Abstract
The phylogenetic relationships among the three orders of modern amphibians (Caudata, Gymnophiona, and Anura) have been estimated based on both morphological and molecular evidence. Most morphological and paleontological studies of living and fossil amphibians support the hypothesis that salamanders and frogs are sister lineages (the Batrachia hypothesis) and that caecilians are more distantly related. Previous interpretations of molecular data based on nuclear and mitochondrial rRNA sequences suggested that salamanders and caecilians are sister groups to the exclusion of frogs. In an attempt to resolve this apparent conflict, the complete mitochondrial genomes of a salamander (Mertensiella luschani) and a caecilian (Typhlonectes natans) were determined (16,656 and 17,005 bp, respectively) and compared with previously published sequences from a frog (Xenopus laevis) and several other groups of vertebrates. Phylogenetic analyses of the mitochondrial data supported with high bootstrap values the monophyly of living amphibians with respect to other living groups of tetrapods, and a sister group relationship of salamanders and frogs. The lack of phylogenetically informative sites in the previous rRNA data sets (because of its shorter size and higher among-site rate variation) likely explains the discrepancy between our results and those based on previous molecular data. Strong support of the Batrachia hypothesis from both molecule- and morphology-based studies provides a robust phylogenetic framework that will be helpful to comparative studies among the three living orders of amphibians and will permit better understanding of the considerably divergent vertebral, brain, and digit developmental patterns found in frogs and salamanders.
Keywords: Lissamphibia, Caudata, Gymnophiona, Anura, mtDNA
Living amphibians (Lissamphibia) are highly successful tetrapods that evolved diverse body plans that differ in modes of locomotion, reproductive specializations, and life histories (1, 2). For instance, the slender body of living salamanders (Caudata) has a well developed tail and proportionally paired limbs, whereas modern caecilians (Gymnophiona) are completely limbless, and are adapted to a fossorial lifestyle, with elongated bodies, protrusible tentacles, and reduced eyes. Extant frogs (Anura) lack tails, and evolved powerful hind limbs and a shortened, stiffened vertebral column (the urostyle)—a unique adaptation for jumping. The earliest fossils currently known of salamanders (Marmorerpeton; ref. 3), caecilians (Eocaecilia; ref. 4), as well as frogs (Prosalirus; ref. 5) all date back to the Jurassic (190–160 million years ago; ref. 6), and demonstrate that all three lineages of extant amphibians acquired their peculiar body plan early on in their evolutionary history. The diversity among amphibians coupled with the lack of shared derived characters plus a poor fossil record complicate assessment of the phylogenetic relationships among the three living orders.
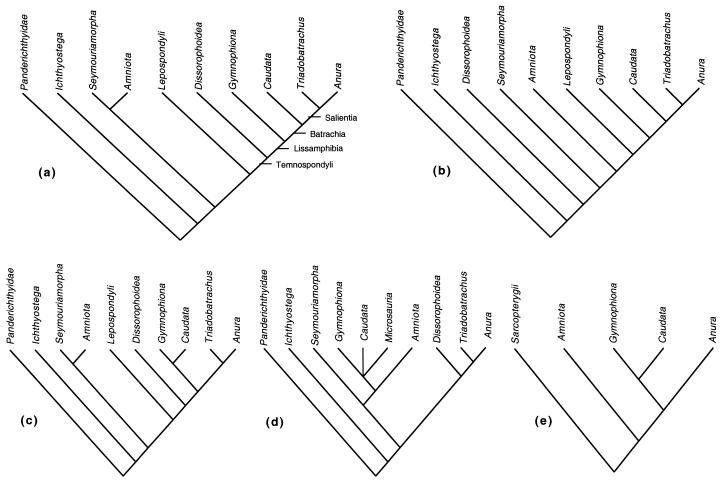
Early workers on amphibian systematics repeatedly rejected the monophyly of Lissamphibia by proposing independent origins of the living orders of modern amphibians (see ref. 7 for a review). However, these studies failed to distinguish between ancestral and derived characters and did not employ any explicit phylogenetic methodology. Szarski (8) and Parsons and Williams (9) integrated previous morphological and paleontological evidence, and concluded that the Lissamphibia were a natural group. The monophyly of Lissamphibia has since gained wide acceptance among researchers (refs. 1 and 10–16; Fig. 1 a–c). A noteworthy exception is Carroll (17–21), who suggests that Lissamphibia are nonmonophyletic because he believes that salamanders and caecilians have affinities with different lineages of microsauria (an extinct group of Lepospondyl amphibians), whereas frogs are related to another extinct group, the temnospondyl amphibians (Fig. 1d).
