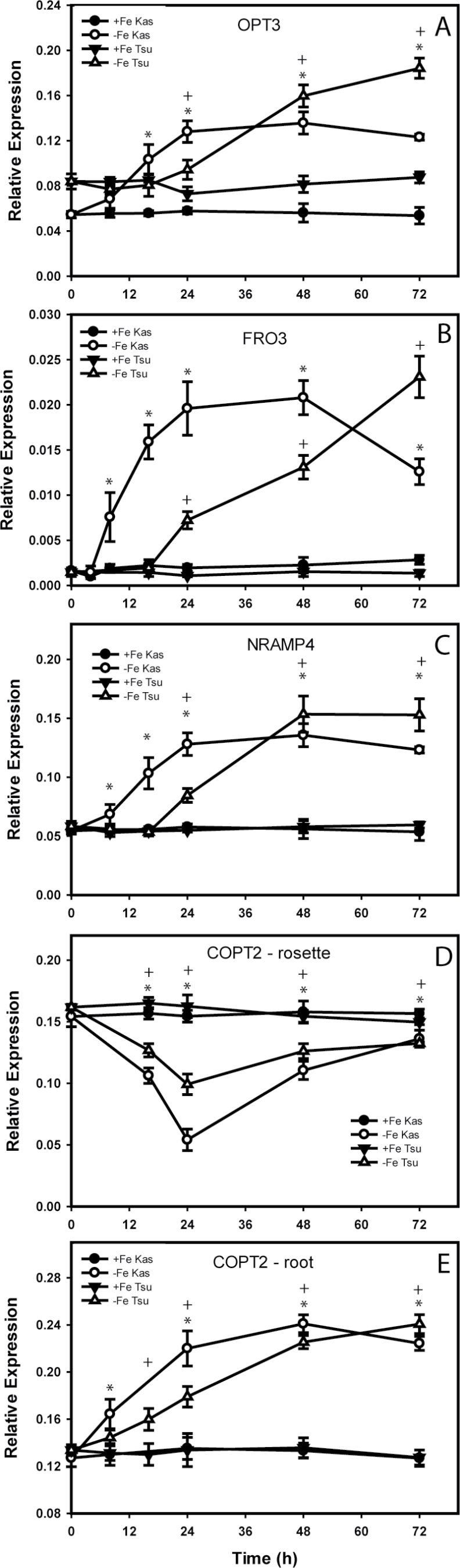
To validate the microarray results, real-time RT-PCR was performed for some of the genes regulated by Fe in both roots and rosettes, using RNA from Kas-1 and Tsu-1 rosettes over a time course. This experiment also served to test whether the difference in timing of response after Fe withdrawal that was observed in roots (Stein and Waters, 2011) was also present in rosettes. Expression of OPT3, FRO3, and NRAMP4 was up-regulated at earlier time points in Kas-1 than in Tsu-1 (Fig. 3). OPT3 was up-regulated on a similar time scale in rosettes to that in roots, whereas NRAMP4 and FRO3 were up-regulated earlier in rosettes than in roots; in Kas-1 at 8h in rosettes for both genes, compared with 16h for NRAMP4 in roots and 48h for FRO3 in roots. As indicated by the array results, COPT2 expression showed opposite responses in roots and rosettes. In roots, COPT2 was up-regulated in Kas-1 by 8h and in Tsu-1 by 16h, whereas in rosettes expression of this gene decreased in both ecotypes from 16h onwards, with a maximum decrease at 24h before recovering to nearly normal levels by 72h.
Fig. 3.
Time course of expression of metal homeostasis-related genes in Kas-1 and Tsu-1 ecotypes. (A) OPT3, (B) FRO3, (C) NRAMP4, (D) COPT2 in rosettes, and (E) COPT2 in roots. n=3 ±SD. *Denotes statistical significance for Kas-1, + denotes statistical significance for Tsu-1 (P < 0.05) between treatments at each time point. +Fe, 50 µM Fe; –Fe, no added Fe.
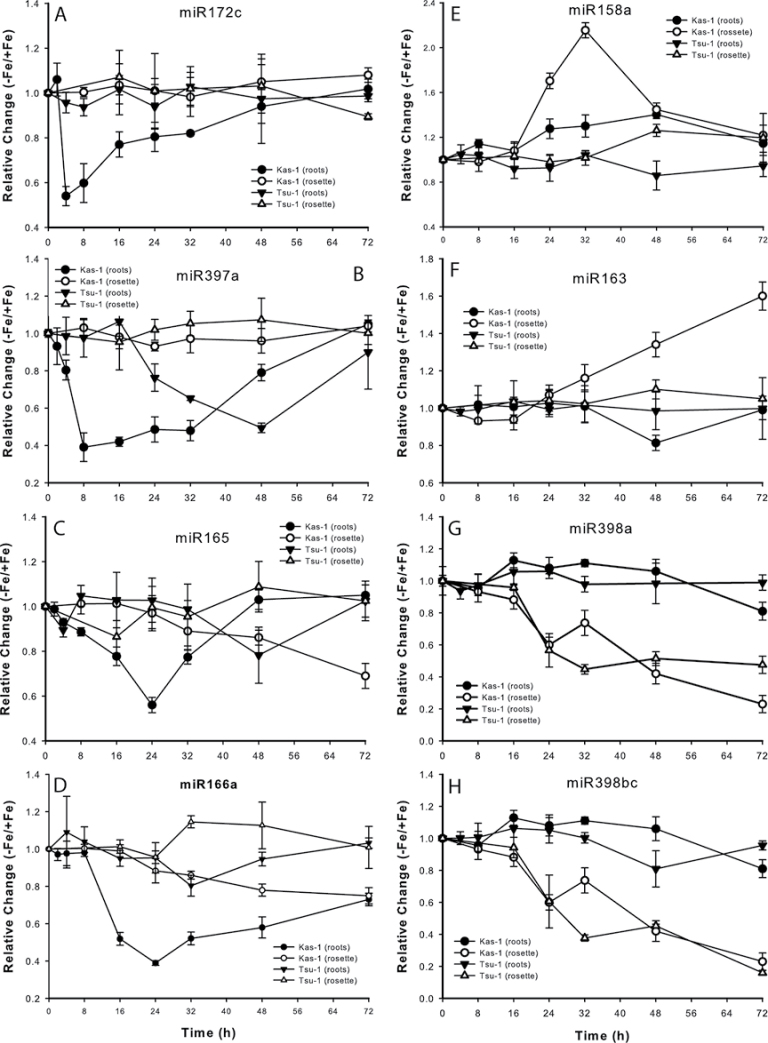
To define further the molecular activity of early responses to Fe deficiency, miRNA expression differences were profiled in roots and rosettes of the early-responding Kas-1 ecotype, at 24h after removal of Fe as compared with expression in Fe-replete plants. These miRNA microarrays indicated that eight miRNAs were significantly differentially regulated after 24h of Fe deficiency. The abundance of these eight miRNAs was quantified by real-time RT-PCR in both Kas-1 and Tsu-1 roots and rosettes over a time course beginning at 2h after Fe removal from the nutrient solution (Fig. 4). Four miRNAs, 172c, 397a, 165a, and 166a, had altered levels in the roots. miR172c decreased in Kas-1 at 4h and then gradually increased to normal levels, while there was no change in expression in Tsu-1. miR397a decreased in both ecotypes, but had an earlier decline then recovery in Kas-1 than in Tsu-1, reaching minimal values in Kas-1 at 8h and in Tsu-1 at 48h. miR165a and 166a had similar expression patterns, in that they decreased then recovered in Kas-1 while decreasing more slowly and steadily in Tsu-1.
