Abstract
Summary: There is a growing need in the research community to apply the molecular evolutionary genetics analysis (MEGA) software tool for batch processing a large number of datasets and to integrate it into analysis workflows. Therefore, we now make available the computing core of the MEGA software as a stand-alone executable (MEGA-CC), along with an analysis prototyper (MEGA-Proto). MEGA-CC provides users with access to all the computational analyses available through MEGA’s graphical user interface version. This includes methods for multiple sequence alignment, substitution model selection, evolutionary distance estimation, phylogeny inference, substitution rate and pattern estimation, tests of natural selection and ancestral sequence inference. Additionally, we have upgraded the source code for phylogenetic analysis using the maximum likelihood methods for parallel execution on multiple processors and cores. Here, we describe MEGA-CC and outline the steps for using MEGA-CC in tandem with MEGA-Proto for iterative and automated data analysis.
Availability: http://www.megasoftware.net/
Contact: s.kumar@asu.edu
1 INTRODUCTION
The molecular evolutionary genetics analysis (MEGA) software is an integrated suite of tools for statistics-based comparative analysis of molecular sequence data based on evolutionary principles (Kumar et al. 2008; Tamura et al. 2011). MEGA is being used by biologists in a large number of laboratories for reconstructing the evolutionary histories of species and inferring the extent and nature of selective forces shaping the evolution of genes and species (see www.megasoftware.net). Additionally, MEGA is used in many classrooms as a tool for teaching the methods used in evolutionary bioinformatics.
One of MEGA’s key features is its graphical user interface (GUI), which facilitates detailed visualization and interactive exploration of sequence data, phylogenetic trees and analysis results. However, over time the needs of MEGA users have expanded due to increasing availability of multi-gene and genome-scale data, which necessitate iterative, high-throughput analysis. In order to address this need, we have re-engineered the MEGA source code so that the computational core, which implements the computation algorithms for all analyses in MEGA, can be used as a stand-alone application and executed through command-line, other applications or scripting languages. We now make available MEGA-CC (computing core) that implements all the computational analyses available in the latest GUI-based edition; for a list of analysis, see Table 1 in Tamura et al. 2011.
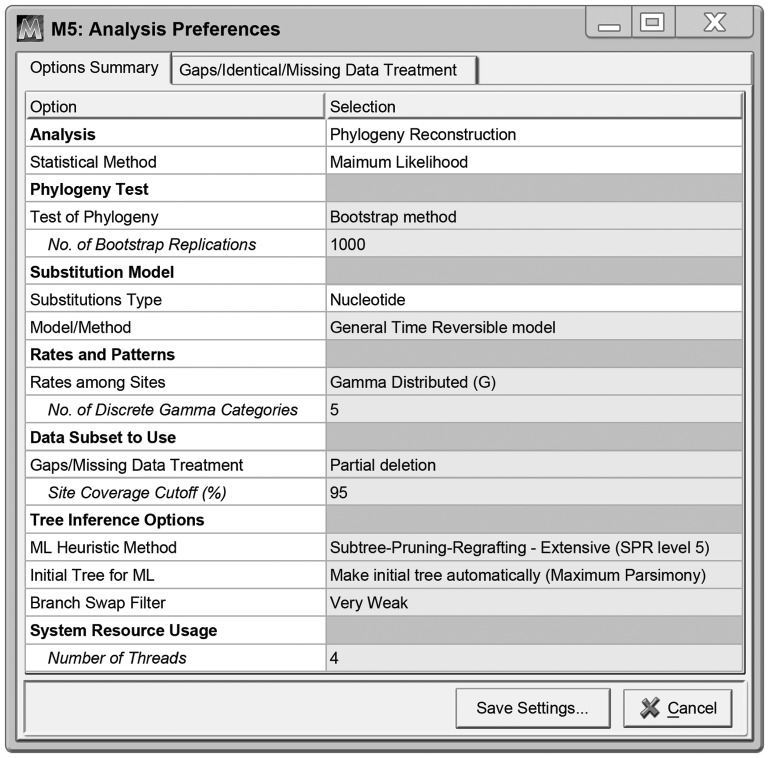
MEGA-CC comes with MEGA-Proto, which provides an easy way to generate a configuration file specifying analysis options for the desired analysis (Fig. 1). The use of MEGA-Proto ensures that the analysis selections are fully validated and that they mirror those in the GUI-based edition of MEGA. This leads to a consistent usage experience across different releases of MEGA and enables the user to test drive their analysis in the GUI-based edition of MEGA 5.0. It also makes it possible for the programming team to more easily keep all versions of MEGA completely synchronized.