Figure 1.
Phylogenetic relationships among recent and fossil amphibians. (a) Temnospondyli as sister group of Lissamphibia (Gymnophiona basal to Caudata and Anura) (13, 40). (b) Lepospondyli (including microsauria and Nectridea) as sister group of Lissamphibia (14, 16). (c) Temnospondyli as sister group of Lissamphibia (Anura basal to Gymnophiona and Caudata) (12). (d) Lissamphibia are not monophyletic. Gymnophiona and Caudata are related to Microsauria, whereas the Anura are related to Temnospondyli (20). (e) Phylogenetic relationships of the living orders of amphibia based on nuclear and mitochondrial rRNA data. Gymnophiona is the sister group of Caudata to the exclusion of Anura (15, 25, 26).
Although monophyly of the Lissamphibia is widely accepted, it is still controversial whether the extinct temnospondyls (Fig. 1a) or the lepospondyls (Fig. 1b) are the closest sister group of Lissamphibia. Moreover, there is no generally accepted consensus regarding the phylogenetic relationships among salamanders, caecilians, and frogs. There are three alternative hypotheses to explain such relationships. (i) Salamanders are the closest living relatives of frogs (and form the clade Batrachia) to the exclusion of caecilians. This hypothesis is the most favored by morphological evidence (refs. 1, 10, 11, and 13; Fig. 1 a and b). This hypothesis has also been suggested recently, based on the phylogenetic analysis of mitochondrial rRNA sequence data, albeit only tentatively (22). (ii) Salamanders are the sister group of caecilians to the exclusion of frogs. Most previous molecular studies support this hypothesis (based on both nuclear and mitochondrial rRNA data) (refs. 15 and 23–26; Fig. 1e). There is also morphological evidence supporting this hypothesis [ref. 12; this topology is also recovered in the analysis of Laurin (16), but he suggests that it may be a spurious result; Fig. 1c]. (iii) Frogs are the sister group of caecilians to the exclusion of salamanders. This hypothesis has apparently never been proposed in print.
To address the questions regarding the monophyly of Lissamphibia with respect to other living groups of tetrapods and the phylogenetic relationships among the three orders of living amphibians (Caudata, Gymnophiona, and Anura) we have sequenced the entire mitochondrial genomes of a salamander (Mertensiella luschani) (R.Z., E. Malaga-Trillo, M. Veith, M. Garcia-Paris, and A.M., unpublished data) and a caecilian (Typhlonectes natans) (22). By analyzing these mitochondrial genomes together with previously published mitochondrial sequence data of a frog (Xenopus laevis) (27) and other selected tetrapods, we provide the most comprehensive molecular data set to date that bears on this question.
Materials and Methods
Sequence Alignment and Phylogenetic Reconstruction.
