Abstract
Functional MRI (fMRI) is a widely used tool for non-invasively measuring correlates of human brain activity. However, its use has mostly been focused upon measuring activity on the surface of cerebral cortex rather than in subcortical regions such as midbrain and brainstem. Subcortical fMRI must overcome two challenges: spatial resolution and physiological noise. Here we describe an optimized set of techniques developed to perform high-resolution fMRI in human SC, a structure on the dorsal surface of the midbrain; the methods can also be used to image other brainstem and subcortical structures.
High-resolution (1.2 mm voxels) fMRI of the SC requires a non-conventional approach. The desired spatial sampling is obtained using a multi-shot (interleaved) spiral acquisition1. Since, T2* of SC tissue is longer than in cortex, a correspondingly longer echo time (TE ~ 40 msec) is used to maximize functional contrast. To cover the full extent of the SC, 8-10 slices are obtained. For each session a structural anatomy with the same slice prescription as the fMRI is also obtained, which is used to align the functional data to a high-resolution reference volume.
In a separate session, for each subject, we create a high-resolution (0.7 mm sampling) reference volume using a T1-weighted sequence that gives good tissue contrast. In the reference volume, the midbrain region is segmented using the ITK-SNAP software application2. This segmentation is used to create a 3D surface representation of the midbrain that is both smooth and accurate3. The surface vertices and normals are used to create a map of depth from the midbrain surface within the tissue4.
Functional data is transformed into the coordinate system of the segmented reference volume. Depth associations of the voxels enable the averaging of fMRI time series data within specified depth ranges to improve signal quality. Data is rendered on the 3D surface for visualization.
In our lab we use this technique for measuring topographic maps of visual stimulation and covert and overt visual attention within the SC1. As an example, we demonstrate the topographic representation of polar angle to visual stimulation in SC.
Keywords: Neuroscience, Issue 63, fMRI, midbrain, brainstem, colliculus, BOLD, brain, Magentic Resonance Imaging, MRI
Protocol
1. Polar-angle Topography Stimulus and Psychophysics
To obtain a polar-angle retinotopic map in the SC, we use a 90° wedge of moving dots as the stimulus (eccentricity 2-9° of visual angle, mean dot-speed 4° / sec) (Fig. 1). It is known that activity in the SC is enhanced by applying covert attention5, hence we use an attention task in our paradigm to increase available signal. On each 2 s trial, subjects are instructed to covertly attend to the entire wedge and perform a speed discrimination task while maintaining fixation. The wedge is divided into 2 × 3 virtual sectors with dots in one of the sectors, randomly chosen on each trial, moving slower or faster than all other dots. After each trial, the wedge is rotated 30° around fixation so that the stimulus rotates fully with a 24 sec period. Each run consists of 9.5 rotations of the stimulus (228 sec), and experimental sessions include 16-18 runs.
To maintain subject's performance on this task throughout the duration of each run, the difficulty of the task is adjusted using two randomly interleaved two-up-one-down staircases. After every two consecutive correct trials the speed difference is reduced by 8%, and for every incorrect trial, the difference is increased by 8%.
Prior to scanning, all subjects practice the visual task outside the scanner until they achieve a stable level of performance; this requires 3-4 20 minute duration practice sessions. Typical discrimination thresholds are in the range of 1°-1.5° / sec.
2. Subject Preparation
Subjects heads are secured with pads to minimize head motion before they are positioned inside the scanner bore. At these high spatial resolutions, fMRI is particularly sensitive to motion artifacts, so head stabilization is critical.
Subjects are given an MRI-compatible button pad in one hand and instructed about which button to press to indicate their judgments about dot speed.
3. Localizing and Prescribing the SC
Human SC is a small but distinct structure, ~9 mm in diameter, located on the roof of the midbrain. When using a small number of fMRI slices, multiple localizer imaging series are required for its precise localization. We run these series along sagittal, axial, and coronal planes.
We then use these localizer images to precisely prescribe the SC with 8-10 contiguous slices, 1.2 mm thick, field-of-view (FOV), 170 mm in an oblique, quasi-axial plane.
