Abstract
In the last decade, genome-wide transcriptome analyses have been routinely used to monitor tissue-, disease- and cell type-specific gene expression, but it has been technically challenging to generate expression profiles from single cells. Here we describe a novel and robust mRNA-Seq protocol (Smart-Seq) that is applicable down to single cell levels. Compared with existing methods, Smart-Seq has improved read coverage across transcripts, which significantly enhances detailed analyses of alternative transcript isoforms and identification of SNPs. We have determined the sensitivity and quantitative accuracy of Smart-Seq for single-cell transcriptomics by evaluating it on total RNA dilution series. Applying Smart-Seq to circulating tumor cells from melanomas, we identified distinct gene expression patterns, including new candidate biomarkers for melanoma circulating tumor cells. Importantly, our protocol can easily be utilized for addressing fundamental biological problems requiring genome-wide transcriptome profiling in rare cells.
Introduction
Analyses of transcriptomes through massively parallel sequencing of cDNAs (mRNA-Seq) generates millions of short sequence fragments that can be analyzed to accurately quantify expression levels1, assemble new transcripts2,3 and investigate alternative RNA processing4,5. These techniques have been consistently pushed towards development of methods that require lower starting amounts of RNA, ideally down to single cells. A protocol initially developed for single-cell microarray studies6 was adapted for mRNA-Seq and used to generate transcriptome data for individual mouse oocytes and early embryonic cells7,8. The method successfully detected thousands of genes expressed in mouse oocytes, and showed increased sensitivity compared with microarrays7. However, this first single-cell mRNA-Seq experiment lacked technical controls, making it impossible to distinguish biological variation between different cells from the technical variation that is intrinsic to cDNA amplification protocols when starting with low amounts of RNA. Therefore, the question remained whether single-cell transcriptomes faithfully represent the RNA population before amplification and how technical variation limits the power to find differential expression. This initial mRNA-Seq method also preferentially amplified the 3′ ends of mRNAs, and hence the data could only be used to identify distal splicing events. Recently, a method for multiplexed single-cell RNA-Seq was introduced that quantifies transcripts through reads mapping to mRNA 5′ ends9. Neither of these methods generates read coverage across full transcripts. Since most mammalian multi-exons genes are subject to alternative RNA processing4,5, there is a need for a single-cell transcriptome method that can both quantify gene expression and provide the coverage for efficient detection of transcript variants and alleles.
In this study, we introduce a single-cell RNA-Sequencing protocol with markedly improved transcriptome coverage, which samples cDNAs from more than just the ends of mRNAs. Using this protocol, we have sequenced the mRNAs from a large number of individual mammalian cells, as well as well-defined dilution series of purified total RNAs, to comprehensively assess how sensitivity, variability and detection of differential expression vary with different amounts of starting material. Our results demonstrate the power of single-cell RNA-Seq for both transcriptional and post-transcriptional studies, and provide valuable insights into the design of experiments that start from few or single cells. To demonstrate the biological importance of this method, we have applied this new assay to putative circulating tumor cells (CTCs) captured from the blood of a melanoma patient to demonstrate how Smart-Seq enables high-quality transcriptome mapping in individual, clinically important cells.
Results
Efficient and robust single-cell RNA-Sequencing using Smart-Seq
For Smart-Seq, first we lysed each cell in hypotonic solution and converted poly(A)+ RNA to full-length cDNA using oligo(dT) priming and SMART template switching technology, followed by 12‐18 cycles of PCR preamplification of cDNA. To enable gene and mRNA isoform expression analyses in single cells, a novel full-transcriptome mRNA-Seq protocol (Smart-Seq) was developed. Smart-Seq makes use of SMART™ template switching technology for the generation of full-length cDNAs and only 12 to 18 cycles of PCR following the initial cDNA synthesis steps. The amplified cDNA was used to construct standard Illumina sequencing libraries using either Covaris shearing followed by ligation of adaptors (PE) or Tn5-mediated “tagmentation” using the Nextera technology (Tn5). Both of these library preparation methods enable random shotgun sequencing of cDNAs (Supplementary Fig. 1). We successfully generated Smart-Seq libraries from 42 individual human or mouse cells, and in addition we generated 64 libraries from dilution series of total RNA derived from human brain (16 samples), mouse brain (28 samples) and universal human reference RNA (UHRR, 20 samples). Each sequencing library was sequenced on the Illumina platform, typically generating over 20 million uniquely mapping reads (Supplementary Table 1). For comparison, several standard mRNA-Seq libraries were also made from 100 ng to a few micrograms of total RNA.
Smart-Seq improves coverage across transcripts
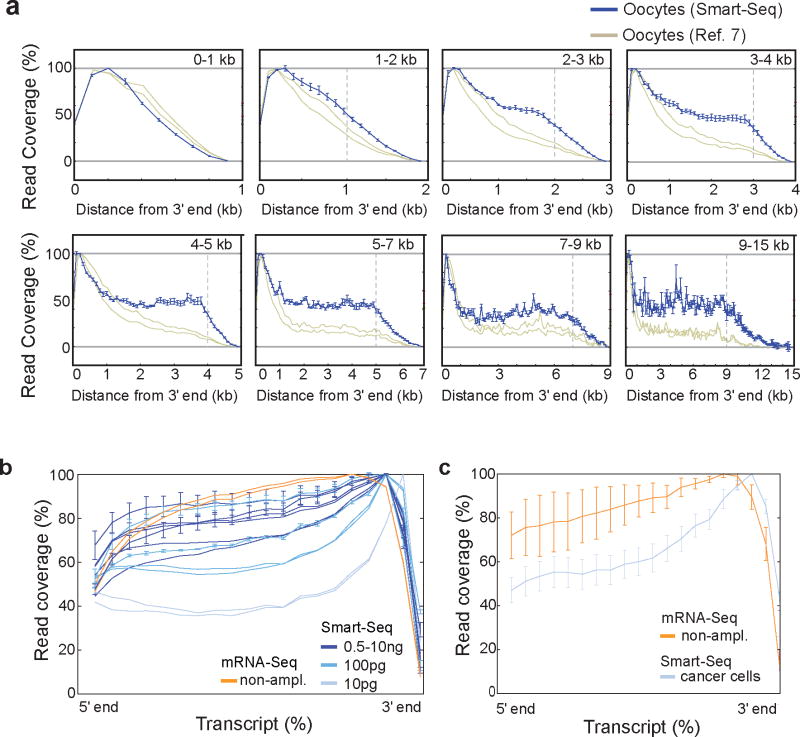
In previous single-cell mRNA-Sequencing studies7,8, the data suffered from a pronounced 3′-end bias that limited analysis across full-length transcripts. We sequenced single-cell transcriptomes from mouse oocytes to enable a direct comparison with published mouse oocyte single-cell data7. Analyses of read coverage across transcripts demonstrated that Smart-Seq has significantly improved full-length coverage of all transcripts longer than 1kb (Fig. 1a and Fig. S2d–k). Smart-Seq analyses of mouse brain RNA at different dilutions showed that even better coverage was obtained with increased starting amounts, with nanogram dilutions reaching close to the coverage observed using standard mRNA-Seq from 100 ng to 1 ug total RNA (Fig. 1b). From only 10 pg input amounts (the estimated amount of RNA in a small eukaryotic cell, Supplementary Table 2) we achieved close to 40% coverage at the 5′ end. Analyses of single-cell transcriptomes from cancer cell lines (four cells each from LNCaP, PC3 and T24) obtained equally good read coverage (Fig. 1c) and, indeed, for 25% of all expressed, multi-exon genes our read coverage enabled full-length transcript reconstruction (exemplified in Fig. S3). We conclude that Smart-Seq has significantly improved read coverage compared to previous single-cell transcriptome methods.