Fig. 4.
Relative changes (–Fe/+Fe) in expression for miRNAs in Kas-1 and Tsu-1 roots and rosettes in response to Fe deficiency. (A) miR172c, (B) miR397a, (C) miR165, (D) miR166a, (E) miR158a, (F) miR163, (G) miR398a, and (H) miR398b/c. n=3 ±SD. 50 µM Fe; –Fe, no added Fe.
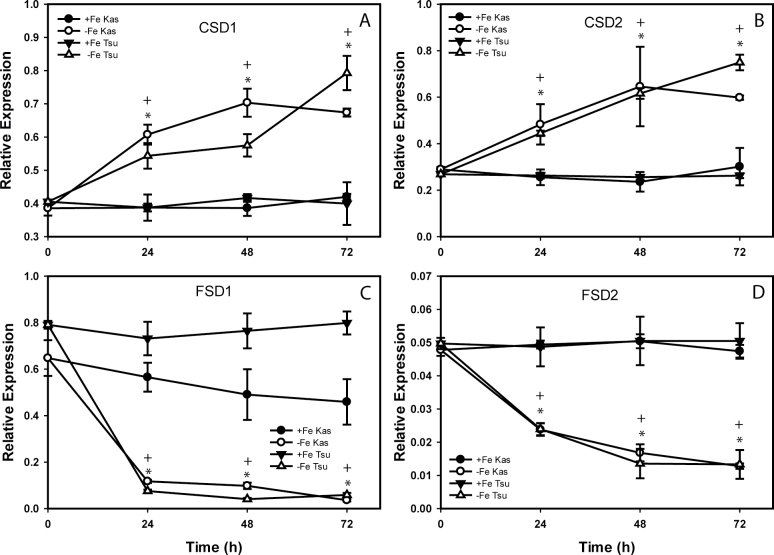
The remaining four miRNAs had altered levels in Fe-deficient rosettes (Fig. 4). MiR158a had a transient increase at 24h and 32h in Kas-1, and gradually increased by 26% in Tsu-1. MiR163 increased at later time points in Kas-1, while this miRNA did not have strong changes in Tsu-1. The miR398s, a and b/c (which are identical at maturity but are encoded by different genes), decreased beginning at 16h in both ecotypes, on a similar time frame, ending at ~20% of the starting abundance. The microarray results were next checked for expression differences for known or predicted (Bonnet et al., 2010) targets of the miRNAs. Several of the target genes were Fe regulated either in roots or in rosettes (Table 4). Some of the most well characterized of these miRNAs, miR398a and b/c, decreased in abundance in rosettes, and had several targets (CSD1, CSD2, and CCS1) that were up-regulated in Fe-deficient rosettes. The known targets of miR398 and miR397 include transcripts for Cu-containing proteins (Sunkar et al., 2006; Yamasaki et al., 2007; Beauclair et al., 2010). Expression of the SOD genes CSD1 and CSD2, and FSD1 and FSD2, were then measured over a time course in Kas-1 and Tsu-1 rosettes in response to withdrawal of Fe (Fig. 5). Unlike OPT3, NRAMP4, and FRO3, the SOD genes responded on similar time scales in both Kas-1 and Tsu-1. Both CSD1 and CSD2 increased to nearly 2-fold in both ecotypes, while FSD1 and FSD2 decreased in both ecotypes. FSD1 decreased to very low levels by 24h, while FSD2 decreased more slowly.
Table 4.
Expression differences of known or potential miRNA targets on Kas-1 and Tsu-1 microarrays Numbers represent fold chage under –Fe relative to +Fe. Root expression data are from Stein and Waters (2011).
| miRNA | Roots | Roots | Rosettes | Rosettes | Roots | Roots | Rosettes | Rosettes | Rosettes | Locus | Description |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tsu-1 24 h | Tsu-1 48 h | Tsu-1 24 h | Tsu-1 48 h | Kas-1 24 h | Kas-1 48 h | Kas-1 24 h | Kas-1 48 h | Col-0 | |||
| miR172c | –1.7 | –2.2 | 1.9 | At3g14770 | Nodulin MtN3 family protein | ||||||
| miR397a | 2.0 | At2g29130 | LAC2; laccase | ||||||||
| miR397a | –1.7 | At5g60020 | LAC17; laccase | ||||||||
| miR158a | 2.4 | At1g64100 | Pentatricopeptide repeat-containing protein | ||||||||
| miR158a | 2.0 | 3.3 | 6.8 | At1g49910 | Pentatricopeptide repeat-containing protein | ||||||
| miR163 | 22.5 | At3g44860 | Farnesoic acid O-methyltransferase | ||||||||
| miR163 | 2.3 | 3.2 | –2.0 | At1g66690 | S-Adenosyl-l-methionine-dependent methyltransferases superfamily protein | ||||||
| miR398a, b/c | 4.4 | 3.4 | At2g28190 | CSD2; copper/zinc superoxide dismutase | |||||||
| miR398a, b/c | 2.5 | 2.5 | At1g08830 | CSD1; copper/zinc superoxide dismutase | |||||||
| miR398a, b/c | 3.3 | 3 | 1.9 | 1.7 | 2.7 | At1g12520 | CCS1; copper/zinc superoxide dismutase copper chaperone | ||||
Fig. 5.
Fe deficiency regulation of SOD genes in Kas-1 and Tsu-1 rosettes. (A) CSD1, (B) CSD2, (C) FSD1, and (D) FSD2. n=3 ±SD. *Denotes statistical significance for Kas-1, + denotes statistical significance for Tsu-1 (P < 0.05) between treatments at each time point. +Fe, 50 µM Fe; –Fe, no added Fe.
