Abstract
Background/Aims
Apolipoprotein E (ApoE) plays an important role in regulating lipid and lipoprotein metabolism, and ApoE genotypes are known to affect plasma lipoprotein concentrations. We investigated whether ApoE genotype determines the disease outcome in hepatitis B virus (HBV)-infected individuals, and verified the association between ApoE genotype and the occurrence of hepatocellular carcinoma (HCC) in patients with chronic liver diseases of various etiologies.
Methods
This hospital-based, case-controlled study enrolled 156 subjects (47 healthy controls, 50 HBV-related liver cirrhosis patients, and 59 HCC patients). ApoE genotypes were determined using PCR-based ApoE genotyping kits. The biological significance of ApoE genotype was verified by measuring serum ApoE levels using an ELISA kits.
Results
The ε3 allele was the most common allele, with allele frequencies among the entire cohort of 5.8%, 84.3%, and 9.9% for the ε2, ε3, and ε4 alleles, respectively. Significantly more of those patients carrying the ε3/3 genotype had developed liver cirrhosis compared to the control subjects. Being an ApoE4 carrier was associated with a lower probability of developing liver cirrhosis. The allele frequencies and genotype distribution of ApoE did not differ significantly between the liver cirrhosis and HCC patients. The serum level of ApoE was significantly higher in patients with liver cirrhosis than in the healthy controls, but did not differ significantly with the ApoE genotype.
Conclusions
The ApoE ε3/3 genotype frequency was higher in patients with HBV-associated liver cirrhosis than in the controls.
Keywords: Apolipoprotein E, Hepatitis B virus, Genotype, Liver cirrhosis
INTRODUCTION
Hepatitis B virus (HBV) infection is now the leading cause of chronic hepatitis, cirrhosis and hepatocellular carcinoma (HCC) in the Asian countries.1 Acute self-limiting HBV infection usually has little impact on morbidity, whereas persistent HBV infection is a major public health problem in Korea.2 The natural history of HBV infection varies, according to the age at which the infection was contracted. Perinatal transmission of HBV, particularly mother-to-child transmission of the virus during the perinatal period, is a common source of chronic infection.1
Host genetic factors may significantly influence the ability to clear HBV following infection. In recent years, increasing attention has been drawn to the role of host genetic factors in the natural course of viral hepatitis. Previously, we demonstrated that polymorphisms in TNF-α, IL-10, IFN-γ genes were correlated with the susceptibility to chronic HBV infection.3,4 However, the exact role of host genetic factors in the clearance of HBV and the risk of HCC occurrence remain uncertain.
Apolipoprotein (ApoE) is mainly found in the lipoprotein bound form, present in chylomicrons, very low density lipoproteins, and high density lipoproteins, as well as a key regulator of lipid and cholesterol metabolism.5,6 ApoE is a polymorphic protein, which arises from three alleles at a single gene locus. The three major isoforms, apo-ε2, apo-ε3, and apo-ε4, differ from one another by single amino acid substitutions, a change, which has profound functional consequences at both the cellular and molecular levels.7 ApoE has raised particular concern since ApoE polymorphisms are related to several chronic disorders, such as Alzheimer's disease and cardiovascular disease.8,9
ApoE genotype could be an important host genetic factor affecting disease progression in chronic liver disease. ApoE isoform appears to have a hepatitis C virus (HCV)-specific protective effect on liver disease, and the outcome of chronic HCV infection is better among the ε4 carriers, due to slow fibrosis progression.10,11 Since HBV may use apolipoprotein pathways to recycle and infect hepatocytes, it is possible that apolipoprotein polymorphism may play a role in the natural history of HBV infection. However, evidence regarding the contribution of ApoE genotype to outcome of HBV infection is limited.12
The aims of this study were to assess if ApoE functional polymorphisms determine disease outcome in HBV infected individuals, and to verify the association with the occurrence of HCC in patients with chronic liver diseases of various etiologies.