Figure 1. Smart-Seq read coverage across transcripts.
(a) Comparison of read coverage over transcripts for Smart-Seq analyzed mouse oocytes (green, n=3) and previously published mouse oocyte transcriptome data (red, n=2)7. Transcripts were grouped according to annotated lengths and analyzed separately, with the transcript lengths indicated in the top right corner of each panel. In each panel, we display the read coverage over the transcripts as a distance from the 3′ end (x-axis), with the vertical dashed gray line showing the length of the shortest included transcripts after which a decline in read coverage is expected. Error bars represent standard deviations among biological replicates. (b) Mean read coverage over transcripts for Smart-Seq data generated from diluted amounts of mouse brain RNA. Independent dilution series (including data from different labs) are shown as separate data sets. For comparison, we included data from standard mRNA-Seq on 100 ng of mouse brain RNA (black). Errors bars represent standard deviations. (c) Read coverage (as in b) for twelve individual human cells of prostate and bladder cancer line origin, analyzed using Smart-Seq (purple) and for prostate cell line LNCaP analyzed with standard mRNA-Seq (black).
Quantitative assessment of single-cell transcriptomics
Analyses of gene expression from millions of cells using mRNA-Seq is highly reproducible and has low technical variation1,4. So far, no single-cell mRNA-Seq study has measured the technical variation intrinsic to the cDNA pre-amplification components of single-cell methods. We therefore diluted microgram amounts of reference total RNA down to nano- and picogram levels and applied Smart-Seq to assess sensitivity, technical variability and detection of differentially expressed transcripts of Smart-Seq on low amounts of total RNA. For comparison, standard mRNA-Seq libraries were generated from 100 ng to microgram levels of reference total RNA.
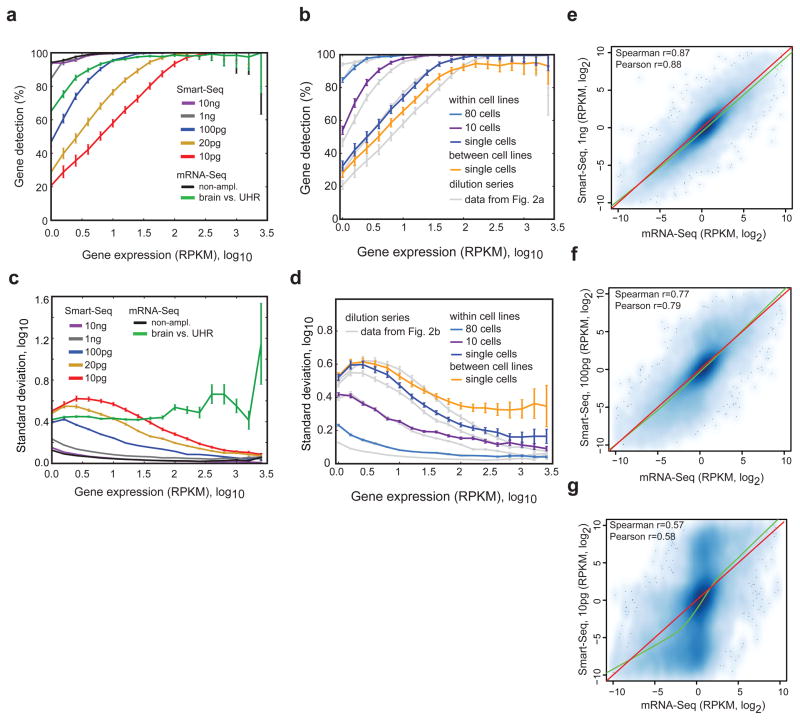
First, we addressed the sensitivity of the method in detecting transcripts present at different expression levels. Starting with 10 ng or 1 ng of total RNA, we found no or minimal decline in sensitivity compared with standard mRNA-Seq. However, lowering the starting amounts to single-cell levels decreased the detection rate of less abundant transcripts (Fig. 2a). Analyses of the twelve cancer cell line cells (four cells each from the LNCaP, PC3 and T24 lines) showed that ~76% of transcripts expressed at 10 RPKM (reads per kilobase exon model and million mappable reads), an expression level that roughly equals the median expression level for detected transcripts, were reproducibly detected in all single-cell profiles (Fig. 2b). We found that the sensitivity of gene detection for the individual cancer cells was similar to that of about 20 pg of starting total RNA (Fig. 2b). Furthermore, we observed that the starting amount of total RNA had a larger impact on sensitivity than the number of PCR cycles used (Supplementary Fig. 5) and that the sequence depth had little effect on transcript detection at levels above a million uniquely mapping reads per cell, with expression levels stabilizing after 3 million uniquely mapped reads (Supplementary Fig. 4b,c). Comparisons of Smart-Seq and previous mouse oocyte data7 demonstrated similar sensitivity (Supplementary Fig. 2a,b). We conclude that transcript detection sensitivity is affected by limiting starting amounts of RNA that lead to random loss of low abundance transcripts, but still the majority of low abundance and the vast majority of highly expressed transcripts are reliably detected even in single cells.
