Abstract
Empirical force fields commonly used to describe the condensed phase properties of complex systems such as biological macromolecules are continuously being updated. Improvements in quantum mechanical (QM) methods used to generate target data, availability of new experimental target data, incorporation of new classes of compounds and new theoretical developments (eg. polarizable methods) make force-field development a dynamic domain of research. Accordingly, a number of improvements and extensions of the CHARMM force fields have occurred over the years. The objective of the present review is to provide an up-to-date overview of the CHARMM force fields. A limited presentation on the historical aspects of force fields will be given, including underlying methodologies and principles, along with a brief description of the strategies used for parameter development. This is followed by information on the CHARMM additive and polarizable force fields, including examples of recent applications of those force fields.
Introduction
Empirical force fields (FF) have become integral tools in the study of complex phenomena in numerous fields such as materials science and biomolecular simulations. The first protein molecular dynamics (MD) simulation was performed in 1970 on a system of 458 atoms over a length of 9.2 ps on an IBM 370, a top-of-the-line computer at the time, using precursor versions of the program CHARMM and CHARMM force field.(1) From its initial conception in the late 1970s to the present time, CHARMM and the associated force field have evolved to allow for MD simulation studies to be performed on a host of systems of different sizes, functions, and complexity. The current version of the CHARMM program, which has been extensively reviewed by Brooks et al (2), is compatible with and has been optimized for multiple types of computer platforms capable of running either in serial or in parallel ranging from single processors PCs to clusters of multi-core processor machines to shared-memory supercomputer installations.
The CHARMM additive force field, one of the most successful force fields in the study of biomolecules, was first described in the seminal paper by Karplus and co-workers in 1983(3). Despite being an integral part of the CHARMM program, it can also be used with a number of MD programs such as AMBER (4), NAMD (5) and GROMACS (6). It has been the force field of choice in many landmark MD simulations, having been used, for example, by Shulten and co-workers in 2006 for the first MD simulation that broke the one million atom MD simulation barrier. Their work on the satellite tobacco mosaic virus, which marked the first all-atom computational model of a full life form, was published two years later.(7) The CHARMM force field was also used in MD simulations performed with ANTON, a specialized supercomputer that executes MD simulations orders of magnitude faster than was previously possible.(8) These advances put additional demands on the force field with respect to accuracy requiring continuous evolution of the parameters comprising the force fields. Indeed, the ongoing evolution of the additive CHARMM force field is what allows it to continue to be of utility for the study of molecular systems of more complex nature and under a longer simulation time frame.
The purpose of the current review is to highlight recent developments in the CHARMM force fields and with emphasis on work originating from our laboratory. Topics covered include strategies employed in parameter optimization, limitations of the force field, and their potential application in the discovery of new therapeutic agents. While the earliest additive CHARMM force field focused on proteins and nucleic acids it has been since extended to lipids (9, 10), carbohydrates (11–15), and to small, typically drug-like, molecules (16) as well as more specialized chemicals, such as quartz (17). In addition, recent enhancements in the protein (18, 19), lipid (9) and nucleic acid (20) force fields have been presented. Current progress in the treatment of induced polarization in the context of the Drude-based polarizable CHARMM force field will also be discussed.
Molecular Mechanics and the CHARMM potential energy function
The molecular mechanics approach was originally pioneered by Alder and Wainwright in the late 1950’s(21). It consists of using a “balls-on-springs” scheme to represent molecules where each ball corresponds to an atom and each spring a covalent bond. This representation allows molecules to be modeled through Newtonian mechanics. Calculation of the forces and integration of the Newton’s equation of motion allows the time evolution of a given system to be calculated, thus giving birth to the field of MD simulations. In molecular mechanics (MM) the potential energy of a system of particles is determined using a defined functional form and associated parameter set, the combination of which is called a force field. In MM force fields the properties of a system of particles are defined by the elasticity of the covalent bonds, the suppleness of two or three adjacent bonds (valence and dihedral angles), as well as the interactions that arise between non-bonded atoms (van der Waals “vdW” and electrostatic interactions). These elements represent the individual contributions to the energy of the system, which are ultimately combined allowing for the energy and forces of a system to be calculated as a function of geometry.
The potential energy function comprising the CHARMM additive force field is
| (1) |
where R⃗ is the ensemble of coordinates of atoms in a system. Internal or intramolecular parameters include bonds (b), angles (θ), Urey-Bradley (UB, S), dihedrals (χ), and impropers (□) where K corresponds to their associated force constants and the symbols having subscript “0” corresponding to the equilibrium values. Non-bonded terms include Lennard-Jones (LJ) parameters εij (well-depth) and Rmin (distance of minimum interaction energy), which are used to define the vdW interactions, and partial charges q, used in Coloumb’s law to describe electrostatic contributions. Force fields based on this type of energy function are referred to as “additive” as they don’t allow the electrostatic parameters to change as a function of environment, such that the total electrostatic energy is simply a sum of all the individual atom-atom terms. Though a large number of force fields have been developed based on the additive model and successfully applied to the study of biomolecular systems, this representation does not account for the phenomenon of electronic polarization and may be considered a significant approximation in these types of force fields.
Explicit inclusion of electronic polarization is expected to represent an important improvement in empirical force fields. For example, in proteins, it was shown that non-polarizable force fields share a systematic error in the calculated electrostatic potential due to a lack of treatment of electronic polarizability.(22) To compensate for the lack of polarizability the additive CHARMM FF, as well as most other macromolecular force fields, deliberately overestimates molecular dipoles of compounds in the gas phase in order to be able to reproduce electrostatic interactions that occur in condensed phase environments. In other words, the parameters are optimized to better reproduce condensed-phase properties at the expense of agreement with gas-phase data. As a basic illustration of the importance of polarization, a water molecule has a gas-phase dipole moment of 1.85 D (23) but an average dipole moment of 2.1 D in a dimer and even higher value in large water clusters (24). The value for the dipole moment in condensed or liquid phase is indicated to be as high as 2.9 D in X-ray and neutron diffraction experiments (25) and ab initio MD simulations have predicted values around 3.0 D (26). Although systematic overestimation of molecular dipoles has worked well in calculating structure and energetics of molecules in the condensed phase, it fundamentally lacks the ability to describe locally induced polarization events, an essential element in intermolecular interactions.
Several of the major energy functions for biological macromolecules, including CHARMM, have been extended to include polarization effects with varying degrees of success. The efforts have been centered around using three methods including induced dipoles, fluctuating point charges, and classical Drude oscillators to describe polarizability.(27) Details on the various implementations have been previously discussed in other reviews, (27, 28) an issue of the Journal of Chemical Theory and Computation that specially addressed polarization (29) and in a more recently published review (30). The classical Drude oscillator is currently being implemented in the context of CHARMM by the MacKerell and Roux groups.(31).
The implementation of molecular polarizability in CHARMM, based on the classical Drude oscillator formalism, involves a massless point charge (ie. the Drude oscillator) being attached to each non-hydrogen atom via a harmonic spring (32)(Figure 1). To model polarization, the position of the additional point charge reacts to the surrounding electrostatic environment, thereby effectively shifting the electron density of the system leading to a change in the atomic and molecular dipoles. To include this effect the electrostatic part of the potential energy function (Eq. 1) is extended by the following equation
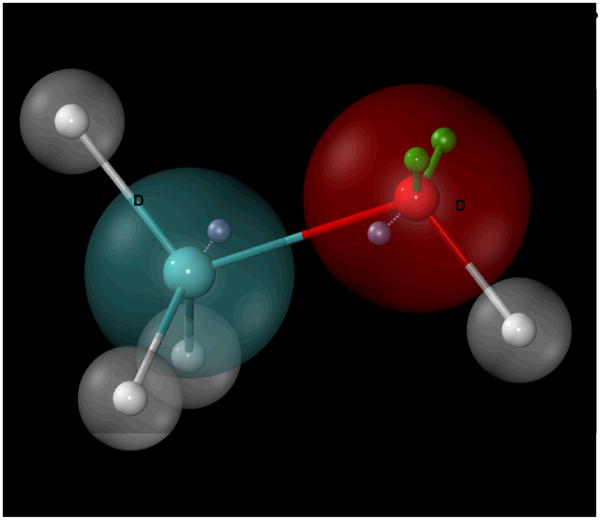
Figure 1.
Schematic representation of how atomic polarizability is treated in the CHARMM polarizable force field using methanol as an example. Drude oscillators (or particles)(blue, “D”) are attached to non-hydrogen parent atoms through harmonic bonds (dashed lines). Oxygen lone-pairs (green, “LP”) are connected with constrained bonds, angles, and dihedrals relative to the C-O-H plane. Hydrogens are not considered as polarizable entities in this model.
| (2) |
for a molecule of N non-hydrogen atoms, where qD and qC are the charges on the Drude particle and its parent atom, respectively; rD and rC are their respective positions, and kD is the force constant of the harmonic spring between the Drude oscillator and its parent atom. In this model, the atomic polarizability (α) of an atom A is related to the charge of the attached Drude particle and the associated harmonic spring through the following equation:
| (3) |
Electrostatic interactions between the Drudes and other charged centers for the 1,4 atom pairs (i.e. atoms separated by 3 covalent bonds) and beyond are treated through Coulomb’s Law the same way as in the additive force field (33). However, 1,2 and 1,3 electrostatic interactions (i.e. atoms separated by 1 or 2 covalent bonds, respectively), which are ignored in the CHARMM additive force field, are reintroduced for Drude particles. These interactions, corresponding to the 1,2 qD(A)-qC(A), or “Drude-parent”, and the 1,3 qD(A)-qC(B), or Drude-adjacent parent, interactions described in Equation 2, are required to create the correct dipole response in a molecule. However, since these electrostatic interactions occur at close proximities, the point charge approximation is unsuitable. To circumvent this issue, the CHARMM Drude force field introduces a damping function initially developed by Thole (34). The Thole screening function
| (4) |
is applied to interactions between two atom centers with charges qi and qj, where rij is the distance between the atom centers and the normalized distance is defined as
| (5) |
such that αi and αj are the polarizabilities of respective charge centers and α, the Thole parameter.