MATERIALS AND METHODS
Study patients
This study is a retrospective case-control study, in which ApoE genotyping and measuring of serum ApoE were performed on the enrolled subjects who were randomly selected. A total of 156 subjects were enrolled in this study, which included 109 case subjects who were diagnosed with liver cirrhosis (LC) or HCC at Ajou University Hospital, between September 2007 and May 2009. Diagnosis of HCC was based on imaging findings of nodules larger than 1 cm, which showed an intense arterial uptake, followed by a washout of contrast in the venous-delayed phases in 4-phase multi-detector CT scan or dynamic contrast enhanced MRI and/or biopsy. Cirrhosis of the liver, on the other hand, was diagnosed pathologically or based on the clinical evidence of portal hypertension, such as visible collateral vessels on the abdominal wall, esophageal varices on esophagogastroscopy, palpable splenomegaly, and sonographically definite findings of cirrhotic liver or ascites. Samples from 47 healthy volunteer, without history of liver disease, were collected as a control. A total of 156 participants underwent Apo E genotyping and of those, 136 participants (87.1%) underwent serum ApoE measurements. All specimens for this study were provided by the Ajou Human Genome Research & Bio-Resource Center, a member of the National Biobank of Korea, which is supported by the Ministry of Health and Welfare. All samples derived from the National Biobank of Korea were obtained with informed consent under institutional review board-approved protocols.
ApoE genotyping and serum ApoE measurements
ApoE genotyping was performed by extracting DNA from whole blood samples. Genotyping for ApoE polymorphisms was carried out by PCR. Amplification reactions were performed in a thermal cycler (Veriti, Life technologies co., CA, USA), using ApoE genotyping PrimerMix Kit (Genotech co., Daejeon, Korea) as a reaction mix. Each reaction mixture was heated at 94℃ for 10 minutes, followed by 30 cycles of amplification (94℃ for 45 s, 60℃ for 45 s, and 72℃ for 45 s). Upon completion of PCR, the products were analyzed by electrophoresis on a 2% ethidium bromide-stained agarose gel.
Of the 156 subjects, serum was available in 136 subjects. Serum ApoE levels were measured in study subjects with the use of an ELISA Kit (ApoE4/Pan-ApoE ELISA Kit; minimum detection limit=4 ng/mL; MLB international co., Woburn, MA, USA) on the absorbance reader (Sunrise, Tecan Group Ltd., Männedorf, Switzerland).
Statistical analysis
All categorical variables are reported as counts and percentages, and comparisons were conducted using a chi-square or Fisher's exact test, as appropriate. Continuous variables are reported as the means±standard deviation with its ranges, and comparisons were conducted using ANOVA with Bonferroni-Dunn test. The homogeneity of variance was tested by Levene's test. Two-sided P values <0.05 were considered statistically significant. All statistical analyses were performed with SPSS software version 16.0.
RESULTS
Patient characteristics
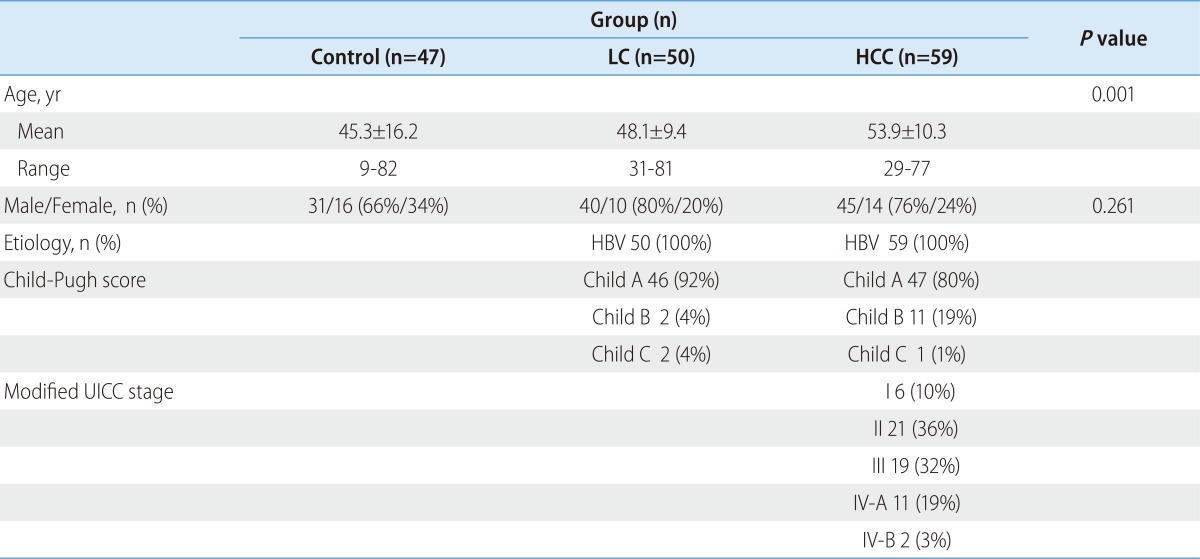
The comparisons of the baseline characteristics, between the three groups who had assessed their ApoE genotypes, are summarized in Table 1. A total of 156 subjects were included (116 men and 40 women), and their ages ranged from 9 to 82 years. The mean age of patients with LC and HCC was higher than that of the healthy controls. The cause of LC and HCC in the enrolled subjects was exclusively HBV infection. Of the 59 HCC patients, 49 patents had liver cirrhosis.