Figure 2. Sensitivity and variability in Smart-Seq from few or single cells.
(a) Percentage of genes reproducibly detected within replicate pairs, binned according to expression level. We performed all pair-wise comparisons within groups of replicates and report the mean and 90% confidence interval. We used Smart-Seq data generated from diluted amounts of human UHR total RNA as indicated. As controls, we added both a comparison of technical replicates of human UHRR analyzed using standard mRNA-Seq protocols with 100 ng input RNA (black line), as well as a comparison of human UHRR and brain RNA from standard mRNA-Seq data (green line). (b) Percentage of genes reproducibly detected within replicate pairs, binned according to expression level (as in a) for human LNCaP, PC3 and T24 cells. We show pair-wise comparisons among single cells from the same cancer cell line (blue line), among multiple cells of the same cell line (purple and blue lines), and comparisons among single cells from different cancer cell lines (yellow line). (c) Standard deviation in gene expression estimates within replicates in bins of genes sorted according to expression levels. Comparisons and colors coded in the same way as in (a) and error bars represents standard error of the mean. (d) Standard deviation in gene expression estimates within replicates (as in c, with comparisons and colors coded in the same manner as in b). (e–g) Scatter plots showing the relative differences between human UHRR and brain gene expression levels estimated from standard mRNA-Seq data on 100 ng input RNA (x-axis) and Smart-Seq generated data (y-axis) starting from 1 ng total RNA (e), 100 pg total RNA (f) and 10 pg total RNA (g). Correlation coefficients computed from log2 transfomed relative gene expression profiles, together with non-linear loess regression curves (green) and y=x lines (red).
Second, we determined the reproducibility in expression levels generated from diluted RNA and individual cells. Comparison of Smart-Seq and previous mouse oocyte data7 demonstrated improved expression level estimation with Smart-Seq (lower oocyte to oocyte variability) across the whole range of expression levels (Supplementary Fig. 2c). Correlation analyses between technical replicates of diluted RNA showed increasing concordance with larger amounts of RNA. Comparing the single cells against the RNA dilution, we observed higher correlations (Pearson correlations of 0.75–0.85) among individual cells of the same type than among dilution replicates at 10 pg (Pearson correlations of 0.65–0.75) (Supplementary Fig. 6). Since variability in measurements of expression levels depends on transcript expression levels, we computed the variability as a function of the expression level (Fig. 2c,d). This analysis showed that Smart-Seq on 10 ng total RNA had the same technical variability as standard mRNA-Seq and that Smart-Seq on 1 ng total RNA showed only a modest increase in technical noise (Fig. 2c). When lowering input amounts down to picogram levels, there was a clear increase in technical variability, particularly for less abundantly expressed transcripts (Fig. 2c). The levels of technical variability at picogram levels of total RNA were compared to the biological variation found in comparisons of human brain and UHRR using standard mRNA-Seq (Fig. 2c, green line). Interestingly, analyses of variation between individual cancer cells of different origin revealed extensive biological variation in highly expressed genes (Fig. 2d).
Finally, we assessed whether pre-amplified single-cell expression profiles were representative of the original expression profiles. Comparing relative gene expression levels (UHRR - brain) estimated using standard mRNA-Seq to those estimated from Smart-Seq with different amounts of input RNA, we again found a high concordance (Fig. 2e–g). Starting with 1 ng or 100 pg total RNA, the relative expression in Smart-Seq and standard mRNA-Seq respectively had Spearman correlations of 0.87 and 0.77 (Fig. 2e,f). Comparisons with 10 pg input RNA showed overall good correlation (Fig. 2g), but identified two populations of transcripts with distorted expression in Smart-Seq data from either human brain or UHRR, reflecting stochastic losses, mostly of low abundance transcripts when starting with such minute RNA of levels (Fig. 2g and Fig. 2a). Pre-amplification of cDNA could also lead to disproportionate amplification of short transcripts, but we found no systematic bias (Supplementary Fig. 7). A previous microarray study analyzed PCR amplified cDNA (from picogram levels) and found the transcriptome overall preserved, but skewed10. Our data from 1 ng and 100 pg total RNA show no skewing, i.e. the loess slopes estimated from the data approximated 1 (Fig. 2e–g). Together, these results demonstrated that transcriptome analyses from few or single cells, in general, preserved relative expression level differences for detected transcripts.
Analyses of transcriptional and post-transcriptional differences from single-cells
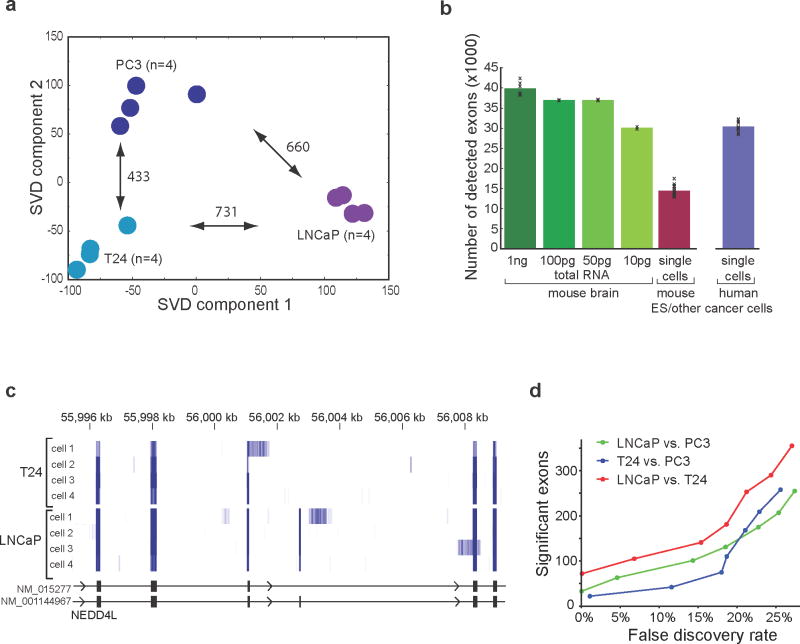
Having demonstrated the improved performance of Smart-Seq on low amounts of RNA compared to previously published methods, we focused our analyses on single-cell transcriptomes from prostate (PC3, LNCaP) and bladder (T24) cancer cell line cells. The twelve individual cells (four from each cell line) clustered according to cell line of origin using their global gene expression levels and we identified hundreds of differentially expressed genes among the three cell lines (Fig. 3a; q<0.05 ANOVA; p<0.05 post-hoc test).