Fig. 1.
The analysis preferences dialog box in MEGA-Proto. It is displayed by the MEGA-Proto application after the user specifies the data type (protein-coding DNA in the present case) and chooses to infer maximum likelihood trees. The user selects the desired settings in this dialog box and saves them into the analysis options file in an appropriate directory for use with MEGA-CC.
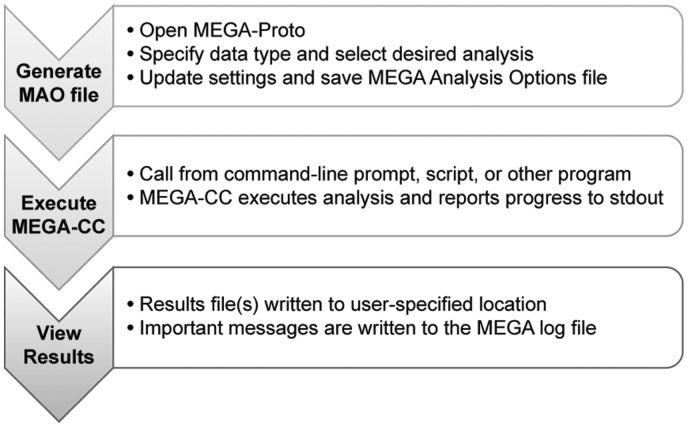
We illustrate the use of MEGA-CC and MEGA-Proto together in an example case where one wishes to infer maximum likelihood (ML) phylogenetic trees for many genes in series and the user has obtained multiple sequence alignments comprised of coding sequence data for each gene. Using the MEGA-Proto application, the user first generates an analysis options file that specifies the chosen settings for the substitution model, genetic code table, gaps/missing data treatment, distribution of rates, topology search approach, source for the initial tree, size of the execution thread pool and optionally the number of replications for carrying out the bootstrap test of phylogeny (Fig. 2).
Fig. 2.
Steps in using MEGA-Proto and MEGA-CC for iterative analysis.
Once the analysis options file has been saved to disk, the user is now ready to execute MEGA-CC to generate an ML phylogeny for each sequence alignment. In this scenario, the user has multiple options for processing the alignment files. The simplest method is to use the ‘File Iterator’ system in MEGA-CC which will search in a user-specified directory for all input data files that are compatible with the active analysis and process them iteratively. In our example, MEGA-CC would process all FastA and MEGA formatted sequence data files found in the specified directory. Alternatively, the ‘File Iterator’ system can operate from a list of files that the user provides in a text file. To use this feature, the user can launch the MEGA-CC executable from a command prompt along with the appropriate parameter flag, and pass it a directory name or the name of a text file which lists all target input files. Alternatively, the user can generate their own script that iterates over the names of the alignment files and launches the MEGA-CC executable for each file. Supported input data files for MEGA-CC are FastA and MEGA files for sequence data, Newick files for phylogenies and MEGA files for distance matrices.
During execution, the progress of the analysis is displayed in the shell window and upon completion, the results files are written to the specified path. The format of the output files depends on the analysis done. For example, in the case of ML phylogeny reconstruction, the resulting phylogeny is written to a Newick formatted text file. Additionally, an optional analysis summary file, which includes information regarding the analysis such as log likelihood and estimated parameter values, is saved alongside the Newick file.
In addition to batch processing of single analyses, MEGA-CC gives users the ability to construct automated workflows that chain together multiple analyses. For instance, starting with a FastA file of DNA sequence data, a multiple sequence alignment can be generated using either MEGA’s native implementation of ClustalW or the integrated MUSCLE alignment program. Next, this alignment can serve as input data for a number of phylogenetic inference methods (e.g. ML, Maximum Parsimony and Neighbor Joining), all of which can be statistically tested by the bootstrap method. Using the sequence alignment and phylogenetic tree that have been created, a user may perform further analyses such as inference of ancestral sequences, tests of natural selection, molecular clock test and estimation of evolutionary rates. Alternatively, MEGA-CC can be easily integrated into pipelines constructed with other bioinformatics applications, as virtually all programming languages facilitate execution of external processes.