Table 1.
Baseline characteristics of the enrolled subjects*
*Values are expressed as the means±SD for continuous variables and n (%) for categorical variables.
HCC, hepatocellular carcinoma; HBV, hepatitis B virus; HCV, hepatitis C virus.
ApoE genotype in case-control population
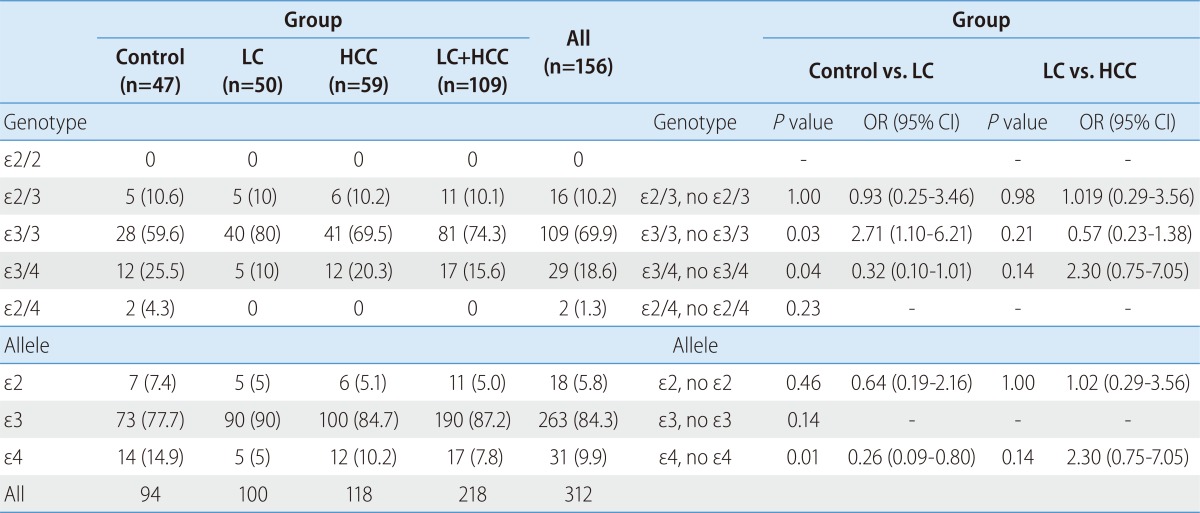
The ApoE genotype distribution and allele frequencies of the subjects are shown in Table 2. Among the 156 subjects, the most common genotype was ε3/3, accounting for 69.9%, followed by ε3/4 and ε2/3, accounting for 15.6% and 10.1%, respectively. The genotypes of ε2/4 and ε4/4 were minor genotypes, which only accounted for 1.3% together. The ε3 allele was the most common allele, with allele frequencies of 5.8% for ε2 allele, 84.3% for the ε3 allele and 9.9 % for ε4 allele in all participants.
Table 2.
Distributions of apolipoprotein E (ApoE) genotypes and alleles in the enrolled patients
*Values are expressed as n (%) for categorical variables.
HCC, hepatocellular carcinoma; LC, liver cirrhosis.
To examine the effect of ApoE genotype on the progression to LC or occurrence of HCC, we analyzed the genotype frequencies among the control, the LC and the HCC subjects. The ApoE genotype was associated with the progression to liver cirrhosis. The odds of developing LC associated, with the carrying of the ε3/3 genotype, were significantly increased (OR 2.71, CI 1.10-6.21) (Table 2). Compared with the subjects, who were ε4 carrier, ε4 non-carrier had an increased susceptibility to LC development (OR 0.26, CI 0.09-0.80). There was no significant difference in the allele frequencies or genotype distribution of ApoE, between liver cirrhosis and HCC patients (Table 2). There is no difference of genotype frequencies according to Child-Pugh score in cirrhotic patients.
