Abstract
Objectives
Aztreonam for inhalation solution (AZLI) was recently approved by the FDA for treating cystic fibrosis (CF) patients infected with Pseudomonas aeruginosa. Here we investigated the effect of aztreonam alone or in combination with tobramycin on P. aeruginosa biofilms grown on CF airway epithelial cells.
Methods
P. aeruginosa biofilms, produced by laboratory strains or clinical isolates, were formed on confluent CF airway cells before treatment overnight with aztreonam or tobramycin alone or in combination. Alternatively, antibiotics were added 1 h after bacterial inoculation to assess their ability to impair biofilm formation at 5 h. Bacterial cfu remaining after treatment were then determined by plate counting.
Results
In the absence of antibiotics, all strains developed biofilms that disrupted CF airway epithelial monolayers overnight. Tobramycin reduced the cfu of all strains grown as biofilms. Aztreonam reduced the cfu of some strains by ∼1 log unit without preserving the integrity of cystic fibrosis airway cell monolayers, while decreasing the biofilms of other clinical isolates by ∼4 log units and protecting the monolayers from being compromised. The combination of aztreonam and tobramycin reduced the cfu of two strains by an additional 0.5 and 2 log units, respectively. Of all the mechanisms explored, Psl exopolysaccharide production might explain the variations in biofilm tolerance to aztreonam in some of the strains.
Conclusions
Effects of aztreonam on P. aeruginosa biofilms in the in vitro co-culture model are strain-dependent. The simultaneous application of aztreonam and tobramycin may be beneficial for a subset of CF patients by eliminating susceptible P. aeruginosa strains.
Keywords: tolerance, AZLI, co-culture, monotherapy
Introduction
Cystic fibrosis (CF) is an inherited disease caused by mutations in the cystic fibrosis transmembrane conductor regulator (CFTR).1,2 In the absence of a functional CFTR, the CF lung becomes chronically infected with bacteria, leading to respiratory failure and death. According to the 2010 Cystic Fibrosis Foundation Patient Registry, Staphylococcus aureus is the dominant (∼67%) microorganism infecting CF patients of age 18–24,3 while Pseudomonas aeruginosa is the dominant microorganism infecting CF patients in older populations (found in ∼80% of adults with CF) and contributes to most morbidity and mortality in these patients.4,5
During early infections, P. aeruginosa is typically in a planktonic, non-mucoid form and can be eradicated by repeated antibiotic treatment.6 However, evidence suggests that during chronic infections P. aeruginosa forms biofilms and becomes more tolerant to antibiotics.7,8 In the biofilm mode of growth, bacteria are encased in an exopolysaccharide (EPS) matrix, which could act as a barrier that prevents antibiotics from penetrating the biofilm.9
The life expectancy of CF patients has greatly improved in the past few decades due to the development and aggressive application of antibiotics.10 Until the inhaled form of aztreonam (AZLI; Cayston®) was approved as an antipseudomonal antibiotic in 2010, the aminoglycoside antibiotic tobramycin for inhalation (TIS, TOBI®), approved for use in 1997, was the only inhaled antibiotic approved for the treatment of pulmonary infections with P. aeruginosa in CF.
Aztreonam is a monobactam antibiotic that exerts its effect by inhibiting bacterial cell wall synthesis and has broad-spectrum activity against Gram-negative bacteria, including P. aeruginosa.11,12 The effectiveness and safety of inhaled aztreonam for CF patients have been investigated in three multinational, multicentre Phase III studies (AIR-CF1 to AIR-CF3).13–15 Regular administration of AZLI to CF patients effectively decreased P. aeruginosa density in their sputum by ∼0.6–1.5log10cfu/g in both short-term (28 days) and long-term (18 months) studies.16
Despite the growing use of TOBI and AZLI by CF patients, few data are available in the literature on the comparative and combination efficacy of these two drugs. To better understand the effect of aztreonam and tobramycin against P. aeruginosa in the context of CF, we performed a comprehensive study to test the antibiotic susceptibility of P. aeruginosa laboratory strain PAO1 and six CF clinical isolates grown as biofilms on CF airway epithelial cells. We used an in vitro co-culture model developed previously by our group,5,17 wherein P. aeruginosa biofilms form on human CF airway epithelial cells. This model recapitulates several aspects of CF lung disease, including more robust biofilm formation on CF airway cells than non-CF control lines and high-level biofilm-mediated antibiotic tolerance akin to what is observed in the CF lung.5,17,18 We also explored the possible additive effect of aztreonam and tobramycin by applying these two drugs simultaneously to the co-culture model. Finally, possible mechanisms for our susceptibility findings were explored. Our study not only provides new insights into P. aeruginosa susceptibility to aztreonam and tobramycin in this co-culture model, but can also be used as a reference for future preclinical study designs.