Figure 3. Transcriptional and post-transcriptional analyses of cancer cell line cells using Smart-Seq.
(a) Categorization of individual cells according to cell line of origin using single-cell Smart-Seq transcriptomes. Singular-value decomposition analysis was conducted for 12 individual cancer cells (4 cells each from the PC3, LNCaP and T24 cancer cell lines) based on global gene expression profiles. Projections are shown based on the first two dimensions that capture most of the variance. The numbers of significantly differentially expressed genes per pairwise cell line comparison are shown next to the arrows (p<0.05, 1-way ANOVA and Tukey post-hoc test). (b) Mean number of alternatively spliced exons with sufficient read coverage for MISO analyses in sequence-depth matched single-cell mRNA-Seq data. Smart-Seq data from diluted mouse brain RNA (green) compared with previously published mouse ES cells8 (red) and twelve Smart-Seq analyzed individual prostate and bladder cell line cells (purple). The error bars represent standard deviation. (c) Single-cell Smart-Seq reads mapping to a portion of the NEDD4L gene locus from four individual T24 and LNCaP cells. Read coverage is shown as a heatmap with darker blue indicating higher read coverage. (d) Number of differentially included exons identified among the PC3, LNCaP and T24 cell lines from single-cell Smart-Seq analysis on four cells per cell line as a function of estimated false discovery rate.
The pronounced 3′-end bias of previous single-cell mRNA-Seq studies has hampered the ability to identify significant alternative splicing differences in single cells. We used the Bayesian mixture of isoforms framework (MISO)11 to infer exon inclusion levels for known alternatively spliced exons in the twelve individual cells. The improved read coverage with Smart-Seq resulted in a two-fold increase in the number of alternatively spliced exons that could be assessed, compared to previously published single-cell mRNA-Seq data (Fig. 3b), significantly improving our ability to detect alternative splicing. Cell-type specific alternative splicing could be inferred from single-cell transcriptomes, as seen in read coverage across the differentially included exon 13 of the NEDD4L gene (Fig. 3c). This exon was frequently included in LNCaP cells (93% mean inclusion level) but was included at much lower levels in T24 cells (15% mean inclusion levels) whereas low expression of NEDD4L in PC3 cells precluded inclusion level estimation. In total, in this comparison of three cancer cell lines, we found one hundred exons with differential exon inclusion levels among the three cell lines, with a less than 1% false discovery rate (Fig. 3d; Supplementary Table 3). We conclude that Smart-Seq significantly improves our ability to detect alternative RNA processing in single cells.
Analyses of circulating tumor cell transcriptomes
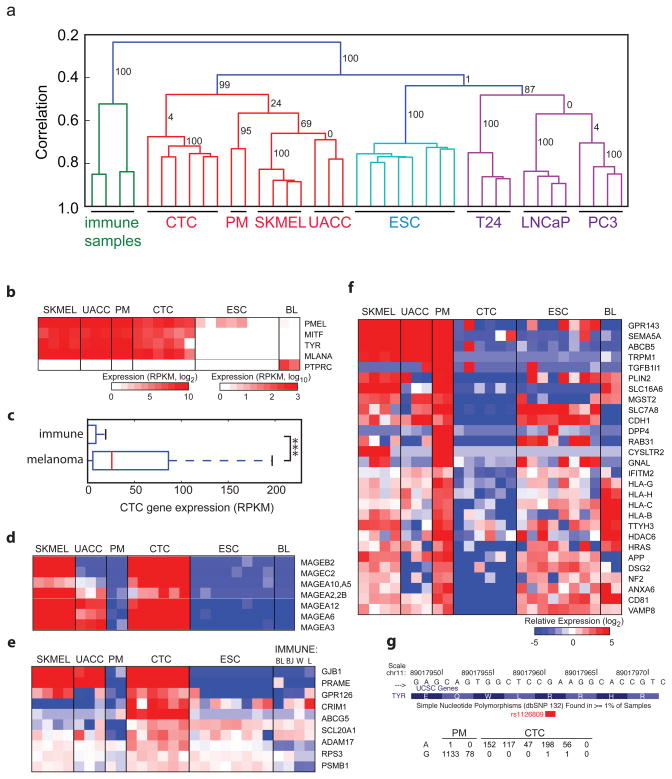
Having demonstrated that Smart-Seq generates quantitative and reproducible single-cell transcriptomes, we asked whether global transcriptome analyses of putative circulating tumor cells (CTCs) could reveal their tumor of origin and provide data to support the use of this method for unbiased cancer-specific biomarker identification. To this end, we generated transcriptomes from NG2+ putative melanoma circulating tumor cells (CTCs) isolated from peripheral blood drawn from a patient with recurrent melanoma using immunomagnetic purification with a MagSweeper instrument (Illumina Inc.)12. For comparison, we also generated Smart-Seq libraries from single cells derived from primary melanocytes (PMs, n=2), melanoma cancer cell line (SKMEL5, n=4 and UACC257, n=3) cells and from human embryonic stem cells (ESCs, n=8). Since the NG2+ putative CTCs were isolated from blood, it was important to compare them to blood cells. The putative CTCs were distinct from lymphoma cell lines (BL41 and BJAB)13 and immune tissues (lymphnode and white blood cell samples), as well as embryonic stem cells, and instead showed high similarity to PMs and melanoma cell line cells. Unsupervised hierarchical clustering and correlation analyses of gene expression levels showed a clear clustering of cells according to cell type of origin (Fig. 4a and Supplementary Fig. 8), and separation from the human brain RNA samples that were previously analyzed with Smart-Seq or mRNA-Seq (data not shown). Further support for the melanocytic origin of the putative melanoma CTCs came from analyses of melanocyte lineage specific markers, as all NG2+ cells expressed high levels of MLANA14, TYR15 and the melanocyte specific m-form of MITF16 but not immune markers such as PTPRC (Fig. 4b), in contrast to peripheral blood lymphocytes (Supplementary Fig. 9). Furthermore, NG2+ cells expressed high levels of melanoma-associated genes (based on our unbiased selection of the 100 transcripts most strongly associated with melanoma, see Methods), but not immune cell-associated genes selected in a similar manner (Fig. 4c, p<3.7e-15, Wilcoxon rank sum test). Thus, both their global transcriptomes and expression patterns of melanoma-associated transcripts clearly support a melanocyte origin for the NG2+ cells putative melanoma CTCs.