Serum ApoE levels in study subjects
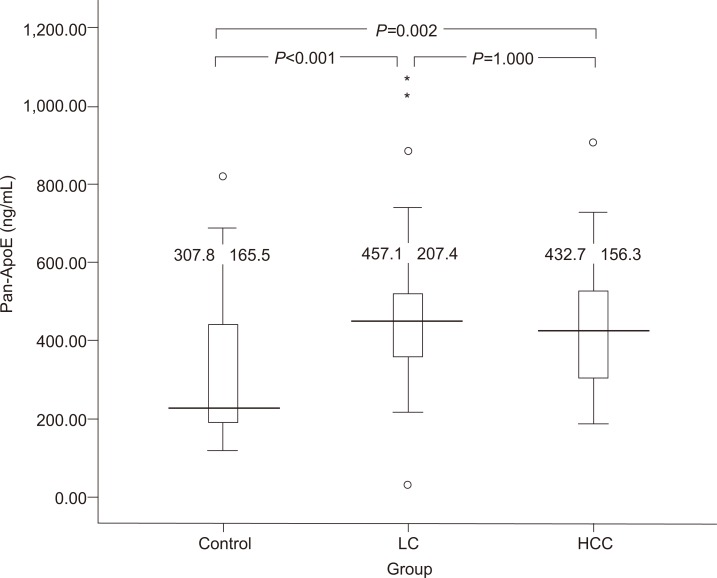
Among to investigate the functional significance at the protein level, we had performed serum ApoE measurements among the 136 individuals in the same set of subjects. The level of serum ApoE was significantly higher in the LC group (P<0.001) and the HCC group (P=0.002) compared with that of the control group (Fig. 1). The mean ApoE level of the LC group tended to be higher than that of the HCC group, but significant differences between the two groups were not observed (Fig. 1).
Figure 1.
Apolipoprotein E (ApoE) levels in the sera of healthy control subjects (n=47), patients with hepatitis-B-virus-related liver cirrhosis (n=50), and patients with hepatocellular carcinoma (HCC; n=59). Serum ApoE levels were significantly higher in the liver cirrhosis (P<0.001) and HCC groups (P=0.001) than in the control group.
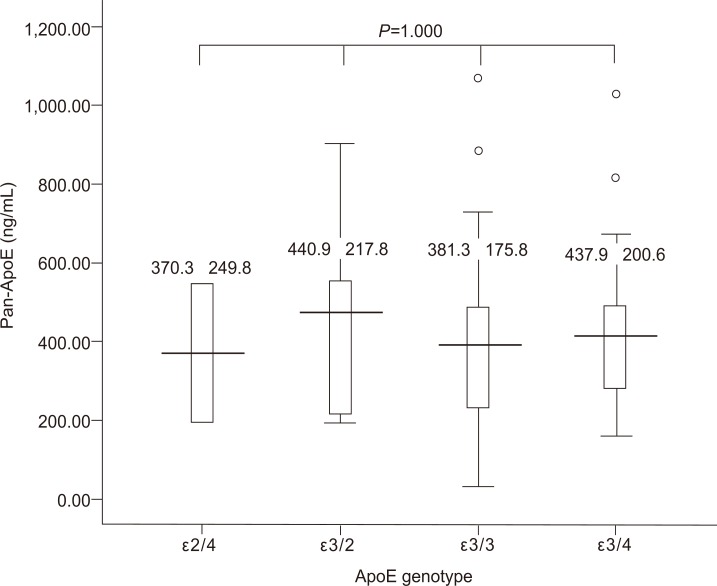
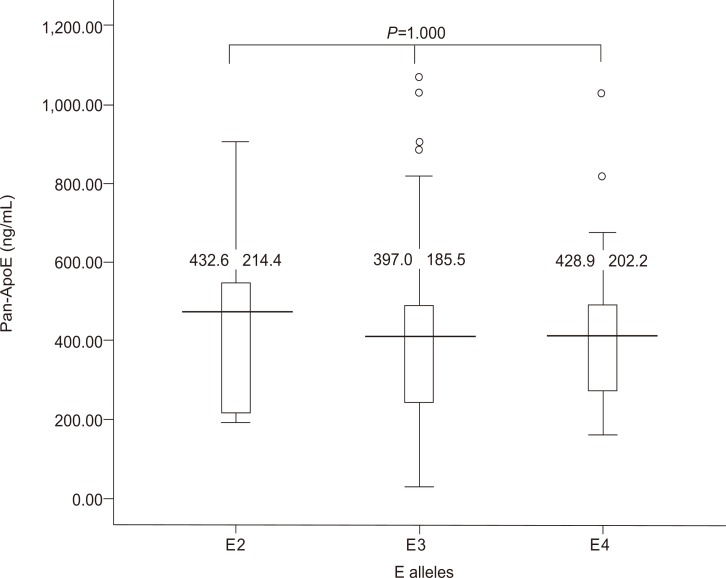
We analyzed whether the serum levels of ApoE correlate with that of ApoE genotypes. Figure 2 demonstrates the serum ApoE levels with regard to ApoE genotype. There was no significant difference in the serum ApoE levels among the different ApoE genotypes. Serum ApoE levels in subjects with different ApoE alleles showed no significant difference (Fig. 3).