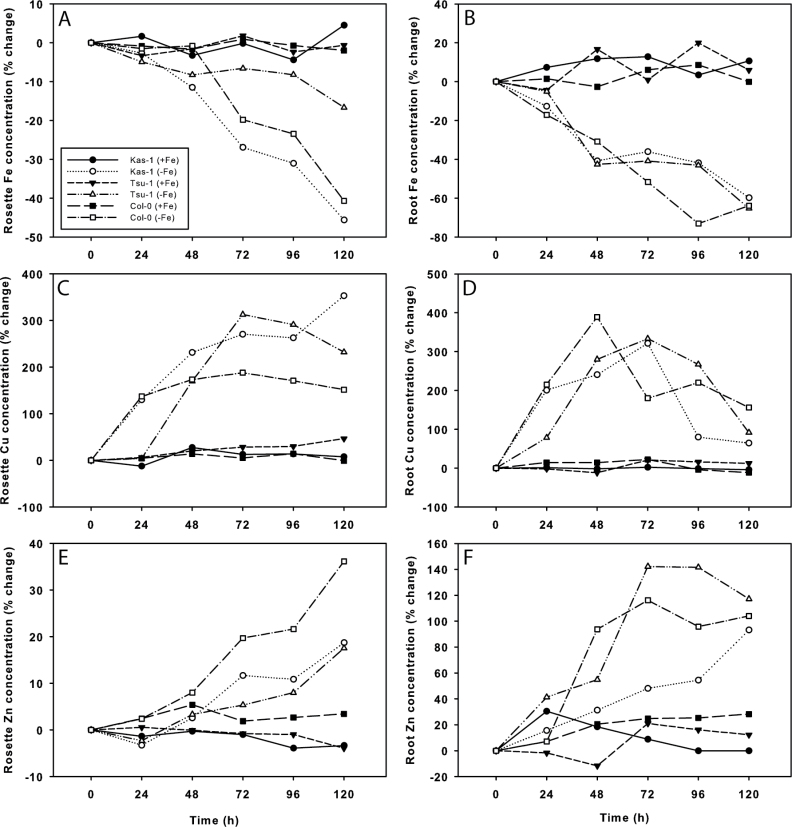
Since CSD1 transcript levels increased and FSD1 levels decreased, a regulation pattern previously shown to be mediated by miR398s in response to high Cu (Yamasaki et al., 2007), the concentrations of the metal micronutrients Cu, Fe, and Zn were measured over a time course in Kas-1, Tsu-1, and Col-0 rosettes and roots (Fig. 6). After Fe was withdrawn, there was no change in bulk rosette or root Fe concentration until 48h. Rosette Fe concentration in Kas-1 and Col-0 decreased rapidly from 48h to 120h, while Tsu-1 concentrations declined more slowly and were not significantly lower until 120h. Root Fe concentrations declined similarly in the three ecotypes. However, the Cu concentration rapidly increased in both roots and rosettes. In Kas-1 and Col-0, the rosette Cu concentration more than doubled within the first 24h, whereas Cu increased in Tsu-1 later, at 48h. Root Cu concentration rapidly increased in all three ecotypes until 48h or 72h, then afterwards declined, but remained elevated compared with +Fe roots. Root Zn concentration gradually doubled, and also increased in rosettes of all three ecotypes, but by much lower percentages than the increase in Cu concentration.
Fig. 6.
Time course changes in metal concentration of Arabidopsis Kas-1, Tsu-1, and Col-0 ecotypes in response to Fe deficiency. Iron concentration in (A) rosettes, (B) roots; Cu concentration in (C) rosettes, (D) roots; Zn concentration in (E) roots, and (F) rosettes. n=3; +Fe, 50 µM Fe; –Fe, no added Fe.
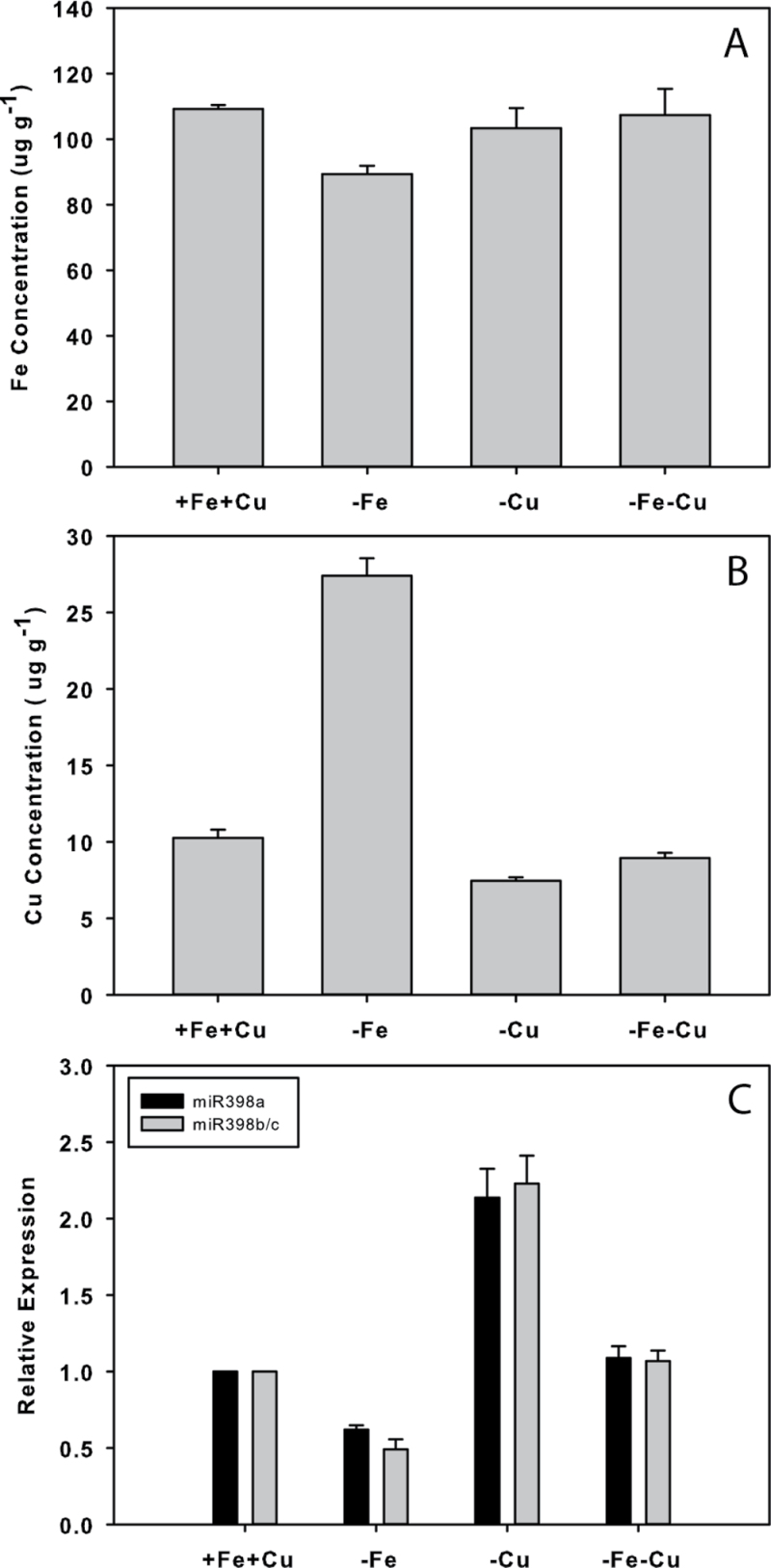
To test whether the changes in miR398 were driven by Fe deficiency or by accumulation of Cu in response to Fe status, Fe, Cu, or both metals were withheld for 3 d before measuring the abundance of miR398s and Fe and Cu concentrations in Col-0 rosettes (Fig. 7). Similar to the time course, under Fe deficiency rosette Fe concentration decreased by 18%, while Cu increased by 166%. Under this treatment, miR398a decreased by >30%, and miR398b/c decreased by >50%. Withholding Cu had no effect on Fe concentration, but led to a decrease in Cu concentration of 27%, while miR398s increased by >100%. Removing both Fe and Cu resulted in no change in Fe and a small decrease in Cu concentration, and a very slight increase in miR398a and miR398b/c (9% and 7%).