Figure 4. Single-cell transcriptomes of circulating tumor cells.
(a) Hierarchical clustering of human samples based on gene expression of highly expressed genes (>100 RPKM). Coloring indicates high-order clusters and the confidence in clusters are indicated with bootstrap values (percentage). Samples analyzed include human immune samples (Burkitt’s lymphoma cell lines BL41 and BJAB, and white blood cells and lymphnode samples) and cells from putative melanoma circulating tumor cells (CTC), primary melanocytes (PM), melanoma cell lines SKMEL5 (SKMEL) and UACC257 (UACC), prostate cancer cell lines (LNCaP, PC3), bladder cancer cell line (T24) and human embryonic stem cells (ESC). (b) Expression of melanocyte makers (PMEL, MITF, TYR, MLANA) and immune marker PTPRC in single-cell transcriptomes from (a) with Burkitt’s lymphoma cell lines BL41 and BJAB (BL). (c) Gene expression levels in CTCs for an unbiased set of 100 immune and melanoma markers. (d–f) Heatmaps showing relative expression of melanoma associated tumor antigens (d), up-regulated plasma-membrane proteins (e), and down-regulated plasma-membrane proteins (f) in single-cell transcriptomes as in (b) with the addition of more immune samples (W: white blood cells, L: lymphnode). (g) Number of reads from individual PMs and putative CTCs that support the reference (G) or risk (A) allele for the melanoma-associated SNP (rs1126809).
We next investigated whether the NG2+ putative CTCs showed signs of originating from a melanoma tumor. Comparison of their gene expression profiles with those of individual PMs identified 289 genes with higher expression in the putative CTCs than the PMs, and 436 genes with significantly lower levels (Supplementary Table 4). The up-regulated genes were significantly enriched (FDR<0.05) for melanoma-associated antigens (Fig. 4d and Supplementary Fig. 10) that have been repeatedly found to be upregulated in cancer17, mitotic cell cycle genes and additional categories (Supplementary Table 5). Down-regulated genes showed enrichment for regulators of cell death and MHC class I genes. Interestingly, the preferentially expressed antigen in melanoma (PRAME) was found highly expressed in NG2+ cells, which together with elevated expression of known melanoma tumor antigens (MAGEs), provides strong support for the conclusion that the NG2+ cells were CTCs that have originated from a melanoma.
In recent years, there has been a strong interest in identifying CTCs from different tumors using the a priori assumption that plasma-membrane proteins would be good diagnostic biomarkers. We used the CTC transcriptome analysis to screen for membrane proteins selectively expressed in melanoma-derived CTCs compared to PMs and immune cells. We identified 9 up-regulated plasma membrane-associated transcripts in the CTCs compared with primary melanocytes (q<0.05 ANOVA; p<0.05 post-hoc test), many of which are not expressed in immune cells and have not been previously associated with melanomas (Fig. 4e). Similarly, screening for loss of expression of plasma-membrane proteins identified 37 genes with significantly lower expression in the CTCs than PMs (Fig. 4f). Of note, epithelial Cadherin 1 (CDH1) showed no expression in the CTCs, and loss of CDH1 is thought to contribute to cancer progression by increasing proliferation, invasion and metastasis18. We also found downregulation of genes associated with the escape from immune surveillance, including five HLA genes (Fig. 4f), and TRPM1, suggesting that these gene expression changes might enable the CTCs to escape from immune surveillance. Notably, low expression of TRPM1 has been shown to correlate with melanoma aggressiveness and metastasis19. Future studies of these membrane proteins will likely enhance our understanding of CTC migration and invasiveness, and these results highlight the utility of studying single CTC cells with RNA-Seq.
Lastly, we investigated whether Smart-Seq transcriptome data could be mined for SNPs and other genetic variants associated with melanomas or other cancers. With the improved read coverage provided by the Smart-Seq method, we were able to identify 4,312 high-confidence genomic sites with support for an alternative allele in at least two CTCs, whereas genotype calls only supported by a single cell showed an excess of novel, likely artifactual, sites (Supplementary Fig. 11) together with a smaller subset (9%) of A-to-G RNA editing sites (data not shown). Ninety-two percent of the high-confidence sites coincided with documented SNPs, for example the melanoma-associated SNP in the TYR gene (rs1126809)20 (Fig. 4g). We conclude that Smart-Seq enables screening for SNPs and mutations in transcribed regions using only few cells.
Discussion
Generating high-coverage transcriptomes from single cells and small numbers of cells will have many applications for studying rare cells; such cells can be either individually picked or identified through cell sorting or laser capture techniques. Our results demonstrate that using Smart-Seq on 10 ng of total RNA is practically indistinguishable from a standard mRNA-Seq, whereas starting with 1 ng (corresponding roughly to 50–100 cells) shows only a minor increase in expression level variability. Therefore, this method could easily be applied to studies on homogeneous cell populations. However, many biologically and clinically important cell types exist in rare quantities and often in heterogeneous milieus, which necessitates single-cell approaches. Smart-Seq generates robust and quantitative transcriptome data from single cells. We found hundreds of differentially expressed genes using only a few individual cells per cell type, e.g. comparing only two primary melanocytes to six melanoma CTCs identified biologically meaningful differences. Even sequencing of a single cell yields useful information, as we, in each cell, detected most of the genes active in a culture of LNCaP cells. Importantly, Smart-Seq has significantly improved read coverage across transcripts, which enables detailed analyses of alternative splicing and identification of SNPs and mutations. Based on our CTC transcriptome results, single-cell analyses are highly informative for identifying candidate biomarkers, SNPs and mutations. In conclusion, datasets obtained with the Smart-Seq protocol provide significantly improved representation of the transcriptomes of individual cells, with relevance for both basic and clinical studies.