Figure 2.
Serum ApoE levels in subjects with different ApoE genotypes. The serum ApoE level did not differ significantly with the ApoE genotype.
Figure 3.
Serum ApoE levels in subjects with different ApoE alleles. The serum ApoE level did not differ significantly with the ApoE allele.
DISCUSSION
Increasing evidence indicates that genetic factors influence the natural history of chronic liver disease. In this study, we have determined whether the ApoE genotypes can predict the outcome of HBV infection and the occurrence of HCC originated from various etiologies. The carriers of apoE4 allelic variant had lower probability of disease progression to liver cirrhosis, but the occurrence of HCC was not affected by ApoE genotypes. Although there is insufficient data to show a direct functional effect of ApoE genotype on the progression to liver cirrhosis, our results suggest a possible genetic association between ApoE genotype and viral clearance in HBV infected patients.
In the current study, ApoE3 allele was associated with a higher probability of progression to HBV-related liver cirrhosis, and ApoE4 allele was a protective factor for developing liver cirrhosis. This result is an agreement with previous report, that a strong association was observed between the apoE3/E3 genotype and the occurrence of more severe liver disease.12 Another recent investigation of the influence of ApoE polymorphism and outcome of HCV infection also suggested that the apoE4 allele protected against severe liver diseases.10
The role of ApoE in the pathogenesis and eradication of chronic HCV is becoming clearer. It is well known that HCV is linked to lipoproteins and mainly uses the LDL receptor pathway to colonize liver cells.13,14 A strong interaction has been postulated to exist between ApoE and hepatitis C virus infection.15 Taken together, ApoE4 seems to be protective against CHC infection and retards the progression of fibrosis. In contrast, HBV interacts with ApoH and possibly interacts with other lipoproteins.16,17 By modulating the transport and the entry of HBV into hepatocytes, apolipoproteins may influence the course of infection, although the virological effects of HBV and HCV are different.
Polymorphisms should have a functional significance. It is essential to show that the change in the gene causes a relevant alteration in the function or level of gene product to establish medically useful links between polymorphism and disease.18 In this study, we assessed both the ApoE genotype and the serum ApoE levels in the same set of patients and sought to confirm the relationship between the serum ApoE levels and liver cirrhosis progression in the enrolled subjects. Higher ApoE levels are observed in patients with liver cirrhosis than in the healthy controls. These results suggest that genetic factor and its gene product may affect the natural course of viral infection. It is not clear that allelic polymorphisms, in the ApoE, can affect the capacity to product ApoE. We assessed whether ApoE production varies, according to the genetic composition of the ApoE. According to our results, ApoE production was not affected by ApoE genotype.
The serum ApoE predominantly originates from the liver and the macrophages.19 ApoE has additional roles, as a modulator of the immune function. Proliferation of both CD4 and CD8 lymphocytes is suppressed by ApoE, which reduces the production of biologically active IL-2.20 In addition, ApoE can have a direct effect on tissue macrophage recruitment, independently, of the lipoprotein metabolism.21 Several data on the role of ApoE in the regulation of inflammation were reported. ApoE has been observed to increase in the serum of patients with sepsis.22 Recently, serum proteomic profiling in patients with drug-induced liver injury demonstrated that elevation of the serum ApoE had the diagnostic power for differentiating patients with drug-induced liver injury from that of the controls.23 In our study, patients with liver cirrhosis showed higher ApoE levels than that of the controls. This result suggests that the elevated ApoE levels can be caused by repeated hepatic inflammation seen in patients with liver cirrhosis.
