Abstract
Background
Non-Hodgkin lymphoma (NHL) is a malignancy of lymphocytes, and there is growing evidence for a role of germline genetic variation in immune genes in NHL etiology.
Methods
To identify susceptibility immune genes, we conducted a 2-stage analysis of single nucleotide polymorphisms (SNPs) from 1,253 genes using the Immune and Inflammation Panel. In Stage 1, we genotyped 7,670 SNPs in 425 NHL cases and 465 controls, and in Stage 2 we genotyped the top 768 SNPs on an additional 584 cases and 768 controls. The association of individual SNPs with NHL risk from a log-additive model was assessed using the Odds Ratios (ORs) and 95% confidence intervals (CI).
Results
In the pooled analysis, only the TAP2 coding SNP rs241447 (MAF=0.26; Thr655Ala) at 6p21.3 (OR=1.34, 95%CI 1.17-1.53) achieved statistical significance after accounting for multiple testing (p=3.1 × 10−5). The TAP2 SNP was strongly associated with follicular lymphoma (FL, OR=1.82, 95%CI 1.46-2.26; p=6.9 × 10−8), and was independent of other known loci (rs10484561 and rs2647012) from this region. The TAP2 SNP was also associated with diffuse large B-cell lymphoma (DLBCL, OR=1.38, 95% CI 1.08-1.77; p=0.011), but not chronic lymphocytic leukemia (OR=1.08; 95% CI 0.88-1.32). Higher TAP2 expression was associated with the risk allele in both FL and DLBCL tumors.
Conclusion
Genetic variation in TAP2 was associated with NHL risk overall, and FL risk in particular, and this was independent of other established loci from 6p21.3.
Impact
Genetic variation in antigen presentation of HLA class I molecules may play a role in lymphomagenesis.
Keywords: genetics, non-Hodgkin lymphoma, immune function, single nucleotide polymorphisms
Introduction
Non-Hodgkin lymphoma (NHL) is a group of heterogeneous malignancies of B and T lymphocytes, as well as other immune cells, although in western populations, B-cell malignancy predominates. Immune dysfunction is an established risk factor for NHL (1), and there is accumulating evidence from multiple independent candidate gene studies that genetic variation in genes involved in immune function and inflammation is associated with NHL risk (2-12). Genome-wide association studies have also identified several loci in and around the HLA region on chromosome 6p21.32-33 (13-16). We previously conducted and published an analysis of NHL risk (425 cases, 465 controls) using the ParAllele (now Affymetrix) Immune and Inflammation Panel, which included 1253 genes that were tagged with 9412 single nucleotides polymorphisms (SNPs) (17). Here, we report the results for a second stage validation of the top 10% of SNPs from that analysis in a new set of 584 NHL cases and 768 controls, and then a pooled analysis on all 1009 cases and 1233 controls. We also formally assessed associations within the most common NHL subtypes: chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL), follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCL).
Methods
Study population and data collection
This study was reviewed and approved by the Human Subjects Institutional Review Board at the Mayo Clinic, and all participants provided written informed consent. Full details on this case-control study have been previously published (17, 18). Briefly, starting on 9/1/02, we offered enrollment to all consecutive cases of newly diagnosed, pathologically-confirmed lymphoma (including CLL) who were age 20 years and older and a resident of Minnesota, Iowa or Wisconsin at the time of diagnosis except cases with a history of HIV infection or who did not speak English. A Mayo Clinic hematopathologist reviewed all materials for each case to verify the diagnosis and to classify each case according to the World Health Organization Classification of Neoplastic Diseases of the Hematopoietic and Lymphoid Tissues (19). This analysis included all subjects enrolled into the study from 9/1/02 through 2/29/08. Of the 1798 eligible patients identified, 1236 (69%) participated, 183 (10%) refused, 39 (2%) were lost to follow-up (i.e., we were unable to contact after multiple attempts), and 340 (19%) did not complete all data collection within 12 months of diagnosis.
Clinic-based controls were recruited from Mayo Clinic Rochester patients under evaluation for a pre-scheduled medical examination in the general medicine divisions of the Departments of Medicine or Family Medicine from 9/1/02 through 2/29/08. Controls had to be at least 20 years old, a resident of Minnesota, Iowa or Wisconsin at time of appointment, and no history of lymphoma or leukemia; controls with a history of HIV infection or who did not speak English were not eligible. Controls were frequency matched to the case distribution on 5-year age group, sex, and geographic location of residence using a computer program that randomly selects subjects from eligible patients. Of the 1899 eligible subjects identified, 1315 (69%) participated, 548 (29%) refused and 36 (2%) did not complete data collection within 12 months of selection.
Participants completed a self-administered risk-factor questionnaire and provided a peripheral blood sample for serum and DNA studies. DNA was extracted from blood samples using a standard procedure (Gentra, Inc).
Genotyping
All participants who had an adequate DNA sample were genotyped as part of a larger genotyping project on a custom Illumina GoldenGate 1536 SNP OPA. Individuals included in the original case-control series (17) were defined as Stage 1 (discovery set); the remainder of the participants were defined as Stage 2 (replication set). Cases diagnosed with Hodgkin lymphoma were excluded from this analysis. A total of 1050 cases and 1274 controls were randomly arranged on 96-well plates, with 50 samples plated in duplicate. One of the CEPH family trios was included on every plate and was also duplicated across each of the plates. The inclusion of these three samples aided in genotyping concordance calculations as well as determination of non-Mendelian inheritance patterns. For duplicated samples, the sample with the higher call rate was used for analysis.
