Abstract
Sequence comparisons of genomes or expressed sequence tags (ESTs) from related organisms provide insight into functional conservation and diversification. We compare the sequences of ESTs from the male accessory gland of Drosophila simulans to their orthologs in its close relative Drosophila melanogaster, and demonstrate rapid divergence of many of these reproductive genes. Nineteen (∼11%) of 176 independent genes identified in the EST screen contain protein-coding regions with an excess of nonsynonymous over synonymous changes, suggesting that their divergence has been accelerated by positive Darwinian selection. Genes that encode putative accessory gland-specific seminal fluid proteins had a significantly elevated level of nonsynonymous substitution relative to nonaccessory gland-specific genes. With the 57 new accessory gland genes reported here, we predict that ∼90% of the male accessory gland genes have been identified. The evolutionary EST approach applied here to identify putative targets of adaptive evolution is readily applicable to other tissues and organisms.
Keywords: positive selection, accessory glands, seminal fluid, peptide hormones, sexual conflict
The Drosophila male accessory gland is a highly specialized reproductive organ. Its function is to secrete seminal-fluid proteins. Therefore, it may be relatively easy to identify many of the proteins found in seminal fluid by sequencing expressed sequence tags (ESTs) from the accessory gland. Secreted accessory gland proteins (Acps) have diverse and important reproductive roles and interesting patterns of evolutionary change. Acps are transferred along with sperm to the female's reproductive tract and have a variety of effects on the female's reproductive physiology (1). Acps increase the egg-laying rate of mated females by inducing oogenesis (2, 3) and ovulation (4), decrease the female's propensity to remate (5), are required for sperm storage (6, 7), and influence egg hatchability (8, 9). Also, Acps may play a role in cryptic female choice (10), sperm competition (11), and intersexual genomic conflict (12)—three evolutionary scenarios thought to promote the divergence of reproductive proteins. The unique role of Acps has made them the focus of much interest by cell and evolutionary biologists, because they seem to be a currency of chemical communication between males and females (1).
Two-dimensional protein electrophoresis has been used to show that male reproductive proteins (including Acps) are twice as diverse as nonreproductive proteins (13), but because the nucleotide sequences encoding these proteins remained unidentified, it was impossible to determine whether positive selection or low constraint on amino acid sequence led to the apparent high divergence of this large class of proteins. Identification of the nucleotide sequences encoding these highly variable proteins will allow for evolutionary inferences of the magnitude of forces affecting their evolution (14) and provide tools for determining the molecular function of the selected gene (2–6, 15).
Conservation of primary sequence has been applied widely as a criterion for functionally important genes or gene regions. For example, the primary amino acid sequence of each core histone gene is >90% identical between plants and animals, presumably because of the conserved role of these proteins in chromatin structure. However, functionally important regions also can be revealed in divergent genes if positive selection is involved in their adaptive divergence (16–18). High levels of amino acid polymorphism within a species also may be a sign that natural selection is favoring high levels of allelic diversity. This pattern is illustrated well by genes involved in the immune response, such as the gene encoding the MHC class I protein, where the region encoding the antigen-binding cleft shows high amino acid diversification (19). A strong signature of positive selection for change is that the number of nonsynonymous substitutions per nonsynonymous site (amino acid altering; dN) significantly exceeds the number of synonymous substitutions per synonymous site (silent; dS; ref. 20). Such analyses also have identified positive selection in genes involved in host–parasite interaction (21), reproduction (22–27), and adaptation to specific environments (28–30).