In addition to the Drude model two other polarizable models have been introduced into CHARMM. The polarizable fluctuating charge model has been implemented by Patel, Brooks and co-workers.(35, 36) This method allows the values of the partial charges to respond to the electric field of their environment effectively altering the molecular dipole using an electronegativity or chemical potential equalization scheme. The CHARMM implementation of the fluctuating charge model has been applied to study small-molecule liquid-vapor interfaces, solvated proteins/peptides, and physiological membrane systems, in which calculation of free energies of solvation was made possible.(37) In addition, Gao and coworkers have introduced the induced dipole model into CHARMM (38). In this model a tensor is applied to each atom in the system to define polarization. Implementation of multiple methods to treat electronic polarization in CHARMM is a feature that will facilitate future force field development.
Parameter Optimization: Strategies and Considerations
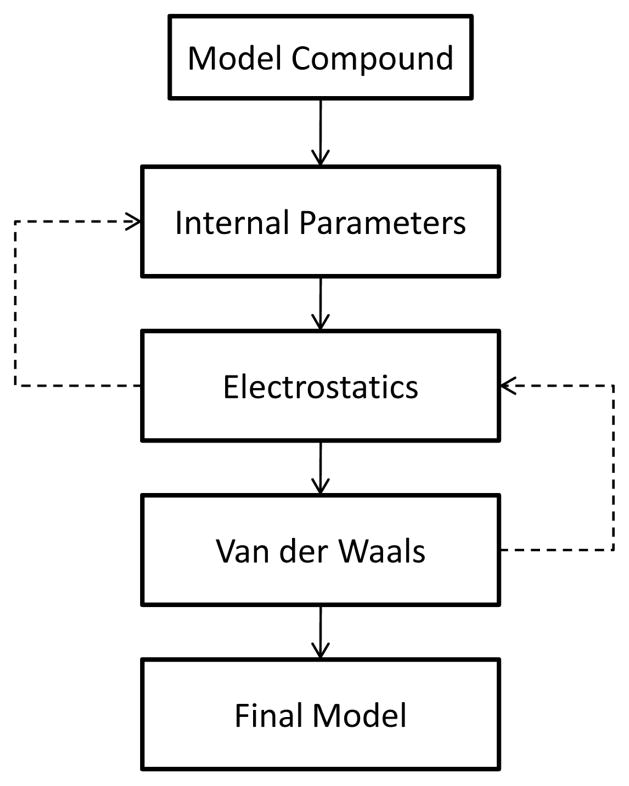
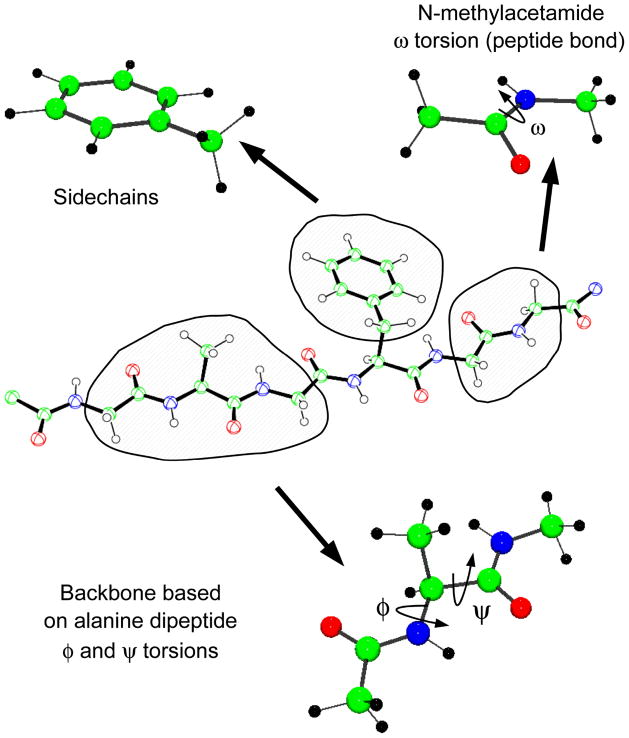
Independent of the form of the potential energy function, a force field is only as good as the quality of the associated parameters. To obtain parameters of sufficient accuracy a procedure is followed in which the parameters in Equation 1 are optimized to reproduce a variety of target properties such as molecular geometries and vibrations, pure solvent properties, and free energies of solvation, among others. The general outline of the parametrization process has been described for the CHARMM additive force field in earlier publications (33, 39) and is depicted in Figure 2. Note that parameter optimization is an iterative approach and several rounds of parametrization are typically required until convergence is achieved. A common strategy in parameter optimization of biological macromolecules is decomposition of the large molecules into smaller model compounds. The advantages are twofold. First, it breaks complex systems into smaller models that are easier to treat using both MM and QM methods. Second, for smaller systems more experimental data are available for smaller systems, including thermodynamic properties of condensed phases, such as heats of vaporization or free energies of aqueous solvation, much of which is essential for accurate optimization of the nonbond portion of the force field. This approach assumes that parameters from the smaller models are transferable to the macromolecules, an assumption that is valid to a limited extent. Figure 3 illustrates how proteins can be decomposed into smaller model compounds; N-methyl acetamide describes the peptide bond, the alanine dipeptide models the protein backbone, and each sidechain is modeled by a corresponding valence-saturated model compound. Similar decompositions are employed for nucleic acids, carbohydrate, lipids, as well as in the construction of larger drug-like molecules from fragments. A well-thought optimization strategy is not only the key to obtain accurate reproduction of QM and experimental target data but also to reduce the labor required to parametrize a molecule.
Figure 2.
A simplified parametrization flow chart.
Figure 3.
Illustration of model compounds used in the parametrization of the CHARMM additive protein force field. (a) N-methyl acetamide is used to model the peptide bond; (b) sidechains, such as in PHE, are modeled by analogous compounds that include terminating methyl or ethyl groups; (c) alanine dipeptide is the model compound for optimization of the ϕ/ψ torsional parameters including CMAP corrections.
Selection of Target Data
A very important step in parameter optimization is the selection of target data. Force field parameter determination is an under-determined and nontrivial problem difficult to automate and very much dependent on human intervention. For the CHARMM force fields a combination of QM data and experimentally determined properties are considered. Although increases in computer power have allowed bigger systems to be studied with more accurate methods, sole reliance on QM methods is inappropriate. This is particularly important for optimization of the nonbond parameters, where dispersion interactions play an important role. However, it should be emphasized that the availability of high quality experimentally determined structural properties (e.g. geometries, vibrations, and conformations) is limited to small molecules, including constituents of nucleic acids and proteins, leading to a reliance on QM data. Macroscopic condensed phase properties such as heats of vaporization or sublimation and free energies of hydration are available for many of the building blocks from which the force field is hierarchically constructed. Although many of these data do not provide an atomic level picture of molecular interactions, when combined with the atomic details provided by QM and structure-based experimental methods (ex. X-ray crystallography and neutron diffraction) they are ideally able to produce a balanced force field, i.e. one that performs well at the atomic level but is also able to reproduce condensed phase experimental properties. The difficulty of attaining this was noted in a 1995 review paper by Halgren (40) in which he commented on the future of force fields:
“The past two years have seen a veritable beehive of activity in force field development. Remarkably, this period has seen the (…) completion of significant re-parameterizations of two currently widely applied force fields - CHARMM and AMBER. That these force fields differ substantially in form and in manner of derivation serves to emphasize that force field development is still as much a matter of art as of science. Someday, consensus on the form and manner of parameterization of molecular force fields may exist, but for now much remains to be learned. This is as it should be, for the problem being addressed is a hard one: to capture faithfully in a computationally tractable model enough of the real, quantum-mechanical physics to insure that a bio-molecular simulation, properly carried out, will yield a correct answer to useful precision.”
Fifteen years later the vision of Halgren has yet to materialize. Force field development is still as much of an art as it is a science. Improvements in QM methods alone have yet to solve the problem and human intervention continues to be extremely important to properly weight the various QM and experimental data being considered during parameters optimization. Finally, problems associated with parameter correlation continue to be a problem.
Optimization procedure
Parametrization of the CHARMM force field consists of obtaining appropriate intramolecular (bond, angle, dihedral, Urey-Bradley and improper, or “out-of-plane”, terms), vdW, and electrostatic parameters that adequately reproduce the selected target data. Determination of the electrostatic parameters differs between the additive and the Drude polarizable force fields in that the polarizabilities (Eq. 3) and Thole factors (Eq. 4) need to be optimized in addition to the point charges in the additive force field. Here, we describe the strategies behind the various steps depicted in Figure 2 that are involved in parameter optimization for the additive and Drude polarizable force fields. As was suggested earlier, parametrization is a tedious process that directly affects the outcome of subsequent usage. It is therefore a requirement that this task be performed in a careful and consistent fashion.