We selected the top 800 (approximately 10%) of the 7670 SNPs that were successfully genotyped in Stage 1 (i.e., passed quality control and were not monomorphic) to genotype in Stage 2; selection was based on the minor allele frequency (MAF) > 5% in control subjects and the trend p-value from the analyses of all Caucasian NHL cases and controls. Of these SNPs, 23 failed Illumina design for this round of genotyping, while four others were no longer mapped uniquely to the same location on the genome. The remaining 773 were genotyped. Using Plink software, we evaluated the genotyping quality. We dropped SNPs with call rates <95% (N=33), SNPs that were monomorphic (N=2), and SNPs that had poor genotype clustering (N=1). After dropping 82 subjects (41 cases and 41 controls) with call rates <90%, we had 1009 cases and 1233 controls in the combined analyses of Stage 1 and 2. Concordance amongst duplicate samples was >99.9%. Hardy-Weinberg equilibrium (HWE) was evaluated among the control subjects for each SNP using an exact test. SNPs with an HWE p-value less than 1×10−3 (N=19) were deemed questionable and were examined further by examining cluster plots. All plots appeared reasonable and no further exclusions were made. Thus, there was a final total of 737 SNPs available for analysis.
Gene expression analysis
Whole exome sequencing (on paired tumor/normal) and gene expression levels from initial (frozen) diagnostic specimens of 36 DLBCL tumors were available (20), of which 11 were also genotyped in this study. Affymetrix HG-U133 plus2.0 microarray chips were used for gene expression profiling and the data were preprocessed using the RMA method (21). We also had whole exome (paired tumor/normal) and RNA next generation sequencing (RNAseq) from initial (frozen) diagnostic specimens of 8 FL tumors (unpublished data); none of these specimens overlapped this study. We compared the gene expression levels from the Affymetrix chip by SNP genotype based on the Illumina OPA genotype call for DLBCL, and the RNAseq levels by SNP genotype based on the tumor exome genotype for FL.
Gene regulatory network analysis
The MetaCore’s auto-expand algorithm (GeneGo Inc. San Diego CA) was used for regulatory gene network analysis. The genes implied by the SNPs were used as the input genes to build the network using the canonical pathways. The auto-expand algorithm draws sub-networks around the input genes and the expansion halts when the sub-networks intersect.
Statistical analysis
Unconditional logistic regression was used to estimate odds ratios (OR) and 95% confidence intervals (CI) for the association between NHL case status and each SNP. Analyses were adjusted for age (including its functional form) and gender and the most common homozygous genotype was treated as the referent category for each of the SNPs. Each SNP was modeled in a log-additive manner in the regression model and the Wald p-value was used to assess significance. Analyses were conducted for Stage 1 Stage 2, and then combined.
The primary analysis focused on all NHL and p<0.001 for the log-additive model in the combined analyses. To determine the proper multiple-comparisons correction for this two-stage design, we used PLINK to subset our original discovery-phase 7670 SNPs, into a set of independent SNPs (R2=0) using the variance inflation factor sliding window approach. The number of independent SNPs (n=352) was then used as a Bonferroni correction for our pooled analyses of Stage 1 and 2 subjects. SNPs with a trend p-value below 1.4×10−4 (=0.05/352) were considered of interest for associations with NHL overall. For SNPs meeting this criterion, we further evaluated other available SNPs from the local region as well as the association with major NHL subtypes (CLL/SLL, DLBCL, FL). The multiple testing threshold for SNPs associated with NHL subtypes was a trend p-value below 4.7×10−5 (0.05/(352*3)). Statistical analyses utilized SAS software (SAS Institute, Inc., Cary, NC).
Results
Cases and controls were well balanced on the study design factors of age, sex and state of residence in each stage (Table 1). The pooled dataset had 1009 NHL cases and 1233 controls, and the most common NHL subtypes were CLL/SLL (N=327), FL (N=238) and DLBCL (N=189).
Table 1.
Characteristics of Stage 1 and Stage 2, Mayo Clinic case-control study of NHL, 2002-2008.