The recently completed genomic sequence of Drosophila melanogaster (31) provides a superb resource against which to perform a comparative EST analysis. Although estimates from differential cDNA hybridization (32–35) and protein electrophoresis studies (36) estimate the number of accessory gland genes in the genome to be ≈25–100 (1, 32–36), only 18 have been isolated to date (32–35). Sequence divergence studies of five Acp genes have revealed two rapidly evolving genes (37–40) and three other Acp genes that are conserved fairly well (40–42). A recent report identified one additional Acp gene subjected to selection (43). The strategy we used in the present study was to isolate and sequence accessory gland ESTs from Drosophila simulans, a close relative of D. melanogaster. There is, on average, 11% silent site difference between these two species (44–46), providing sufficient numbers of substitutions for reliable estimates of dN and dS. More divergent comparisons would introduce the problem of multiple substitutions, which can obscure signs of selection. Comparing sequences from D. simulans to the D. melanogaster genome simultaneously identifies the D. melanogaster gene sequence for further functional studies and provides an estimate of divergence for evolutionary inferences. To this end, we prepared an oligo(dT)-primed cDNA library from dissected D. simulans accessory glands. To enrich for male-specific ESTs, we performed a differential hybridization step in which we probed the cDNA library with 32P-labeled adult female D. simulans cDNA. Only colonies hybridizing weakly or not at all to the female cDNA were selected for further analysis. Thus, our collection for analysis is enriched for accessory gland genes expressed only in males, although it is possible that genes expressed at low levels in females might still be present in our EST collection.
Materials and Methods
cDNA Library Preparation and Screening.
Total RNA was purified from 500 dissected D. simulans accessory glands by the guanidinium thiocyanate/CsCl method (47), yielding 2 μg of RNA. mRNA was isolated by using the Qiagen Oligotex kit. Oligo(dT)-primed cDNA was prepared and cloned directionally into pSport (BRL) following the manufacturer's directions. Because many of the known Acps are small genes, size selection allowed for small genes to be included. The resulting library was 98% recombinant, containing 225,000 colony-forming units (cfus) with an average insert size of 1.3 kb. Three thousand cfus were plated at low density, and colony lifts were prepared and probed. Oligo(dT)-primed first-strand female cDNA was prepared by using BRL superscript II reverse transcriptase incorporating 32P-labeled dCTP and then denatured at 65°C for 30 min in 0.3 M NaOH. Hybridization was for 18 h at 65°C in 5 × standard saline phosphate (SSPE)/5 × Denhardt's solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA)/0.5% SDS/0.2 mg/ml salmon sperm DNA. Final washes were at 65°C, 0.1 × SSPE for 10 min. Three hundred fifty nonhybridizing colonies were selected for further analysis. Plasmid DNA was purified by using the Qiagen miniprep kit and spotted onto Hybond-XL membranes (Amersham Pharmacia). Clones were rechecked for male specificity by dot-blot hybridization of purified plasmid DNA. Additionally, eight previously identified Acps (Acp32CD, Acp33A, Acp62F, Acp63F, Acp76A, Acp98AB, Acp36DE, and Acp26Aa; refs. 32 and 33) were screened out by probing with 32P-labeled PCR products. Three hundred twenty-eight clones remained male predominant.
To identify which of the ESTs are likely to be expressed specifically in the main cells of the accessory glands, we performed a second differential hybridization screen. Transgenic D. melanogaster flies expressing wild-type diphtheria toxin in their accessory glands (DTA-E) under the control of an accessory gland main cell-specific promoter (48) make no detectable main cells Acps and show degeneration of their accessory gland main cells. The secondary cells of their accessory glands (4% of the accessory gland; ref. 49) are unaffected and still synthesize proteins (48). We hybridized the accessory gland ESTs to 32P-labeled cDNA made from wild-type males or from DTA-E males. Hybridization conditions were identical to those described above. Hybridization to a cDNA probe from wild-type D. melanogaster males but not to DTA-E D. melanogaster males defined those ESTs that are accessory gland main cell-specific in their expression and thus a portion of the Acp genes. However, those Acp genes transcribed in secondary cells (or elsewhere in the body) still will be expressed in DTA-E flies and hybridize to the DTA-E cDNA probe.
Sequencing was from Qiagen purified plasmid DNA performed on a Beckman CEQ-2000 automated sequencer. Only 5′ ends were sequenced. Thirty-seven inserts were less than 50 bp and were not analyzed further. In total, 212 independent genes were identified. Of these genes, 176 matched putative protein-coding regions in the annotated D. melanogaster genome, whereas the remaining 36 matched 5′ or 3′ noncoding regions.