A total of 12 complete mitochondrial genomes representing the major groups of tetrapods were analyzed (GenBank accession nos. for African lungfish, Protopterus dolloi, L42813; coelacanth, Latimeria chalumnae, U82228; clawed frog, Xenopus laevis, M10217; caecilian, Typhlonectes natans, AF154051; Lusehan's salamander, Mertensiella luschani, AF154053; skink, Eumeces egregius, AB016606; painted turtle, Chrysemys picta, AF069423; alligator, Alligator mississippiensis, Y13113; chicken, Gallus gallus, X52392; opossum, Didelphis virginiana, Z29573; blue whale, Balaenoptera musculus, X72204; human, Homo sapiens, D38112). Tetrapod mtDNAs were selected so that their molecular evolutionary rates were not statistically different and so that long-branch attraction effects could be avoided (e.g., the snake mtDNA shows an unusually accelerated evolutionary rate and was omitted; ref. 28). Nucleotide sequences were aligned by using clustal x (29) and refined by eye. Gaps resulting from the alignment were treated as missing data. Ambiguous alignments, mainly in 5′ and 3′ ends of protein coding genes in the DHU and TψC arms of the tRNAs and in several highly variable regions of the rRNA genes were excluded from the phylogenetic analyses. Maximum parsimony (MP) analyses were performed by using heuristic searches [tree bisection and reconnection (TBR) branch swapping; multiple trees (MULTREE) option in effect] with 10 random stepwise additions of taxa. Transitions (Ti) and transversions (Tv) were given equal weight according to the empirical Ti:Tv ratio (1:1). The neighbor-joining (NJ) (30) analysis of the alignment was based on an HKY85 (31) distance matrix. In the maximum likelihood (ML) analyses [Hasegawa, Kishino, Yano (HKY)85 model], Ti:Tv ratios were optimized to maximize the likelihood analysis; empirical base frequencies were used. Robustness of the phylogenetic results was tested either by bootstrap analyses (32) with 500 pseudoreplications (MP and NJ) or by the quartet puzzling method (33) with 1,000 puzzling steps (ML). All phylogenetic analyses were performed by using paup* Version 4.0b4a (34).
Results and Discussion
Complete Mitochondrial DNA Evidence Supports the Batrachia Hypothesis.
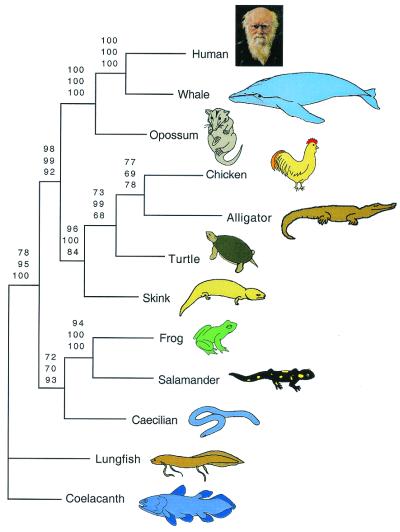
Mitochondrial protein-coding, tRNA, and rRNA gene sequences were combined into a single data set that produced an alignment of 15,686 positions; of those positions, 2,635 were excluded because of ambiguity, 4,825 were constant, and 6,472 were parsimony-informative. All three commonly used methods of phylogenetic inference (MP, NJ, and ML) arrived at the same tree topology (Fig. 2). This fully resolved tree supports the monophyly of living amphibians with respect to other living tetrapods (1, 8–16) and favors a sister group relationship for salamanders and frogs (the Batrachia hypothesis; refs. 1, 10, 11, 13, and 22). Robustness of our results was confirmed by high bootstrap support of all nodes in the tree (Fig. 2). This result contradicts the hypothesis of a salamander + caecilian grouping (12) that was suggested by previous molecular work based on rRNA data sets (15, 23, 24, 26).
Figure 2.
Majority rule (50%) consensus trees depicting living amphibian relationships. Mitochondrial protein-coding, tRNA, and rRNA gene sequences were combined into a single data set that was subjected to MP (bootstrap values based on 500 bootstrap pseudoreplications; upper value of each triplet of numbers), NJ (bootstrap values based on 500 bootstrap pseudoreplications; middle value of each triplet of numbers), and ML (quartet puzzling support values based on 1,000 puzzling steps; lowest value of each triplet of numbers) analyses. Lobe-finned fishes (lungfish and coelacanth) were used as outgroup taxa.
Several recent studies have pointed out the risk of recovering misleading phylogenetic relationships among distantly related taxa when only short sequence data sets are analyzed (35–38). Larger or relatively heterogenous data sets, i.e., complete mitochondrial genomes with many phylogenetically informative sites, are required to confidently resolve most questions dealing with major phylogenetic events (35–38). Our results would seem to support the conclusions of these studies.