Materials and methods
Bacterial strains, growth medium and antibiotics
P. aeruginosa strain PAO1 used in the confocal microscopy studies carries a plasmid (pSMC21) that constitutively expresses green fluorescent protein (GFP) and is stable without antibiotic selection.19 P. aeruginosa strain PAO1 is a well characterized and broadly used laboratory strain. The strain ZK2870 and its mutant counterpart ZK2870ΔpelA were kindly provided by Roberto Kolter.20 The P. aeruginosa strain PAO1 Δpsl mutant, mucoid laboratory strain FRD1 and its non-mucoid derivative FRD1algT::Tn501 were obtained from Dan Wozniak.21,22 The strains SMC715, SMC725 and SMC738 and their corresponding Δpsl mutants were provided by Michael Zegans.23 Clinical P. aeruginosa isolates SMC1585, SMC1587, SMC1595, SMC1596, SMC5450 and SMC5451 were collected from the sputa of CF patients by the clinical microbiology laboratory at the Dartmouth-Hitchcock Medical Center (DHMC). Each strain was collected from a different patient and previous antibiotic application of the patients was not recorded. The mucoid phenotype of these clinical isolates was determined by the standard clinical test,24 in which the bacteria were grown on sheep blood agar, MacConkey's agar and Mueller–Hinton agar at 35°C in 5% CO2 or in room air. All P. aeruginosa strains were cultured in lysogeny broth (LB) medium (10 g of tryptone, 5 g of yeast extract and 10 g of NaCl dissolved in 1 L of water) overnight at 37°C before further dilution and application to the airway cells. The antibiotics tobramycin and aztreonam were obtained from Sigma (St Louis, MO, USA) and MP Biomedicals (Solon, OH, USA), respectively. Clinically relevant concentrations of tobramycin (1000 mg/L) and aztreonam (700 mg/L) were chosen for this study, which are equivalent to the peak concentrations measured in the sputum of CF patients after aerosolized drug application.14,25
Cell lines and cell culture
Human bronchial epithelial cells (CFBE41o−, homozygous for the ΔF508-CFTR mutation) were isolated from a CF patient.26,27 CFBE41o− cells overexpressing ΔF508-CFTR (CFBE cells hereafter), a gift from Dr J. P. Clancy,28 were used throughout the study as host cells. CFBE cells were maintained in minimal essential media (MEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 50 U/mL penicillin, 2 mg/mL puromycin, 50 mg/mL streptomycin and 5 mg/mL plasmocin in a 5% CO2/95% air incubator at 37°C. CFBE cells were grown at a seeding density of 0.2 × 106/well in a 12-well plate (Corning Incorporated, Corning, NY, USA) or at 2 × 106/coverslip on a 40 mm diameter glass coverslip (Bioptechs, Butler, PA, USA) for 7–9 days to establish confluent monolayers. The regular cell growth medium was switched to a modified medium (MEM without phenol red plus 2 mM l-glutamine; referred to as ‘imaging medium’ hereafter) on the day the experiment was performed.
Cytotoxicity assay
To examine whether aztreonam alone or in combination with tobramycin causes cytotoxicity in CFBE cells, we measured the levels of lactate dehydrogenase (LDH) released upon exposure to these antibiotics for 16 h using the CytoTox 96 non-radioactive cytotoxicity assay kit from Promega, as described previously.17
Static co-culture assay
The static co-culture assay was used to form P. aeruginosa biofilms on CFBE cells in a standard multiwell tissue culture plate, as described previously.17 Briefly, CFBE cells were grown in 12-well plates for 7–9 days as described above. The cells were washed twice with imaging medium before P. aeruginosa was inoculated into each well at a multiplicity of infection (MOI) of ∼30 (∼6 × 106 cfu/well). The plates were incubated at 37°C, 5% CO2 for 1 h to allow bacterial attachment to the airway cells. The supernatant was then replaced with imaging medium containing 0.4% arginine and incubated for 5 h to form biofilms on CFBE cells. To assess the efficacy of antibiotic treatment in preformed biofilms, the plates were washed twice with imaging medium and antibiotics were applied at designated concentrations (tobramycin, 1000 mg/L; aztreonam, 700 mg/L) to disrupt established biofilms for 16 h. The supernatant was then removed and washed twice with imaging medium. CFBE cells were lysed with 0.1% Triton X-100 for ∼15 min. The lysate was vortexed for 3 min before serial dilution and spot titration onto LB plates to determine the cfu/well. The bacterial strain was defined as ‘susceptible’ to the antibiotic treatment in the static co-culture model if the CFBE monolayers were not disrupted after overnight antibiotic treatment and there was more than a 2 log10 difference in cfu recovery between no antibiotic treatment and antibiotic treatment.
To test the ability of antibiotics to prevent biofilm formation, these compounds were applied after the 1 h period for bacterial attachment, the plates were incubated for 5 h, and cfu/well was determined as described above. The detection limit of the static co-culture assay was ∼200 cfu/well. All experiments were performed at least three times.
Flow chamber imaging assay
We used the flow chamber imaging assay to assess antibiotic susceptibility visually. In this assay, GFP-labelled P. aeruginosa were used to form biofilms on human airway cells as described previously.5,29 Briefly, a Focht Chamber System 2 (FCS2) from Bioptechs was assembled and mounted on the stage of an inverted Nikon microscope. CFBE cells were grown on a 40 mm diameter glass coverslip for 7–9 days to form a confluent polarized monolayer and washed twice with imaging medium before assembly into the FCS2 system. Bacteria were injected on the apical side of the CFBE cells in the chamber and allowed to attach for 2 h without flow. Subsequently the flow was started at the rate of 20 mL/h for 4 h to allow the formation of biofilms. Antibiotics (tobramycin, aztreonam or their combination) were then added to the imaging medium reservoir and applied with the flow overnight (16 h). Alternatively, antibiotics were added 2 h after the inoculation of bacteria and applied with the flow for 4 h to examine the prevention of biofilm formation by the antibiotics. The integrity of the CFBE monolayers was assessed visually by differential interference contrast microscopy.
Confocal microscopy imaging was performed on a Nikon TE2000 Livescan Swept Field Confocal microscope equipped with a QuantEM:512SC EMCCD camera (Photometrics, Tucson, AZ, USA). A ×60 oil-immersion, 1.3 numerical aperture objective was used for this study. The acquired z-stack images (with 1 μm interval) were compressed to one image layer using maximum intensity projection with the Nikon NIS-Elements software.
The flow cell and static biofilm methods are both based on the concept of allowing bacteria to incubate on epithelial cells to allow initial attachment before removing the unattached planktonic bacteria and allowing the remaining attached bacteria to form biofilms. The static co-culture assay has a higher throughput since it uses multiwell culture plates, while the flow chamber assay is ideal for high-resolution imaging. Several studies from our group show corresponding similar behaviours between organisms grown in these two systems.5,17,29
RT–PCR analysis for bacterial biofilm markers
The protocol for RT–PCR studies of the bacterial biofilm marker tolA was described in detail previously.29 The rplU gene, which is constitutively expressed under the conditions examined here, was used as a control. Briefly, P. aeruginosa was grown on CFBE cells for 6 h using the static co-culture assay as described above. Total bacterial RNA was harvested, purified with the RNeasy kit from Qiagen (Valencia, CA, USA) and treated with DNase using the DNA-free kit from Ambion (Austin, TX, USA). The First-Strand Synthesis Kit for RT–PCR (Ambion) was used to synthesize the complementary sequence and the RNA/cDNA hybrids were used as templates for PCR reactions using the annealing conditions and primer sets reported previously.29,30 Samples were normalized to the quantity of RNA used in the reverse transcriptase reaction.