Statistical Analyses.
The genomic region (including 500 bp upstream and downstream) of the genes identified by our ESTs were compared by using blast to the Berkeley EST database. This strategy assures that our ESTs and previous ESTs did not just identify different regions of the same gene or missed because of annotation errors. Genes with an E value > 10−10 were considered not found in the Drosophila EST database. Signal sequences were predicted by using the program signalp (http://www.cbs.dtu.dk/services/SignalP-2.0/).
To determine the number of Acp genes in the D. melanogaster genome, we analyzed the frequency of multiply hit genes identified in our EST analysis. We can get a bound on the number of genes expressed in the accessory gland by assuming that they are all equally likely to be recovered in the cDNA clones. In fact, there is wide variation in expression levels (5, 32–35), thus any estimate that we make assuming equal expression will be lower than the true number. If there were equal expression levels, the numbers of genes recovered in clones 1, 2, and 3 times, etc., are expected to have a Poisson distribution. We can estimate the number of distinct genes expressed in the accessory gland by estimating the Poisson parameter by maximum likelihood, then using this parameter to estimate the size of the zero class (those genes present in the genome that were not observed). The library was prescreened with eight different Acp genes, thus we can apply this estimation procedure on what is left after screening and simply add the count of screened genes back at the end. We consider only those clones that are main cell accessory gland-enriched (by virtue of the differential screening described below) or those that encode a protein with a predicted signal peptide.
Assessment of the significance of excess of dN over dS was determined as follows. dN and dS were estimated as two free parameters by maximum likelihood (L1) by using paml (50, 51). The likelihood was calculated also for the null model, having dN equal to dS (L0). The negative of twice the difference in the logarithm (log) likelihood obtained from these two models (−2[log(L0)−log(L1)]) was compared with the χ2 distribution with 1 degree of freedom. Although the likelihood ratio test is a large-sample test (20), computer simulations have shown it to be reliable for short sequences for this type of comparison (52). Similar results are obtained by the method of Nei and Gojobori (53), which uses a Z test to determine whether dN − dS is significantly different from zero.
ANOVA was used to test the null hypothesis that the Acp genes have the same dN and dS values as non-Acp genes. We analyzed the raw data and log-transformed data. Log transformation improved the fit of the data to the normal distribution in a few cases; however, analyses on both the raw data and log-transformed data yielded the same conclusions.
Results and Discussion
Two hundred and eighty five male-predominant clones were sequenced, identifying 212 independent genes (Table 1, which is published as supplemental material on the PNAS web site, www.pnas.org). Thirty-five percent of the genes (75 independent genes) matching these ESTs did not have matches from the 91,833 ESTs that have been generated as part of the Drosophila genome project (54), probably because previous EST libraries did not use adult male accessory glands. Remarkably, 47% of the genes showed no evidence of homology to non-Drosophila sequences in GenBank. This proportion is higher than the overall average for the D. melanogaster genome (29%; ref. 31). Consistent with the accessory gland being a secretory tissue, 24% of the predicted genes (51 independent genes) encode proteins with putative signal sequences. Eight percent of the genes (18 independent genes) correspond to genomic regions of the D. melanogaster genome in which no genes have been predicted (31). The annotation of these genes was missed by gene-prediction algorithms (55), most likely because accessory gland genes generally have low codon bias, short ORFs, and tend to diverge rapidly enough to make ortholog identification difficult.
Identification of Acp Genes.
We performed a second differential hybridization step to identify Acp genes, using probes from D. melanogaster flies lacking Acp transcripts, because of expression of diphtheria toxin in the main cells of their accessory glands (DTA-E; ref. 48), and wild-type flies. ESTs that hybridize strongly to the wild-type probe but not to the DTA-E probe are candidates for encoding accessory gland main cell-specific transcripts (Fig. 1). Approximately 15% of the ESTs (32 independent genes) appear to be accessory gland main cell-specific by this stringent criterion. Of these, 81% (26 independent genes) are novel candidate Acps (Table 1). This probing will not identify all potential Acps, because accessory gland-specific genes that are expressed at low levels and accessory gland genes expressed also, or exclusively, in secondary cells, will not be detected as accessory gland-specific by such an analysis (48). In fact, many of the ESTs that encode proteins with predicted signal sequences (and therefore are probably secreted proteins) are not identified as accessory gland main cell-specific by these criteria. Nonetheless, the two differential-hybridization steps demonstrate that the EST sequences are enriched for male-specific accessory gland genes.