Fig. 7.
Responses to Fe and/or Cu deficiency in Col-0 rosettes. Changes in (A) Fe and (B) Cu concentration. (C) Changes in miR398a and miR398b/c abundance. – Fe, no added Fe; –Cu no added Cu; –Fe–Cu, omission of both.
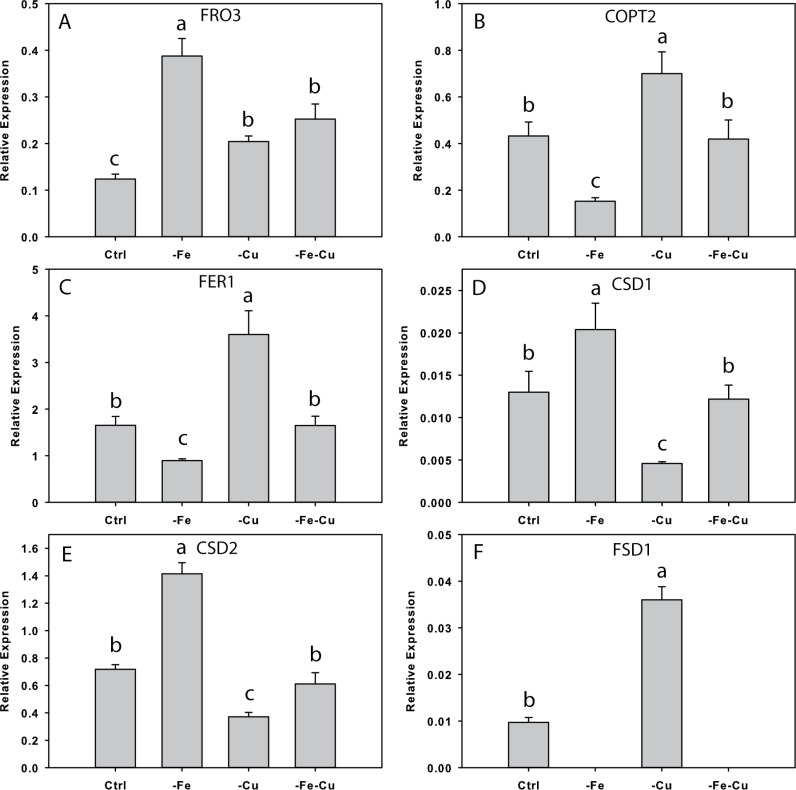
The effect of Fe and Cu deficiency treatments on genes that responded to Fe deficiency that are or may also be regulated by Cu was then investigated (Fig. 8). After 3 d, FRO3 was up-regulated 3-fold under Fe deficiency, 1.7-fold under Cu deficiency, and 2-fold under both Cu and Fe deficiency. COPT2 transcript levels decreased under Fe deficiency and increased under Cu deficiency, similar to FER1, whereas CSD1 and CSD2 increased under Fe deficiency and decreased under Cu deficiency. Transcript levels of these three genes were unchanged when both Fe and Cu were withheld. FSD1 decreased to undetectable levels under both –Fe and –Fe–Cu treatments, and increased under Cu deficiency.
Fig. 8.
Fe and/or Cu regulation of gene expression in Col-0 rosettes. (A) FRO3, (B) COPT2, (C) FER1, (D) CSD1, (E) CSD2, and (F) FSD1; n=3 ±SD. Different letters denote statistical significance by ANOVA (P < 0.05), followed by Duncan’s test. Ctrl, 50 µM Fe and 0.5 µM Cu; – Fe, no added Fe and 0.5 µM Cu; –Cu, 50 µM Fe and no added Cu; –Fe–Cu, omission of both.
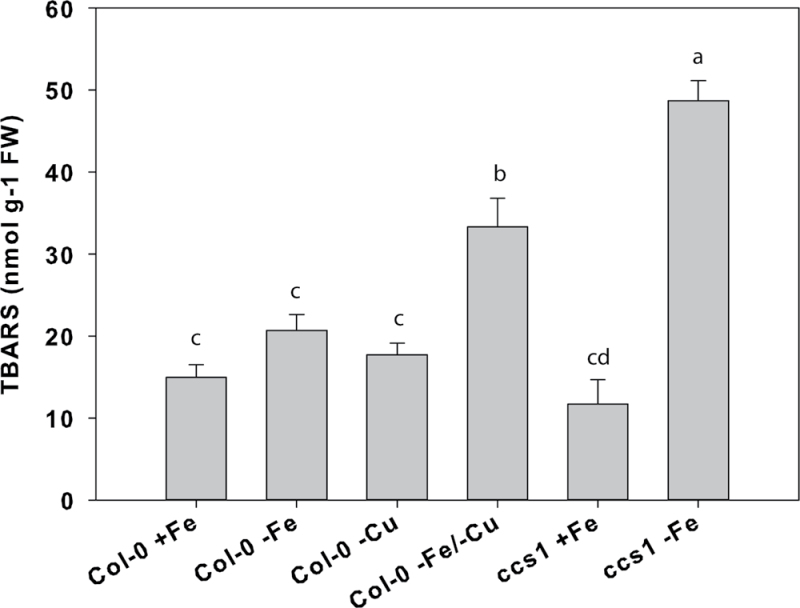
Since SODs function to protect against oxidative stress, a lipid peroxidation assay was used following treatment of rosettes with methyl viologen to measure the capacity of plants to scavenge reactive oxygen species when grown under deficiencies of Fe, Cu, or both metals (Fig. 9). Deficiencies of Fe or Cu resulted in slight increases in formation of TBARS, indicating a compromise in reactive oxygen species protection. However, deficiencies of both metals resulted in a >2-fold increase in formation of TBARS in Col-0 rosettes. The effect of Fe deficiency on reactive oxygen species scavenging was then tested in the ccs1 mutant, which is defective in the copper chaperone for SODs that is essential for the insertion of Cu into the apoproteins to form functional CuSOD proteins (Chu et al., 2005). Under Fe-replete conditions, this mutant had no increase in formation of TBARS relative to Col-0, but under Fe deficiency the plants were less able to scavenge reactive oxygen, as lipid peroxidation was 4-fold greater.
Fig. 9.
Lipid peroxidation in rosettes of Arabidopsis plants after 3 d growth on –Fe–Cu, or –Fe–Cu solution. n=3 ±SD. Different letters denote statistical significance by ANOVA (P < 0.05), followed by Duncan’s test. +Fe, 50 µM Fe and 0.5 µM Cu; – Fe, no added Fe and 0.5 µM Cu; –Cu, 50 µM Fe and no added Cu; –Fe–Cu, omission of both.