Sequencing of the mucA gene
To sequence the mucA locus of the bacterial strains, the region was first amplified by PCR, using the forward primer 5′-GTGAAGCAATCGACAAAGCTCTGCAG-3′, which anneals within algU, 51 bp upstream of mucA, and the reverse primer 5′-CTGCCAAGCAAAAGCAACAGGGAGG-3′, which anneals within mucB, 18 bp downstream of mucA, to produce a 706 bp product. The PCR products were then purified using the Qiagen PCR Purification kit, and sequenced at the Dartmouth College Molecular Biology Core Facility using the primer 5′-CTTTGTTGCGAGAAGCCTGACACAGC-3′, which anneals 24 bp upstream of mucA. The mucA locus of each strain was analysed in quadruplicate, and the sequences were aligned and compared with the wild-type P. aeruginosa PAO1 mucA sequence using ClustalX software. The open reading frames were converted into codons and the respective amino acids using the program GCK, and ClustalX was used again for the alignment and comparison of amino acid sequences.
Results and discussion
Aztreonam alone or in combination with tobramycin was not toxic to CFBE cells
Peak concentrations of TOBI and AZLI measured in sputum isolates from CF patients have been reported to be ∼1000 and 700 mg/L, respectively,14,25 thus these concentrations were used throughout the study. We performed an LDH assay to examine whether the concentrations of tobramycin and aztreonam used in this study would be cytotoxic to CFBE cells. Approximately 6% LDH release was observed from CFBE cells treated with aztreonam alone (700 mg/L) or in combination with tobramycin (1000 mg/L) overnight, which was not statistically significant from the medium control (data not shown). Tobramycin (1000 mg/L) alone also showed no cytotoxicity.5
Biofilm-grown P. aeruginosa PAO1 is tolerant to aztreonam, but not tobramycin, using the co-culture model
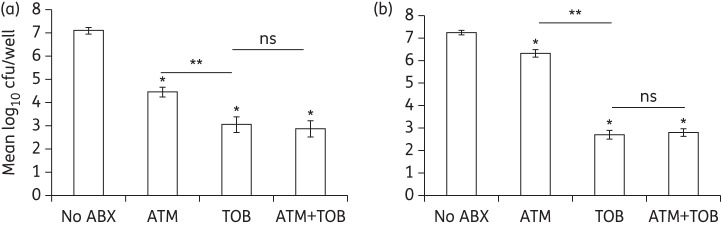
We previously showed that tobramycin at a clinically relevant concentration was able to prevent the formation of biofilms and disrupt established biofilms on CF-derived airway cells,5 likely by limiting the growth of P. aeruginosa as well as reducing the number of viable organisms in a preformed biofilm. In this study, we first investigated the ability of aztreonam to prevent formation of biofilms formed by the laboratory strain P. aeruginosa PAO1 on CFBE cells, using the static co-culture assay (Figure 1a). We found that aztreonam reduced P. aeruginosa PAO1 cfu by ∼3 log units/well compared with no antibiotic treatment. Tobramycin reduced cfu by ∼1 log unit more than aztreonam treatment. The combination of tobramycin and aztreonam, however, was no more effective than tobramycin alone. CFBE monolayers remained intact with or without antibiotic treatment in the prevention assay.
Figure 1.
Preformed biofilms of the P. aeruginosa laboratory strain PAO1 showed antibiotic tolerance to aztreonam, but not tobramycin, using the static co-culture assay. (a) Static co-culture prevention assay. Aztreonam (700 mg/L), tobramycin (1000 mg/L) or their combination was applied 1 h after bacterial inoculation to prevent formation of biofilms. The cfu were measured 5 h after drug treatment. Following treatment with tobramycin alone or tobramycin + aztreonam ∼103 cfu were recovered, while ∼104 cfu were recovered following treatment with aztreonam alone. No additive effect of tobramycin and aztreonam was revealed for prevention of P. aeruginosa PAO1 biofilm formation. CFBE monolayers remained intact with or without antibiotic application. (b) Static disruption assay. Biofilms were formed on CFBE cells 6 h post-inoculation, followed by treatment with antibiotics as indicated for 16 h. Following treatment with tobramycin alone or the combination of tobramycin and aztreonam ∼103 cfu were recovered, while ∼106 cfu were recovered following treatment with aztreonam alone. No additive effect of tobramycin and aztreonam was revealed for disruption of PAO1 biofilms. The detection limit for cfu recovery in the static assay was 200cfu/well (i.e. 2.3 logcfu/well). *P < 0.05 versus no antibiotic. **P < 0.05. ns, not significantly different (one-way ANOVA and Tukey's test). CFBE monolayers were disrupted if no antibiotics or only aztreonam was applied. ABX, antibiotic; ATM, aztreonam; TOB, tobramycin.
In the absence of antibiotics, P. aeruginosa PAO1 developed biofilms that destroyed the integrity of CFBE monolayers overnight. Aztreonam reduced the cfu of preformed P. aeruginosa PAO1 biofilms by ∼1 log unit compared with no antibiotic treatment (Figure 1b), but did not preserve the integrity of CFBE monolayers (not shown). Based on the definition outlined in the Materials and methods section, in which a >2 log reduction in cfu and disruption of the airway epithelial monolayer is used as a cut-off for susceptibility, our data indicate that biofilm-grown P. aeruginosa PAO1 is tolerant to aztreonam. It should be noted that the counted cfu are likely an underestimation in the no antibiotic and aztreonam treatments due to the loss of unattached CFBE monolayers during final washing steps in the static co-culture assay (refer to the Materials and methods section). By contrast, tobramycin was effective in killing established P. aeruginosa PAO1 biofilms in the static co-culture model and reduced the cfu by >4 log units (Figure 1b). CFBE monolayers with preformed biofilms were also protected by tobramycin treatment. The combination of tobramycin and aztreonam showed efficacy similar to that of tobramycin alone.