Figure 1.
Differential hybridization of the EST library with 32P-labeled cDNA from wild-type males (WT) and DTA-E males (both D. melanogaster), the latter having no main cell accessory gland genes expressed. Row 1 shows a sample EST clone that hybridizes strongly with the male wild-type probe but not the DTA-E probe, and is thus a candidate Acp. Rows 2 and 3 show example EST clones with equal (strong or weak, respectively) hybridization to wild type and DTA-E, and thus cannot be identified as potential Acps by this method.
If we consider as Acps either those genes that are accessory gland main cell-specific by our differential hybridization or those male genes expressed in the accessory gland and encoding proteins with a predicted signal sequence (indicating they encode secreted proteins), we can estimate the total number of Acp proteins from the distribution of multiple isolations of the same genomic sequence. The data contained 46 putative Acp genes that were represented 1 time, 6 that were represented 2 times, 3 that were found in 3 clones, and 4 that were found in 4 clones. The 4 remaining distinct genes were present in 6, 8, 9, and 17 clones. Fitting this data to a Poisson distribution yields a maximum likelihood estimate of 75 genes. To this number we must add the 8 genes that were prescreened initially, giving an estimate of 83 genes expressed in the accessory gland. Because of variation in gene expression level, this number should be viewed as a rough lower bound. Additional screening of the original library will quantify the variation in expression levels.
With the 18 previous genes (32–35) and the 57 new independent Acp genes identified here (26 by loss of expression in DTA-E flies and an additional 31 with signal sequences), we estimate that 90% of the 83 accessory gland-specific genes in the D. melanogaster genome now have been identified. Interestingly, there is a significant absence of Acp genes on the X chromosome. Although theoretical models have suggested that there are grounds to expect the opposite result, namely a clustering of male fertility genes on the X chromosome (56), that the Drosophila X is dosage-compensated in males by hypertranscription on a gene-by-gene basis (57, 58) may explain the deficit of X-linked Acps (ref. 32 and this study). Transposition of an autosomal Acp to the X chromosome would result initially in reduced expression in males, which may be disadvantageous (1).
The types of genes identified as encoding Acps are consistent with what is known about accessory gland function. Previous studies identified 13 genes encoding small novel secreted peptides (32–35), some of which have been demonstrated to have reproductive functions (2–5). Our analysis identified 12 additional peptides in this class. Previous studies also identified secreted proteins that undergo regulated proteolysis (32, 33, 57, 58), and two putative protease inhibitors (refs. 32 and 59–61 and O. Lung, U. Tram, C. Finnerty, M. Epper-Mains, J. Kalb, and M.F.W., unpublished data). Here, we identified nine additional proteases and six additional protease inhibitors that may be involved in proteolytic cascades. Additionally, we identified six separate lipase genes. Lipase activity has been detected in D. melanogaster male reproductive organs (62), but no accessory gland-expressed lipase genes had been identified previously. Possible functional roles of lipases include providing nutritional value for females, alteration of sperm membranes to facilitate fertilization, and modification of female reproductive tract for efficient sperm and Acp usage. Nearly 50% of the new genes have no known motifs, suggesting our EST collection to be a rich source of new protein motifs.
Evolutionary Analyses of Divergence.
To gain insight into the evolutionary forces affecting the divergence of these reproductive proteins, we calculated the levels of synonymous and nonsynonymous substitution (20, 50, 51) for those D. simulans ESTs that match protein-coding region sequences in D. melanogaster (176 total independent comparisons; the remaining 36 ESTs matched 5′ or 3′ noncoding regions). The goal here was to identify potential candidate genes whose divergence was promoted by positive Darwinian selection, as indicated by a high dN/dS ratio. Below, we demonstrate that different evolutionary forces affect the Acp genes compared with non-Acp genes, and provide evidence that the high dN/dS ratios do not result from the Acp genes being pseudogenes or from sequencing errors.