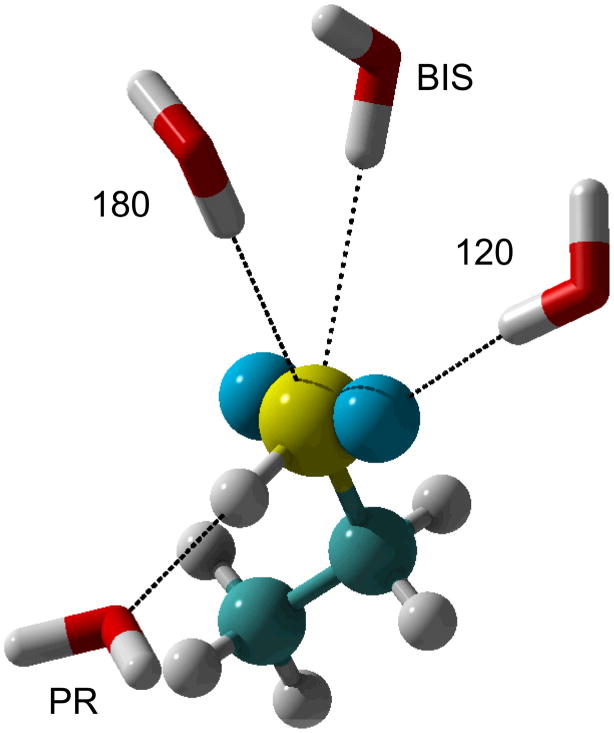
The development of an accurate electrostatic model is one of the more tedious steps in the parametrization workflow, laying the foundation for the model to reproduce the proper electronic response as a function of environment. In the additive CHARMM force field optimization of point charges is based on a supramolecular approach where the charges are adjusted to reproduce QM HF/6-31G(d) interaction energies and geometries of the model compound with, typically, individual water molecules as illustrated in Figure 4. Placement of water molecules at different orientations around the molecule has the distinct advantage of implicitly accounting for local electronic polarization, a desirable feature for accurate reproduction of condensed phase properties. In addition, interactions with water are often supplemented with dimers of the model compounds and with dipole moments of the models, with the dipole moments overestimated to account for condensed phase effects.(39, 41). For a number of biomolecular force fields the most common optimization methods are based on QM electrostatic potential (ESP) approaches. ESP methods consist of determining atomic charges based on reproduction of QM ESP evaluated on a grid surrounding the model compound.(42–44) The advantage of these methods is of convenience since charges can be determined quickly for any compound for which the QM ESP can be determined. An extension of ESP methods is the inclusion of restraints during fitting, referred to as the restrained ESP (RESP) approach,(45) which overcomes limitations on the determination of charges on buried atoms.(46) It is important to take note that solutions from both supramolecular and ESP approaches are conformation dependent, requiring care in selecting the appropriate conformation or conformations when performing the charge optimization.
Figure 4.
Orientation of test water molecules around the polarizable atom of interest exemplified through ethanethiol. Carbon is in cyan, sulfur in yellow and LP in blue. “120” is probing for interactions along the S-LP axis. “180” is oriented towards the C–S bond. “BIS” water points at the bisection of the C-S-H valence angle. “PR” is pointed directly at the sulfur proton and along the S–H bond.
Electrostatic parameters in the Drude polarizable force field are obtained from restrained fitting to perturbed QM ESP maps calculated on grid points located on concentric Connolly surfaces around the molecule. In some cases fitting is supplemented with reproduction of the molecular dipole moment and diagonal elements of the polarization tensor.(31) The determination of the atomic polarizabilities and Thole factors (34) requires multiple perturbed ESPs, typically calculated at the B3LYP/aug-cc-pVDZ level, each giving the electronic response of the molecule to a point charge. Perturbing ions are placed mainly along chemical bonds and lone pairs. To more accurately reproduce hydrogen-bond interactions as a function of orientation lone pairs (LPs) are added to hydrogen bond acceptors. LPs typically carry the charge of the atom to which they are attached (N, O or S) while the associated polarizability and Thole factor are assigned to the parent atom. In addition, anisotropic polarizability of hydrogen bond acceptors was found to be required to reproduce interactions with ions as a function of orientation (see reference (47) for an example on sulfurs and Figure 5 for an illustration). Although gas-phase dipoles are readily reproducible with full atomic polarizabilities, scaling of the polarizabilities was shown to be necessary to reproduce condensed-phase properties as seen in the SWM4-DP and SWM4-NDP water models.(48) A scaling factor of approximately 0.7 was found appropriate for the water models but for other classes of molecules scaling factors ranging from 0.6 to 1.0 were shown to be appropriate with 1.0 being full polarizability. For instance, a scaling factor of 0.7 was used in thiols and a factor of 0.85 was needed for dimethyl disulfide and 0.6 for ethylmethyl sulfide (47). Other scaling factors are 0.7 for primary and secondary alcohols (49), 0.85 for aromatics (50), N-containing heterocycles (51), nucleic acid bases and ethers (52) and 1.0 for alkanes (53). A value of 0.724, identical to the water model, was recently used in ion parameters(54). Final optimization of the polarizabilities consists of testing the model for reproduction of the pure solvent dielectric and adjusting the polarizability scaling if necessary. A generalization can be made here such that, although scaling factors will differ from one class of model compounds to another, they will remain in the aforementioned range.
Figure 5.
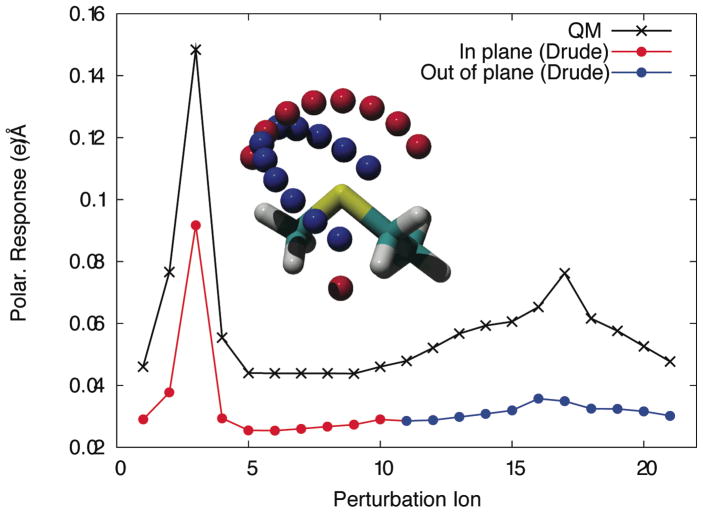
The anisotropic nature of polarizability for a sulfur atom in ethylmethylsulfide is probed using +0.5e point charges placed on two perpendicular arcs (inset). Differences between QM perturbed and unperturbed electrostatic potentials are used to determine polarization response as a function of orientation. Polarizability anisotropy parameters are fitted to reproduce relative response curves obtained through quantum mechanical calculations.
Determination of the vdW terms is one of the most important aspects in the development of a force field for condensed phase simulations and is indeed the most tedious step in the process. Jorgensen pioneered the use of condensed phase simulations, usually pure liquids, as the basis for optimization of the LJ parameters.(55, 56) Typically, once partial atomic charges are assigned via the supramolecular approach, the LJ parameters for a model compound can be adjusted to reproduce the experimentally determined pure solvent heat of vaporization and density, as well as isothermal compressibility and heat capacity, crystal heat of sublimation and lattice geometry and/or free energy of aqueous solvation, as available. Although this is an effective method for the fine-tuning of the parameters, the solution is by far trivial. A major issue is the parameter correlation paradigm, such that LJ parameters for different atoms in a molecule (e.g., H and C in ethane) as well as between the εij and Rmin terms on an individual atom can compensate for changes in each other, making it difficult to pinpoint the “correct” LJ parameters of a molecule based on reproduction of condensed phase properties alone.(57) To overcome this problem a method has been developed to determine the relative value of the LJ parameters based on high level QM data (58) and the absolute values based on localized εij and Rmin scans for the reproduction of experimental data.(59, 60) This approach requires supramolecular interactions between rare gases and the model compound. However, once satisfactory LJ parameters are optimized for atoms in a class of functional groups they can often be directly transferred to other molecules with those functional groups without further optimization.
Reproduction of experimental hydration free energies reflects how well the electrostatic and vdW parameters reproduce interactions with bulk water. This property is especially important in the determination of the free energy of ligand binding, where the solvation free energy of the ligand plays a major role in the overall binding affinity. Recently, in the context of the polarizable Drude force field, it has been shown that atom-pair specific LJ parameters need to be used in order to minimize discrepancies between calculated and experimental hydration free energies while simultaneously reproducing pure solvent heats of vaporization and molecular volumes. This can be achieved through the NBFIX facility in CHARMM, and consists of overwriting well-depths and vdW radii based on the standard combination rules with atom-pair specific Lennard-Jones parameters (e.g. atom-Ow) that are optimized to reproduce the aqueous free energies (19). As shown in Table I this has led to considerably better agreement with experimental values for free energies of hydration calculated with the Drude polarizable force field versus those calculated with the additive force field. The need for atom-pair specific LJ parameters is due to limitations in the combining rules currently use; (19) efforts are ongoing in our laboratory to identify new LJ combining rules such that atom-pair specific parameters will not be required.
Optimization of the internal parameters in the CHARMM force field relies heavily on QM calculations and data from the Cambridge Crystallographic Database (61). The parametrization effort consists of determining appropriate force constants and equilibrium values for the bond, angle, improper and Urey-Bradley terms and force constants, multiplicities and phase angles for the dihedral term. Target data includes geometrical information, supplemented by vibrational spectra, including assignments of the normal mode contributions in terms of primitive modes.(62, 63) In compounds with rotatable bonds, conformational energies as a function of rotation around those bonds are used as additional target data for the optimization of the associated dihedral parameters (64, 65). Examples include the peptide bond and the glycosidic linkage (11, 12). An automated fitting program is currently available (66) and is especially useful for large multidimensional target data sets such as those generated for glycosidic linkages between monosaccharides (11).
It should again be noted that the parameter optimization process follows an iterative approach (Figure 2). Thus, if changes are made to one aspect of the parameters for a model compound, other parameters have to be re-evaluated with respect to reproduction of the target data and optimized as required. While this may be considered tedious, given the now extensive nature of the CHARMM force fields, this procedure typically only requires two or three cycles to reach an adequate level of convergence.
Recent developments and applications of the CHARMM additive force field
While a range of parameters in the context of the additive force field are available, continuous optimization and development efforts have improved the accuracy and expanded the coverage of the CHARMM additive force field allowing more accurate simulations on heterogeneous molecular systems. Current versions of the additive force field include CHARMM22/CMAP (39, 67) for proteins, CHARMM27(68) for nucleic acids, CHARMM36(9, 10) for lipids, CHARMM37(11–15) for carbohydrates, and CGenFF(16) for small molecules. As discussed below, updates of the CHARMM22/CMAP protein and CHARMM27 nucleic acid parameters are nearing completion as of the writing of this article. The parameters are available at the MacKerell group web page at (http://mackerell.umaryland.edu/CHARMM_ff_params.html) as well as being included with the CHARMM package. It should be emphasized that these different parameter sets are subgroups of the unified CHARMM additive force field as all the contained parameters are compatible, a prerequisite for calculations of complex heterogeneous systems. Recent adjustments to the organization of the individual files will facilitate the application of the CHARMM force field to such systems. This section will provide information complementary to previous published reviews (33, 39) on recent improvements and extension in the additive force field in CHARMM. Discussions about the current progress in the CHARMM polarizable force field will ensue.