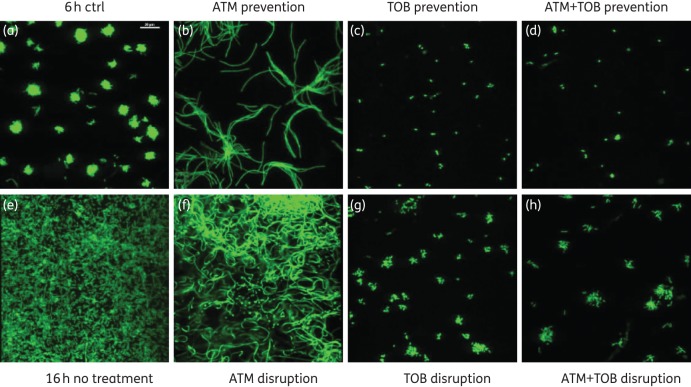
We next used confocal microscopy with GFP-labelled P. aeruginosa PAO1 grown on CFBE cells to assess the impact of different antibiotic treatment regimens on established biofilms formed by this microbe (Figure 2), using the flow chamber co-culture assay. A mushroom-like biofilm structure was formed on CFBE cells 6 h post-inoculation in the absence of treatment (Figure 2a), as reported previously.5 Without antibiotic treatment, this biofilm became a homogeneous sheet-like flat structure overnight (Figure 2e). This flat biofilm phenotype of P. aeruginosa has been reported previously.31 The bacteria became filamentous when aztreonam was applied alone either to prevent biofilm formation (Figure 2b) or to disrupt established biofilms (Figure 2f), indicating possible metabolic stress of P. aeruginosa PAO1 induced by the drug.32 In contrast, P. aeruginosa PAO1 treated with tobramycin (Figure 2c and g) or the combination of tobramycin and aztreonam (Figure 2d and h) resulted in killing of most of the bacteria, with only small clusters of bacteria remaining on the epithelial cells. From the confocal images and the data presented in Figure 1, it is apparent that aztreonam was less effective in preventing P. aeruginosa PAO1 biofilm formation and disrupting preformed P. aeruginosa PAO1 biofilms than was tobramycin or the combination of the two antibiotics.
Figure 2.
Confocal images of biofilms formed by P. aeruginosa PAO1 on CFBE cells with application of aztreonam (700 mg/L), tobramycin (1000 mg/L) or their combination using the flow cell assay. (a) P. aeruginosa PAO1 biofilms on CFBE cells 6 h post-inoculation. (b–d) Antibiotics were applied 1 h after PAO1 inoculation, and the co-culture incubated for 5 h to prevent biofilm formation. (e–h) Biofilms were preformed for 6 h, then medium (no treatment control) or antibiotics were applied for 16 h to disrupt established biofilms. Aztreonam alone caused filamentation of P. aeruginosa PAO1. Scale bar = 20 μm. ATM, aztreonam; ctrl, control; TOB, tobramycin.
CF clinical isolates grown as biofilms on airway cells show a variable response to aztreonam
To extend our findings beyond a single laboratory strain of P. aeruginosa, the effects of aztreonam and tobramycin were further evaluated on six clinical isolates using the static co-culture model. We used three clinical isolates of P. aeruginosa characterized as non-mucoid (SMC1587, SMC1595 and SMC1596) and three characterized as mucoid (SMC5450, SMC5451 and SMC1585), as judged by a standard clinical laboratory test described in the Materials and methods section. The formation of biofilms was confirmed by measuring the expression of the gene preferentially expressed in biofilm-grown P. aeruginosa, tolA, in these clinical isolates 6 h post-inoculation on CFBE cells. All clinical isolates demonstrated a visible PCR product derived from the tolA transcript (Figure S1, available as Supplementary data at JAC Online). As expected, there was no detectable tolA expression in P. aeruginosa PAO1 grown planktonically, but this gene was expressed in the PAO1 strain grown as a biofilm on airway cells (Figure S1, available as Supplementary data at JAC Online). These data indicate that clinical strains grown on CF airway cells adopt properties consistent with biofilm-grown bacteria, a result confirming our previous findings.5,29
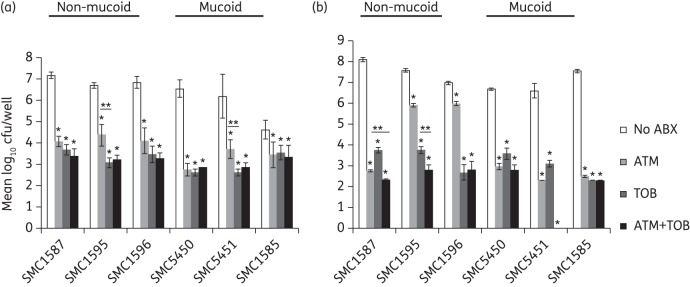
Both the prevention and disruption assays were performed on these clinical isolates to assess their susceptibility to aztreonam and tobramycin alone, or in combination, when grown on CFBE cells. In the prevention assay, both aztreonam and tobramycin prevented biofilm formation on CFBE cells for all six clinical isolates and reduced the number of cfu up to 4 log units (Figure 3a). Tobramycin-mediated inhibition was more robust than aztreonam-mediated inhibition for two of the strains (SMC1595 and SMC5451) and similar to aztreonam-mediated inhibition in the other four strains (SMC1587, SMC1596, SMC5450 and SMC1585). The combination of aztreonam and tobramycin worked no better to prevent biofilm formation than tobramycin alone for the six clinical P. aeruginosa strains tested here.
Figure 3.