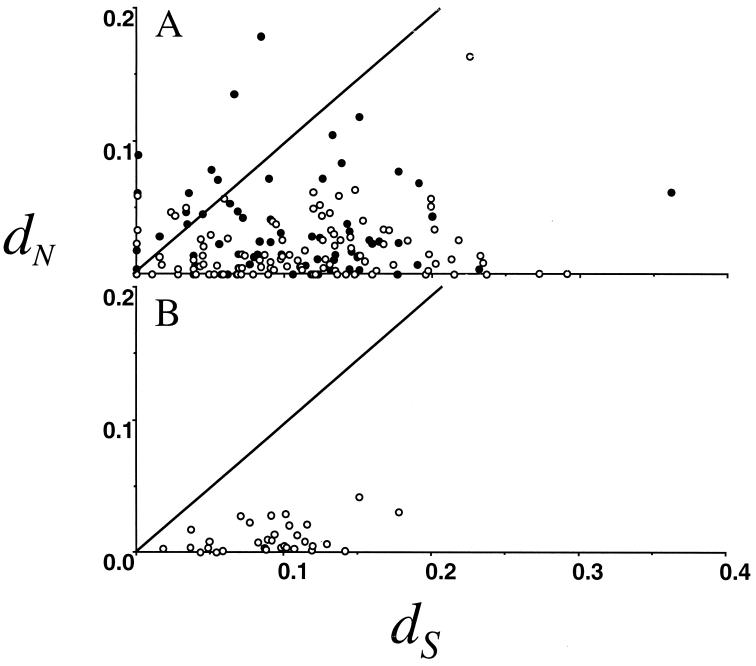
Fig. 2 shows dN plotted against dS for all of the independent genes from our EST study; the values for all pairwise comparisons are presented in Supplementary Material. For 140 comparisons, the dN/dS ratio was significantly different from 1, as assessed by a likelihood ratio test. Most of these cases (n = 134) have dN/dS ratios significantly less than 1, suggesting purifying selection associated with functional constraint. In six independent cases, the dN/dS ratio was significantly greater than 1, suggesting positive selection for amino acid diversification rather than relaxation of constraint. In total, 11% (19 independent genes) of the ESTs have dN/dS ratios exceeding 1. As a class, the Acp genes had a level of nonsynonymous substitution (dN = 0.052 ± 0.009) that was more than twice that of non-Acp genes (dN = 0.024 ± 0.002), a result that was highly significant by ANOVA (F1, 180 = 62.22, P < 0.0001). For comparison, we plotted dN and dS for 32 nonreproductive genes in D. melanogaster and D. simulans (44–46) and found that on average dN is substantially less than dS (Fig. 2). Importantly, the range of dS values for the ESTs and nonreproductive proteins is similar, and an ANOVA test failed to reject the null hypothesis of equal levels of divergence. This is evidence that the high dN/dS ratios observed in our EST sequences are not caused by sequencing errors, because such errors would inflate both dS and dN values. Several of the ESTs were distinct clones of the same gene. Alignment of these repeats revealed 56 mismatches in 7,800 bp for a maximum overall sequencing error rate of 0.7%, a figure that is far too small to be responsible for the excess nonsynonymous differences seen (particularly considering that many of these differences represent true polymorphisms and not sequencing errors). Additionally, the average dS of our ESTs (dS = 0.11) is identical to the average dS (dS = 0.11) calculated from complete genes between D. simulans and D. melanogaster (44–46), indicating that our estimates are reliable despite the short length of the sequences (average alignment length between coding regions was 285 bp). Similarly, a contrast of dN of our non-Acp ESTs to the nonreproductive genes of refs. 44–46 revealed no difference (F1, 179 = 0.53, P = 0.467). The regulated expression and lack of frame-shift mutations between species argues against our Acp ESTs being pseudogenes. Finally, equivalence of dS in these two samples is consistent also with homogeneity of mutation rates and other factors influencing the levels of sequence divergence.