| Stage 1 (425 cases, 465 controls) |
Stage 2 (584 cases, 768 controls) |
Pooled Estimate (1009 cases, 1233 controls) |
||||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |
| Age | ||||||
| <40 | 21 (4.9%) | 29 (6.2%) | 25 (4.3%) | 61 (7.9%) | 46 (4.6%) | 90 (7.3%) |
| 40-49 | 73 (17.2%) | 53 (11.4%) | 69 (11.8%) | 117 (15.2%) | 142 (14.1%) | 170 (13.8%) |
| 50-59 | 80 (18.8%) | 95 (20.4%) | 138 (23.6%) | 161 (21%) | 218 (21.6%) | 256 (20.8%) |
| 60-69 | 135 (31.8%) | 138 (29.7%) | 185 (31.7%) | 223 (29%) | 320 (31.7%) | 361 (29.3%) |
| 70+ | 116 (27.3%) | 150 (32.3%) | 167 (28.6%) | 206 (26.8%) | 283 (28%) | 356 (28.9%) |
| Sex | ||||||
| Male | 252 (59.3%) | 262 (56.3%) | 349 (59.8%) | 411 (53.5%) | 601 (59.6%) | 673 (54.6%) |
| Female | 173 (40.7%) | 203 (43.7%) | 235 (40.2%) | 357 (46.5%) | 408 (40.4%) | 560 (45.4%) |
| Residence | ||||||
| Minnesota | 276 (64.9%) | 313 (67.3%) | 399 (68.3%) | 509 (66.3%) | 675 (66.9%) | 822 (66.7%) |
| Iowa | 86 (20.2%) | 85 (18.3%) | 110 (18.8%) | 159 (20.7%) | 196 (19.4%) | 244 (19.8%) |
| Wisconsin | 63 (14.8%) | 67 (14.4%) | 75 (12.8%) | 100 (13%) | 138 (13.7%) | 167 (13.5%) |
| Family History of NHL | ||||||
| No | 327 (95.1%) | 387 (96.7%) | 467 (94.9%) | 644 (97.4%) | 794 (95.0%) | 1031 (97.2%) |
| Yes | 17 (4.9%) | 13 (3.3%) | 25 (5.1%) | 17 (2.6%) | 42 (5.0%) | 30 (2.8%) |
| NHL Subtype | ||||||
| CLL/SLL | 123 (30.8%) | 204 (37.6%) | 327 (34.7%) | |||
| FL | 113 (28.3%) | 125 (23%) | 238 (25.2%) | |||
| DLBCL | 65 (16.3%) | 124 (22.8%) | 189 (20%) | |||
| MZL | 30 (7.5%) | 29 (5.3%) | 59 (6.3%) | |||
| MCL | 26 (6.5%) | 27 (5%) | 53 (5.6%) | |||
| TCL | 16 (4%) | 19 (3.5%) | 35 (3.7%) | |||
| Other/NOS | 27 (6.8%) | 15 (2.8%) | 42 (4.5%) | |||
SNPs in the pooled analysis with a p<0.001 are shown in Table 2. Only the top ranked SNP from TAP2 met the corrected p-value threshold of 1.4×10−4. This TAP2 SNP is common (minor allele frequency 0.26) and leads to a coding change at position 665 (Thr→Ala). Compared to the GG genotype, there was an increased risk of NHL with the GA (OR=1.30; 95% CI 1.09-1.55) and the AA (OR=1.89; 95% CI 1.33-2.68) genotypes.
Table 2.
SNPs ranked by p-value (p<0.001) from the pooled analysis of a 2-stage evaluation of the ParAllele (Affymetrix) Immune and Inflammation Panel, Mayo Clinic case-control study of NHL, 2002-2008.
| Stage 1 (425 cases, 465 controls) |
Stage 2 (584 cases, 768 controls) |
Pooled Estimate (1009 cases, 1233 controls) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP rsID | Gene | chr | Minor Allele |
OR* (95% CI) | p- value |
OR* (95% CI) | p-value | MAF† | OR* (95% CI) | p-value |
| rs241447 | TAP2 | 6 | G | 1.30 (1.05, 1.60) | 0.015 | 1.36 (1.13, 1.63) | 0.00099 | 0.24 | 1.34 (1.17, 1.53) | 0.000031 |
| rs2857597 | AIF1 | 6 | A | 0.78 (0.63, 0.98) | 0.029 | 0.76 (0.63, 0.91) | 0.0031 | 0.28 | 0.77 (0.67, 0.88) | 0.00019 |
| rs754505 | NFATC1 | 18 | A | 0.65 (0.44, 0.95) | 0.028 | 0.62 (0.44, 0.86) | 0.0045 | 0.08 | 0.63 (0.49, 0.81) | 0.00032 |
| rs6746608 | BCL2L11 | 2 | A | 0.86 (0.71, 1.04) | 0.13 | 0.77 (0.66, 0.90) | 0.0012 | 0.46 | 0.81 (0.71, 0.91) | 0.00044 |
| rs3819545 | VDR | 12 | G | 1.25 (1.03, 1.52) | 0.023 | 1.25 (1.07, 1.46) | 0.0059 | 0.36 | 1.24 (1.10, 1.40) | 0.00048 |
| rs1894408 | HLA-DOB | 6 | G | 1.15 (0.95, 1.39) | 0.14 | 1.29 (1.10, 1.51) | 0.0018 | 0.36 | 1.23 (1.09, 1.39) | 0.00076 |
| rs7425883 | ZAP70 | 2 | C | 0.88 (0.70, 1.10) | 0.26 | 0.71 (0.59, 0.86) | 0.00062 | 0.22 | 0.78 (0.67, 0.90) | 0.00077 |
| rs2365736 | PLXNC1 | 12 | G | 1.24 (1.02, 1.50) | 0.034 | 1.25 (1.06, 1.46) | 0.0065 | 0.35 | 1.23 (1.09, 1.40) | 0.00078 |
| rs4764191 | PTPRO | 12 | A | 0.63 (0.43, 0.92) | 0.017 | 0.70 (0.51, 0.95) | 0.024 | 0.08 | 0.66 (0.52, 0.84) | 0.00084 |
Odds Ratio (OR) and 95% confidence interval (CI), adjusted for age and sex.
Minor allele frequency (MAF) among controls.