P. aeruginosa clinical isolates from the sputum of the CF patients (SMC1585, SMC1587, SMC1595, SMC1596, SMC5450 and SMC5451) showed diverse antibiotic susceptibility to aztreonam, and a possible additive effect of tobramycin and aztreonam using the static co-culture assay. (a) Static co-culture prevention assay. Aztreonam (700 mg/L), tobramycin (1000 mg/L) or the combination of both was applied 1 h after bacterial inoculation to prevent formation of biofilms. cfu were measured 5 h after drug treatment. There was no additive effect of tobramycin and aztreonam in prevention of biofilm formation. (b) Static co-culture disruption assay. Biofilms were formed on CFBE cells 6 h post-inoculation. Aztreonam (700 mg/L), tobramycin (1000 mg/L) or the combination of both was then applied for 16 h. No cfu were recovered for aztreonam + tobramycin treatment for SMC5451. In all cases, CFBE monolayers were disrupted without antibiotic treatment. Clinical isolates SMC1595 and SMC1596 showed limited susceptibility to aztreonam when grown as a biofilm on CFBE cells. The monolayers were also disrupted when treated with aztreonam alone for isolates SMC1595 and SMC1596, while the CFBE monolayers remained intact for the other isolates. CFBE monolayers were protected from disruption for all six isolates when treated with tobramycin or the combination of tobramycin and aztreonam. Aztreonam and tobramycin had an additive effect for SMC1587 and SMC1595. For statistical analysis, all comparisons were done within each strain. *P < 0.05 versus no antibiotic. **P < 0.05 (one-way ANOVA and Tukey's test). ABX, antibiotic; ATM, aztreonam; TOB, tobramycin.
In the disruption assay CFBE monolayers were compromised, as judged by visual inspection after overnight incubation, for all six clinical isolates in the absence of antibiotic treatment. For two of the six clinical isolates (SMC1595 and SMC1596), overnight application of aztreonam did not protect the CFBE monolayer from being disrupted and only reduced the bacteria by ∼1 log10 cfu/well (Figure 3b). Aztreonam demonstrated much better killing of biofilms formed by the other four clinical isolates (SMC1585, SMC1587, SMC5450 and SMC5451), with an ∼4–5 log10 cfu/well reduction (Figure 3b) and protection of the CFBE monolayers. In contrast, tobramycin yielded an ∼4 log unit killing of all six clinical strains and preserved the integrity of CFBE monolayers in all cases.
Interestingly, despite its inability to effectively reduce the biofilms of some isolates, aztreonam was more effective against half of the clinical isolates (SMC1587, SMC5450 and SMC5451) than was tobramycin. It was also noted in our study that the P. aeruginosa isolates tolerant to aztreonam were all non-mucoid.
Finally, the combination of aztreonam and tobramycin only had an additive effect on biofilm disruption of strains SMC1587 and SMC1595 (P < 0.05) in the co-culture model. Previous research has shown that the combination of antibiotics with different modes of action against P. aeruginosa might be more efficient than monotherapy for eliminating those isolated from CF lungs.33–36 Since aztreonam and tobramycin belong to β-lactam and aminoglycoside antibiotic families, respectively, we hypothesized that the combination of aztreonam and tobramycin might be more efficient than monotreatment to kill biofilms on CFBE cells. Our results partially supported this hypothesis and demonstrated that the additive effect of combined antibiotic treatment was strain-dependent. Thus, our results indicated that combined treatment with aztreonam and tobramycin might be beneficial for a subset of CF patients only. However, because most CF patients are infected with multiple bacterial strains with varying degrees of susceptibility to these antibiotics, it is difficult to anticipate how the data presented here for single isolates in vitro would relate to authentic clinical situations.
Overproduction of alginate does not mediate aztreonam tolerance
As shown above, aztreonam had a variable effect on P. aeruginosa biofilms formed on CFBE cells. Therefore, we decided to explore several hypotheses to explain these observed differences in response to aztreonam. We noted that all three mucoid clinical strains were susceptible to aztreonam in the co-culture model (SMC5450, SMC5451 and SMC1585; Figure 3b). Therefore, mucoidy was tested as a possible mechanism of aztreonam susceptibility. The mucoid phenotype is caused by the overproduction of alginate, which in clinical strains is often regulated by the activation of the algT gene, which encodes an alternative sigma factor.21,37
The isogenic strains FRD1 (mucoid) and its well-characterized, non-mucoid mutant FRD1algT::Tn501 were used to examine the response to aztreonam with the static co-culture assay (Figure S2, available as Supplementary data at JAC Online). The wild-type FRD1 and algT mutant (FRD1algT::Tn501) both showed a decrease of ∼2 log units when tested for the prevention of biofilm formation by aztreonam (Figure S2a, available as Supplementary data at JAC Online). We also found that both strains were susceptible to aztreonam when grown as established biofilms, showing a reduction of ∼4 log units after treatment (Figure S2b, available as Supplementary data at JAC Online). These two strains were also equally susceptible to tobramycin in the co-culture model. For both strains, antibiotic treatment also protected the CFBE monolayers from being compromised. Therefore, the hyperproduction of alginate (mucoidy) does not appear to alter the tolerance of P. aeruginosa strain PAO1 to aztreonam, and indicates that the basis of tolerance to aztreonam by the mucoid clinical isolates is unlikely to be overproduction of alginate.