Figure 2.
The number of nonsynonymous substitutions per nonsynonymous site (dN) plotted against the number of synonymous substitutions per synonymous site (dS) for the D. simulans accessory gland EST library (A), and random nonreproductive proteins (B) compared with D. melanogaster genomic sequence. ● are putative Acps (by differential hybridization or containing signal sequences), whereas ○ cannot be identified as putative Acps by these methods. The line shows the neutral expectation of dN = dS. Data for the lower panel are from Moriyama and Powell (44). dN and dS were estimated by the maximum likelihood method (50, 51). Similar results are obtained by using the method of Nei and Gojobori (53).
The average dN of the accessory gland-specific ESTs (dN = 0.052) is 2.1 times that of the nonaccessory gland-specific genes (dN = 0.024), a result that stands in stark contrast to the virtually identical levels of synonymous divergence quantified above. This high level of amino acid sequence divergence for the male accessory gland-specific genes is similar to the 2-fold higher level of divergence of male reproductive proteins analyzed by two-dimensional gel protein electrophoresis (13). A dN/dS ratio of approximately 1 can result from either positive selection or lack of functional constraint. The criterion for positive selection that requires a dN/dS ratio across the entire gene being greater than 1 to identify positive selection is extremely stringent. It is possible that the genes identified here with a dN/dS ratio of approximately 1 will show signs of positive selection with additional tests. For example, genes such as the MHC glycoprotein HLA-A (19) and mammalian zona pellucida glycoproteins (64) have pairwise dN/dS ratios less than 1. However, comparisons of these genes among several species, taking into account variation in the dN/dS ratio between sites and lineages, has detected the action of positive selection on portions of these genes (19, 63). Similar analysis of the putative targets of positive selection identified in this study will be informative, but will require additional taxon sampling.
The identification of the large group of male reproductive genes presented here opens the door to molecular tests of cryptic female choice (10), sperm competition (11), and intersexual genomic conflict (12). The general approach we describe here, blasting EST sequences from a closely related species to one whose full genome has been sequenced, promises to be a powerful approach for identifying potential targets of positive selection at a genomic scale and is readily applicable to any tissue or organism.
Supplementary Material
Acknowledgments
We thank J. Calkins, R. Nielsen, V. Bauer DuMont, Y. Lei, E. N. Moriyama, M. Dermitzakis, B. Lazzaro, K. Montooth, S. Kresovich, S. Schloss, S. Howell, J. Mueller, and S. Tanksley for comments or discussion. W.J.S. was supported by a National Science Foundation/Alfred P. Sloan postdoctoral fellowship and a National Institutes of Health National Research Service Award postdoctoral fellowship. Support was provided to A.G.C. and H.M.W.-D. from National Science Foundation Grant DEB 9527592; to M.F.W. from National Science Foundation Grant IBN97−23356 and National Institutes of Health Grant HD38921; and to C.F.A. from National Institutes of Health Grant GM36431.