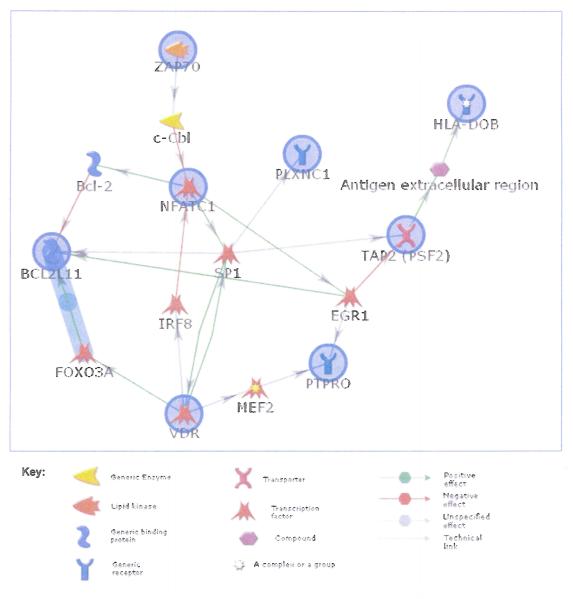
Besides the SNP from TAP2, there were two other SNPs, rs2857597 from AIF1 and rs1894408 from HLA-DOB, that were from the 6p21.3 region, while the other top SNPs were from genes on chromosome 18 (NFATC1), 2 (ZAP70) and 12 (VDR, PLXNC1, PTPRO). When we performed a regulatory network analysis of these 9 genes using MetaCore’s auto-expand algorithm, 8 out of 9 genes (excluding AIF1) were functionally connected with only 1 node (gene) away from each other (Figure 1), suggesting that virtually all of the top hits from the study are closely related from regulatory perspective.
Figure 1.
Regulatory network analysis of the top genes with SNPs with p<0.001 using MetaCore’s shortest pathway algorithm (GeneGo Inc.); the nodes that are circled are the genes of interest.
We next evaluated the chromosome 6p21 region with all SNPs available from the replication phase along with results for the major NHL subtypes (Table 3). There were several additional nominally significant (p<0.01) SNPs between TAP2 and AIF1, including SNPs in BAT3, C2, and HLA-DRA. In NHL subtype analyses, the strongest associations for SNPs from this region were for FL: rs241447 (TAP2), rs1894408 (HLA-DOB), rs7192 (HLA-DRA) and rs7746553 (C2), and of these three SNPs, all exceeded our multiple testing p-value for the subtype analyses (i.e., 4.7 × 10−5). For CLL/SLL and DLBCL, SNPs from the 6p21.3 region did not meet the multiple testing threshold p-value of 4.7 × 10−5, but they did show similar, albeit slightly weaker, ORs for CLL/SLL (except for TAP2 and C2 SNPs) and DLBCL (except for the HLA-DR SNPs).
Table 3.
Results for all NHL and NHL subtypes for 6p21.3 region, pooled results, Mayo Clinic case-control study of NHL, 2002-2008.
| All NHL |
CLL/SLL |
Follicular Lymphoma |
DLBCL |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID | Gene | Position | Minor Allele |
MAF | OR* (95% CI) | p-trend | OR* (95% CI) | p-trend | OR* (95% CI) | p-trend | OR* (95% CI) | p-trend |
| rs3093986 | LOC† | 31601400 | A | 0.21 | 0.80 (0.69, 0.93) | 0.0045 | 0.84 (0.67, 1.04) | 0.11 | 0.57 (0.43, 0.75) | 8.5E-05 | 0.80 (0.60, 1.07) | 0.13 |
| rs2857597‡ | AIF1 | 31692979 | A | 0.28 | 0.77 (0.67, 0.88) | 0.00019 | 0.85 (0.69, 1.04) | 0.11 | 0.64 (0.50, 0.82) | 0.00035 | 0.63 (0.48, 0.83) | 0.0013 |
| rs2242656 | BAT3 | 31722081 | G | 0.21 | 0.79 (0.68, 0.91) | 0.0017 | 0.74 (0.59, 0.92) | 0.0081 | 0.63 (0.48, 0.82) | 0.00068 | 0.80 (0.60, 1.06) | 0.12 |
| rs7746553 | C2 | 32003952 | C | 0.14 | 1.31 (1.11, 1.54) | 0.0011 | 1.13 (0.89, 1.43) | 0.31 | 1.68 (1.32, 2.14) | 2.3E-05 | 1.59 (1.21, 2.10) | 0.00090 |
| rs8084 | HLA-DRA | 32519013 | A | 0.47 | 0.85 (0.76, 0.96) | 0.0073 | 0.80 (0.67, 0.95) | 0.012 | 0.67 (0.55, 0.82) | 7.9E-05 | 0.92 (0.73, 1.14) | 0.44 |
| rs7192 | HLA-DRA | 32519624 | A | 0.43 | 0.84 (0.75, 0.95) | 0.0040 | 0.80 (0.67, 0.95) | 0.012 | 0.57 (0.46, 0.70) | 1.1E-07 | 0.94 (0.75, 1.17) | 0.57 |
| rs1894408 | HLA-DOB | 32894811 | G | 0.36 | 1.23 (1.09, 1.39) | 0.00076 | 1.17 (0.98, 1.39) | 0.090 | 1.62 (1.33, 1.98) | 1.5E-06 | 1.14 (0.91, 1.43) | 0.27 |
| rs241447‡ | TAP2 | 32904728 | G | 0.24 | 1.34 (1.17, 1.53) | 3.1E-05 | 1.08 (0.88, 1.32) | 0.48 | 1.82 (1.46, 2.26) | 6.9E-08 | 1.38 (1.08, 1.77) | 0.011 |
| rs714289 | HLA-DMB | 33013789 | G | 0.07 | 0.99 (0.78, 1.25) | 0.93 | 1.02 (0.73, 1.43) | 0.90 | 0.69 (0.44, 1.08) | 0.11 | 1.00 (0.63, 1.56) | 0.98 |
Ordinal OR and 95% confidence interval from the log additive model, adjusted for age and sex.