Discussion
Rosette Fe deficiency-regulated genes
To date, little was known about alterations in gene expression in Fe-deficient Arabidopsis leaves and rosettes. The first of such studies used a custom array to study changes in expression of metabolism-related genes (Thimm et al., 2001). Later, the 8300 gene Affymetrix Arabidopsis GenChip was used for profiling transporter genes in Fe-, Cu-, and Zn-deficient shoots and roots of Col-0 (Wintz et al., 2003), and these authors reported three genes that were up-regulated and three genes that were down-regulated in leaves in response to Fe deficiency. Using the Affymetrix ATH1 microarray, 108 and 446 up-regulated, and 22 and 244 down-regulated genes were identified in Kas-1 and Tsu-1, respectively (P ≤ 0.05, absolute fold change ≥2) (Supplementary Fig. S1 at JXB online). Clearly, there are differences in transcriptome-level responses between these ecotypes, and also between Kas-1, Tsu-1, and Col-0 grown by another research group (Schuler et al., 2011), as was observed in Fe-deficient roots (Stein and Waters, 2011). Kas-1 and Tsu-1 grown side by side had largely non-overlapping sets of Fe-regulated genes in rosettes (Fig. 1), and these sets also mostly did not overlap with those of Fe-deficient Col-0. This study seeks to take advantage of the diverse responses of multiple ecotypes to identify robustly Fe-regulated genes that respond similarly in multiple ecotypes. These common genes represent responses that are conserved across disparate genotypes and are likely to be the most important universal responses to Fe deficiency (Stein and Waters, 2011). The Kas-1 and Tsu-1 ecotypes are known to have constitutive differences in expression of nearly 6000 genes in leaves, and >350 genes responded differently to soil drying between these ecotypes (Juenger et al., 2010). Another cause of the differences in Fe-regulated genes in Kas-1 and Tsu-1 may be the delayed responses to Fe deficiency observed in Tsu-1, for example in root ferric reductase activity and expression of several Fe uptake genes (Stein and Waters, 2011). In rosettes, delayed expression of several genes (Fig. 2), a delayed decrease in Fe concentration after Fe withdrawal, and delayed Cu accumulation were also observed (Fig. 6). Thus, it is possible that primarily earlier responding genes were captured in the Tsu-1 data set compared with Kas-1 or Col-0.
Comparison of shoot and root Fe regulons
The genes listed in Table 1 represent key Fe deficiency response genes in Arabidopsis rosettes, and indicate that a major theme in this response is alteration of metal homeostasis. Comparing the Kas-1, Tsu-1, and Col-0 data, there were several genes that had altered expression in response to Fe deficiency in all three ecotypes, namely FRO3, OPT3, BTS, and the ‘unknown protein’ gene At1g47400. Interestingly, several of the Fe-regulated genes in rosettes were also regulated by Fe deficiency in roots (Table 3). It is not clear at this point whether genes that are Fe regulated in both organs have identical roles in roots and rosettes, but for many of these genes that would not be unexpected. Genes such as FRO3, OPT3, NRAMP4, ZIF1, and BTS were all associated with the basic helix–loop–helix (bHLH) transcription factor PYE in roots (Long et al., 2010), suggesting that the PYE network may carry out similar functions in leaves. The OPT3 gene is likely to be involved in phloem transport of Fe in various tissues including roots and leaves, as the knockdown line opt3-2 had increased Fe localization in vascular tissues (Stacey et al., 2008). ZIF1 and NRAMP4 are both involved in vacuolar function and metal compartmentalization. ZIF1 has been implicated in transport of NA into vacuoles, which leads to Zn sequestration, while knockouts or overexpression lines of ZIF1 had disrupted distribution of Fe between roots and shoots (Haydon et al., 2012). NRAMP4 is involved in transporting Fe out of vacuoles (Lanquar et al., 2005), which may be a source of stored Fe that is utilized when Fe supply from roots is no longer available.
The roles of certain consistently Fe-regulated rosette genes, such as At1g47400, At5g05250, and ORG1, in Fe deficiency are not known, but these genes were among the small percentage of those with highly conserved expression changes in response to Fe deficiency in all of the three diverse ecotypes (Table 1). This suggests that important new processes for adaptation to Fe deficiency remain to be discovered, and the method used here for filtering for genes that respond to stimuli in multiple diverse ecotypes may help focus future research on genes with universal responses.
Other notable gene expression responses in rosettes and roots were down-regulation of ferritins FER1 and FER4, and FeSOD FSD1. These changes should result in decreased synthesis and subsequent sequestration of Fe into these proteins. Ferritin proteins decrease in low-Fe leaves (Ravet et al., 2009), which would presumably release stored Fe to replace Fe that is no longer being taken up by roots. FSD1 is known to be up-regulated by Cu, but results in Fig. 8 suggest that Fe may also regulate expression of this gene.
Differences in response time for Kas-1 and Tsu-1 were observed in both tissues for certain genes. In the present real-time RT-PCR time course, up-regulation of OPT3 and NRAMP4 occurred more rapidly in both roots (Stein and Waters, 2011) and rosettes (Fig. 3) of Kas-1 than Tsu-1. Comparing rosette and root tissues, expression levels of some genes respond to Fe deficiency in rosettes more rapidly than they do in roots. NRAMP4 was up-regulated in Kas-1 rosettes within 8h (Fig. 3), while it was not up-regulated in Kas-1 roots until 16h after Fe withdrawal (Stein and Waters, 2011). This more rapid up-regulation in rosettes was most dramatic for FRO3, which was up-regulated in rosettes by 8h (Fig. 3), but not in roots until 48h (Stein and Waters, 2011). This rapid response occurred prior to any decrease in bulk rosette Fe concentration (Fig. 6), and raises interesting questions, such as do Fe perception mechanisms in shoots detect Fe inside of cells or organelles, or is it the response to some other parameter that is sensed, such as the influx of Fe or some other molecule into leaves as it is delivered from roots in the xylem? Do leaves respond to Fe deficiency prior to roots and send a phloem-mobile signal to up-regulate root gene expression? These have been long-running questions in the field, but an interorgan signalling molecule has not been identified. However, miR158a levels were increased in rosettes of both ecotypes. The abundance of this miRNA was strongly increased (log2 2.4-fold) by Fe deficiency in Brassica napus phloem sap (Buhtz et al., 2010). This raises the intriguing possibility that miR158 could be a long-distance phloem-mobile signal between shoots and roots, as is the case for miR395 for sulphur status (Jones-Rhoades and Bartel, 2004; Buhtz et al., 2010) and miR399 for phosphate status (Chiou and Lin, 2011). Increased abundance of miR158a peaked at >2-fold in whole rosettes of Kas-1, while this miRNA increased by ~30% in Tsu-1. The bulk rosette samples may dilute the true levels if this miRNA is primarily localized to the phloem, and more detailed experiments are necessary to address this question.