Interestingly, morphology- and molecule-based studies have disagreed profoundly regarding extant amphibian relationships. Most morphological studies have consistently concluded that salamanders and frogs are sister groups (1, 10, 11, 13), whereas molecular studies based on nuclear and mitochondrial rRNA data have supported caecilians as the sister taxon of salamanders (15, 23, 24, 26). Our analyses based on complete mitochondrial sequence data may contribute to a resolution of the apparent conflict between morphological and molecular evidence. In agreement with previous morphology-based hypotheses, our molecular data overwhelmingly support monophyly of living amphibians and the hypothesis that salamanders and frogs are more closely related to each other than to caecilians (Fig. 2). There are several reasons why we believe that our extensive molecular data set may be more reliable than the smaller rRNA data sets on this question. It is well known that rRNA data sets show a high degree of among-site rate variation, and consequently, few sites are phylogenetically informative at any given level of divergence (39). This pattern can lead to the recovery of spurious phylogenies based on homoplasious rather than phylogenetically informative sites, and likely explains why previous molecular studies based on rRNA data were prone to recover a salamander + caecilian clade. In fact, a recent reanalysis of the mitochondrial rRNA evidence was able to moderately support a salamander + frog sister group relationship (22).
Identity of the Sister Group of Lissamphibia Can Be Gleaned Only from Paleontological Data.
Of course, molecular data cannot directly assess the monophyly of Lissamphibia because it also includes fossil groups. However, as indicated by our molecular data (Fig. 2), a sister group relationship between salamanders and frogs can indirectly reject those hypotheses based on morphological and paleontological data that suggest nonmonophyly of living amphibians because of an independent origin of salamanders (refs. 18–21; Fig. 1d). Moreover, the monophyly of living amphibians with respect to living amniotes (Fig. 2) can indirectly reject those hypotheses that relate caecilians and salamanders to microsaurs, and they suggest a closer relationship of the latter to amniota rather than to Temnospondyli (refs. 17–21; Fig. 1d). In this regard, the molecular phylogeny presented further supports the hypothesis that lissamphibians (including fossil groups) constitute a single clade with respect to extinct amphibian groups.
Unfortunately, molecular data cannot provide information on the question of the closest extinct relative of the Lissamphibia. Only paleontological evidence can determine whether the extinct Temnospondyli (e.g., refs. 13 and 40) or the Lepospondyli (14, 16, 20) are the closest sister group to modern amphibians (but see below).
Congruent Molecular and Morphological Evidence Provides a Robust Phylogenetic Framework That Is Necessary for Comparative Studies Among Amphibians.
The hypothesis presented here implies that frogs provide the ideal extant outgroup to analyze phylogenetic relationships within salamanders (including, e.g., the open question of the relative positions of the families Sirenidae and Proteidae; ref. 41); inversely, salamanders are likely the best living outgroup to study anuran systematics (including the contentious monophyly of archaeobatrachia; ref. 42). The use of the closest living relative as outgroup is relevant both in morphological (because it could directly affect the polarity of several characters) and molecular (e.g., because it could prevent the attraction of long branches to basal positions) studies. Moreover, our results have implications for the decision to choose between alternative positions of several fossil taxa that are closely related to the origin of the three lineages of living amphibians. For instance, the recent placement of the albanerpetontids (a group of salamander-like fossil amphibians) as sister group of the Batrachia (43) is supported by the molecular phylogeny (a putative caecilian + salamander clade would have contradicted it). Furthermore, by providing an independent source of phylogenetic information, our results confirm the authenticity of some of the putative morphological synapomorphies (up to nine unique shared-derived characters; see ref. 13) that diagnose the Batrachia, and might encourage the search for new shared-derived traits between salamanders and frogs. Moreover, our results invite further research on the causes of the now seemingly homoplasious characters that group caecilians with salamanders.
The geographic argument based on the current world distribution of amphibians and rRNA evidence that salamanders and caecilians originated from a common ancestor because of the breakup of the supercontinent Pangea (15) is invalidated by our data. Furthermore, the recent discovery of Czatkobatrachus from the early Triassic of Poland (44) suggests that, at least, Salientia (the stem-group of anurans) occurred in all of Pangea (Triadobatrachus is from early Triassic of Madagascar; ref. 45), and that the separation of salamanders and frogs predated the breakup of Pangea.