LOC100129921
Top hit from Table 2.
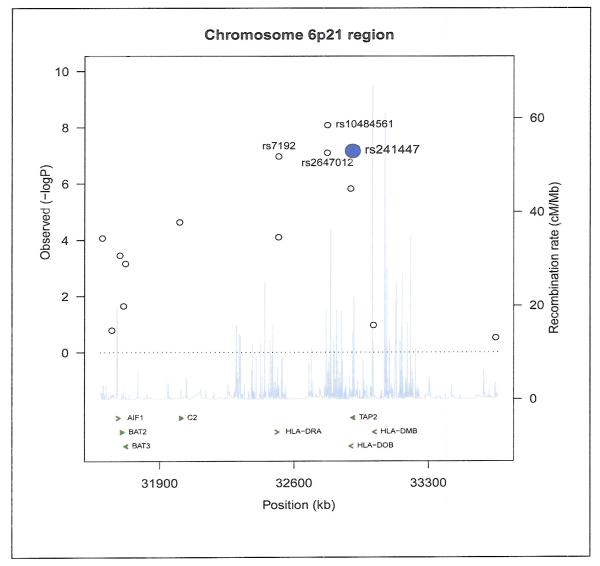
Also available from the larger genotyping project were two GWAS SNPs previously identified in the 6p21.3 region for FL but which were not on the Immune and Inflammation SNP platform: rs10484561 published by Conde et al. (14) and rs2647012 published by Smedby et al. (15); the Mayo Clinic study contributed primary data to the latter study for rs2647012. Figure 2 shows our results for this region for FL. There were strong associations for both of these FL GWAS SNPs: rs10484561 (allelic OR=2.23, 95% CI 1.70-2.92; p-trend=8.26 × 10−9) and rs2647012 (OR=0.56, 95% CI 0.45-0.69; 8.03 × 10−8). Our top FL SNP rs241447 (TAP2) was not in strong LD with the FL GWAS SNPs rs10484561 (r2=0.16; D’=0.67) or rs2647012 (r2=0.014; D’=0.25) based on genotypes in our 1233 controls. After simultaneous adjustment for all three SNPs in a logistic regression analysis, rs10484561 (allelic OR=2.16, 95% CI 1.66-2.81; p-trend=1.11 × 10−8), rs2647012 (OR=0.57, 95% CI 0 0.46-0.70; 1.04 × 10−7), and rs241447 (OR=1.81; 95% CI 1.46-2.24; p-trend=6.89 × 10−8) remained significant, supporting independent effects. We observed no interactions between our top hit rs241447 and either FL GWAS SNPs rs10484561 or rs2647012 (data not shown). We did not genotype the third GWAS SNP rs6457327; however, in HapMap data, this SNP was not in LD with rs241447 (r2=0.012).
Figure 2.
Plot of observed p-values for 6p21 region for follicular lymphoma, highlighting the top hits from this study (rs241447 and rs7192) and the two published GWAS hits (rs10484561 and rs2647012).
The other SNP strongly associated with FL, rs7192 from HLA-DRA, was not in strong LD with our top FL SNP rs241447 (; r2=0.037; D’=0.40) nor with rs10484561 identified by the Conde et al. (r2=0.077; D’=0.96), and rs7192 remained significant after adjustment for our top FL SNP rs241447 (OR=0.61; 95% CI 0.49-0.75; p=7.9 × 10−6) and for the Conde et al (14) FL GWAS SNP rs10484561 (OR=0.64; 95% CI 0.51-0.79; 5.1 × 10−5). In contrast, rs7192 was in stronger LD with the Smedby et al SNP rs2647012 (r2=0.52; D’=0.73), and after adjustment for the latter SNP, rs7192 remained marginally statistically significant (OR=0.71; 95% CI 0.53-0.96; p=0.028).
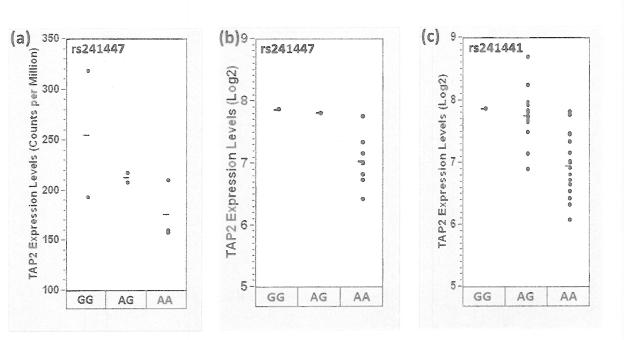
Finally, we explored whether rs241447 genotype was associated with TAP2 mRNA expression. From the set of 8 FLs with paired tumor-normal exome and RNAseq data, there was a trend of higher TAP2 expression in patients with the GG or GA compared to the AA genotype (p=0.14) (Figure 3a). In the case-control study, the dominant model OR for TAP2 in FL (GG or GA versus AA genotype) was 2.01 (95% CI 1.51-2.68). From the set of 11 DLBCL cases genotyped in the case-control study that also had tumor gene expression measured using the Affymetrix HG-U133 plus2.0 microarray chips, there was higher TAP2 expression in patients with the GG or GA compared to the AA genotype (p=3.3 × 10−6) (Figure 3b). In the case-control study, the dominant model OR for TAP2 in DLBCL (GG or GA versus AA genotype) was 1.39 (95% CI 1.01-1.91). For DLBCL, we also assessed genotype based on the exome sequencing available on 36 tumors. Unfortunately, the exome sequencing data did not have sufficient coverage at the rs241447 position to make a reliable genotype call, but a SNP in perfect LD (rs241441) was well covered, and there was higher TAP2 expression in patients with the GG or GA compared to the AA genotype (p=0.0030) (Figure 3c).