Next, high-resolution T1-weighted structural images are obtained using a three-dimensional (3D) RF-spoiled GRASS (SPGR) sequence (15° flip angle, 1.2 mm voxels) once prior to collecting the functional data and once after. These images are used as a reference to align the fMRI data to a high-resolution structural reference volume obtained in a separate session that we describe later.
4. Functional MRI Parameters
All imaging was performed on a GE Signa HD12 3T MRI scanner using the GE-supplied 8-channel, head coil. Excitation was a 6.4 ms windowed-sinc pulse applied using the scanner body coil.
To obtain 1.2 mm sampling in human SC, we use a three-shot spiral-trajectory acquisition6,7. Three shots are needed for several reasons. In particular, single-shot acquisition for our scanner and FOV requires > 77 msec, too long to be practical. The multiple shots are combined together after correction by subtracting the initial value and linear trend of the phase. TE is incremented by 2 ms on the first frame to estimate a field map from the first two volumes acquired, and this map is used for linear correction. Reconstructed images had an SNR of ~20. Temporal power spectra in SC voxels typically showed little of the structure associated with physiological noise; the use of a 3-shot acquisition had a strong filtering effect on the comparatively high-frequency effects of cardiac pulse and respiration. Other noise-reduction techniques are problematic in this high-resolution context. For example, retrospective correction methods such as RETROICOR6 are not applicable to multi-shot data, and cardiac gating introduces noise and artifact associated with disturbance of T1 equilibrium.
Echo time, TE = 40 msec, is longer than typically used in cortex (30 msec) because we measured a correspondingly longer T2* in SC tissue (~60 msec) than observed in cortical gray matter (~45 msec).
Acquisition bandwidth is limited to 62.5 kHz to reduce peak gradient current that causes unwanted heating on our scanner. We choose TR = 1 sec, so with three shots a volume is acquired every 3 sec.
5. Structural MRI and 3D Modeling
In a single separate session for each subject, we obtain a high-resolution (0.7 mm sampling) reference volume using a T1-weighted sequence that gives good tissue contrast (3D SPGR, 15° flip angle, inversion prepared with TI = 450 msec, 2 excitations, ~28 minute duration, 0.7 mm voxels).
In this reference volume, we segment the tissue of the midbrain, brainstem, and portions of the thalamus (Fig. 2A) using a combination of automatic and manual techniques provided by the ITK-SNAP application2. In particular, we use an automatic segmentation tool in which the user seeds multiple points within each brainstem; the software then automatically expands the segmentation around the seed points constrained within a region defined by contrast and intensity criteria. This automatic segmentation is then adjusted, if necessary, using manual, "paint-like" voxel tools.
The cerebrospinal fluid-tissue interface of the SC is interpolated from the segmentation using isodensity surface tessellation, and this initial surface is refined to reduce aliasing artifacts (Fig. 2, B and D) using a variational deformable-surface algorithm3. This surface provides vertices and outward normal vectors to be used as a reference for the laminar calculations (described below) as well as a means to visualize functional data.
6. Image Analysis
To analyze the fMRI data, we use the mrVista software package (available for download at http://white.stanford.edu/mrvista.php) as well as tools developed on the mrVista framework in our lab. In the next few steps we use standard mrVista package tools:
Initialize the session in mrVista, choosing the option to spatially normalize the intensity of the averaged data to reduce the effects of coil inhomogeneity. The normalization uses a homomorphic method, that is, dividing by a low-pass filtered version of the temporally averaged volume image intensities with an additive robust correction for estimated noise. The first half cycle of images (12 sec) is discarded to avoid transient MR equilibrium and hemodynamic effects.
Correct within-run motion. Motion is estimated from the time series of volumes. However, because of the relatively low signal-to-noise ratio (SNR) of the images, we first perform a 5-sample boxcar smoothing upon the time series. Each volume is then registered to the average of the last 5 samples. Note that smoothing is only used to estimate the motion, and the actual data is not smoothed.
Form temporal averages of each run's motion-corrected image data, and use these averages to correct the between-run motion using the last run as reference.
Perform a slice-timing correction. We use sequential slice acquisition, so errors due to performing timing correction after the motion correction will be small (~125 msec).
Average the multiple runs recorded within each session to improve SNR.