Mutations in the mucA gene do not correlate with aztreonam tolerance
In CF isolates of P. aeruginosa, activation of algT often results from the loss-of-function mutation in mucA, which codes for an anti-sigma factor.37,38 In addition, mucA mutations also lead to reduced expression of bacterial virulence factors,39 which could be associated with the reduced cytotoxic effect on airway cells in our co-culture model among mucoid strains. That is, we observed that the disruption of CFBE monolayers was always associated with P. aeruginosa tolerance to aztreonam in the co-culture model, indicating a possible link between tolerance to this antibiotic and the functional status of MucA. To test this idea, the mucA locus from each of the clinical strains was sequenced and compared with the 584 bp wild-type mucA gene of P. aeruginosa PAO1. The results are summarized in Table 1 (columns 1–3). Strains SMC1587, SMC1596, SMC5450 and SMC5451 each had wild-type mucA alleles. Both FRD1 strains and strain SMC1585 had a single nucleotide deletion at position 430, resulting in a frameshift mutation and premature stop codon at position 440. The non-mucoid SMC1595 had a nucleotide insertion between bases 190 and 191, resulting in a frameshift mutation and premature stop codon at base 500. Given that one of the four strains carrying mucA mutations and two of the five strains carrying wild-type mucA alleles showed aztreonam tolerance, there was no strict correlation between mucA alleles and tolerance to this antibiotic.
Table 1.
Summary of characteristics of P. aeruginosa strains tested in this study
| Strain | Mucoidy | mucA allele | ATM susceptibility of biofilmsa | Additive effect of ATM + TOB | MICP TOB (mg/L) | MICP ATM (mg/L) |
|---|---|---|---|---|---|---|
| PAO1 | no | WT | tolerant | no | 0.38 | 1.0 |
| SMC1585 | yes | mutant | susceptible | no | 0.125 | 0.19 |
| SMC1587 | no | WT | susceptible | yes | 8.0 | 0.38 |
| SMC1595 | no | mutant | tolerant | yes | 1.5 | 2.0 |
| SMC1596 | no | WT | tolerant | no | 1.0 | 3.0 |
| SMC5450 | yes | WT | susceptible | no | 1.0 | 0.25 |
| SMC5451 | yes | WT | susceptible | no | 4.0 | >256 |
| FRD1 | yes | mutant | susceptible | ND | 1.0 | 0.38 |
| FRD1algT::Tn501 | no | mutant | susceptible | ND | 0.75 | 0.25 |
| ZK2870 | no | ND | tolerant | ND | 1.0 | 4.0 |
| ZK2870ΔpelA | no | ND | tolerant | ND | 1.0 | 4.0 |
ATM, aztreonam; MICP, planktonic MIC; ND, not determined; TOB, tobramycin; WT, wild-type.
aP. aeruginosa strains were scored as tolerant to aztreonam when the CFBE monolayers were disrupted after overnight aztreonam treatment using the static co-culture assay and there was <2 log10 difference in cfu recovery between no antibiotic treatment and aztreonam treatment.
Mutation in pelA does not mediate aztreonam tolerance
To investigate whether other EPS components besides alginate may be involved in mediating aztreonam tolerance, we tested the effect of a mutation in the pelA gene on the background of a clinical P. aeruginosa isolate. The pel locus is responsible for synthesis of a glucose-rich component of EPS in non-mucoid P. aeruginosa.20 Recently, our group discovered that the minimal bactericidal concentrations of tobramycin and gentamicin were 4-fold higher for a biofilm of the P. aeruginosa ΔpelA mutant grown on an abiotic surface than for its wild-type counterpart.40
To test whether the Pel EPS impacts antibiotic tolerance when bacteria are grown as biofilms on airway cells, P. aeruginosa strain ZK2870 and its ΔpelA variant were grown on CFBE cells to examine their susceptibility to aztreonam. We found a decrease in cfu recovery for both ZK2870 and ZK2870ΔpelA strains by ∼1000-, 10 000- and 40 000-fold with aztreonam, tobramycin and the combined antibiotic treatment, respectively [Figure S3a (Figure S3 is available as Supplementary data at JAC Online)]. Moreover, the preformed biofilms on CFBE cells of both strains demonstrated high tolerance to aztreonam treatment (Figure S3b, available as Supplementary data at JAC Online) and CFBE cells were disrupted overnight. We have also tested the ΔpelA mutant on the P. aeruginosa PA14 strain background using the static co-culture assay and found that both PA14 wild-type and PA14 ΔpelA mutant demonstrated tolerance to aztreonam treatment, but not tobramycin or the combination treatment (Figure S3c,d, available as Supplementary data at JAC Online), which is consistent with what we found in the ZK2870 strain. We conclude that differential ability to produce the Pel EPS could not explain the aztreonam tolerance for the P. aeruginosa strains tested.
Mutation in psl might mediate aztreonam tolerance in some P. aeruginosa strains
Like pel, the psl locus is also responsible for the synthesis of an EPS component in non-mucoid P. aeruginosa strains and appears to contribute to a mannose-rich EPS component.20 Hence, we investigated the effect of a mutant in the psl gene on the background of several P. aeruginosa isolates, including P. aeruginosa laboratory strain PAO1 and clinical isolates SMC715, SMC725 and SMC738, using our static co-culture assay. Interestingly, we found that, unlike wild-type PAO1, the preformed biofilm of the PAO1 Δpsl mutant exhibited increased susceptibility to overnight aztreonam treatment [Figure S4b (Figure S4 is available as Supplementary data at JAC Online)]. The CFBE monolayers were protected from being compromised after overnight treatment with aztreonam for the mutant, while they were disrupted for wild-type P. aeruginosa PAO1. Moreover, there was an ∼2 log10 decrease in the remaining cfu for the mutant compared with the wild-type PAO1 when treated overnight with aztreonam. However, for the other three clinical strains tested (SMC715, SMC725 and SMC738), both the wild-type and psl mutants demonstrated tolerance to aztreonam treatment using the static co-culture assay (Figure S4d, f and h, available as Supplementary data at JAC Online) and the CFBE monolayers were disrupted overnight when treated alone with aztreonam. All four strains demonstrated susceptibility to tobramycin or the combination treatment, although tobramycin treatment was significantly more effective for the SMC715 psl mutant than wild-type SMC715 (Figure S4d, available as Supplementary data at JAC Online). These data indicated that the Psl EPS might play a role in antibiotic tolerance for some, but not all, of the P. aeruginosa strains.