Figure 3.
Comparison of the TAP2 genotype and expression levels: (a) FL, rs241447 genotype (from exome sequencing) and TAP2 RNAseq levels (N=8); (b) DLBCL, rs241447 genotype (based on Illumina genotyping) and TAP2 expression level (N=11) and (c) DLBCL, rs241441 genotype (based on exome sequencing) and TAP2 expression level (N=36).
Discussion
We have conducted follow-up analyses of the top 10% of SNPs from the ParAllele (Affymetrix) Immune and Inflammation panel in a new set of 584 cases and 768 controls, for a combined sample of 1009 cases and 1233 controls. We found that the common SNP rs241447 (minor allele frequency 0.26) in TAP2 from the 6p21.3 region showed a significant association with risk of NHL overall after correcting for multiple testing; the association was particularly strong for FL, but was also apparent for DLBCL. Higher TAP2 expression was associated with the risk allele in both FL and DLBCL tumors.
The 6p21.3 region is a large, complex and immune gene-rich region that has been previously implicated as a susceptibility locus for overall NHL risk (5, 10-12, 15). Further, this region has been flagged as a region of interest for not only for NHL, but also for the specific NHL subtypes of FL (10, 11, 13, 14), DLBCL (5, 10, 15), and familial CLL/SLL (16). In NHL subtype analyses, we found genome-wide significance for the TAP2 SNP rs241447 with FL risk, as well as a weaker but still evident association with DLBCL but no association with CLL/SLL. TAP2 was not in the top 40 stage 1 SNPs for FL in either of the published GWA studies (14, 15). The TAP2 SNP rs241447 is predicted to be “damaging” by SIFT (22), and is located in an evolutionary conserved domain across 28 species based on multiz (23) and phastCon (24) calculations. In FL, rs241447 was not in LD with either of the previously identified FL GWAS SNPs rs10484561 (14) and rs2647012 (15), and all three SNPs remained significant in a multivariate model. Our results independently replicate rs10484561 in FL (14), and identify TAP2 as a novel and independent risk loci for FL and perhaps DLBCL.
TAP2 (transporter 2, ATP-binding cassette, sub-family B) is a member of the multidrug resistance protein (MRP)/TAP subfamily of ATP-binding cassette (ABC) transporters, and is involved in both multidrug resistance and antigen presentation (25, 26). TAP2 forms a heterodimer with TAP1 in order to transport peptides (ranging from ions to large proteins) from the cytoplasm to the endoplasmic reticulum (25, 26), and is essential for loading of antigen on HLA class I protein on the cell surface (27). TAP2 and TAP1 are located in the MHC II locus of chromosome 6, between HLA-DOB and HLA-DMB, and genetic variation in these genes has been associated with type 1 diabetes, systemic lupus erythematosus (SLE), and celiac disease (26), conditions that have been associated with overall NHL risk in some studies (1). The TAP2 SNP rs241447 specifically has been positively associated with SLE (OR=1.46 per allele, 95% CI 1.14-1.88) (28) and inversely associated with type 1 diabetes (OR=0.43; 95% CI 0.35-0.52) (29), and these associations were not due to LD with HLA-DRB1 or DR-DQ, respectively. SLE has more consistently been associated with NHL risk, including DLBCL ad FL risk, while the association for type 1 diabetes with NHL overall or for NHL subtypes has been mixed (30).
Some studies have reported LD between TAP2 and HLA class II alleles (31, 32), while others have not (28, 29, 33). While HLA class II alleles (HLA-DRB1*0101 and *13) were associated with follicular lymphoma in one recent study (10), we did not have genotyping for class II alleles and so could not address LD with TAP2, and this remains an important future research question. The TAP2 SNP is also in a region of high LD with several other coding SNPs, including rs241448 (ter687Q) and rs241449 (a synonymous SNP), and haplotypes formed by these alleles leads to alternative splicing and different isoforms of the protein known to have different peptide selectivity (29). Down-regulation or a loss of TAP expression (by mutation or other mechanisms) leads to loss of surface HLA class I expression, allowing tumors to escape immune recognition (25). Our data suggests that common genetic variation in the TAP2 gene is associated with TAP2 expression and increased risk of NHL, particularly FL, raising the hypothesis that TAP2 may predispose to lymphomagenesis, perhaps by influencing antigen presentation of HLA class I molecules.
While no other genes met our multiple testing threshold for all NHL, AIF1 (12), BCL2L11 (34, 35), and VDR (6) have previously been implicated in either NHL overall or one of the common subtypes. Germline genetic variation in ZAP70 has not been associated with NHL risk, but ZAP70 expression has been associated with prognosis in CLL (36). While NFATC1, PLXNC1 and PTPRO have not been associated with NHL, NFATC1 is known to regulate the expression of growth and survival genes including MYC, TNF, CD40L, and BAFF, all of which have also been linked to lymphomagenesis (5, 11, 37, 38). However, given the high potential for false positive results in this setting, our results will need to be replicated in other studies or through pooled analyses.