Align the structural data from the fMRI session to the reference structural volume using a robust intensity-based registration algorithm8. Load the alignment and segmentation into mrVista.
Transform the functional time series data to the segmented reference volume. In the following steps we use tools developed in our lab to perform further analysis.
Compute a distance map by calculating the distance between each SC tissue voxels and its nearest vertex on the SC surface. These distances are used to measure laminar position within the reference volume.
Perform a laminar segmentation process to enable depth averaging of time-series data to improve the SNR. Small (1.6 mm diameter) disks of tissue are associated with each vertex of the surface model along the entire superficial SC surface, and each disk is then extended both inward and outward from the SC tissue using the local surface normals to form an individual laminar neighborhood4.
For each point on the SC surface, we use these laminar associations to average the time series over a specified depth range. Since, visually responsive neurons are primarily present in the superficial layers of the SC, for the visual stimulation experiment we used a depth range of 0-1.8 mm.
7. Topographic Mapping Analysis
For analyzing the topographic representation of the data, Coherence analysis is performed on the depth-averaged time series by fitting a sinusoid at the stimulus repetition frequency (i.e., 1/24 Hz for the 24 sec stimulus described above) for each voxel. From this fit, we derive surface maps of response amplitude, coherence, and phase. This analysis is performed in the frequency domain, and is a common technique to quantify retinotopic maps in visual cortex9,10.
The phase of the sinusoidal fit measures the position of the stimulus. Zero-phase corresponds to the upper vertical meridian (Fig. 3). The stimulus then rotates clockwise, so a π/2 phase corresponds to when the stimulus has rotated to the horizontal meridian in the right visual field. After π radians of phase, the stimulus crosses into the left visual field, and so forth.
We also obtained boundaries of the entire superficial extent of the SC using manual inspection of the high-resolution T1-weighted volume anatomy. These boundaries are marked in Fig. 3 by red dashed lines.
fMRI does not directly measure neural activity, but rather a blood-flow response that is tightly but sluggishly coupled to neural activity. This adds a hemodynamic delay to the phase response. The delay is estimated by taking all voxels above our coherence threshold from each SC region-of-interest, and centering their means around π in the complex plane. In our SC data, these delays are quite small, of the order of 2-4 sec. We remove this delay by rotating the colormap counter-clockwise by 45 degrees (corresponding to 3 sec) (Fig. 3).
8. Representative Results
Phase data overlaid on a 3D surface of the SC (Fig. 3) shows that the response to visual stimulation is represented contralaterally in the SC, i.e., the left visual field is represented on the right SC and vice-a-versa.
There is also a retinotopic organization of the activity. The right upper visual field is represented medially on the left colliculus (blue-magenta), and the lower field is represented laterally (red-yellow). Similarly, the left upper visual field is represented medially on the right colliculus (blue-cyan) and lower is represented laterally (green-yellow).
This topography is consistent with the results of non-human primate studies of SC responses: 1) to visual stimulation using single-unit electrophysiology, and 2) the mapping of microstimulation induced saccadic eye movements11,12.
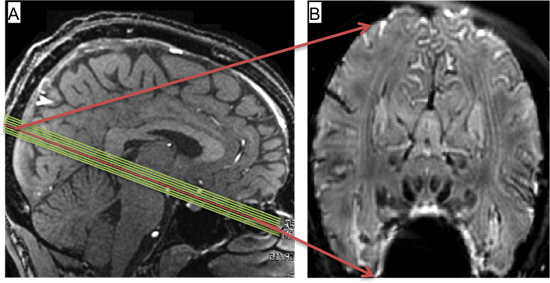 Figure 1. A) Slice prescription viewed on mid-sagittal image. B) Temporal mean (from 1-run) of functional images obtained from central slice marked in red in A).
Figure 1. A) Slice prescription viewed on mid-sagittal image. B) Temporal mean (from 1-run) of functional images obtained from central slice marked in red in A).