Planktonic MIC does not correlate with aztreonam tolerance in biofilms
Finally, we examined the in vitro susceptibility of these P. aeruginosa isolates grown planktonically by measuring the MIC of tobramycin and aztreonam for the clinical strains analysed here. The experimentally determined MIC of aztreonam did not always reflect the antibiotic susceptibility of the same strain grown in the co-culture model (Table 1, columns 4, 6 and 7). For example, clinical isolate SMC5451 had a planktonic MIC of aztreonam of >256 mg/L yet was susceptible to aztreonam treatment when grown as a preformed biofilm on airway cells. The other strains tested, however, consistently demonstrated tolerance to aztreonam in the co-culture model when MIC was ≥1 mg/L. Tobramycin MICs for all tested strains ranged from 0.125 to 8 mg/L, but did not reflect the similar antibiotic tolerance measured in the co-culture model for these isolates (Figures 1 and 3). Therefore, the response of planktonic bacteria to antibiotics in liquid culture did not effectively predict the responsiveness of biofilm-grown bacteria on a biotic surface. Thus, while reported susceptibility tests measure the MICs for planktonic bacteria or biofilm inhibitory concentrations on abiotic surfaces,34,41 our in vitro co-culture model allows for formation of bacterial biofilms on airway epithelial cells and more closely resembles the form of bacteria in CF lungs.
Summary
Here we studied the effects of two clinically available inhaled antibiotics, tobramycin and aztreonam, as well as their combination on P. aeruginosa biofilms formed on CF-derived human airway cells. We found that the effect of aztreonam on this co-culture model was variable and strain-dependent. These data indicate that only a subset of CF patients might respond to aztreonam therapy, and for some patients a combination of aztreonam and tobramycin may be more effective than monotherapy with either antibiotic. In contrast, all clinical isolates tested showed a significant reduction (3–5 log) in biofilm cfu when treated with tobramycin, a finding consistent with the efficacy of this antibiotic as a monotherapy in clinical studies.42–44 The aztreonam tolerance of some P. aeruginosa strains does not appear to be due to Pel EPS production, mucoidy, mucA allele status or planktonic resistance. However, the Psl EPS production appears to relate to antibiotic tolerance in some of the strains studied. In addition to the potential modes of aztreonam tolerance tested here, other mechanisms, such as lower bacterial outer membrane permeability, antibiotic efflux pump and β-lactamase resistance, may play a role in P. aeruginosa tolerance to aztreonam.45 Further investigations are needed to fully understand the selective tolerance of CF patient-derived P. aeruginosa isolates to aztreonam.
Funding
This work was supported by the National Institutes of Health R01-HL074175 (B. A. S.), P20 RR-018787 (B. A. S.), the Cystic Fibrosis Foundation Research Development Program (B. A. S.), the Hitchcock Foundation PPG (B. A. S. and G. A. O.) and Novartis/CTBSMC545000DUSNC01 (G. A. O.).
Transparency declarations
All authors: none to declare.
This work was supported in part by Novartis, the company that makes the product tobramycin.
Author contributions
Q. Y., S. M.-M., B. A. S. and G. A. O. conceived and designed the experiments, Q. Y., E. F. G., S. M.-M. and J. D. S. performed the experiments, Q. Y., E. F. G., S. M.-M., B. A. S. and G. A. O. analysed the data, and Q. Y., E. F. G., B. A. S. and G. A. O. wrote the paper.
Supplementary data
Figures S1 to S4 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
Preliminary results of this paper were presented at the Twenty-fifth Annual North American Cystic Fibrosis Conference, Anaheim, CA, USA, 2011 (Abstract #293).
References
- 1.Riordan JR, Rommens JM, Kerem B, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–73. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Kerem B, Rommens JM, Buchanan JA, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–80. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 3.Cystic Fibrosis Foundation. Patient Registry Annual Data Report 2010. http://www.cff.org/UploadedFiles/LivingWithCF/CareCenterNetwork/PatientRegistry/2010-Patient-Registry-Report.pdf. (2 July 2012, date last accessed)
- 4.Heijerman H. Infection and inflammation in cystic fibrosis: a short review. J Cyst Fibros. 2005;4(Suppl 2):3–5. doi: 10.1016/j.jcf.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Moreau-Marquis S, O'Toole GA, Stanton BA. Tobramycin and FDA-approved iron chelators eliminate Pseudomonas aeruginosa biofilms on cystic fibrosis cells. Am J Respir Cell Mol Biol. 2009;41:305–13. doi: 10.1165/rcmb.2008-0299OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geller DE. Aerosol antibiotics in cystic fibrosis. Respir Care. 2009;54:658–70. doi: 10.4187/aarc0537. [DOI] [PubMed] [Google Scholar]
- 7.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–74. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh PK, Schaefer AL, Parsek MR, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–4. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 9.Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–9. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 10.Hoiby N. Recent advances in the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. BMC Med. 2011;9:32. doi: 10.1186/1741-7015-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elborn JS, Henig NR. Optimal airway antimicrobial therapy for cystic fibrosis: the role of inhaled aztreonam lysine. Expert Opin Pharmacother. 2010;11:1373–85. doi: 10.1517/14656566.2010.482102. [DOI] [PubMed] [Google Scholar]