Polypeptides and Proteins (CMAP)
The protein φ/ψ parameters are crucial for accurate simulation and sampling of the conformational properties of polypeptides and protein. Studies from around the turn of the millennium indicated that the intrinsic form of the dihedral term in the potential energy function limits the ability of force fields in reproducing QM data and experimental structures of proteins (69) as well as the ability to fold small peptides. To address this issue, improvements in the backbone energetics of the CHARMM22 force field were undertaken in the form of a 2D dihedral energy correction map (CMAP) that could be applied to the φ/ψ surface.(67) Since alanine is the common denominator for all natural amino acids with the exception of glycine and proline, CMAP was calculated based on QM φ/ψ energy surfaces of the alanine dipeptide. The correction grid applies to all residues in the CHARMM22 force field except glycine and proline for which separate CMAPs were implemented. This alanine dipeptide-based CMAP was further adjusted empirically through MD simulations of protein crystals and small peptides, with the latter using an implicit solvent model, bringing significant improvements in reproducing crystallographic φ/ψ distributions.(70) Furthermore, a study on hen lysozyme has shown that CMAP corrections significantly reduce the discrepancies in the backbone dynamics between CHARMM22 and NMR observations.(71) The force field containing this grid-based energy correction scheme is referred to as CHARMM22/CMAP.
The CHARMM22/CMAP force field has been used to study the dynamics of various systems in contexts ranging from structure-function analysis of proteins as well as drug development. Guvench et al (72) performed MD simulations to elucidated the structural basis for substrate binding in SHP-2, a non-receptor tyrosine phosphatase. It was shown that substrate pY-peptide binding is modulated by Tyr66, which, through conformational flexibilities in its backbone, acts as a gate for the binding cleft. CHARMM was also used to derive the mechanisms for binding of the anticancer drug imatinib to its receptor.(73) In this study, a combination of CHARMM22/CMAP and newly developed parameters for imatinib were used to conduct free energy calculations yielding good agreement with experimental binding affinities and allowed the researchers to suggest imatinib modifications with improved binding. Long MD simulations on the order of hundreds of nanoseconds are often required to trace subtle structural changes in proteins that could play pivotal roles in activity. Such is the case for the recent structure-activity relationship (SAR) assessment of the naturally occurring small protein PcFK1 that displays antimalarial properties.(74) Correlated movements of the loop, turns, and sheet regions on the protein were identified through 150 ns simulations of wild-type and mutant forms of PcFK1 using the CHARMM22/CMAP force field, from which a SAR model was developed. This diverse set of examples demonstrates the breadth of applications possible with and the fidelity of simulation results offered by the CHARMM22/CMAP force field.
The ability to accurately reproduce experimental structures and energies is essential for successful applications of force fields in MD simulations. One major issue with CHARMM22/CMAP is that although CMAP corrections gave better agreement with NMR measurements for folded proteins, simulations of unstructured peptides such as Ala5 is biased towards α-helical conformations.(75) Efforts to address the problem including revisions of the CMAP and amino acid side-chain torsions are ongoing (R. Best, M. Feig, X. Zhu and A. MacKerell, Personal communication). Tests of the new parameters are currently pending completion and release of these parameters is anticipated by the time this article is published. Improvements of secondary structure and sidechain torsion sampling brought to the CHARMM protein force field will likely improve the quality of MD simulations for the study of both the structural and dynamical properties of proteins.
Nucleic Acids
The CHARMM27 nucleic acid force field(68) was developed to fully support computational analysis of DNA and RNA. It consists of a full reoptimization of an earlier version of the CHARMM additive force field for nucleic acids that was shown to favor the A form of DNA.(76) CHARMM27 has been used in a study by Priyakumar and MacKerell (77) to investigate DNA base-flipping, which has provided a detailed understanding of the mechanisms and free energy profile for the breaking of Watson-Crick base-pairing and base stacking interactions associated with a C:G base flip. In another study, DNA:RNA hybrid systems were simulated and shown to yield good agreement with experimental observations (78). A separate study examined the structural basis of DNA triplex formation (79) and the impact of base modification on triplex sequence specificity. Results from the simulation will aid the design of efficient triple helix-forming oligonucleotides useful for gene targeting. One issue that remains to be addressed is the overestimation of BI/BII ratio for DNA duplexes such that unrestrained simulations fail to reproduce the sequence-dependent BII populations, a problem also seen in other force fields.(80) This offers room for further improvements to the CHARMM27 force field so to expand its utility in the study of the conformation, energy, and sequence-dependent behavior of nucleic acids; efforts to address the BI/BII issue are ongoing (L. Nilsson and A. MacKerell, Personal communication)
CHARMM27 has also been shown to be of utility in the context of RNA. One recent example is a study of the adenine riboswitch (81) as well as the DNA:RNA hybrid study mentioned above. Another study noted the presence of local opening of WC base pairs in duplex RNA, an phenomena indicated to be consistent with experimental data for GC base pairs but not for AU pairs (82). Other studies include MD simulations of a protein-RNA complex (83), structural characterization of the locked nucleic acid-RNA duplexes (84), and free energy profile of base-flipping in dsRNA (85). Two recent studies in which both the CHARMM27 and AMBER (param99) force fields were used are of particular interest. In a study investigating folding of two RNA hairpins based on potential of mean force calculations (86) it was observed that the AMBER force field significantly overestimated the experimental folding energy while CHARMM27 was in significantly better agreement with experiment, though one of the hairpins was indicated to be slightly unstable at room temperature. A second study involved a RNA 18-mer duplex (87) that showed the RMS fluctuations in MD simulations using AMBER to be significantly lower than that of CHARMM27, with the AMBER model staying closer to the canonical starting structure. To understand these phenomena, as well as local base opening seen in these CHARMM27 RNA simulations, systematic analysis was undertaken from which the sampling of the 2′OH moiety was identified as a possible cause of the local opening. Adjustments to the corresponding parameters were made and a number of models were obtained in which base opening was decreased (20). Notably, the new models lead to significantly improved reproduction of a range of experimental data, including better agreement with crystal and NMR structures, while still yielding free energies of folding of the RNA hairpins studied by Deng and Cieplak (86) similar to those calculated with CHARMM27, indicating the adjustments to not over stabilize the folded state. It is anticipated that the optimized parameter set will be referred to as CHARMM36.
Lipids
The CHARMM27r(88) lipid force field has been used for the study of biological systems involving the lipid bilayer. Recent publications include, for example, elucidation of the free energy profile and barriers for cholesterol flip-flops in three different types of lipid environments(89), simulation of a peptide in lipid bilayers(90) to better understand the dynamics of peptides as a function of membrane type and thickness, and exploration of the mechanism behind which a serine protease is anchored to a bilayer(91). Success of these studies is associated, in part, with revisions of the original CHARMM27 force field. In the revision parameters for the aliphatic tails were optimized using high level QM calculations as target data to yield better agreements with NMR order parameters for the aliphatic tails. However, one major flaw exists in the CHARMM27r version where positive surface tension of the bilayer results in shrinking of the membrane to a gel-phase state well above the transition temperature.(92) This flaw has been recently addressed by Klauda et al. (92) where headgroup torsions parameters and LJ and partial atomic charges of the ester linkage were re-optimized to yield the CHARMM36 lipid force field. This version reproduces experimental surface areas for saturated and unsaturated lipid bilayers, which prevents unwarranted shrinking of the lipid bilayer. One issue that remains to be addressed is that of the treatment of long-range LJ interactions, which, as discussed by the authors of the CHARMM36 article, is different for monolayers than with bilayers, with the latter being correctly treated with standard truncation methods common to all simulation programs. Accordingly, the CHARMM36 version of the lipid force field is suitable for the majority of the current research interests involving lipids including heterogeneous systems involving bilayers.
Carbohydrates
The additive CHARMM force field has historically contained a limited number of parameters for carbohydrates(93). It has recently been extended to include a range of carbohydrates, an omnipresent entity in cells that is essential in many cellular processes. Modeling of carbohydrates is further driven by scientific interest in understanding their biological and physiological functions in complex systems involving glycoconjugates and carbohydrate-protein complexes. In contrast to proteins, carbohydrates lack clear structural classifications.(94) While some polysaccharides have a regular repeating unit that favors structural periodicities, many are widely diverse, varying in the context of multiple factors such as the types of constituent monosaccharides and the type of the glycosidic linkages, variations that effect their ability to form regular structures and repeating networks of hydrogen bonds formed by the constituent hydroxyl groups.(95–97) Such structural diversity presents a challenge for the development of reliable parameters.
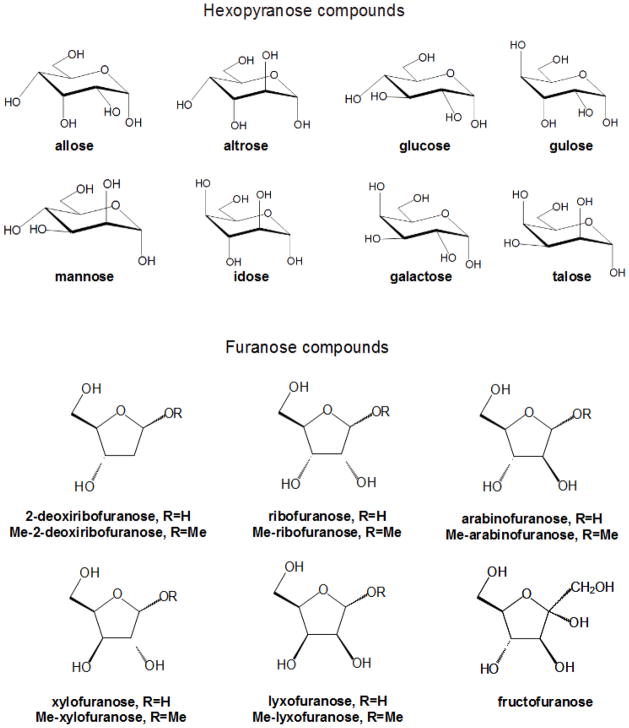
In the past few years, development of a carbohydrate force field compatible with the additive CHARMM force field has taken place in our group. The work has been systematic, covering major classes of mono- and disaccharides. As a result parameters are currently available for hexopyranose (15), aldopentofuranose (14), and fructofuranose (14) monosaccharides and as well as an extensive set of glycosidic linkage types (11, 12) as required to study di- and polysaccharides. In addition, optimization of parameters for non-hydroxyl moieties present in eukaryotic carbohydrates is nearing completion.