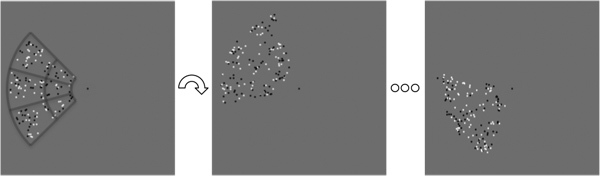 Figure 2. Polar-angle topography stimulus. A 90° wedge of moving black-and-white dots on a gray background rotated slowly around fixation. The wedge was divided into an array of 6 virtual sectors (gray lines added to emphasize sectors) to allow the subject to perform a speed discrimination task in a randomly chosen sector.
Figure 2. Polar-angle topography stimulus. A 90° wedge of moving black-and-white dots on a gray background rotated slowly around fixation. The wedge was divided into an array of 6 virtual sectors (gray lines added to emphasize sectors) to allow the subject to perform a speed discrimination task in a randomly chosen sector.
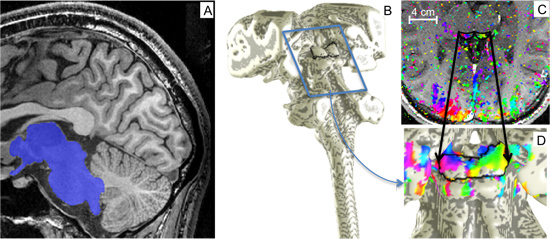 Figure 3. Segmentation and surface modeling. A) The midbrain, brainstem, and portions of the thalamus were segmented from high-resolution MRI anatomy volumes. B) A surface was created at the edge of segmented region. C) Sinusoidal-fit phase data viewed on an inplane slice (coherence > 0.25). D) A rotated and enlarged view of the brainstem surface model was used to visualize phase data on the SC.
Figure 3. Segmentation and surface modeling. A) The midbrain, brainstem, and portions of the thalamus were segmented from high-resolution MRI anatomy volumes. B) A surface was created at the edge of segmented region. C) Sinusoidal-fit phase data viewed on an inplane slice (coherence > 0.25). D) A rotated and enlarged view of the brainstem surface model was used to visualize phase data on the SC.
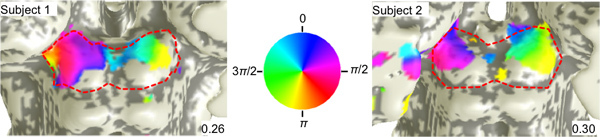 Figure 4. Polar-angle maps. fMRI phase maps in two subjects that encode visual polar angle. Coherence thresholds for each map are provided on the bottom right. The color wheel relates the overlaid phases of the stimulus in polar angle to their visual field positions.
Figure 4. Polar-angle maps. fMRI phase maps in two subjects that encode visual polar angle. Coherence thresholds for each map are provided on the bottom right. The color wheel relates the overlaid phases of the stimulus in polar angle to their visual field positions.
Discussion
Our acquisition and data-analysis techniques enable the measurement of neural activity in subcortical human brain structures at high resolution (1.2 mm voxels). The 3-shot spiral acquisition reduces the physiological noise that is particularly detrimental to fMRI measurements around the midbrain. In addition, our laminar segmentation of the tissue allows us to perform depth averaging of the data that helps improve the SNR. We have used these methods to show precise polar-angle topographic maps of visual stimulation and covert visual attention in the human SC1. The laminar segmentation also enables the analysis of depth profiles of functional activity that vary under experimental control1.
Our imaging methods open up new avenues for neuroscience experimentation in human subcortical structures. These methods can enable translation of fine-scale research done on animals in subcortical areas to humans, e.g., investigating the organization of auditory responses in structures such as the inferior colliculus and the cochlear nucleus13-15, or visual and multisensory responses in thalamic nuclei such as pulvinar16,17. Finally, these techniques could provide functional localization for small structures such as the subthalamic nucleus and globus pallidus, which are often targets for deep brain stimulation in patients with Parkinson's disease, dystonia or chronic pain18-21.
Disclosures
No conflicts of interest declared.
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant BCS 1063774.