Strengths of this study include the use of carefully designed case-control study (18); central pathology review and classification; a well characterized, comprehensive panel of immune and inflammation genes based on HapMap SNPs; a two-stage design; and relatively large sample size. Limitations include lower power to assess NHL subtypes and use of a white population, although this enhances internal validity in the setting of a genetic association study. We have previously published data from this study showing lack of population stratification in this study population (17). Finally, we were also able to adjust for the two strongest GWAS SNPs. In summary, TAP2 appears to be a strong candidate susceptibility gene for NHL, particularly FL. Further genetic and protein are needed to confirm abnormalities or aberrant function of TAP2 are warranted.
Acknowledgment
We thank Sondra Buehler for her editorial assistance.
Grant Support: National Cancer Institute/NIH grant R01 CA92153.
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Alexander DD, Mink PJ, Adami HO, Chang ET, Cole P, Mandel JS, et al. The non-Hodgkin lymphomas: A review of the epidemiologic literature. Int J Cancer. 2007;120:1–39. doi: 10.1002/ijc.22719. [DOI] [PubMed] [Google Scholar]
- 2.Forrest MS, Skibola CF, Lightfoot TJ, Bracci PM, Willett EV, Smith MT, et al. Polymorphisms in innate immunity genes and risk of non-Hodgkin lymphoma. Br J Haematol. 2006;134:180–3. doi: 10.1111/j.1365-2141.2006.06141.x. [DOI] [PubMed] [Google Scholar]
- 3.Lan Q, Zheng T, Rothman N, Zhang Y, Wang SS, Shen M, et al. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107:4101–8. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang SS, Cerhan JR, Hartge P, Davis S, Cozen W, Severson RK, et al. Common genetic variants in proinflammatory and other immunoregulatory genes and risk for non-hodgkin lymphoma. Cancer Res. 2006;66:9771–80. doi: 10.1158/0008-5472.CAN-06-0324. [DOI] [PubMed] [Google Scholar]
- 5.Rothman N, Skibola CF, Wang SS, Morgan G, Lan Q, Smith MT, et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol. 2006;7:27–38. doi: 10.1016/S1470-2045(05)70434-4. [DOI] [PubMed] [Google Scholar]
- 6.Purdue MP, Lan Q, Kricker A, Grulich AE, Vajdic CM, Turner J, et al. Polymorphisms in immune function genes and risk of non-Hodgkin lymphoma: findings from the New South Wales non-Hodgkin Lymphoma Study. Carcinogenesis. 2007;28:704–12. doi: 10.1093/carcin/bgl200. [DOI] [PubMed] [Google Scholar]
- 7.Cerhan JR, Liu-Mares W, Fredericksen ZS, Novak AJ, Cunningham JM, Kay NE, et al. Genetic variation in tumor necrosis factor and the nuclear factor-{kappa}B canonical pathway and risk of non-Hodgkin’s Lymphoma. Cancer Epidemiol Biomarkers Prev. 2008;17:3161–9. doi: 10.1158/1055-9965.EPI-08-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skibola CF, Bracci PM, Halperin E, Nieters A, Hubbard A, Paynter RA, et al. Polymorphisms in the estrogen receptor 1 and vitamin C and matrix metalloproteinase gene families are associated with susceptibility to lymphoma. PLoS One. 2008;3:e2816. doi: 10.1371/journal.pone.0002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerhan JR, Novak AJ, Fredericksen ZS, Wang AH, Liebow M, Call TG, et al. Risk of non-Hodgkin lymphoma in association with germline variation in complement genes. Br J Haematol. 2009;145:614–23. doi: 10.1111/j.1365-2141.2009.07675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang SS, Abdou AM, Morton LM, Thomas R, Cerhan JR, Gao X, et al. Human leukocyte antigen class I and II alleles in non-Hodgkin lymphoma etiology. Blood. 2010;115:4820–3. doi: 10.1182/blood-2010-01-266775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skibola CF, Bracci PM, Nieters A, Brooks-Wilson A, de Sanjose S, Hughes AM, et al. Tumor necrosis factor (TNF) and lymphotoxin-alpha (LTA) polymorphisms and risk of non-Hodgkin lymphoma in the InterLymph Consortium. Am J Epidemiol. 2010;171:267–76. doi: 10.1093/aje/kwp383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang SS, Menashe I, Cerhan JR, Cozen W, Severson RK, Davis S, et al. Variations in chromosomes 9 and 6p21.3 with risk of non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2011;20:42–9. doi: 10.1158/1055-9965.EPI-10-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skibola CF, Bracci PM, Halperin E, Conde L, Craig DW, Agana L, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41:873–5. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conde L, Halperin E, Akers NK, Brown KM, Smedby KE, Rothman N, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet. 2010;42:661–4. doi: 10.1038/ng.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smedby KE, Foo JN, Skibola CF, Darabi H, Conde L, Hjalgrim H, et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21.32 and suggests shared genetic susceptibility with diffuse large B-cell lymphoma. PLoS Genet. 