Development of CHARMM force field parameters for cyclic hexopyranoses and furanoses (Figure 6) and acyclic polyalcohols, acyclic carbohydrates, and inositol followed a common approach, including the usual array of target data: QM optimized molecular geometries, experimental geometries derived from small crystals, vibrational frequencies, solute-water interaction energies, molecular volumes, and heats of vaporization or sublimation. One of the more time consuming stages is the extensive QM sampling of conformational space of the furanose monosaccharides was for the two-dimensional ring pucker energy surfaces. Parametrization of acyclic polyalcohols, acyclic carbohydrates, and inositol had been complicated by the intramolecular hydrogen bonds and by the increased flexibility of the compounds versus their cyclic counterparts. The strategy employed to solve this problem was to perform constrained optimizations, with a total of ~1,500 conformations considered. In all cases, simultaneous consideration of a vast amount of QM data presents a challenge. To deal with that an automated scheme(66), based on Monte Carlo simulated annealing (MCSA), was used to accommodate the large amount of data. The performance of the newly developed force field parameters was evaluated by comparing experimental and calculated condensed-phase properties from MD simulations. Good agreement was found for crystal lattice parameters and densities, diffusion coefficients as well as NMR proton-proton couplings in aqueous solution in a subsequent study(98) and is indicative of the quality of the CHARMM force field parameters for carbohydrates.
Figure 6.
Cyclic hexopyranose and furanose compounds parametrized for the CHARMM carbohydrate force field.
Parameters for Small Molecules: Applications in Drug Design
An important utility of empirical force fields is to aid in the design and development of new therapeutic agents.(99, 100) Because of the extent of structural and chemical diversity in drug molecules, parameters geared towards transferability across a wide range of compounds is required and sought after by many force field development communities. Fulfilling this purpose are specialized force field parameters including MMFF (101), AMBER GAFF (102), CVFF (103), COMPASS (104) and the commercial version of CHARMm (105). These force fields are appropriate, for example, for methods that identify biologically active or “lead” compounds from a large database of small molecules as well as computer-based de novo molecular design of drug-like molecules. Using the aforementioned force fields new drug entities can be readily assembled, analyzed, and subjected to computational predictions of activity. However, it should be emphasized that reliable calculations such as those required for target-based structure optimization of lead compounds require the use of a well-balanced force field that properly treats both the drug and target (e.g. protein) molecules. In addition, to accurately capture the subtle differences across a series of test compounds it is generally necessary that at least some level of parameter optimization of the molecules of interest be performed, due to the inherent limited transferability of parameters within a force field. With sufficient optimization, a force field can provide superior accuracy in conformational sampling and energy evaluations, leading to improved results from a variety of techniques such as 4D-QSAR(106), conformationally-sampled pharmacophore (CSP) (107) and related methods, as well as free energy perturbation calculations (108).
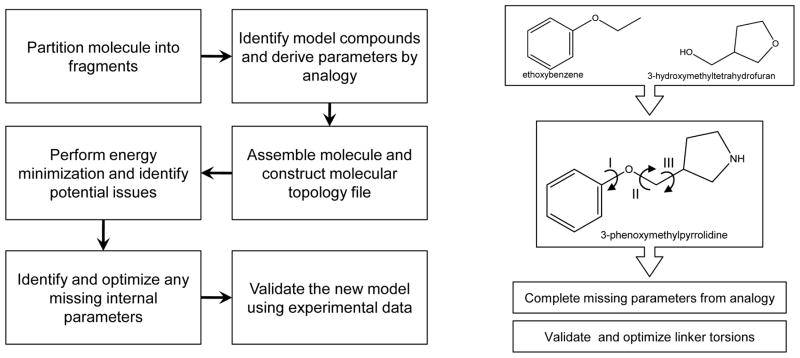
The increasing need for computational analysis of drug-like molecules was the driving force behind the development of the CHARMM General Force field (CGenFF) (16). Over 100 common and medicinally useful molecular scaffolds and linkers have been formally added to the existing collection of model compounds in the CHARMM force field. From this pool of model compounds, researchers can intuitively assemble compounds of interest, followed by evaluation of the quality of the resulting model and optimization of selected parameters to improve the accuracy of the treatment of the molecule of interest. Typically, one would only need to optimize the linker regions between ring systems. A summary of the steps involved in creating a new molecule is shown in Figure 7. CGenFF was conceived using a methodology consistent with the parametrization philosophy behind the biological CHARMM additive force field. As a result, CGenFF is fully compatible with the biological portions of the force field, which makes possible simulations of heterogeneous systems such as protein-ligand interactions. This concept is similar to the philosophy used in OPLS (109, 110). For complex systems or cases where a large number of molecules need to be studied, scripts that generate initial guesses of the atom type and parameters are available on the MacKerell group web page (http://mackerell.umaryland.edu/) and efforts are ongoing with respect to the development of a web server that automatically assigns atom types, charges and parameters (https://www.paramchem.org/). The ultimate goal of that server will be the ability to validate and optimize selected parameters, a capability that is anticipated to facilitate the expansion of CGenFF to cover a larger range of chemical space. The availability of CGenFF will likely spur additional interest in computer-aided drug design using the CHARMM force field.
Figure 7.
Flowchart for the determination of parameters for drug-like molecules as described in reference 16. 3-phenoxymethylpyrrolidine is assembled from two of its constituents, ethoxybenzene and 3-hydroxymethyltetrahydrofuran, available from the CGenFF. Parameters were identified for the pyrrolidine group by analogy. Optimization of the dihedrals I, II, and III is required to produce an accurate computational model for this molecule.
Recent advances in the CHARMM polarizable force field
The initial step in developing the polarizable force field based on the Drude oscillator was the parametrization of water. A four-site polarizable water model SWM4-NDP was developed(48). In this model, Drude particles have negative charges rather than positive, as was in the previous SWM4-DP model (111). Such representation more accurately reflects the shifting of electron distributions associated with changes in the environment. The convention of using negatively charged Drude particles has been maintained in subsequent development efforts. SWM4-NDP yields a clear improvement over the additive TIP3P (112) model in that both gas-phase dipole moments and condensed-phase dielectric constant are simultaneously reproduced with high accuracy. The ability of reproducing the orientation and extent of quadrupoles is also a clear indication of the superiority of the polarizable water model over that of the additive.
From the time the 4-point polarizable water model SWM4-NDP was published (48) until now, significant progress has been made in the development of CHARMM polarizable force fields for proteins, lipids and nucleic acids. Current available model compounds for proteins include the backbone N-methylacetamide (113) and amino acid sidechain models such as alkanes (53), alcohols (49), aromatics (50, 51), amides (113), and sulfur-containing compounds (47). Compared to the additive force field, polarizable models display increased accuracy in gas-phase dipole moments, dielectric constants of bulk solvents, and free energies of solvation. Finalization of both the peptide backbone and sidechain parameters is ongoing and preliminary simulations of small polypeptides as well as full proteins have been performed. A full polarizable model of DPPC lipids has been created and successfully applied to simulate a bilayer (114); additional optimization of that model is ongoing. In addition, parameters for nucleic acid bases have recently been reported (115) and preliminary simulations of DNA and RNA have been performed. Finally, optimization of Drude parameters for carbohydrates has been initiated, such that the availability of a comprehensive Drude polarizable force field for biological molecules in the next several years is anticipated.
Development efforts have also extended to ions, which are not only an integral part of the cellular environment but also an important component of metalloproteins (54, 116). One major issue encountered during this optimization was that of polarization catastrophe, as seen in other polarizable models (38, 117, 118). This primarily results from strong Coulombic interactions that overcome LJ repulsion and lead to uncontrolled drifting of the Drude particle away from the atomic center when atoms approach to closely, a problem enhanced most notably in the case of dications. To address this issue the Drude energy function was extended to include an anharmonic hyperpolarizability damping term
| (6) |
where a force constant Khyp is applied once the Drude displacement, Δr, is a distance greater than Δrcut. This term limits the maximum displacement of Drude particles with negligible effects on the overall electrostatic properties. The force constant of 40,000 kcal/mol/Å4 was selected based on QM-estimated maximum induced dipole of halide anions such as Br− and I− (54). A Δrcut of 0.2 Å is currently used for simulations of biomolecules using the Drude polarizable force field in CHARMM. Implementation of the damping function provides an effective way to minimize the polarizability catastrophe issue commonly seen in polarizable force fields.
Conclusion and Perspectives
Molecular mechanics as a tool to investigate experimentally difficult to access properties of molecular systems has gained wide acceptance within the scientific community. After years of effort, the CHARMM additive force field has evolved into a full featured force field capable of treating biologically relevant macromolecules: proteins, lipids nucleic acids and carbohydrates as well as a range of small drug-like molecules. Improvements in this force field, which relies on fine-tuning of parameters to reproduce an ever increasing collection of QM and experimental data, are ongoing.
Aside from fundamental algorithmic changes and advances in computer power, improvements in empirical force field can be made in three tiers. These include modifications to the underlying form of the potential energy function on which the force field is based, expansion of the number of explicitly parametrized model compounds, and additional refinements of available force fields. In this review all three of the issues have been addressed in the context of the CHARMM force field development efforts. Moreover, as computational power continues to increase and conformational sampling methodologies improves, it will be possible to both improve present force fields as well as extend empirical force fields to more complex energy functions, as evidenced by ongoing efforts in the development of polarizable force fields. It is anticipated that the inherent advantage of polarizable force fields over current additive models will allow them to be of significant utility in the application of molecular mechanics approaches to solving biological questions.