Methods
Generation and amplification of Smart-Seq cDNA
We have developed a new method for the generation of cDNA from total RNA or from single cells, called Smart-Seq. Briefly, polyA+ RNA was reverse transcribed through tailed oligo-dT priming directly in total RNA or a whole cell lysate using Moloney Murine Leukemia Virus Reverse Transcriptase (MMLV RT). Once the reverse transcription reaction reaches the 5′ end of an RNA molecule, the terminal transferase activity of MMLV adds a few non-templated nucleotides to the 3′ end of the cDNA. The carefully designed SMARTer II A oligo then base-pairs with these additional nucleotides, creating an extended template. The reverse transcriptase then switches templates and continues transcribing to the end of the oligonucleotide. The resulting full-length cDNA contains the complete 5′ end of the mRNA, as well as an anchor sequence that serves as a universal priming site for second strand synthesis. The cDNA is then amplified using 12 cycles for 1 ng of total RNA, 15 cycles for 100 pg of total RNA, and 18 cycles for 10 pg total RNA or from single cells. The exact number of cycles for each dilution replicate or single-cell is detailed in Supplementary Table 1. The Smart-Seq cDNA generation and amplification methods developed for this manuscript have recently become available in a kit marketed by Clontech called the “SMARTer Ultra Low RNA Kit for Illumina sequencing”. Although all the libraries in this manuscript were generated before the kit became commercially available, our protocol is reflected in the detailed instructions for generating cDNA from few cells or 100 pg–10 ng of total RNA that is now included in the manual for this kit. For single cell applications, each cell (or control RNA) was added in max 1 λ of media to 4 λ of hypotonic lysis buffer consisting of 0.2% Triton X-100 and 2 U/μl of ribonuclease (RNase) inhibitors (Clontech, 2313B) in RNase free water. The deposition of an intact cell in the hypotonic lysis buffer leads to immediate lysis and stabilization of the RNA through RNase inhibitors. Then, poly(A)+ RNA was reverse-transcribed through tailed oligo(dT) priming using the CDS primer (5′‐AAGCAGTGGTATCAACGCAGAGTACT(30)VN‐3′, where V represents A, C or G) directly in total RNA or a whole cell lysate using Moloney murine leukemia virus reverse transcriptase (MMLV RT).
Construction and sequencing of Smart-Seq sequencing libraries
Amplified cDNA (~5ng cDNA) was used to construct Illumina sequencing libraries using either Illumina’s “Ultra Low Input mRNA-Seq Guide” (the “PE” protocol) or a modification of Epicentre’s Nextera DNA sample preparation protocol (the “Tn5” protocol). With the PE protocol, the amplified cDNA was fragmented using a Covaris acoustic shearing instrument. The resulting fragments were end-repaired, followed by the addition of a single A base, ligation to Illumina PE adaptors, and then amplification with 12–18 cycles of PCR (depending on starting amounts of RNA, see Supplementary Table 1 for detailed instructions of all libraries generated). With the Tn5 protocol, the amplified cDNA was “tagmentated” at 55°C for 5 min in 20μl reaction with 0.25μl of transposase and 4μl of 5x HMW Nextera reaction buffer. 35μl of PB was added to the tagmentation reaction mix to strip the transposase off the DNA, and the tagmentated DNA was purified with 88μl of SPRI XP beads (sample to beads ratio of 1:1.6). Purified DNA was then amplified by 9 cycles of standard Nextera PCR. The libraries were sequenced on either Illumina’s HiSeq 2000, GAIIx or MiSeq instruments and all clusters that passed filter were exported into fastq files. Details on the sequence depth, sequencing platform and library construction method for each dilution replicate and single cell are included in Supplementary Table 1. All data shown in the figures of this manuscript were generated using the PE protocol unless otherwise specified in the figure legend.
Construction and sequencing of standard mRNA-Seq libraries
We generated mRNA-Seq transcriptome data following the Illumina mRNA-Seq kit from 100 ng and 1 microgram of total RNA, as detailed in Supplementary Table 1.
Isolation of individual CTCs from peripheral blood
10 ml of peripheral blood was collected from a male patient with recurrent, metastatic melanoma using K2 EDTA blood collection tubes (Becton Dickinson, NJ, USA). The blood sample was processed within 3 hours of collection. The erythrocytes in 4.5 ml of the blood sample were lysed with BD Pharm Lyse™ lysing solution (Becton Dickinson, NJ, USA) for 10 minutes at room temperature. The nucleated cells were pelleted, resuspended in HBSS containing 1% BSA and 5mM EDTA, pelleted, resuspended in 1ml of HBSS containing 1% BSA and 5mM EDTA. The nucleated cells were stained with PE-conjugated anti-human CD45 IgG to label leukocytes. The cells were subsequently reacted with biotinylated anti-human CSPG4 (a.k.a. NG2) mouse IgG at 4 °C for 2 hours, washed with HBSS, and reacted with streptavidin-conjugated MG980A magnetic beads at 4 °C for 2 hours. The cells were captured based on magnetic sweeping to harvest the beads from cell suspension using the MagSweeper instrument (Illumina Inc.) as previously described12. The harvested cells were stained with 5 μg/ml Calcein AM (Life Technologies, Carlsbad, CA) in HBSS for 20 minutes to identify viable cells. Manual picking of viable cells showing desired Calcein-positive/CD45-negative/bead-attached profile was performed to isolate cells for molecular profiling. The individual cells were placed into 2.5 μl of Superblock (Thermo Scientific, Waltham, MA) containing 4000 Unit/ml RNase inhibitor (New England Biolabs, Ipswich, MA) and stored at −80 °C until preparation of Smart-Seq libraries.
Isolation of mouse oocytes and human lymphocytes
MII oocytes were isolated from 4-week old CAST/EiJ female mice. Mice were superovulated by injection of 5 IU PMSG, followed by injection of 5 IU of hCG 48 hrs later. MII oocytes were isolated 14–15 hrs after hCG treatment by dissection of the ampulla of the oviduct and cumulus cells were removed by hyaluronidase digestion. Single oocytes were manually picked, lysed in dilution buffer, and cDNA constructed as described above. Peripheral blood lymphocytes from healthy human volunteers were isolated on Ficoll gradients using LymphoPrep (Fresenius Kabi, Norway). Individual cells were manually picked into lysis buffer and cDNA constructed as described above.