Our results will also be helpful in polarizing the divergent developmental patterns exhibited by Caudata, Anura, and Gymnophiona (46, 47). For instance, vertebral development in frogs is interpreted as being rather similar to that inferred for Temnospondyli (21), which may represent the primitive condition for all tetrapods. However, the pattern of vertebral centra ossification in salamanders (48) and caecilians (49) is more similar to that inferred for Lepospondyli (representing a derived condition). If lissamphibians evolved from lepospondyls (14, 16), and accepting that salamanders and frogs are sister group taxa, the most parsimonious explanation to the observed patterns is that the type of vertebral development in salamanders and caecilians is primitive for lissamphibians, and that the ancestor of frogs reelaborated the developmental pattern found in the extinct temnospondyls (this pattern may be a constraint associated with metamorphosis in frogs; ref. 21). Alternatively, whether temnospondyls are the sister group of modern amphibians, the early ossification of vertebral centra evolved independently in caecilians and salamanders, whereas frogs would have retained the primitive condition of all tetrapods.
Brain development of frogs and salamanders differs considerably. The brains of frogs are morphologically much more complex than those of salamanders (50). On the other hand, salamanders and caecilians show similarity in many features of brain development (51). These patterns seemingly would contradict the Batrachia hypothesis. However, the simple brain morphology of salamanders may be a secondarily derived condition associated with an increase in genome, cell size, and pedomorphic evolution (51, 52). Our phylogeny supports this hypothesis of a secondary simplification in salamanders.
The three orders of living amphibians differ markedly in their limb morphologies (6). By providing a robust phylogenetic framework, our results may also aid in polarizing the divergent limb development patterns found in amphibians (53). For instance, frogs and salamanders follow divergent pathways of digital formation (e.g., refs. 46 and 54). Frogs develop their digits in a posterior to anterior sequence (55). However, salamanders show the opposite pattern (46). Based on strict ontogenetic criteria, these patterns were used in the past as evidence of amphibian (and tetrapod) polyphyly (e.g., ref. 56; see ref. 47 for a review). Yet, based on the robust and congruent morphological and molecular evidence that supports the Batrachia hypothesis, the interpretation of these developmental patterns would need to be quite different. Digit formation in frogs is considered to be the ancestral condition for all tetrapods, whereas the salamander pattern represents a derived condition (46, 55, 57, 58). Recent molecular developmental studies demonstrate that expression patterns of Hoxa-11 in the limb buds of frogs are similar to those found in amniota (59), whereas salamanders show a derived pattern.
The interpretation of many comparative studies among the three orders of living amphibians largely depends on whether the salamanders are the closest living relative of frogs or the living sister group of caecilians (60). In contrast to previous molecular analyses based on nuclear and mitochondrial rRNA data, our results, based on the analysis of complete mitochondrial genome data, support the Batrachia (salamander + frog) hypothesis and are thereby in agreement with most morphological evidence. Our analyses demonstrate that some phylogenetic controversies between morphology and molecules that exist in the literature are only apparent rather than real and frequently caused by a lack of phylogenetically informative characters in one of the two (morphological or molecular) relatively small data sets analyzed. General support of the Batrachia hypothesis allows a confident polarization of characters in many comparative studies that explore the astonishing biological diversity within and among extant orders of amphibians.
Acknowledgments
We thank Borja Sanchiz, David B. Wake, and three anonymous reviewers for insightful comments on the manuscript. We dedicate this paper to the memory of Pere Alberch, who brilliantly combined development and evolution, taking amphibians as a case model. R.Z. was sponsored by a postdoctoral contract of the Ministerio de Educacion y Cultura of Spain. This work received partial financial support from grants from the Lion Foundation, the Deutsche Forschungsgemeinschaft, the University of Konstanz, and Grant DEB-9615178 from the U.S. National Science Foundation (to A.M.).
Abbreviations
- MP
maximum parsimony
- NJ
neighbor joining
- ML
maximum likelihood
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF154053).