References
- Katyal S, Zughni S, Greene C, Ress D. Topography of covert visual attention in human superior colliculus. Journal of Neurophysiology. 2010;104:3074–3083. doi: 10.1152/jn.00283.2010. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Xu G, Pan Q, Bajaj CL. Discrete Surface Modeling Using Partial Differential Equations. Computer Aided Geometric Design. 2006;23:125–145. doi: 10.1016/j.cagd.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ress D, Glover GH, Liu J, Wandell B. Laminar profiles of functional activity in the human brain. NeuroImage. 2007;34:74–84. doi: 10.1016/j.neuroimage.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Schneider KA, Kastner S. Effects of sustained spatial attention in the human lateral geniculate nucleus and superior colliculus. J. Neurosci. 2009;29:1784–1795. doi: 10.1523/JNEUROSCI.4452-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH. Simple analytic spiral K-space algorithm. Magn. Reson. Med. 1999;42:412–415. doi: 10.1002/(sici)1522-2594(199908)42:2<412::aid-mrm25>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Glover GH, Lai S. Self-navigated spiral fMRI: interleaved versus single-shot. Magn. Reson. Med. 1998;39:361–368. doi: 10.1002/mrm.1910390305. [DOI] [PubMed] [Google Scholar]
- Nestares O, Heeger DJ. Robust multiresolution alignment of MRI brain volumes. Magn. Reson. Med. 2000;43:705–715. doi: 10.1002/(sici)1522-2594(200005)43:5<705::aid-mrm13>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Engel SA, Glover GH, Wandell BA. Retinotopic organization in human visual cortex and the spatial precision of functional MRI. Cereb. Cortex. 1997;7:181–192. doi: 10.1093/cercor/7.2.181. [DOI] [PubMed] [Google Scholar]
- Schneider KA, Kastner S. Visual responses of the human superior colliculus: a high-resolution functional magnetic resonance imaging study. Journal of Neurophysiology. 2005;94:2491–2503. doi: 10.1152/jn.00288.2005. [DOI] [PubMed] [Google Scholar]
- Cynader M, Berman N. Receptive-field organization of monkey superior colliculus. Journal of Neurophysiology. 1972;35:187–201. doi: 10.1152/jn.1972.35.2.187. [DOI] [PubMed] [Google Scholar]
- Robinson DA. Eye movements evoked by collicular stimulation in the alert monkey. Vision Research. 1972;12:1795–1808. doi: 10.1016/0042-6989(72)90070-3. [DOI] [PubMed] [Google Scholar]
- Schreiner CE, Langner G. Laminar fine structure of frequency organization in auditory midbrain. Nature. 1997;388:383–385. doi: 10.1038/41106. [DOI] [PubMed] [Google Scholar]
- Baumann S. Orthogonal representation of sound dimensions in the primate midbrain. Nature Neuroscience. 2011;14:423–425. doi: 10.1038/nn.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmierca MS. A discontinuous tonotopic organization in the inferior colliculus of the rat. J. Neurosci. 2008;28:4767. doi: 10.1523/JNEUROSCI.0238-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender D. Retinotopic organization of macaque pulvinar. Journal of Neurophysiology. 1981;46:672. doi: 10.1152/jn.1981.46.3.672. [DOI] [PubMed] [Google Scholar]
- Grieve KL, Acuña C, Cudeiro J. The primate pulvinar nuclei: vision and action. Trends in Neurosciences. 2000;23:35–39. doi: 10.1016/s0166-2236(99)01482-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Oroz MC. The subthalamic nucleus in Parkinson's disease: somatotopic organization and physiological characteristics. Brain. 2001;124:1777. doi: 10.1093/brain/124.9.1777. [DOI] [PubMed] [Google Scholar]
- Romanelli P. Microelectrode recording revealing a somatotopic body map in the subthalamic nucleus in humans with Parkinson disease. Journal of Neurosurgery. 2004;100:611–618. doi: 10.3171/jns.2004.100.4.0611. [DOI] [PubMed] [Google Scholar]
- DeLong MR, Crutcher MD, Georgopoulos AP. Primate globus pallidus and subthalamic nucleus: functional organization. Journal of Neurophysiology. 1985;53:530. doi: 10.1152/jn.1985.53.2.530. [DOI] [PubMed] [Google Scholar]
- Houeto JL. Acute deep-brain stimulation of the internal and external globus pallidus in primary Dystonia functional mapping of the pallidum. Archives of Neurology. 2007;64:1281–1286. doi: 10.1001/archneur.64.9.1281. [DOI] [PubMed] [Google Scholar]