Alignment of short reads to genome and transcriptome
Reads were independently aligned using Bowtie21 against the respective genome assembly (hg19 or mm9) and transcriptome sequences (Ensembl, human and mouse annotations downloaded 16th May 2011 and 13th Dec 2010, respectively). Transcriptome mapped reads were converted from transcriptome coordinates to genomic coordinates and thereafter compared with the genome mapped reads to identify reads that map to a unique genomic location. This procedure ensured that mapped reads were unique across both the genome and transcriptome, while allowing for reads to map to different transcripts of the same gene in the initial transcriptome mapping. The uniquely mapped reads were converted to binary BAM files using Samtools22. The resulting transcriptome data was visualized using the Integrated Genome Viewer (IGV, Broad Institute) using the histogram visualization for Fig. 1a,d and heatmap visualization for Supplementary Fig. 3.
Expression level estimation and technical comparisons of sensitivity and variation
Gene expression levels for Refseq transcripts were summarized as RPKM values and read counts using rpkmforgenes23. RefSeq annotations for human and mouse were downloaded on the 31st August 2011 and 13th Dec 2010 respectively. RPKM calculations only considered uniquely mappable positions for transcript length normalizations using the ENCODE Mappability track (wgEncodeCrgMapabilityAlign50mer.bigWig) for human and in-house computed uniqueness files for mouse. Overlapping RefSeq transcripts were collapsed giving one expression value per gene locus. Only 10 million randomly selected mapped reads were used per sample in order to compare sensitivity and variation in gene and exon levels. Samples with fewer than 10 million uniquely mappable reads (a few ES cells8) were therefore discarded from analyses. Samples with 20 pg of total RNA (used in Fig. 2b,d) were simulated by using 5 million reads each of two different 10 pg samples. Analyses of gene detection (Fig. 2a,b and Supplementary Fig. 4a,b) were calculated over pairs of technical replicates or individual cells. Genes were binned by the highest expression level of the two samples, and was considered detected if it had an RPKM above 0.1 in both samples. The mean for all possible pairs of technical replicates within a group was used together with standard deviations using the adjusted Wald method. Analyses of variation (Fig. 2b,d) were also calculated on pairs of samples, binning genes by the mean of log expression, excluding genes below 0.1 RPKM in either sample. Since gene expression levels across single cells are often log normally distributed24, we calculated absolute difference in log10 expression values and standard deviations by multiplying mean variation in a bin with 0.886. Scatter plots were generated in R using smoothScatter (geneplotter package) and loess nonlinear regression using the graphics package. Pearson and Spearman correlations were computed using absolute or relative expression levels as log2 RPKM values. We included publicly available human UHRR, brain and LNCaP data for comparison4,25–27. To analyze how sensitivity and variation improve with a larger numbers of cells, we used Smart-Seq data generated from 10 LNCaP cells (Supplementary Table 1). In order to obtain estimates for the effect for larger numbers of cells (used in Fig. 2b,d), we created two combined samples using 25, 10 and 3 cell samples from picked LNCaP cells, 25, 10 and 5 cell samples from LNCaP cells spiked into healthy donor’s blood and isolated using the EPCAM marker, and 2 single-cell LNCaP samples, achieving a total of 80 cells per each of the two sample pools. These were sequence-depth matched to 10 millions reads, by using 125,000 random reads from single-cell samples, 375,000 from 3-cell samples etc.
Analyses of read coverage across transcriptome
The read coverage analyses were based on human and mouse RefSeq transcripts. Reads were mapped to RefSeq transcripts directly rather than to the genome, using Bowtie allowing for up to 10 hits per read. Each transcript was divided into forty equally sized bins and the number of reads was counted for each bin and gene. The read count per bin for each gene was divided by total read count for that gene before the bins for all the different genes were summed up. The calculated read coverage per bin was later normalized through the division by the bin with the largest read coverage. The mean and standard deviation over replicates were shown in (Fig. 1 and Supplementary Fig. 2) including all transcripts with at least 10 mapped reads. Analyses of full-length transcript reconstructions were based on RefSeq annotations and we defined full-length reconstructed genes as those for which we obtained correct exon-intron structure throughout all annotated exons of at least one isoform. We limited the analyses to expressed (≥ 0.1 RPKM) and multi-exon (≥ 2 exons) genes.
Singular value decomposition
The global transcript expression values for cancer cells were analyzed using singular value decomposition (SVD) to determine the fundamental patterns in the transcriptomes. The expression levels in RPKM were normalized to unit length and the SVD computed using SVDMAN28. Each cell was then projected onto the two strongest SVD components to visualize the overall similarity in gene expression (Fig. 3a).
Analyses of differential expression
One-way analysis of variance (ANOVA) was performed on expression levels (RPKM, log2) followed by Tukey post-hoc test in R/Bioconductor. Only genes significant after multiple testing corrections (5% FDR, Benjamini-Hochberg) were evaluated with post-hoc test (p-values < 0.05). Lists of significantly differential expressed genes are found in Supplementary Table 4 for CTC, PM and melanoma cell line comparisons, and in Figure 3a for comparisons between prostate and bladder cancer cell line cells.
Selection of marker genes for melanoma and immune cells
In order to identify the 100 transcripts most strongly associated with melanoma and immune cells, respectively, we initially calculated the difference in mean gene expression between melanoma samples29 and a combination of monocytes30, T-cells31, white blood cells and lymphnode samples (from Fig. 4a). The differences were divided by the highest expression value in any of the samples, to avoid differences driven by outlier expression values in one replicate only. We ranked genes according to this metric and selected the 100 strongest markers for the melanoma and for the immune cell combination. We then evaluated the mean expression values of each gene in the individual putative CTCs. To include the monocyte SAGE data, we converted 1.5 RPM to 1 RPKM, assuming an average transcripts length of 1.5 kb23.