Abbreviations
- EST
expressed sequence tag
- Acps
accessory gland protein
- DTA-E
D. melanogaster males expressing wild-type diphtheria toxin in their accessory glands
- log
logarithm
Footnotes
References
- 1.Wolfner M F. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- 2.Soller M, Bownes M, Kubli E. Dev Biol. 1999;208:337–351. doi: 10.1006/dbio.1999.9210. [DOI] [PubMed] [Google Scholar]
- 3.Heifetz Y, Tram U, Wolfner M F. Proc R Soc London. 2001;268:175–180. doi: 10.1098/rspb.2000.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heifetz Y, Lung O, Frongillo E A, Wolfner M F. Curr Biol. 2000;10:99–102. doi: 10.1016/s0960-9822(00)00288-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen P S, Stumm-Zollinger E, Aigaki T, Balmer J, Bienz M, Bohlen P. Cell. 1988;54:291–298. doi: 10.1016/0092-8674(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 6.Neubaum D M, Wolfner M F. Genetics. 1999;153:845–857. doi: 10.1093/genetics/153.2.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tram U, Wolfner M F. Genetics. 1999;153:837–844. doi: 10.1093/genetics/153.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prout T, Clark A G. Proc R Soc London Ser B. 2000;267:201–203. doi: 10.1098/rspb.2000.0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, T., Herndon, L., Heifetz, Y., Partridge, L. & Wolfner, M. F. Proc. R. Soc. London, in press. [DOI] [PMC free article] [PubMed]
- 10.Eberhard W G, Cordero C. Trends Ecol Evol. 1995;10:493–496. doi: 10.1016/s0169-5347(00)89205-8. [DOI] [PubMed] [Google Scholar]
- 11.Clark A G, Aguadé M, Prout T, Harshman L G, Langley C H. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice W R. Nature (London) 1996;381:232–234. doi: 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- 13.Civetta A, Singh R S. J Mol Evol. 1995;41:1085–1095. doi: 10.1007/BF00173190. [DOI] [PubMed] [Google Scholar]
- 14.Aquadro C F. Curr Opin Gen Dev. 1997;7:835–840. doi: 10.1016/s0959-437x(97)80048-2. [DOI] [PubMed] [Google Scholar]
- 15.Rong Y S, Golic K G. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen R, Yang Z. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki Y, Gojobori T. Mol Biol Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Nielsen R, Goldman N, Pedersen A M. Genetics. 2000;155:471–439. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes A L, Nei M. Nature (London) 1992;355:402–403. doi: 10.1038/355402b0. [DOI] [PubMed] [Google Scholar]
- 20.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ. Press; 2000. [Google Scholar]
- 21.Hughes A L. Mol Biol Evol. 1992;9:381–393. doi: 10.1093/oxfordjournals.molbev.a040730. [DOI] [PubMed] [Google Scholar]
- 22.Clark A G, Kao T H. Proc Natl Acad Sci USA. 1991;88:9823–9227. doi: 10.1073/pnas.88.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y H, Ota T, Vacquier V D. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- 24.Swanson W J, Vacquier V D. Proc Natl Acad Sci USA. 1995;92:4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metz E C, Palumbi S R. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 26.Ferris P J, Pavlovic C, Fabry S, Goodenough U W. Proc Natl Acad Sci USA. 1997;94:8634–8639. doi: 10.1073/pnas.94.16.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyckoff G J, Wang W, Wu C-I. Nature (London) 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 28.Stewart C-B, Schilling J W, Wilson A C. Nature (London) 1987;330:401–404. doi: 10.1038/330401a0. [DOI] [PubMed] [Google Scholar]
- 29.Bargelloni L, Marcato S, Patarnell T. Proc Natl Acad Sci USA. 1998;95:8670–8675. doi: 10.1073/pnas.95.15.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoyama S, Zhang H, Radlwimmer F B, Blow N S. Proc Natl Acad Sci USA. 1999;96:6279–6284. doi: 10.1073/pnas.96.11.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 32.Wolfner M F, Harada H A, Bertram M J, Stelick T J, Kraus K W, Kalb J M, Lung Y O, Neubaum D M, Park M, Tram U. Insect Biochem Mol Biol. 1997;27:825–834. doi: 10.1016/s0965-1748(97)00056-8. [DOI] [PubMed] [Google Scholar]