As a final word, we must bear in mind that even the most accurate force field is only as good as the computational design with which it is applied. Carefully crafted calculations require an understanding of the strengths and weaknesses of the applied force field. These considerations are also important for interpreting results from the calculations. With this review we hope to provide a basis towards an expanded understanding of force fields with the purpose of facilitating proper implementation of the CHARMM force fields.
Table 1.
Comparison of experimental/QM and empirical gas phase dipole moments, μ, pure solvent dielectric constants, ε, and free energies of hydration (kcal/mol) for various model compounds using the additive and Drude polarizable CHARMM force fields.
| Exp. | Additive | Drude | ΔexpAdditive | ΔexpDrude | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Molecule | μ | ε | ΔGhydr | μ | ε | ΔGhydr | μ | ε | ΔGhydr | μ | ε | ΔGhydr | μ | ε | ΔGhydr |
| Ethanea | 1.76 | 1.77 | 0 | 1.01 | 2.17 | 0 | 1.71 | 1.84 | −0.75 | 0.4 | −0.05 | 0.07 | |||
| Propanea | 0.084 | 1.8 | 1.98 | 0.002 | 1.02 | 1.98 | 0.108 | 1.8 | 1.63 | −0.082 | −0.78 | 0 | 0.024 | 0 | −0.35 |
| Isobutanea | 0.132 | 1.82 | 2.28 | 0.012 | 1.02 | 1.86 | 0.226 | 1.9 | 2.28 | −0.12 | −0.8 | −0.42 | 0.094 | 0.08 | 0 |
| MeOHb | 1.7 | 32.61 | −5.11 | 2.38 | 17.2 | −4.98 | 1.83 | 30.1 | −4.64 | 0.68 | −15.41 | 0.13 | 0.13 | −2.51 | 0.47 |
| EtOHb | 1.71 | 24.85 | −5.01 | 2.36 | 18.8 | −5.34 | 1.81 | 21.4 | −4.97 | 0.65 | −6.05 | −0.33 | 0.1 | −3.45 | 0.04 |
| 1-PrOHb | 1.55 | 20.52 | −4.83 | 2.35 | 15.2 | −5.33 | 1.82 | 19.5 | −4.85 | 0.8 | −5.32 | −0.5 | 0.27 | −1.02 | −0.02 |
| 1-BuOHb | 1.66 | 17.33 | −4.72 | 2.36 | 10.8 | −5.6 | 1.8 | 21.2 | −4.67 | 0.7 | −6.53 | −0.88 | 0.14 | 3.87 | 0.05 |
| EtSHc | 1.58 | 6.67 | −1.3 | 1.44 | 4.3 | −0.86 | 1.78 | 7.79 | −1.37 | −0.14 | −2.37 | 0.44 | 0.2 | 1.12 | −0.07 |
| 1-PrSHc | 1.6 | 5.94 | −1.05 | 1.62 | 5.94 | −1.14 | 1.05 | 0.02 | 0 | −0.09 | |||||
| EMSc | 1.85 | −1.83 | 1.73 | 4.98 | −1.09 | 1.89 | 6.91 | −1.88 | −0.12 | 0.74 | 0.04 | −0.05 | |||
| DMDSc | 1.56 | 9.6 | −1.46 | 1.13 | 3.03 | −0.02 | 1.66 | 8.44 | −1.37 | −0.43 | −6.57 | 1.44 | 0.1 | −1.16 | 0.09 |
| CPENd | 0.02 | 1.96 | 1.2 | 0.05 | 1.02 | 0.73 | 0.02 | 1.63 | 0.81 | 0.03 | −0.94 | −0.47 | 0 | −0.33 | −0.39 |
| CHEXd | 0 | 2.02 | 1.23 | 0 | 1.02 | 0.89 | 0 | 1.66 | 1.42 | 0 | −1 | −0.34 | 0 | −0.36 | 0.19 |
| THFd | 1.75 | 7.43 | −3.47 | 1.97 | 5.42 | −3.34 | 1.69 | 6.8 | −3.78 | 0.22 | −2.01 | 0.13 | −0.06 | −0.63 | −0.31 |
| THPd | 1.58 | 5.54 | −3.12 | 2.03 | 4.97 | −3.35 | 1.58 | 5.03 | −3.2 | 0.45 | −0.57 | −0.23 | 0 | −0.51 | −0.08 |
| DEEd | 1.15 | 4.24 | −1.76 | 1.81 | 4.96 | −2 | 1.2 | 3.53 | −1.6 | 0.66 | 0.72 | −0.24 | 0.05 | −0.71 | 0.16 |
| DMOEd | 2.19 | 7.22 | −4.84 | 3.1 | 6.76 | −4.52 | 2.12 | 5.61 | −3.78 | 0.91 | −0.46 | 0.32 | −0.07 | −1.61 | 1.06 |
| DMEd | 1.3 | 6.53 | −1.92 | 1.86 | 9.71 | −1.44 | 1.3 | 6.36 | −1.25 | 0.56 | 3.18 | 0.48 | 0 | −0.17 | 0.67 |
|
| |||||||||||||||
| Average | 0.3 | −2.9 | 0.1 | 0.1 | −0.4 | 0.1 | |||||||||
| SD | 0.4 | 4.3 | 0.6 | 0.1 | 1.5 | 0.4 | |||||||||
Acknowledgments
We would like to thank Dr. Sairam S. Mallajosyula, Dr. Christopher Baker, as well as everyone in the MacKerell group for help with various references, active discussions and moral support. This work was supported by a departmental fellowship from the School of Pharmacy, University of Maryland, Baltimore to XZ, the University of Maryland Computer-Aided Drug Design Center and NIH grants GM070855, GM015101 and GM0772558 and NSF grant CHE-0823198.
References
- 1.McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 2.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 4.Case DA, TAD, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, RCW, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra GIK, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, HG, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, MGS, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER. University of California; San Francisco: 2010. p. 11. [Google Scholar]
- 5.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjelkmar Pr, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. Journal of Chemical Theory and Computation. 2010;6(2):459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 7.Chandler DE, Hsin J, Harrison CB, Gumbart J, Schulten K. Intrinsic curvature properties of photosynthetic proteins in chromatophores. Biophys J. 2008;95(6):2822–2836. doi: 10.1529/biophysj.108.132852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, et al. Anton, a special-purpose machine for molecular dynamics simulation. SIGARCH Comput Archit News. 2007;35(2):1–12. [Google Scholar]
- 9.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, et al. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. The Journal of Physical Chemistry B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feller SE, Yin D, Pastor RW, MacKerell AD., Jr Molecular Dynamics Simulation of Unsaturated Lipid Bilayers at Low Hydration: Parametrization and Comparison with Diffraction Studies. Biophy J. 1997;73:2269–2279. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raman EP, Guvench O, MacKerell AD., Jr CHARMM additive all-atom force field for glycosidic linkages in carbohydrates involving furanoses. J Phys Chem B. 2010;114(40):12981–12994. doi: 10.1021/jp105758h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guvench O, Hatcher ER, Venable RM, Pastor RW, Mackerell AD. CHARMM Additive All-Atom Force Field for Glycosidic Linkages between Hexopyranoses. J Chem Theory Comput. 2009;5(9):2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatcher E, Guvench O, Mackerell AD. CHARMM Additive All-Atom Force Field for Acyclic Polyalcohols, Acyclic Carbohydrates and Inositol. J Chem Theory Comput. 2009;5(5):1315–1327. doi: 10.1021/ct9000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatcher E, Guvench O, Mackerell AD. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J Phys Chem B. 2009;113(37):12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, et al. Additive empirical force field for hexopyranose monosaccharides. J Comput Chem. 2008;29(15):2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopes PE, Murashov V, Tazi M, Demchuk E, Mackerell AD., Jr Development of an empirical force field for silica. Application to the quartz-water interface. J Phys Chem B. 2006;110(6):2782–2792. doi: 10.1021/jp055341j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacKerell AD, Jr, Feig M, Brooks CL., 3rd Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc. 2004;126(3):698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 19.Baker C, Lopes P, Zhu X, Roux Bt, MacKerell A. Accurate Calculation of Hydration Free Energies using Pair-Specific Lennard-Jones Parameters in the CHARMM Drude Polarizable Force Field. Journal of Chemical Theory and Computation. 2010;6(4):1181–1198. doi: 10.1021/ct9005773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denning EJ, Priyakumar DU, Nilsson L, Mackerell AD., Jr Impact of hydroxyl sampling on the conformational properties of RNA: Update of the CHARMM all-atom additive force field for RNA. 2011 doi: 10.1002/jcc.21777. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alder BJWTE. Phase Transition for a Hard Sphere System. J Chem Phys. 1957;27(1208) [Google Scholar]
- 22.Hernandez G, Anderson JS, LeMaster DM. Polarization and polarizability assessed by protein amide acidity. Biochemistry. 2009;48(27):6482–6494. doi: 10.1021/bi900526z. [DOI] [PubMed] [Google Scholar]
- 23.David R, Lide E. Handbook of Chemistry and Physics. 84. CRC Press; Boca Raton, Florida: 2003. [Google Scholar]
- 24.Gregory JK, Clary DC, Liu K, Brown MG, Saykally RJ. The Water Dipole Moment in Water Clusters. Science. 1997;275(5301):814–817. doi: 10.1126/science.275.5301.814. [DOI] [PubMed] [Google Scholar]
- 25.Badyal YS, Saboungi ML, Price DL, Shastri SD, Haeffner DR, Soper AK. Electron distribution in water. The Journal of Chemical Physics. 2000;112(21):9206–9208. [Google Scholar]
- 26.Silvestrelli PL, Parrinello M. Water Molecule Dipole in the Gas and in the Liquid Phase. Physical Review Letters. 1999;82(16):3308. [Google Scholar]
- 27.Rick SW, Stuart SJ. Potentials and algorithms for incorporating polarizability in computer simulations. In: Lipkowitz KB, Boyd DB, editors. Reviews in Computational Chemistry. Wiley-VCH; Hoboken, NJ: 2002. pp. 89–146. [Google Scholar]
- 28.Halgren TA, Damm W. Polarizable force fields. Curr Opin Struct Biol. 2001;11(2):236–242. doi: 10.1016/s0959-440x(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen WL. Special Issue on Polarization. Journal of Chemical Theory and Computation. 2007;3(6):1877–1877. doi: 10.1021/ct700252g. [DOI] [PubMed] [Google Scholar]
- 30.Lopes P, Roux B, MacKerell A. Molecular modeling and dynamics studies with explicit inclusion of electronic polarizability: theory and applications. Theoretical Chemistry Accounts: Theory, Computation, and Modeling (Theoretica Chimica Acta) 2009;124(1):11–28. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anisimov VM, Lamoureux G, Vorobyov IV, Huang N, Roux B, MacKerell AD. Determination of Electrostatic Parameters for a Polarizable Force Field Based on the Classical Drude Oscillator. J Chem Theory Comput. 2005;1(1):153–168. doi: 10.1021/ct049930p. [DOI] [PubMed] [Google Scholar]