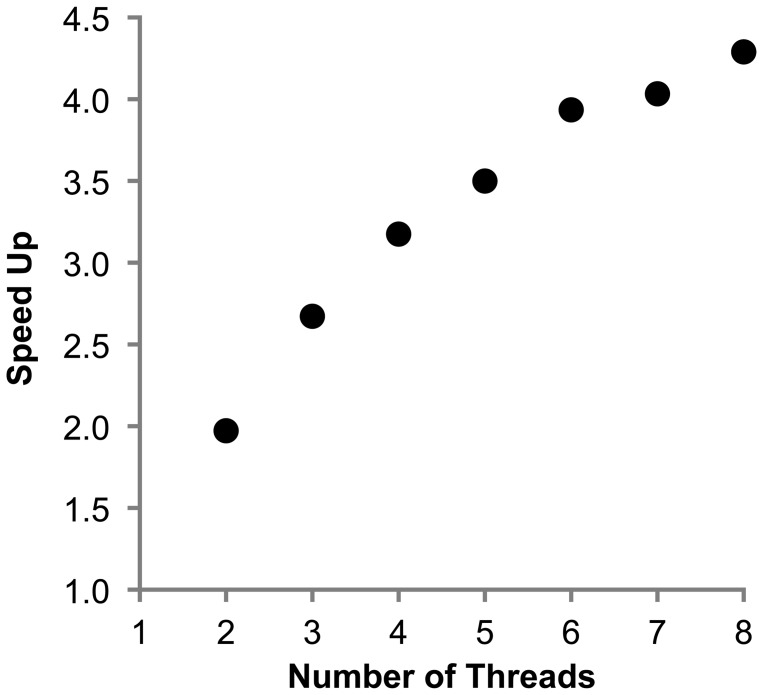
We have also re-programmed ML analyses to efficiently utilize multiple processors and cores on the same machine for increased performance, which will greatly reduce the run time for computationally demanding ML inferences. For example, MEGA-CC reduces the time taken to complete ML tree search by almost half (49%) when using two threads (speed-up = 1.97) for an alignment containing 500 sequences, each having 2000 base pairs (Fig. 3). Addition of the third thread reduces the time further (speed up = 2.73), a similar performance was achieved by RAxML version 7.3.2 (Stamatakis et al. 2006) for two and three threads. (More detailed comparison of algorithms and speed-ups will be published elsewhere.) Because virtually all new desktop workstations are equipped with at least a few cores (or processors), many will see significant performance increases when opting to use multiple threads and cores.
Fig. 3.
Speed-up achieved in MEGA-CC when inferring ML phylogeny using multiple cores. Each dot shows the time taken by using only one thread divided by the time taken by multiple threads on the same machine. Results are from an analysis using a simulated sequence alignment containing 500 sequences that were 2000 base pairs each (see Tamura et al. 2011). The Subtree-Pruning-and-Regrafting procedure under a GTR + G substitution model (four discrete categories) was used on an Intel Xeon processor with eight hyper-threaded processing cores on Windows 7.
In summary, MEGA-CC will address the needs of biologists interested in iterative and automated analyses. Currently, MEGA-CC and MEGA-Proto are available for the Windows 32-bit computing platform. We plan to release the 64-bit version of MEGA-CC in the near future that will be natively compiled for all major operating systems and come with a web-based version of MEGA-Proto.
ACKNOWLEDGEMENTS
We thank Carol Williams and Sudhindra Gadagkar for editorial comments, and all the members of our laboratories who tested and provided feedback on early releases of this software.
Funding: Research grants from the US National Institutes of Health (HG002096-11 and GM081066-04 to SK) and Japan Society for the Promotion of Science (KT).
Conflict of Interest: none declared
REFERENCES
- Kumar S., et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., et al. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed model. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tamura K., et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]