- 33.Monsma S A, Wolfner M F. Genes Dev. 1988;2:1063–1073. doi: 10.1101/gad.2.9.1063. [DOI] [PubMed] [Google Scholar]
- 34.DiBenedetto A J, Harada H A, Wolfner M F. Dev Biol. 1990;139:134–148. doi: 10.1016/0012-1606(90)90284-p. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer U. Mol Gen Genet. 1986;202:219–225. [Google Scholar]
- 36.Chen P S. Ann Soc Entomol Fr. 1991;27:231–244. [Google Scholar]
- 37.Tsaur S C, Wu C-I. Mol Biol Evol. 1997;14:544–549. doi: 10.1093/oxfordjournals.molbev.a025791. [DOI] [PubMed] [Google Scholar]
- 38.Aguadé M. Genetics. 1999;152:543–551. doi: 10.1093/genetics/152.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguadé M. Genetics. 1998;150:1079–1089. doi: 10.1093/genetics/150.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguadé M, Miyashita N, Langley C. Genetics. 1992;132:755–770. doi: 10.1093/genetics/132.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cirera S, Aguadé M. Genetics. 1997;147:189–197. doi: 10.1093/genetics/147.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt T, Choffat Y, Schneider M, Hunziker P, Fuyama Y, Kubli E. Insect Biochem Mol Biol. 1993;23:571–579. doi: 10.1016/0965-1748(93)90030-v. [DOI] [PubMed] [Google Scholar]
- 43.Begun D J, Whitley P, Todd B L, Waldrip-Dail H M, Clark A G. Genetics. 2000;156:1879–1888. doi: 10.1093/genetics/156.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriyama E N, Powell J R. J Mol Evol. 1997;45:378–379. doi: 10.1007/pl00006243. [DOI] [PubMed] [Google Scholar]
- 45.Bauer V L, Aquadro C F. Mol Biol Evol. 1997;14:1252–1257. doi: 10.1093/oxfordjournals.molbev.a025734. [DOI] [PubMed] [Google Scholar]
- 46.Begun D J, Whitley P. Proc Natl Acad Sci USA. 2000;97:5960–5965. doi: 10.1073/pnas.97.11.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacDonald R J, Swift G H, Przybyla A E, Chirgwin J M. Methods Enzymol. 1987;152:219–227. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]
- 48.Kalb J M, DiBenedetto A J, Wolfner M F. Proc Natl Acad Sci USA. 1993;90:8093–8097. doi: 10.1073/pnas.90.17.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertram M J, Akerkar G A, Ard R L, Gonzalez C, Wolfner M F. Mech Dev. 1992;38:33–40. doi: 10.1016/0925-4773(92)90036-j. [DOI] [PubMed] [Google Scholar]
- 50.Goldman N, Yang Z. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z. paml, Phylogenetic Analysis by Maximum Likelihood. England: Univ. College London; 2000. , Version 3.0. [Google Scholar]
- 52.Yang Z, Swanson W J, Vacquier V D. Mol Biol Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]
- 53.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 54.Rubin G M, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, Harvey D A. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- 55.Reese M G, Kulp D, Tammana H, Haussler D. Genome Res. 2000;10:529–538. doi: 10.1101/gr.10.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rice W R. Evolution (Lawrence, Kans) 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 57.Franke A, Baker B S. Curr Opin Cell Biol. 2000;12:351. doi: 10.1016/s0955-0674(00)00099-5. [DOI] [PubMed] [Google Scholar]
- 58.Meller V H. Trends Cell Biol. 2000;10:54–59. doi: 10.1016/s0962-8924(99)01693-1. [DOI] [PubMed] [Google Scholar]
- 59.Monsma S A, Harada H A, Wolfner M F. Dev Biol. 1990;142:465–475. doi: 10.1016/0012-1606(90)90368-s. [DOI] [PubMed] [Google Scholar]
- 60.Park M, Wolfner M F. Dev Biol. 1995;171:694–722. doi: 10.1006/dbio.1995.1315. [DOI] [PubMed] [Google Scholar]
- 61.Bertram M J, Neubaum D M, Wolfner M F. Insect Biochem Mol Biol. 1996;26:971–980. doi: 10.1016/s0965-1748(96)00064-1. [DOI] [PubMed] [Google Scholar]
- 62.Smith G M, Rothwell K, Wood S L, Yeaman S J, Bownes M. Biochem J. 1994;304:775–779. doi: 10.1042/bj3040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swanson W J, Yang Z, Wolfner M F, Aquadro C F. Proc Natl Acad Sci USA. 2001;98:2509–2514. doi: 10.1073/pnas.051605998. . (First Published February 20, 2001; 10.1073/pnas.051605998) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.