ApoE was up-regulated in the tissue of HCC developed in patients with chronic viral hepatitis C and serum of HCC patients.24,25 The course of HCC development is a multistep process, and cirrhosis has been considered as a precancerous stage. Because only a subset of LC patients develops HCC, it is of great interest to identify factors that affect the transition from LC to cancer. Hence, only patients with cirrhosis were enrolled in our study, however, there were no differences between liver cirrhosis and HCC patients in ApoE genotype and serum ApoE levels.
Our study has some limitations in its study design. First, study subjects did not include the patients with chronic hepatitis or inactive HBV carrier. Thus, this study could not adequately evaluate the effects of the ApoE genotype on the progression of fibrosis. As a consequence, ApoE genotype could, in fact, be associated with an increased susceptibility to the HBV persistence, not to fibrosis progression. It would be more reasonable to select non-cirrhotic chronic liver disease as a control to find factors of disease progression. Second, sample size was small. Prediction of patients who will develop HCC in patients with chronic liver disease is an important unmet clinical need. We were unable to assess this issue clearly in this current study because of the sample size being small. Further investigations with a larger sample size are needed.
In conclusion, the ApoE ε3/3 genotype frequencies were higher in patients with HBV associated liver cirrhosis rather than controls. The serum ApoE levels are higher in patients with liver cirrhosis compared with that of the healthy individuals. It appears that the usefulness of ApoE genotype as a predictor of viral clearance after HBV infection and diagnostic value of the serum ApoE levels for the diagnosis of liver cirrhosis need to be defined.
Acknowledgements
This study was supported by a grant from the Korean Health R & D project, Ministry of Health and Welfare, Republic of Korea (A010383).
Abbreviations
- ApoE
apolipoprotein E
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- LC
liver cirrhosis
Footnotes
The authors have no conflicts to disclose.
References
- 1.Kwon SY, Lee CH. Epidemiology and prevention of hepatitis B virus infection. Korean J Hepatol. 2011;17:87–95. doi: 10.3350/kjhep.2011.17.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi HJ, Ko SY, Choe WH, Seo YS, Kim JH, Byun KS, et al. Clinical features of acute viral hepatitis B in Korea: a multi-center study. Korean J Hepatol. 2011;17:307–312. doi: 10.3350/kjhep.2011.17.4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheong JY, Cho SW, Chung SG, Lee JA, Yeo M, Wang HJ, et al. Genetic polymorphism of interferon-gamma, interferon-gamma receptor, and interferon regulatory factor-1 genes in patients with hepatitis B virus infection. Biochem Genet. 2006;44:246–255. doi: 10.1007/s10528-006-9029-y. [DOI] [PubMed] [Google Scholar]
- 4.Cheong JY, Cho SW, Hwang IL, Yoon SK, Lee JH, Park CS, et al. Association between chronic hepatitis B virus infection and interleukin-10, tumor necrosis factor-alpha gene promoter polymorphisms. J Gastroenterol Hepatol. 2006;21:1163–1169. doi: 10.1111/j.1440-1746.2006.04304.x. [DOI] [PubMed] [Google Scholar]
- 5.Minihane AM, Jofre-Monseny L, Olano-Martin E, Rimbach G. ApoE genotype, cardiovascular risk and responsiveness to dietary fat manipulation. Proc Nutr Soc. 2007;66:183–197. doi: 10.1017/S0029665107005435. [DOI] [PubMed] [Google Scholar]
- 6.Jofre-Monseny L, Minihane AM, Rimbach G. Impact of apoE genotype on oxidative stress, inflammation and disease risk. Mol Nutr Food Res. 2008;52:131–145. doi: 10.1002/mnfr.200700322. [DOI] [PubMed] [Google Scholar]
- 7.Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 8.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 9.Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 10.Wozniak MA, Itzhaki RF, Faragher EB, James MW, Ryder SD, Irving WL. Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology. 2002;36:456–463. doi: 10.1053/jhep.2002.34745. [DOI] [PubMed] [Google Scholar]