- 12.Sykes RB, Bonner DP, Bush K, et al. Azthreonam (SQ 26,776), a synthetic monobactam specifically active against aerobic Gram-negative bacteria. Antimicrob Agents Chemother. 1982;21:85–92. doi: 10.1128/aac.21.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCoy KS, Quittner AL, Oermann CM, et al. Inhaled aztreonam lysine for chronic airway Pseudomonas aeruginosa in cystic fibrosis. Am J Respir Crit Care Med. 2008;178:921–8. doi: 10.1164/rccm.200712-1804OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Retsch-Bogart GZ, Quittner AL, Gibson RL, et al. Efficacy and safety of inhaled aztreonam lysine for airway Pseudomonas in cystic fibrosis. Chest. 2009;135:1223–32. doi: 10.1378/chest.08-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oermann CM, Retsch-Bogart GZ, Quittner AL, et al. An 18-month study of the safety and efficacy of repeated courses of inhaled aztreonam lysine in cystic fibrosis. Pediatr Pulmonol. 2010;45:1121–34. doi: 10.1002/ppul.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkins MD, Elborn JS. Aztreonam lysine: a novel inhalational antibiotic for cystic fibrosis. Exp Rev Respir Med. 2010;4:435–44. doi: 10.1586/ers.10.48. [DOI] [PubMed] [Google Scholar]
- 17.Anderson GG, Moreau-Marquis S, Stanton BA, et al. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun. 2008;76:1423–33. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreau-Marquis S, Stanton BA, O'Toole GA. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther. 2008;21:595–9. doi: 10.1016/j.pupt.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloemberg G, O'Toole GA, Lugtenberg B, et al. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–51. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004;186:4457–65. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett ES, Perlegas D, Wozniak DJ. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU) J Bacteriol. 1999;181:7401–4. doi: 10.1128/jb.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, Jackson KD, Landry RM, et al. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–21. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zegans ME, Wozniak D, Griffin E, et al. Pseudomonas aeruginosa exopolysaccharide Psl promotes resistance to the biofilm inhibitor polysorbate 80. Antimicrob Agents Chemother. 2012 doi: 10.1128/AAC.00373-12. doi:10.1128/AAC.00373–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahba AH, Darrell JH. The identification of atypical strains of Pseudomonas aeruginosa. J Gen Microbiol. 1965;38:329–42. doi: 10.1099/00221287-38-3-329. [DOI] [PubMed] [Google Scholar]
- 25.Geller DE, Pitlick WH, Nardella PA, et al. Pharmacokinetics and bioavailability of aerosolized tobramycin in cystic fibrosis. Chest. 2002;122:219–26. doi: 10.1378/chest.122.1.219. [DOI] [PubMed] [Google Scholar]
- 26.Bruscia E, Sangiuolo F, Sinibaldi P, et al. Isolation of CF cell lines corrected at ΔF508-CFTR locus by SFHR-mediated targeting. Gene Ther. 2002;9:683–5. doi: 10.1038/sj.gt.3301741. [DOI] [PubMed] [Google Scholar]
- 27.Cozens AL, Yezzi MJ, Kunzelmann K, et al. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 28.Hentchel-Franks K, Lozano D, Eubanks-Tarn V, et al. Activation of airway Cl− secretion in human subjects by adenosine. Am J Respir Cell Mol Biol. 2004;31:140–6. doi: 10.1165/rcmb.2004-0012OC. [DOI] [PubMed] [Google Scholar]
- 29.Moreau-Marquis S, Bomberger JM, Anderson GG, et al. The ΔF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am J Physiol Lung Cell Mol Physiol. 2008;295:L25–37. doi: 10.1152/ajplung.00391.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mah TF, Pitts B, Pellock B, et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–10. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 31.Kirisits MJ, Parsek MR. Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell Microbiol. 2006;8:1841–9. doi: 10.1111/j.1462-5822.2006.00817.x. [DOI] [PubMed] [Google Scholar]
- 32.Orlicek SL. Aztreonam. Semin Pediatr Infect Dis. 1999;10:45–50. [Google Scholar]
- 33.Ghani M, Soothill JS. Ceftazidime, gentamicin, and rifampicin, in combination, kill biofilms of mucoid Pseudomonas aeruginosa. Can J Microbiol. 1997;43:999–1004. doi: 10.1139/m97-144. [DOI] [PubMed] [Google Scholar]
- 34.Dales L, Ferris W, Vandemheen K, et al. Combination antibiotic susceptibility of biofilm-grown Burkholderia cepacia and Pseudomonas aeruginosa isolated from patients with pulmonary exacerbations of cystic fibrosis. Eur J Clin Microbiol Infect Dis. 2009;28:1275–9. doi: 10.1007/s10096-009-0774-9. [DOI] [PubMed] [Google Scholar]
- 35.Herrmann G, Yang L, Wu H, et al. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J Infect Dis. 2010;202:1585–92. doi: 10.1086/656788. [DOI] [PubMed] [Google Scholar]
- 36.Araoka H, Baba M, Tateda K, et al. In vitro combination effects of aztreonam and aminoglycoside against multidrug-resistant Pseudomonas aeruginosa in Japan. Jpn J Infect Dis. 2012;65:84–7. [PubMed] [Google Scholar]
- 37.Xie ZD, Hershberger CD, Shankar S, et al. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–6. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin DW, Schurr MJ, Mudd MH, et al. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–81. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones AK, Fulcher NB, Balzer GJ, et al. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J Bacteriol. 2010;192:5709–17. doi: 10.1128/JB.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan W, Bernier SP, Kuchma SL, et al. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int Microbiol. 2010;13:207–12. doi: 10.2436/20.1501.01.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King P, Lomovskaya O, Griffith DC, et al. In vitro pharmacodynamics of levofloxacin and other aerosolized antibiotics under multiple conditions relevant to chronic pulmonary infection in cystic fibrosis. Antimicrob Agents Chemother. 2010;54:143–8. doi: 10.1128/AAC.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramsey BW, Dorkin HL, Eisenberg JD, et al. Efficacy of aerosolized tobramycin in patients with cystic fibrosis. N Engl J Med. 1993;328:1740–6. doi: 10.1056/NEJM199306173282403. [DOI] [PubMed] [Google Scholar]
- 43.Ramsey BW, Pepe MS, Quan JM, et al. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. Cystic Fibrosis Inhaled Tobramycin Study Group. N Engl J Med. 1999;340:23–30. doi: 10.1056/NEJM199901073400104. [DOI] [PubMed] [Google Scholar]
- 44.Konstan MW, Flume PA, Kappler M, et al. Safety, efficacy and convenience of tobramycin inhalation powder in cystic fibrosis patients: the EAGER trial. J Cyst Fibros. 2011;10:54–61. doi: 10.1016/j.jcf.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hancock RE. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative Gram-negative bacteria. Clin Infect Dis. 1998;27(Suppl 1):S93–9. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.