The differences in expression of several genes in roots and rosettes also suggest that some Fe-responsive genes are regulated by different mechanisms in different tissues, or by a complex interaction of multiple transcription factors. For example, the transcription factor FIT is thought to regulate FRO3 expression in roots (Wu et al., 2005), but FIT is not expressed in leaves (Colangelo and Guerinot, 2004), so another regulatory system must be in place in leaves. In addition to PYE and BTS, the transcription factors bHLH039 and bHLH101 were up-regulated in both Kas-1 and Tsu-1 (Table 1). The bHLH039 protein has been shown to interact with FIT (Yuan et al., 2008), but expression of bHLH039 and bHLH101 is not FIT dependent (Wang et al., 2007). Thus, it is possible that one or a combination of these proteins regulates the leaf Fe regulon. Another root Fe- and FIT-regulated gene, COPT2 (Colangelo and Guerinot, 2004; Buckhout et al., 2009; Garcia et al., 2010; Yang et al., 2010), was up-regulated in roots in the time course, but down-regulated in rosettes. This may reflect an opposite regulation of COPT2 by Fe deficiency in roots with simultaneous down-regulation in leaves in response to increased Cu accumulation in rosettes, since COPT2 is also regulated by Cu (Sancenon et al., 2003; Yamasaki et al., 2009; del Pozo et al., 2010).
Do microRNAs modulate Fe and Cu cross-talk?
Plants have complex regulation of Cu levels in cells (Burkhead et al., 2009). In Arabidopsis, Cu uptake systems, Cu chaperone proteins, and P-type ATPases that transport Cu into organelles are all highly regulated. Under Cu excess, CSD1 and CSD2 transcript and protein levels are increased, and miR398s are not present. Under Cu deficiency, miR398b and miR398c become abundant, and CSD1 and CSD2 transcripts are down-regulated as miR398s bind to and direct degradation of their transcripts (Sunkar et al., 2006; Yamasaki et al., 2007; Beauclair et al., 2010). At the same time, FSD1 transcript and protein levels increase under Cu deficiency, which allows FeSOD to replace the CuSODs functionally. Under Cu deficiency, increased FeSOD and decreased CuSOD proteins were observed in Arabidopsis, Brassica juncea, tomato, maize, and rice leaves (Cohu and Pilon, 2007). The transcription of miR397a, the miR398s, FSD1, and also COPT2 is dependent on the transcription factor SPL7 (Yamasaki et al., 2009; Bernal et al., 2012). The present results suggest that a similar but opposite Fe–Cu cross-talk system is at work in Arabidopsis. That is, under Fe deficiency, FeSODs are down-regulated and the proteins are functionally replaced by CuSODs by increased expression of CSD1 and CSD2. A model for this interaction is presented as Supplementary Fig. S3 at JXB online. The time course results show that FeSOD transcripts declined to very low levels within 24h of Fe withdrawal (Fig. 5). During the same time period, Cu accumulation had already more than doubled for Kas-1 and Col-0, with Tsu-1 Cu accumulation delayed by 1 d (Fig. 6). In Chlamydomonas reinhardtii, a similar cross-talk was observed (Page et al., 2012), but with FeSODs being replaced by an Mn-containing SOD protein, induced under Fe starvation. No Cu/Zn-containing SODs are present in C. reinhardtii. This could indicate a conserved metal interchange among plants, dependent on the metal availability.
Three of the eight miRNAs that responded to Fe deficiency are known regulators of Cu protein transcript degradation. miR397a decreased rapidly in Kas-1 roots after removal of Fe, reaching its lowest levels at 8h, and also decreased in Tsu-1, but at later time points (Fig. 4). This miRNA is abundant under low Cu conditions, where it targets and directs destruction of transcripts of several laccase proteins (Abdel-Ghany and Pilon, 2008), which are Cu-containing enzymes. Decreased abundance of miR397a, as in the Fe-deficient roots studied here, should relieve this post-transcriptional regulation and result in increased transcript levels. Indeed, the microarray results showed that LAC2, one of the miR397a targets (Abdel-Ghany and Pilon, 2008) and a Cu-regulated gene (Bernal et al., 2012), was increased by 2.1-fold in Fe-deficient Kas-1 roots at 24h. This miRNA was also shown to be down-regulated in Fe-deficient B. napus roots, leaves, and phloem sap, while it was up-regulated under Cu deficiency in phloem and roots (Buhtz et al., 2010). This same study showed that miR398a increased in leaves and phloem sap under Cu deficiency, but decreased in roots, leaves, and phloem sap under Fe deficiency (Buhtz et al., 2010). In the present study, miR398a and miR398b/c showed decreased abundance in rosettes following removal of Fe, consistent with results described above and also with results from Fe-deficient bean leaves (Valdés-López et al., 2010). The miR398s are known to target and direct degradation of transcripts of several Cu-containing proteins, notably CSD1, CSD2, and CCS1 (Sunkar et al., 2006; Yamasaki et al., 2007; Beauclair et al., 2010), which in this study were all up-regulated by microarray and/or RT-PCR (Table 4, Fig. 5). The Cu concentration also increased in the rosettes (Fig. 6), and as such it is possible that the Fe deficiency down-regulation of miR398s was by Cu regulation. This possibility was addressed by withholding Fe and Cu individually or simultaneously. The results in Fig. 7 suggested that miR398a and b/c are regulated by both Fe and Cu in opposition, with Fe deficiency acting to decrease levels of miR398s and Cu deficiency acting to increase levels of miR398s, since there was no net change when Col-0 was made both Fe and Cu deficient. The results of increased or decreased miR398s expression were reflected in transcript levels of CSD1 and CSD2 under these treatments (Fig. 8), while the non-miR398s target FRO3 was up-regulated under Fe, Cu, and simultaneous deficiencies. Additional study will be required to state definitively that Fe directly regulates the abundance of miR398s. In the case that miR398s are not regulated by Fe deficiency per se, but rather indirectly through increased Cu accumulation in rosettes, this still represents a mechanism of Fe–Cu cross-talk, if Cu uptake/accumulation is directly responsive to Fe deficiency (see below).
Although the Kas-1 and Tsu-1 ecotypes used in this study differed widely in their timing of response (including Cu accumulation) and transcriptional profiles in response to Fe deficiency, the regulation of the miR398s, the two CuSODs (CSD1 and CSD2), and the FeSODs was nearly identical. If miR398s are used as a mechanism for increasing CuSODs to replace FeSODs under Fe deficiency, this would probably be conserved across plant species. Nutrient status regulation of miR398 was also observed in Populus trichocarpa by Cu deficiency (Lu et al., 2011) and in Phaseolus vulgaris by Fe deficiency (Valdés-López et al., 2010). In the Sanger miRbase, homologues are present in several other plant species, including: Oryza sativa, Glycine max, Medicago truncatula, Pinus taeda, Vitis vinifera, Brassica juncea, Aquilegia caerula, Zea mays, Sorghum bicolor, Citrus sinensis, Ricinus communis, Gossypium raimondii, Arabidopsis lyrata, Arachis hypogea, Theobroma cacao, Salvia sclarea, and Brachypodium distachyon. It should be noted that miR398s have also been shown to respond to other stimuli that might alter the need to modulate the capacity for oxidative stress tolerance, such as salt stress, water stress, treatment with ultraviolet light, and other stresses (Zhu et al., 2011).
Cu uptake under Fe deficiency: lack of specificity of up-regulated Fe uptake systems?