- 32.Lamoureux G, Roux B. Modeling induced polarization with classical Drude oscillators: Theory and molecular dynamics simulation algorithm. The Journal of Chemical Physics. 2003;119(6):3025–3039. [Google Scholar]
- 33.MacKerell AD., Jr Empirical force fields for biological macromolecules: overview and issues. J Comput Chem. 2004;25(13):1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 34.Thole BT. Molecular polarizabilities calculated with a modified dipole interaction. Chem Phys. 1981;59:341–350. [Google Scholar]
- 35.Patel S, Brooks CL. CHARMM fluctuating charge force field for proteins: I parameterization and application to bulk organic liquid simulations. J Comp Chem. 2004;25(1):1–15. doi: 10.1002/jcc.10355. [DOI] [PubMed] [Google Scholar]
- 36.Patel S, Mackerell AD, Brooks CL. CHARMM fluctuating charge force field for proteins: II -Protein/solvent properties from molecular dynamics simulations using a nonadditive electrostatic model. J Comp Chem. 2004;25(12):1504–1514. doi: 10.1002/jcc.20077. [DOI] [PubMed] [Google Scholar]
- 37.Patel S, Brooks CL. Fluctuating charge force fields: Recent developments and applications fromsmall molecules to macromolecular biological systems. Mol Simul. 2006;32(3–4):231–249. [Google Scholar]
- 38.Xie W, Pu J, Mackerell AD, Gao J. Development of a polarizable intermolecular potential function (PIPF) for liquid amides and alkanes. J Chem Theory Comput. 2007;3(6):1878–1889. doi: 10.1021/ct700146x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Halgren TA. POTENTIAL-ENERGY FUNCTIONS. Current Opinion in Structural Biology. 1995;5(2):205–210. doi: 10.1016/0959-440x(95)80077-8. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. Journalof the American Chemical Society. 1988;110(6):1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 42.Singh UC, Kollman PA. AN APPROACH TO COMPUTING ELECTROSTATIC CHARGES FOR MOLECULES. Journal of Computational Chemistry. 1984;5(2):129–145. [Google Scholar]
- 43.Chirlian LE, Francl MM. ATOMIC CHARGES DERIVED FROMELECTROSTATIC POTENTIALS -A DETAILED STUDY. Journal of Computational Chemistry. 1987;8(6):894–905. [Google Scholar]
- 44.Merz KM. ANALYSIS OF A LARGE DATA-BASE OF ELECTROSTATIC POTENTIAL DERIVED ATOMIC CHARGES. Journal of Computational Chemistry. 1992;13(6):749–767. [Google Scholar]
- 45.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. The Journal of Physical Chemistry. 1993;97(40):10269–10280. [Google Scholar]
- 46.Francl MM, Carey C, Chirlian LE, Gange DM. Charges fit to electrostatic potentials .2. Can atomic charges be unambiguously fit to electrostatic potentials? Journal of Computational Chemistry. 1996;17(3):367–383. [Google Scholar]
- 47.Zhu X, MacKerell AD., Jr Polarizable empirical force field for sulfur-containing compounds based on the classical Drude oscillator model. J Comput Chem. 2010;31(12):2330–2341. doi: 10.1002/jcc.21527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamoureux G, Harder E, Vorobyov IV, Roux B, MacKerell AD., Jr A polarizable model of water for molecular dynamics simulations of biomolecules. Chem Phys Lett. 2006;418(1–3):245–249. [Google Scholar]
- 49.Anisimov VM, Vorobyov IV, Roux B, MacKerell JAD. Polarizable empirical force field for the primary and secondary alcohol series based on the classical Drude model. J Chem Theory Comp. 2007;3:1927–1946. doi: 10.1021/ct700100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopes PE, Lamoureux G, Roux B, Mackerell AD., Jr Polarizable empirical force field for aromatic compounds based on the classical drude oscillator. J Phys Chem B. 2007;111(11):2873–2885. doi: 10.1021/jp0663614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopes PE, Lamoureux G, Mackerell AD., Jr Polarizable empirical forcefield for nitrogen-containing heteroaromatic compounds based on the classical Drude oscillator. J Comput Chem. 2009;30(12):1821–1838. doi: 10.1002/jcc.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker CM, Mackerell AD., Jr Polarizability rescaling and atom-based Thole scaling in the CHARMM Drude polarizable force field for ethers. J Mol Model. 2009 doi: 10.1007/s00894-009-0572-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vorobyov IV, Anisimov VM, MacKerell AD., Jr Polarizable empirical force field for alkanes based on the classical Drude oscillator model. J Phys Chem B. 2005;109(40):18988–18999. doi: 10.1021/jp053182y. [DOI] [PubMed] [Google Scholar]
- 54.Yu H, Whitfield TW, Harder E, Lamoureux G, Vorobyov I, et al. Simulating Monovalent and Divalent Ions in Aqueous Solution Using a Drude Polarizable Force Field. J Chem Theory Comput. 2010;6(3):774–786. doi: 10.1021/ct900576a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorgensen WL. Optimized intermolecular potential functions for liquid alcohols. The Journal of Physical Chemistry. 1986;90(7):1276–1284. [Google Scholar]
- 56.Jorgensen WL, Madura JD, Swenson CJ. Optimized intermolecular potential functions for liquid hydrocarbons. Journal of the American Chemical Society. 1984;106(22):6638–6646. [Google Scholar]
- 57.MacKerell AD., Jr . Atomistic Models and Force Fields. In: Becker OM, MacKerell AD Jr, Roux B, Watanabe M, editors. Computational Biochemistry and Biophysics. Marcel Dekker, Inc; New York: 2001. pp. 7–38. [Google Scholar]
- 58.Yin DX, MacKerell AD. Ab initio calculations on the use of heliumand neon as probes of the van der Waals surfaces of molecules. Journal of Physical Chemistry. 1996;100(7):2588–2596. [Google Scholar]
- 59.Yin DX, Mackerell AD. Combined ab initio empirical approach for optimization of Lennard-Jones parameters. Journal of Computational Chemistry. 1998;19(3):334–348. doi: 10.1002/jcc.1166. [DOI] [PubMed] [Google Scholar]
- 60.Chen IJ, Yin DX, MacKerell AD. Combined ab initio/empirical approach for optimization of Lennard-Jones parameters for polar-neutral compounds. Journal of Computational Chemistry. 2002;23(2):199–213. doi: 10.1002/jcc.1166. [DOI] [PubMed] [Google Scholar]
- 61.Allen F. The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallographica Section B. 2002;58(3 Part 1):380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 62.Bright Wilson JE, Decius JC, Cross PC. Molecular Vibrations: The Theory of Infrared and Raman Vibrational Spectra. Dover Publications; 1980. [Google Scholar]
- 63.Pulay P, Fogarasi G, Pang F, Boggs JE. Systematic ab initio gradient calculation of molecular geometries, force constants, and dipole moment derivatives. Journal Of The American Chemical Society. 1979;101(10):2550–2560. [Google Scholar]
- 64.Beachy MD, Chasman D, Murphy RB, Halgren TA, Friesner RA. Accurate ab Initio Quantum Chemical Determination of the Relative Energetics of Peptide Conformations and Assessment of Empirical Force Fields. Journal of the American Chemical Society. 1997;119(25):5908–5920. [Google Scholar]
- 65.Brooks C, Case DA. Simulations of peptide conformational dynamics and thermodynamics. Chemical Reviews. 1993;93(7):2487–2502. [Google Scholar]
- 66.Guvench O, MacKerell AD., Jr Automated conformational energy fitting for force-field development. J Mol Model. 2008;14(8):667–679. doi: 10.1007/s00894-008-0305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 68.MacKerell AD, Jr, Banavali N, Foloppe N. Development and current status of the CHARMM force field for nucleic acids. Biopolymers. 2000;56(4):257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 69.Feig M, MacKerell AD, Brooks CL. Force Field Influence on the Observation of π-Helical Protein Structures in Molecular Dynamics Simulations. The Journal of Physical Chemistry B. 2003;107(12):2831–2836. [Google Scholar]
- 70.Price DJ, Brooks CL., 3rd Modern protein force fields behave comparably in molecular dynamics simulations. J Comput Chem. 2002;23(11):1045–1057. doi: 10.1002/jcc.10083. [DOI] [PubMed] [Google Scholar]
- 71.Buck M, Bouguet-Bonnet S, Pastor RW, MacKerell AD., Jr Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys J. 2006;90(4):L36–38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guvench O, Qu CK, MacKerell AD., Jr Tyr66 acts as a conformational switch in the closed-to-open transition of the SHP-2 N-SH2-domain phosphotyrosine-peptide binding cleft. BMC Struct Biol. 2007;7:14. doi: 10.1186/1472-6807-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aleksandrov A, Simonson T. A molecular mechanics model for imatinib and imatinib:kinase binding. J Comput Chem. 2010;31(7):1550–1560. doi: 10.1002/jcc.21442. [DOI] [PubMed] [Google Scholar]
- 74.Gleeson MP, Deechongkit S, Ruchirawat S. Molecular dynamics investigation of psalmopeotoxin I. Probing the relationship between 3D structure, anti-malarial activity and thermal stability. J Mol Model. 2010 doi: 10.1007/s00894-010-0732-6. [DOI] [PubMed] [Google Scholar]