2011;7:e1001378. doi: 10.1371/journal.pgen.1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slager SL, Rabe KG, Achenbach SJ, Vachon CM, Goldin LR, Strom SS, et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2011;117:1911–6. doi: 10.1182/blood-2010-09-308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerhan JR, Ansell SM, Fredericksen ZS, Kay NE, Liebow M, Call TG, et al. Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110:4455–63. doi: 10.1182/blood-2007-05-088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerhan JR, Fredericksen ZS, Wang AH, Habermann TM, Kay NE, Macon WR, et al. Design and validity of a clinic-based case-control study on the molecular epidemiology of lymphoma. Int J Mol Epidemiol Genet. 2011;2:95–113. [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffe ES, Harris NL, Stein H, Vardiman JW. World Health Organization Classification of Tumours: Pathology and Genetics, Tumours of Hematopoietic and Lymphoid Tissues. IARC Press; Lyon: 2001. [Google Scholar]
- 20.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphona (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 23.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–15. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lankat-Buttgereit B, Tampe R. The transporter associated with antigen processing: function and implications in human diseases. Physiol Rev. 2002;82:187–204. doi: 10.1152/physrev.00025.2001. [DOI] [PubMed] [Google Scholar]
- 26.Koehn J, Fountoulakis M, Krapfenbauer K. Multiple drug resistance associated with function of ABC-transporters in diabetes mellitus: molecular mechanism and clinical relevance. Infect Disord Drug Targets. 2008;8:109–18. doi: 10.2174/187152608784746510. [DOI] [PubMed] [Google Scholar]
- 27.de la Salle H, Hanau D, Fricker D, Urlacher A, Kelly A, Salamero J, et al. Homozygous human TAP peptide transporter mutation in HLA class I deficiency. Science. 1994;265:237–41. doi: 10.1126/science.7517574. [DOI] [PubMed] [Google Scholar]
- 28.Ramos PS, Langefeld CD, Bera LA, Gaffney PM, Noble JA, Moser KL. Variation in the ATP-binding cassette transporter 2 gene is a separate risk factor for systemic lupus erythematosus within the MHC. Genes Immun. 2009;10:350–5. doi: 10.1038/gene.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu HQ, Lu Y, Marchand L, Bacot F, Frechette R, Tessier MC, et al. Genetic control of alternative splicing in the TAP2 gene: possible implication in the genetics of type 1 diabetes. Diabetes. 2007;56:270–5. doi: 10.2337/db06-0865. [DOI] [PubMed] [Google Scholar]
- 30.Ekstrom Smedby K, Vajdic CM, Falster M, Engels EA, Martinez-Maza O, Turner J, et al. Autoimmune disorders and risk of non-Hodgkin lymphoma subtypes: a pooled analysis within the InterLymph Consortium. Blood. 2008;111:4029–38. doi: 10.1182/blood-2007-10-119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronningen KS, Undlien DE, Ploski R, Maouni N, Konrad RJ, Jensen E, et al. Linkage disequilibrium between TAP2 variants and HLA class II alleles; no primary association between TAP2 variants and insulin-dependent diabetes mellitus. Eur J Immunol. 1993;23:1050–6. doi: 10.1002/eji.1830230511. [DOI] [PubMed] [Google Scholar]
- 32.Cesari M, Hoarau JJ, Caillens H, Robert C, Rouch C, Cadet F, et al. Is TAP2*0102 allele involved in insulin-dependent diabetes mellitus (type 1) protection? Hum Immunol. 2004;65:783–93. doi: 10.1016/j.humimm.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Alvarado-Guerri R, Cabrera CM, Garrido F, Lopez-Nevot MA. TAP1 and TAP2 polymorphisms and their linkage disequilibrium with HLA-DR, -DP, and -DQ in an eastern Andalusian population. Hum Immunol. 2005;66:921–30. doi: 10.1016/j.humimm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Morton LM, Purdue MP, Zheng T, Wang SS, Armstrong B, Zhang Y, et al. Risk of non-Hodgkin lymphoma associated with germline variation in genes that regulate the cell cycle, apoptosis, and lymphocyte development. Cancer Epidemiol Biomarkers Prev. 2009;18:1259–70. doi: 10.1158/1055-9965.EPI-08-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly JL, Novak AJ, Fredericksen ZS, Liebow M, Ansell SM, Dogan A, et al. Germline variation in apoptosis pathway genes and risk of non-Hodgkin’s lymphoma. Cancer Epidemiol Biomarkers Prev. 2011;19:2847–58. doi: 10.1158/1055-9965.EPI-10-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zenz T, Frohling S, Mertens D, Dohner H, Stilgenbauer S. Moving from prognostic to predictive factors in chronic lymphocytic leukaemia (CLL) Best Pract Res Clin Haematol. 2010;23:71–84. doi: 10.1016/j.beha.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Novak AJ, Slager SL, Fredericksen ZS, Wang AH, Manske MM, Ziesmer S, et al. Genetic variation in B-cell-activating factor is associated with an increased risk of developing B-cell non-Hodgkin lymphoma. Cancer Res. 2009;69:4217–24. doi: 10.1158/0008-5472.CAN-08-4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skibola CF, Nieters A, Bracci PM, Curry JD, Agana L, Skibola DR, et al. A functional TNFRSF5 gene variant is associated with risk of lymphoma. Blood. 2008;111:4348–54. doi: 10.1182/blood-2007-09-112144. [DOI] [PMC free article] [PubMed] [Google Scholar]