Several of the present results suggest that Cu accumulation in rosettes is a specific Fe deficiency response, regulated by Fe deficiency but separate from the well-known Fe uptake system (i.e. FRO2 and IRT1). The increase in Cu accumulation in Fe-deficient plants has previously been attributed to non-specific uptake by up-regulation of Fe acquisition systems, and to the fact that ferric-chelate reductase activity can also reduce Cu2+, which may increase Cu uptake (Norvell et al., 1993; Cohen et al., 1997), and some mutants with constitutive Fe demand have increased Cu accumulation in leaves (Welch et al., 1993; Delhaize, 1996; Stacey et al., 2008). However, the present results indicate that the increased uptake of Cu may not be non-specific, but instead is a part of the Fe deficiency response (see below). Additionally, a recent study has indicated that Cu2+ reduction for Cu uptake is carried out not by FRO2, but rather by FRO4 and FRO5, which are regulated by Cu status through the SPL7 transcription factor (Bernal et al., 2012). The time course data indicate that the increase in Cu uptake and accumulation in rosettes had already occurred prior to maximal induction of FIT, FRO2, or IRT1, and before a measurable up-regulation of ferric-chelate reductase activity (Fig. 6; Stein and Waters, 2011). Secondly, altered expression of Cu-transporting proteins or Cu homeostasis genes in roots (CCH, OPT3, COPT2, and ZIP2) and rosettes (CCS1, OPT3, and COPT2) was observed, which suggests that altered Cu metabolism is part of the rosette Fe deficiency response. Thirdly, the results suggest that miR398s and downstream targets are regulated by both Fe and Cu. If miR398s are down-regulated by Fe deficiency, this would have a post-transcriptional effect to increase steady-state levels of CSD1 and CSD2, which happened in both ecotypes. Additionally, when Cu and Fe were withheld simultaneously, FSD1 transcripts remained at levels resembling Fe deficiency alone rather than the elevated levels observed under Cu deficiency alone. Fourthly, a decrease in fitness, as shown by increased oxidative stress damage, was measured when increased Cu uptake under Fe deficiency was blocked by withholding Fe and Cu simultaneously, indicating that there is a physiological role for increased Cu accumulation to supply CuSODs. Lastly, increased oxidative stress damage was also measured in the Fe-deficient ccs1 mutant, which genetically blocked formation of functional CuSOD proteins, indicating that a specific role for accumulation of Cu under Fe deficiency is to supply Cu to CuSODs in the absence of functional FeSODs. Together, these data present a model that suggests that increased Cu uptake into Fe-deficient Arabidopsis is not a result of lack of specificity of Fe uptake, but is an important adaptation to Fe deficiency.
Conclusions and future directions
Two genome-wide transcript profiling techniques, Affymetrix microarray and miRNA microarray, were used to identify Fe-regulated transcripts in rosettes. Together, several of the genes and miRNAs that were identified indicated a link between Fe deficiency and Cu metabolism, which may function to provide protection from oxidative stress. The nature of this link may be miR398. Future studies will determine how Fe deficiency regulates levels of this miRNA. Another future direction is to determine which genes are responsible for regulation and transport of Cu for increased Cu accumulation under Fe deficiency.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Venn diagram of microarray expression of Arabidopsis rosette transcripts in response to Fe deficiency.
Figure S2. Over-represented GO-slim categories for Kas-1, Tsu-1, and Col-0 rosette Fe-regulated genes.
Figure S3. Model of Fe–Cu cross-talk in Arabidopsis thaliana.
Table S1. Primers used in this study.
Acknowledgements
The authors thank Laura Hock for expert technical assistance, and Michael A. Grusak for helpful discussion of this work.
References
- Abdel-Ghany SE, Pilon M. 2008. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis Journal of Biological Chemistry 283 15932––15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauclair L, Yu A, Bouche N. 2010. microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis The Plant Journal 62 454––462 [DOI] [PubMed] [Google Scholar]
- Bernal M, Casero D, Singh V, et al. 2012. Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis The Plant Cell 24 738––761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, He Y, Billiau K, Van de Peer Y. 2010. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics Bioinformatics 26 1566––1568 [DOI] [PubMed] [Google Scholar]
- Buckhout TJ, Yang TJW, Schmidt W. Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics. 2009;10:147. doi: 10.1186/1471-2164-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A, Pieritz J, Springer F, Kehr J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biology. 2010;10:64. doi: 10.1186/1471-2229-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KAG, Abdel-Ghany SE, Cohu CM, Pilon M. 2009. Copper homeostasis New Phytologist 182 799––816 [DOI] [PubMed] [Google Scholar]
- Chaignon V, Di Malta D, Hinsinger P. 2002. Fe-deficiency increases Cu acquisition by wheat cropped in a Cu-contaminated vineyard soil New Phytologist 154 121––130 [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Research. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants Annual Review of Plant Biology 62 185––206 [DOI] [PubMed] [Google Scholar]
- Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, Jinn TL. 2005. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis Plant Physiology 139 425––436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CK, Norvell WA, Kochian LV. 1997. Induction of the root cell plasma membrane ferric reductase. An exclusive role for Fe and Cu Plant Physiology 114 1061––1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohu CM, Pilon M. 2007. Regulation of superoxide dismutase expression by copper availability Physiologia Plantarum 129 747––755 [Google Scholar]
- Colangelo EP, Guerinot ML. 2004. The essential basic helix–loop–helix protein FIT1 is required for the iron deficiency response The Plant Cell 16 3400––3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo T, Cambiazo V, González M. 2010. Gene expression profiling analysis of copper homeostasis in Arabidopsis thaliana Biochemical and Biophysical Research Communications 393 248––252 [DOI] [PubMed] [Google Scholar]
- Delhaize E. 1996. A metal-accumulator mutant of Arabidopsis thaliana Plant Physiology 111 849––855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Poschl Y, Raschke A, Ullrich K, Ettingshausen S, Hauptmann V, Grosse I, Quint M. 2010. Natural variation of transcriptional auxin response networks in Arabidopsis thaliana The Plant Cell 22 2184––2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Quint M. 2011. Expression level polymorphisms: heritable traits shaping natural variation Trends in Plant Science 16 481––488 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. 2008. Cell identity mediates the response of Arabidopsis roots to abiotic stress Science 320 942––945 [DOI] [PubMed] [Google Scholar]
- Doerge RW. 2002. Mapping and analysis of quantitative trait loci in experimental populations Nature Reviews Genetics 3 43––52 [DOI] [PubMed] [Google Scholar]
- Garcia MJ, Lucena C, Romera FJ, Alcantara E, Perez-Vicente R. 2010. Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis Journal of Experimental Botany 61 3885––3899 [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. 2010. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants Plant Physiology and Biochemistry 48 909––930 [DOI] [PubMed] [Google Scholar]
- Hansch R, Mendel RR. 2009. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl) Current Opinion in Plant Biology 12 259––266 [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Kawachi M, Wirtz M, Hillmer S, Hell R, Krämer U. 2012. Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis The Plant Cell 24 724––737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges DM, DeLong JM, Forney CF, Prange RK. 1999. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds Planta 207 604––611 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA Molecular Cell 14 787––799 [DOI] [PubMed] [Google Scholar]