- 11.Mueller T, Gessner R, Sarrazin C, Graf C, Halangk J, Witt H, et al. Apolipoprotein E4 allele is associated with poor treatment response in hepatitis C virus (HCV) genotype 1. Hepatology. 2003;38:1592. doi: 10.1016/j.hep.2003.09.042. author reply 1592-1593. [DOI] [PubMed] [Google Scholar]
- 12.Toniutto P, Fattovich G, Fabris C, Minisini R, Burlone M, Pravadelli C, et al. Genetic polymorphism at the apolipoprotein E locus affects the outcome of chronic hepatitis B. J Med Virol. 2010;82:224–331. doi: 10.1002/jmv.21642. [DOI] [PubMed] [Google Scholar]
- 13.Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pumeechockchai W, Bevitt D, Agarwal K, Petropoulou T, Langer BC, Belohradsky B, et al. Hepatitis C virus particles of different density in the blood of chronically infected immunocompetent and immunodeficient patients: Implications for virus clearance by antibody. J Med Virol. 2002;68:335–342. doi: 10.1002/jmv.10208. [DOI] [PubMed] [Google Scholar]
- 15.Mazumdar B, Banerjee A, Meyer K, Ray R. Hepatitis C virus E1 envelope glycoprotein interacts with apolipoproteins in facilitating entry into hepatocytes. Hepatology. 2011;54:1149–1156. doi: 10.1002/hep.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefas I, Rucheton M, D'Angeac AD, Morel-Baccard C, Seigneurin JM, Zarski JP, et al. Hepatitis B virus Dane particles bind to human plasma apolipoprotein H. Hepatology. 2001;33:207–217. doi: 10.1053/jhep.2001.20531. [DOI] [PubMed] [Google Scholar]
- 17.Mehdi H, Kaplan MJ, Anlar FY, Yang X, Bayer R, Sutherland K, et al. Hepatitis B virus surface antigen binds to apolipoprotein H. J Virol. 1994;68:2415–2424. doi: 10.1128/jvi.68.4.2415-2424.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenthal N, Schwartz RS. In search of perverse polymorphisms. N Engl J Med. 1998;338:122–124. doi: 10.1056/NEJM199801083380210. [DOI] [PubMed] [Google Scholar]
- 19.Wu AL, Windmueller HG. Relative contributions by liver and intestine to individual plasma apolipoproteins in the rat. J Biol Chem. 1979;254:7316–7322. [PubMed] [Google Scholar]
- 20.Kelly ME, Clay MA, Mistry MJ, Hsieh-Li HM, Harmony JA. Apolipoprotein E inhibition of proliferation of mitogen-activated T lymphocytes: production of interleukin 2 with reduced biological activity. Cell Immunol. 1994;159:124–139. doi: 10.1006/cimm.1994.1302. [DOI] [PubMed] [Google Scholar]
- 21.Shiri-Sverdlov R, Wouters K, van Gorp PJ, Gijbels MJ, Noel B, Buffat L, et al. Early diet-induced non-alcoholic steatohepatitis in APOE2 knock-in mice and its prevention by fibrates. J Hepatol. 2006;44:732–741. doi: 10.1016/j.jhep.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Thompson PA, Kitchens RL. Infection induces a positive acute phase apolipoprotein E response from a negative acute phase gene: role of hepatic LDL receptors. J Lipid Res. 2008;49:1782–1793. doi: 10.1194/jlr.M800172-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell LN, Vuppalanchi R, Watkins PB, Bonkovsky HL, Serrano J, Fontana RJ, et al. Serum proteomic profiling in patients with drug-induced liver injury. Aliment Pharmacol Ther. 2012;35:600–612. doi: 10.1111/j.1365-2036.2011.04982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanc JF, Lalanne C, Plomion C, Schmitter JM, Bathany K, Gion JM, et al. Proteomic analysis of differentially expressed proteins in hepatocellular carcinoma developed in patients with chronic viral hepatitis C. Proteomics. 2005;5:3778–3789. doi: 10.1002/pmic.200401194. [DOI] [PubMed] [Google Scholar]
- 25.Ritorto MS, Borlak J. Combined serum and tissue proteomic study applied to a c-Myc transgenic mouse model of hepatocellular carcinoma identified novel disease regulated proteins suitable for diagnosis and therapeutic intervention strategies. J Proteome Res. 2011;10:3012–3030. doi: 10.1021/pr101207t. [DOI] [PubMed] [Google Scholar]