Detection of alternatively spliced exons
We analyzed exon inclusion levels for a collection of alternatively skipped exons previously identified from EST and cDNA data4. We used the Mixture of Isoforms (MISO) framework11 to calculate exon inclusion levels with confidence intervals. We used the default MISO settings, which require at least 20 reads mapping to the alternative exon or the immediate upstream or downstream exon or exon-exon junctions between them. For a fair assessment of coverage across known alternatively spliced exons (Fig. 3b), we matched the sequence-depth by randomly sampling 10 million uniquely mapped reads per sample.
Hierarchical clustering analyses
Genes with average expression above 20 RPKM (3,690 genes) were clustered by Spearman correlation and complete linkage using python scipy (hcluster). To evaluate the significance (or robustness) of each branchpoint, we generated thousand bootstrap gene set replicates that were independently clustered and from these we counted the percentage of times each branch was recovered.
Analyses of differential exon inclusion
To find significant differences in inclusion levels of alternative exons we applied a t-test with variance shrinkage, known to counter-act false positives in microarray analyses32. A variance was calculated for each alternative exon based on the exon inclusion levels across biological replicates. For each sample group (cell line) the 90th percentile of the variation was included in the variance term ((90th percentile variation + gene variation)/2) when calculating the t-statistic. The null distribution of the t-statistic was calculated by shuffling the sample labels (cell to cell line mapping) repeatedly and for each shuffle compute the t-statistics, thus allowing the conversion of t-statistics to p-values for the cancer cell comparison. To estimate false discovery rates, the sample groups were randomly split in half and combined with half from the other sample group, and the number of significant exons was counted using the t-statistics introduced above (repeatedly, to vary the random splitting of sample groups). The false discovery rate was then estimated as the number of significant exons in random shuffles divided by the number of significant events with correct sample groups. The numbers of significant exons at different false discovery rates were presented in Fig. 3d.
SNP and mutation detection
CTC RNA-Seq Fastq files were mapped to transcriptome (Ensembl, annotations downloaded 16th May 2011) and genome with BWA33, allowing for no indels and removing multi-mapping reads. Samtools rmdup22 was used to filter PCR duplicates, and BAM files were reordered by Picard (http://picard.sourceforge.net). Variant sites were called by the Genome Analysis Toolkit34 jointly on reads from all six CTC samples, with a quality score threshold for sites of 500 and requiring detection in two or more samples (see Supplementary Figure 11 for more detailed information on varying these threshold). We limited the analyses to transcribed regions using RefSeq gene models, and the last 35 bp of transcripts were not considered to remove false positives arising from mapping of reads with partial poly(A) tail. Analyses of overlap with known SNPs were bases on dbSNP build 13235.
Supplementary Material
Acknowledgments
We thank Chris Burge and Gösta Winberg for critical reading of the manuscript, Tiffany Juarez and Jenna Cotton at the Rebecca and John Moores Cancer Center at UCSD for their professional help in IRB protocol preparation and aquisition of clinical samples, AmirAli Talasaz and Gordon Cann for assistance with the Magsweeper, Science for Life laboratory (Stockholm) for assistance with MiSeq sequencer. Y.C.W was supported by a fellowship from the Marie Mayer Foundation. L.C.L. was supported by NIH K12HD001259. J.F.L. was supported by NIH R33MH87925 and CIRM (CL1–00502, RT1–01108, TR1–01250, and RN2–00931). R.S. was supported by European Research Council (Starting grant 243066), Swedish Research Council (2008–4562), Foundation for Strategic Research (FFL4) and Åke Wiberg Foundation (756194131).
Footnotes
The reported sequence read data have been deposited in Gene Expression Omnibus at NCBI (GSE38495). S.L., R.L., I.K., and G.P.S. are employees and shareholders of Illumina Inc, but the remaining authors have no competing financial interests.
References
- 1.Mortazavi A, Williams B, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008 doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 2.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 6.Kurimoto K, et al. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 8.Tang F, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell stem cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Islam S, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome research. 2011 doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iscove NN, et al. Nature Biotechnology. Vol. 20. Nature Publishing Group; 2002. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA; pp. 940–943. [DOI] [PubMed] [Google Scholar]
- 11.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talasaz AH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011 doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jungbluth AA, et al. Expression of melanocyte-associated markers gp-100 and Melan-A/MART-1 in angiomyolipomas. An immunohistochemical and rt-PCR analysis. Virchows Arch. 1999;434:429–435. doi: 10.1007/s004280050362. [DOI] [PubMed] [Google Scholar]
- 15.Tomita Y, Montague PM, Hearing VJ. Anti-T4-tyrosinase monoclonal antibodies--specific markers for pigmented melanocytes. J Invest Dermatol. 1985;85:426–430. doi: 10.1111/1523-1747.ep12277121. [DOI] [PubMed] [Google Scholar]
- 16.Fang D, Setaluri V. Role of microphthalmia transcription factor in regulation of melanocyte differentiation marker TRP-1. Biochem Biophys Res Commun. 1999;256:657–663. doi: 10.1006/bbrc.1999.0400. [DOI] [PubMed] [Google Scholar]
- 17.Chomez P, et al. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 18.Tang A, et al. E-cadherin is the major mediator of human melanocyte adhesion to keratinocytes in vitro. Journal of Cell Science. 1994;107(Pt 4):983–992. doi: 10.1242/jcs.107.4.983. [DOI] [PubMed] [Google Scholar]
- 19.Duncan LM, et al. Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis. Cancer Res. 1998;58:1515–1520. [PubMed] [Google Scholar]
- 20.Gudbjartsson DF, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 21.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS computational biology. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengtsson M, Ståhlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Research. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Au KF, Jiang H, Lin L, Xing Y, Wong WH. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38:4570–4578. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sam LT, et al. A Comparison of Single Molecule and Amplification Based Sequencing of Cancer Transcriptomes. PLoS ONE. 2011;6:12. doi: 10.1371/journal.pone.0017305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wall ME, Dyck PA, Brettin TS. SVDMAN--singular value decomposition analysis of microarray data. Bioinformatics. 2001;17:566–568. doi: 10.1093/bioinformatics/17.6.566. [DOI] [PubMed] [Google Scholar]
- 29.Berger MF, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–427. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zawada AM, et al. SuperSAGE evidence for CD14++CD16+ monocytes as a third monocyte subset. Blood. 2011;118:e50–e61. doi: 10.1182/blood-2011-01-326827. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature Biotechnology. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature reviews Genetics. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Research. 1999;9:677–679. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.