References
- 1.Duellman W E, Trueb L. Biology of Amphibians. Baltimore: Johns Hopkins Univ. Press; 1994. [Google Scholar]
- 2.Pough F H, Janis C M, Heiser J B. Vertebrate Life. Englewood Cliffs, NJ: Prentice–Hall; 1998. pp. 297–341. [Google Scholar]
- 3.Evans S E, Milner A R, Mussett F. Geobios (Jodhpur, India) 1988;21:539–552. [Google Scholar]
- 4.Jenkins F A, Walsh D M. Nature (London) 1993;365:246–249. [Google Scholar]
- 5.Shubin N H, Jenkins F A. Nature (London) 1995;377:49–52. [Google Scholar]
- 6.Wake M H. Zoology (Jena) 1997;100:141–151. [Google Scholar]
- 7.Trueb L, Cloutier R. In: Origins of the Major Groups of Tetrapods: Controversies and Consensus. Schultze H P, Trueb L, editors. Ithaca, NY: Cornell Univ. Press; 1991. pp. 175–193. [Google Scholar]
- 8.Szarski H. Q Rev Biol. 1962;37:189–241. [Google Scholar]
- 9.Parsons T S, Williams E E. Q Rev Biol. 1963;38:26–53. [Google Scholar]
- 10.Rage J C, Janvier P. Geobios (Jodhpur, India) 1982;6:65–83. [Google Scholar]
- 11.Milner A R. In: The Phylogeny and Classification of the Tetrapods. Benton M J, editor. Vol. 1. Oxford: Clarendon; 1988. pp. 59–102. [Google Scholar]
- 12.Bolt J R. In: Origins of the Major Groups of Tetrapods: Controversies and Consensus. Schultze H P, Trueb L, editors. Ithaca, NY: Cornell Univ. Press; 1991. pp. 194–222. [Google Scholar]
- 13.Trueb L, Cloutier R. In: Origins of the Major Groups of Tetrapods: Controversies and Consensus. Schultze H P, Trueb L, editors. Ithaca: Cornell Univ. Press; 1991. pp. 223–313. [Google Scholar]
- 14.Laurin M, Reisz R. In: Amniote Origins. Sumida S S, Martin K L, editors. New York: Academic; 1997. pp. 9–59. [Google Scholar]
- 15.Feller A E, Hedges S B. Mol Phylogenet Evol. 1998;9:509–516. doi: 10.1006/mpev.1998.0500. [DOI] [PubMed] [Google Scholar]
- 16.Laurin M. Ann Sci Nat Zool Biol Anim. 1998;1:1–42. [Google Scholar]
- 17.Carroll R L, Currie P J. J Linn Soc London Zool. 1975;57:229–247. [Google Scholar]
- 18.Carroll R L, Holmes R. J Linn Soc London Zool. 1980;68:1–40. [Google Scholar]
- 19.Carroll R L. Vertebrate Paleontology and Evolution. New York: Freeman; 1988. [Google Scholar]
- 20.Carroll R L. Bull Mus Nat Hist Nat (Paris 4eme Série) 1995;17:389–445. [Google Scholar]
- 21.Carroll R L, Kuntz A, Albright K. Evol Dev. 1999;1:36–48. doi: 10.1111/j.1525-142x.1999.t01-2-.x. [DOI] [PubMed] [Google Scholar]
- 22.Zardoya R, Meyer A. Genetics. 2000;155:765–775. doi: 10.1093/genetics/155.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson A, Wilson A C. Mol Biol Evol. 1989;6:131–154. doi: 10.1093/oxfordjournals.molbev.a040539. [DOI] [PubMed] [Google Scholar]
- 24.Hedges S B, Moberg K D, Maxson L R. Mol Biol Evol. 1990;7:607–633. doi: 10.1093/oxfordjournals.molbev.a040628. [DOI] [PubMed] [Google Scholar]
- 25.Hedges S B, Maxson L R. Herpetol Monogr. 1993;7:27–42. [Google Scholar]
- 26.Hay J M, Ruvinsky I, Hedges S B, Maxson L R. Mol Biol Evol. 1995;12:928–937. doi: 10.1093/oxfordjournals.molbev.a040270. [DOI] [PubMed] [Google Scholar]