- 75.Best RB, Buchete NV, Hummer G. Are current molecular dynamics force fields too helical? Biophys J. 2008;95(1):L07–09. doi: 10.1529/biophysj.108.132696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.MacKerell AD, Wiorkiewicz-Kuczera J, Karplus M. An all-atom empirical energy function for the simulation of nucleic acids. Journal of the American Chemical Society. 1995;117(48):11946–11975. [Google Scholar]
- 77.Priyakumar UD, MacKerell AD. Base flipping in a GCGC containing DNA dodecamer: A comparative study of the performance of the nucleic acid force fields, CHARMM, AMBER, and BMS. Journal of Chemical Theory and Computation. 2006;2(1):187–200. doi: 10.1021/ct0501957. [DOI] [PubMed] [Google Scholar]
- 78.Priyakumar UD, Mackerell AD., Jr Atomic detail investigation of the structure and dynamics of DNA.RNA hybrids: a molecular dynamics study. J Phys Chem B. 2008;112(5):1515–1524. doi: 10.1021/jp709827m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Semenyuk A, Darian E, Liu J, Majumdar A, Cuenoud B, et al. Targeting of an interrupted polypurine:polypyrimidine sequence in Mammalian cells by a triplex-forming oligonucleotide containing a novel base analogue. Biochemistry. 2010;49(36):7867–7878. doi: 10.1021/bi100797z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heddi B, Foloppe N, Oguey C, Hartmann B. Importance of accurate DNA structures in solution: the Jun-Fos model. J Mol Biol. 2008;382(4):956–970. doi: 10.1016/j.jmb.2008.07.047. [DOI] [PubMed] [Google Scholar]
- 81.Priyakumar UD, MacKerell AD., Jr Role of the Adenine Ligand on the Stabilization of the Secondary and Tertiary Interactions in the Adenine Riboswitch. Journal of Molecular Biology. 2010;396(5):1422–1438. doi: 10.1016/j.jmb.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pan Y, MacKerell AD., Jr Altered structural fluctuations in duplex RNA versus DNA: a conformational switch involving base pair opening. Nucleic Acids Research. 2003;31(24):7131–7140. doi: 10.1093/nar/gkg941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.MacKerell AD, Jr, Nilsson L. Molecular dynamics simulations of nucleic acid-protein complexes. Current Opinion in Structural Biology. 2008;18(2):194–199. doi: 10.1016/j.sbi.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pande V, Nilsson L. Insights into structure, dynamics and hydration of locked nucleic acid (LNA) strand-based duplexes from molecular dynamics simulations. Nucleic Acids Res. 2008;36(5):1508–1516. doi: 10.1093/nar/gkm1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hart K, Nystrom B, Ohman M, Nilsson L. Molecular dynamics simulations and free energy calculations of base flipping in dsRNA. RNA. 2005;11(5):609–618. doi: 10.1261/rna.7147805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deng N-J, Cieplak P. Free Energy Profile of RNA Hairpins: A Molecular Dynamics Simulation Study. Biophysical Journal. 2010;98(4):627–636. doi: 10.1016/j.bpj.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Faustino I, Pérez A, Orozco M. Toward a Consensus View of Duplex RNA Flexibility. Biophysical Journal. 2010;99(6):1876–1885. doi: 10.1016/j.bpj.2010.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klauda JB, Brooks BR, MacKerell AD, Jr, Venable RM, Pastor RW. An ab initio study on the torsional surface of alkanes and its effect on molecular simulations of alkanes and a DPPC bilayer. J Phys Chem B. 2005;109(11):5300–5311. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 89.Jo S, Rui HA, Lim JB, Klauda JB, Im W. Cholesterol Flip-Flop: Insights from Free Energy Simulation Studies. Journal of Physical Chemistry B. 2010;114(42):13342–13348. doi: 10.1021/jp108166k. [DOI] [PubMed] [Google Scholar]
- 90.Rui HA, Im W. Protegrin-1 Orientation and Physicochemical Properties in Membrane Bilayers Studied by Potential of Mean Force Calculations. Journal of Computational Chemistry. 2010;31(16):2859–2867. doi: 10.1002/jcc.21580. [DOI] [PubMed] [Google Scholar]
- 91.Broemstrup T, Reuter N. How does proteinase 3 interact with lipid bilayers? Phys Chem Chem Phys. 2010;12(27):7487–7496. doi: 10.1039/b924117e. [DOI] [PubMed] [Google Scholar]
- 92.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ha SN, Giammona A, Field M, Brady JW. A revised potential-energy surface for molecular mechanics studies of carbohydrates. Carbohydr Res. 1988;180(2):207–221. doi: 10.1016/0008-6215(88)80078-8. [DOI] [PubMed] [Google Scholar]
- 94.Woods RJ. Three-dimensional structures of oligosaccharides. Curr Opin Struct Biol. 1995;5(5):591–598. doi: 10.1016/0959-440x(95)80049-2. [DOI] [PubMed] [Google Scholar]
- 95.Jeffrey GA, Saenger W. Hydrogen Bonding in Biological Structures. Springer-Verlag; Berlin: 1991. [Google Scholar]
- 96.Schmidt RK, Tasaki K, Brady JW. Computer Modeling Studies of the Interacton of Water with Carbohydrates. Journal of Food Engineering. 1994;22:43–57. [Google Scholar]
- 97.Kirschner KN, Woods RJ. Solvent interactions determine carbohydrate conformation. PNAS. 2001;98:10541–10545. doi: 10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hatcher E, Säwén E, Widmalm G, MacKerell JAD. Conformational properties of methyl β-maltoside and methyl α-and β-cellobioside disaccharides. J Phys Chem B. 2010 doi: 10.1021/jp109475p. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Young DC. Molecular Mechanics. John Wiley & Sons, Inc; 2009. [Google Scholar]
- 100.Wong CF, McCammon JA. Protein simulation and drug design. Adv Protein Chem. 2003;66:87–121. doi: 10.1016/s0065-3233(03)66003-1. [DOI] [PubMed] [Google Scholar]
- 101.Halgren TA. MMFF VII. Characterization of MMFF94, MMFF94s, and Other Widely Available Force Fields for Conformational Energies and for Intermolecular-Interaction Energies and Geometries. J Comp Chem. 1999;20:730–748. doi: 10.1002/(SICI)1096-987X(199905)20:7<730::AID-JCC8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 102.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing ofa general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 103.Ewig CS, Berry R, Dinur U, Hill J-R, Hwang M-J, et al. Derivation of Class II Force Fields. VIII. Derivation of a General Quantum Mechanical Force Field for Organic Compounds. J Comp Chem. 2001;22:1782–1800. doi: 10.1002/jcc.1131. [DOI] [PubMed] [Google Scholar]
- 104.Sun H. COMPASS: An ab Inito Force-Field Optimized for Condensed-Phase Applications-Overview with Details on Alkane and Benzene Compounds. J Phys Chem B. 1998;102:7338–7364. [Google Scholar]
- 105.Momany FA, Rone R. Validation of the General Purpose QUANTA 3.2/CHARMm Force Field. Journal of Computational Chemistry. 1992;13(7):888–900. [Google Scholar]
- 106.Hopfinger AJ, Wang S, Tokarski JS, Jin BQ, Albuquerque M, et al. Construction of 3D-QSAR models using the 4D-QSAR analysis formalism. Journal of the American Chemical Society. 1997;119(43):10509–10524. [Google Scholar]
- 107.Bernard D, Coop A, MacKerell AD., Jr 2D conformationally sampled pharmacophore: a ligand-based pharmacophore to differentiate delta opioid agonists from antagonists. J Am Chem Soc. 2003;125(10):3101–3107. doi: 10.1021/ja027644m. [DOI] [PubMed] [Google Scholar]
- 108.Reddy MR, Erion MD. Free Energy Calculations in Rational Drug Design. Springer -Verlag; [Google Scholar]
- 109.Price MLP, Ostrovsky D, Jorgensen WL. Gas-phase and liquid-state properties of esters, nitriles, and nitro compounds with the OPLS-AA force field. Journal of Computational Chemistry. 2001;22(13):1340–1352. [Google Scholar]
- 110.McDonald NA, Jorgensen WL. Development of an All-Atom Force Field for Heterocycles. Properties of Liquid Pyrrole, Furan, Diazoles, and Oxazoles. The Journal of Physical Chemistry B. 1998;102(41):8049–8059. [Google Scholar]
- 111.Lamoureux G, Jr, Roux B. A simple polarizable model of water based on classical Drude oscillators. Journal of Chemical Physics. 2003;119(10):5185–5197. [Google Scholar]
- 112.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 113.Harder E, Anisimov VM, Whitfield T, MacKerell AD, Jr, Roux B. Understanding the dielectric properties of liquid amides from a polarizable force field. J Phys Chem B. 2008;112(11):3509–3521. doi: 10.1021/jp709729d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Harder E, Mackerell AD, Jr, Roux B. Many-body polarization effects and the membrane dipole potential. J Am Chem Soc. 2009;131(8):2760–2761. doi: 10.1021/ja806825g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baker CM, Anisimov VM, MacKerell AD., Jr Development of CHARMM polarizableforce field for nucleic acid bases based on the classical Drude oscillator model. J Phys Chem B. 2011;115(3):580–596. doi: 10.1021/jp1092338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lamoureux G, Roux B. Absolute Hydration Free Energy Scale for Alkali and Halide Ions Established from Simulations with a Polarizable Force Field. J Phys Chem B. 2006;110:3308–3322. doi: 10.1021/jp056043p. [DOI] [PubMed] [Google Scholar]
- 117.Yu H, van Gunsteren WF. Accounting for polarization in molecular simulation. Computer Physics Communications. 2005;172(2):69–85. [Google Scholar]
- 118.Darley MG, Handley CM, Popelier PLA. Beyond Point Charges: Dynamic Polarization from Neural Net Predicted Multipole Moments. Journal of Chemical Theory and Computation. 2008;4(9):1435–1448. doi: 10.1021/ct800166r. [DOI] [PubMed] [Google Scholar]