- Juenger TE, Sen S, Bray E, Stahl E, Wayne T, Mckay J, Richards JH. 2010. Exploring genetic and expression differences between physiologically extreme ecotypes: comparative genomic hybridization and gene expression studies of Kas-1 and Tsu-1 accessions of Arabidopsis thaliana Plant, Cell and Environment 33 1268––1284 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D. 2009. Quantitative genomics: analyzing intraspecific variation using global gene expression polymorphisms or eQTLs Annual Review of Plant Biology 60 93––114 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, West MAL, van Leeuwen H, Kim K, Doerge RW, Michelmore RW, St Clair DA. 2006. Genomic survey of gene expression diversity in Arabidopsis thaliana Genetics 172 1179––1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Bolte S, et al. 2005. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron EMBO Journal 24 4041––4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. 2010. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots The Plant Cell 22 2219––2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SF, Yang CM, Chiang VL. 2011. Conservation and diversity of MicroRNA-associated copper-regulatory networks in Populus trichocarpa Journal of Integrative Plant Biology 53 879––891 [DOI] [PubMed] [Google Scholar]
- Mukherjee I, Campbell NH, Ash JS, Connolly EL. 2006. Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper Planta 223 1178––1190 [DOI] [PubMed] [Google Scholar]
- Norvell WA, Welch RM, Adams ML, Kchian LV. 1993. Reduction of Fe(III), Mn(III) and Cu(II) chelates by roots of pea (Pisum sativum L.) or soybean (Glycine max) Plant and Soil 156 123––126 [Google Scholar]
- Page MD, Allen MD, Kropat J, Urzica EI, Karpowicz SJ, Hsieh SI, Loo JA, Merchant SS. 2012. Fe sparing and Fe recycling contribute to increased superoxide dismutase capacity in iron-starved Chlamydomonas reinhardtii The Plant Cell 24 2649––2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Ravet K, Tapken W. 2011. The biogenesis and physiological function of chloroplast superoxide dismutases Biochimica et Biophysica Acta 1807 989––998 [DOI] [PubMed] [Google Scholar]
- Puig S, Andres-Colas N, Garcia-Molina A, Penarrubia L. 2007. Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications Plant, Cell and Environment 30 271––290 [DOI] [PubMed] [Google Scholar]
- Raven JA, Evans MCW, Korb RE. 1999. The role of trace metals in photosynthetic electron transport in O2-evolving organisms Photosynthesis Research 60 111––149 [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. 2009. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis The Plant Journal 57 400––412 [DOI] [PubMed] [Google Scholar]
- Sancenon V, Puig S, Mira H, Thiele DJ, Penarrubia L. 2003. Identification of a copper transporter family in Arabidopsis thaliana Plant Molecular Biology 51 577––587 [DOI] [PubMed] [Google Scholar]
- Schuler M, Keller A, Backes C, Philippar K, Lenhof HP, Bauer P. Transcriptome analysis by GeneTrail revealed regulation of functional categories in response to alterations of iron homeostasis in Arabidopsis thaliana. BMC Plant Biology. 2011;11:10. doi: 10.1186/1471-2229-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller S, Terry N. 1980. Limiting factors in photosynthesis. 2. Iron stress diminishes photo-chemical capacity by reducing the number of photosynthetic units Plant Physiology 65 121––125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G. 2008. The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds Plant Physiology 146 589––601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RJ, Waters BM. 2011. Use of natural variation reveals core genes in the transcriptome of iron-deficient Arabidopsis thaliana roots Journal of Experimental Botany 63 1039––1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. 2006. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance The Plant Cell 18 2051––2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Takahashi M, Tsukamoto T, et al. 2006. Biosynthesis and secretion of mugineic acid family phytosiderophores in zinc-deficient barley The Plant Journal 48 85––97 [DOI] [PubMed] [Google Scholar]
- Terry N. 1980. Limiting factors in photosynthesis. 1. Use of iron stress to control photo-chemical capacity in vivo Plant Physiology 65 114––120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry N. 1983. Limiting factors in photosynthesis. 4. Iron stress-mediated changes in light-harvesting and electron-transport capacity and its effects on photosynthesis in vivo Plant Physiology 71 855––860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ. 2001. Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis Plant Physiology 127 1030––1043 [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Yang SS, Aparicio-Fabre R, Graham PH, Reyes JL, Vance CP, Hernández G. 2010. MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity New Phytologist 187 805––818 [DOI] [PubMed] [Google Scholar]
- van Leeuwen H, Kliebenstein DJ, West MAL, Kim K, van Poecke R, Katagiri F, Michelmore RW, Doerge RW, Clair DA. 2007. Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid The Plant Cell 19 2099––2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poecke RMP, Sato M, Lenarz-Wyatt L, Weisberg S, Katagiri F. 2007. Natural variation in RPS2-mediated resistance among Arabidopsis accessions: correlation between gene expression profiles and phenotypic responses The Plant Cell 19 4046––4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Baumlein H, Weisshaar B, Bauer P. 2007. Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana Planta 226 897––908 [DOI] [PubMed] [Google Scholar]
- Waters BM, Chu HH, DiDonato RJ, Roberts LA, Eisley RB, Lahner B, Salt DE, Walker EL. 2006. Mutations in Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiology 141 1446––1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Grusak MA. 2008. Whole-plant mineral partitioning throughout the life cycle in Arabidopsis thaliana ecotypes Columbia, Landsberg erecta, Cape Verde Islands, and the mutant line ysl1ysl3 New Phytologist 177 389––405 [DOI] [PubMed] [Google Scholar]
- Waters BM, Troupe GC. 2012. Natural variation in iron use efficiency and mineral remobilization in cucumber (Cucumis sativus) Plant and Soil 352 185––197 [Google Scholar]
- Welch RM, Norvell WA, Schaefer SC, Shaff JE, Kochian LV. 1993. Induction of iron(III) and copper(II) reduction in pea (Pisum sativum L.) roots by Fe and Cu staus: does the root-cell plasmalemma Fe(III)-chelate reductase perform a general role in regulating cation uptake? Planta 1993 555––561 [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen WQ, Chang HS, Zhu T, Vulpe C. 2003. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis Journal of Biological Chemistry 278 47644––47653 [DOI] [PubMed] [Google Scholar]
- Wu HL, Li LH, Du J, Yuan YX, Cheng XD, Ling HQ. 2005. Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana Plant and Cell Physiology 46 1505––1514 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. 2007. Regulation of copper homeostasis by micro-RNA in Arabidopsis Journal of Biological Chemistry 282 16369––16378 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. 2009. SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis The Plant Cell 21 347––361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJW, Lin W-D, Schmidt W. 2010. Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks Plant Physiology 152 2130––2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YX, Wu HL, Wang N, Li J, Zhao WN, Du J, Wang DW, Ling HQ. 2008. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis Cell Research 18 385––397 [DOI] [PubMed] [Google Scholar]
- Zhu C, Ding Y, Liu H. 2011. MiR398 and plant stress responses Physiologia Plantarum 143 1––9 [DOI] [PubMed] [Google Scholar]