- 27.Roe B A, Din-Pow M, Wilson R K, Wong J F. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- 28.Kumazawa Y, Ota H, Nishida M, Ozawa T. Genetics. 1998;150:313–329. doi: 10.1093/genetics/150.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson J D, Gibson T J, Plewniak F, Jeanmougin J, Higgins D G. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 32.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 33.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 34.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer; 1998. , Version 4.0. [Google Scholar]
- 35.Cummings M P, Otto S P, Wakeley J. Mol Biol Evol. 1995;12:814–822. doi: 10.1093/oxfordjournals.molbev.a040258. [DOI] [PubMed] [Google Scholar]
- 36.Russo C A M, Takezaki N, Nei M. Mol Biol Evol. 1996;13:525–536. doi: 10.1093/oxfordjournals.molbev.a025613. [DOI] [PubMed] [Google Scholar]
- 37.Zardoya R, Meyer A. Mol Biol Evol. 1996;13:933–942. doi: 10.1093/oxfordjournals.molbev.a025661. [DOI] [PubMed] [Google Scholar]
- 38.Takezaki N, Gojobori T. Mol Biol Evol. 1999;16:590–601. doi: 10.1093/oxfordjournals.molbev.a026141. [DOI] [PubMed] [Google Scholar]
- 39.Olsen G J, Woese C R. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 40.Panchen A L, Smithson T R. Biol Rev Camb Philos Soc. 1987;62:341–438. [Google Scholar]
- 41.Larson A, Dimmick W W. Herpetol Monogr. 1993;7:77–93. [Google Scholar]
- 42.Ford L S, Cannatella D C. Herpetol Monogr. 1993;7:94–117. [Google Scholar]
- 43.McGowan G, Evans S E. Nature (London) 1995;373:143–145. [Google Scholar]
- 44.Evans S E, Borsuk-Bialynicka M. Acta Palaeo Polonica. 1998;43:573–580. [Google Scholar]
- 45.Rage J, Rocek Z. Paleontographica Abt A. 1989;206:1–16. [Google Scholar]
- 46.Alberch P, Gale E A. Evolution. 1985;39:8–23. doi: 10.1111/j.1558-5646.1985.tb04076.x. [DOI] [PubMed] [Google Scholar]
- 47.Hanken J. Evol Biol. 1986;20:389–417. [Google Scholar]
- 48.Wake D B, Lawson R. J Morphol. 1973;139:251–300. doi: 10.1002/jmor.1051390302. [DOI] [PubMed] [Google Scholar]
- 49.Wake D B, Wake M H. Mem Soc Zool Fr. 1986;43:67–70. [Google Scholar]
- 50.Roth G, Blanke J, Wake D B. Proc Natl Acad Sci USA. 1994;91:4796–4800. doi: 10.1073/pnas.91.11.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roth G, Maujoks-Manteuffel C, Nishikawa K, Schmidt A, Wake D B. Brain Behav Evol. 1993;42:137–170. doi: 10.1159/000114147. [DOI] [PubMed] [Google Scholar]
- 52.Roth G, Nishikawa K C, Wake D B. Brain Behav Evol. 1997;50:50–59. doi: 10.1159/000113321. [DOI] [PubMed] [Google Scholar]
- 53.Wake D B, Shubin N. J Morphol. 1994;220:407–408. [Google Scholar]
- 54.Shubin N, Alberch P. Evol Biol. 1986;20:318–390. [Google Scholar]
- 55.Shubin N. Evol Biol. 1995;28:39–86. [Google Scholar]
- 56.Holmgren N. Acta Zool (Stockholm) 1933;20:89–124. [Google Scholar]
- 57.Shubin N H. In: Homology: The Hierarchical Basis of Comparative Biology. Hall B K, editor. San Diego: Academic; 1994. pp. 249–271. [Google Scholar]
- 58.Shubin N, Wake D B. Am Zool. 1996;36:51–60. [Google Scholar]
- 59.Wagner G P, Khan P A, Blanco M J, Misof B, Liversage R A. Am Zool. 1999;39:686–694. [Google Scholar]
- 60.Cannatella D C, Hillis D M. Herpetol Monogr. 1993;7:1–7. [Google Scholar]