Abstract
Viral particles are biological machines that have evolved to package, protect, and deliver the viral genome into the host via regulated conformational changes of virions. We have developed a procedure to modify lysine resides with S-methylthioacetimidate (SMTA) across the pH range from 5.5 to 8.5. Lysine residues that are not completely modified are involved in tertiary or quaternary structural interactions, and their extent of modification can be quantified as a function of pH. This procedure was applied to the pH-dependent structural transitions of Brome Mosaic Virus (BMV). As the reaction pH increases from 5.5 to 8.5, the average number of modified lysine residues in the BMV capsid protein increases from six to twelve, correlating well with the known pH dependent swelling behavior of BMV virions. The extent of reaction of each of the capsid protein’s lysine residues has been quantified at eight pH values using coupled liquid chromatography-tandem mass spectrometry. Each lysine can be assigned to one of three structural classes identified by inspection of the BMV virion crystal structure. Several lysine residues display reactivity that indicates their involvement in dynamic interactions that are not obvious in the crystal structure. The influence of several capsid protein mutants on the pH-dependent structural transition of BMV has also been investigated. Mutant H75Q exhibits an altered swelling transition accompanying solution pH increases. The H75Q capsids show increased reactivity at lysine residues 64 and 130, residues distal from the dimer interface occupied by H75, across the entire pH range.
Keywords: Brome Mosaic Virus, virion structure, pH dependence, chemical modification, mass spectrometry, Lysine, Conformational Change
Introduction
Viral particles are biological machines whose structure has evolved to package, protect, and deliver the viral genome into the host.1–2 The close integration of virion structure and function underscores the need to study virion architecture and the structural transitions that regulate the viral life cycle.3–8 There is also considerable recent interest in studying virion structure to advance the nano engineering of viral particles for the delivery of drugs and molecular therapeutics.9–13
Viral particles are dynamic, flexible structures that undergo conformational changes across a wide range of time scales.3–4 The large scale, slow conformational changes often associated with virion assembly, maturation, or infection can be kinetically trapped and characterized crystallograpically or by electron microscopic image reconstruction.4 Virions also show rapid fluctuations around an equilibrium structure that have been described as “breathing” vibrational modes.4,14 Conformational changes on both time scales can be studied using labeling techniques such as hydrogen-deuterium exchange (HDX) and covalent modification.15–19 Covalent modification techniques can either decrease the masses of proteins, as is seen with limited proteolysis to remove flexible “loops and fringes” of a protein 20–22, or increase protein masses, as in selective modification of specific amino acid side chains.23–31 In contrast to HDX, covalent modifications are not labile on the timescale of the analysis, thus minimizing back exchange of the label and reducing complexities in data interpretation. Side chain reactivity can be affected by secondary, tertiary and quaternary structure, and the patterns of reactivity displayed by a protein can be used to infer structural features and conformational changes.23 An additional advantage of covalent modification procedures is that hydrolytic enzymes and side chain modifying reagents have well characterized residue selectivities that allow additional sequence information to be derived from mass spectrometric analysis of modified proteins or peptides.23–26
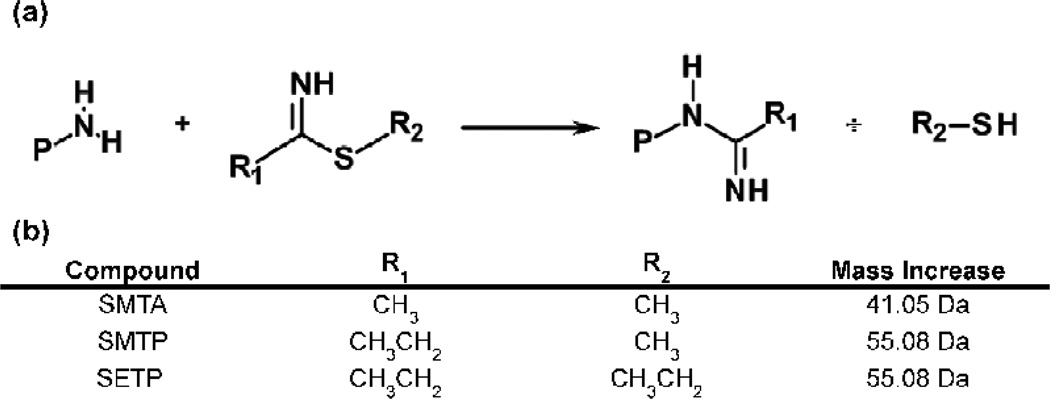
Thioimidates modify amino groups according to Figure 1a, resulting in the mass increases summarized in Figure 1b. We have used thioimidate reagents such as S-methylthioacetimidate (SMTA) to probe the structure of soluble proteins and the proteins of the bacterial ribosome.32–38 Lysines that are protected from SMTA modification are buried at the interface of protein-protein or protein-nucleic acid interactions, or are involved in ionic orhydrogen bonding interactions with other amino acid residues.33–34,36,38 The average extent of modification of proteins shows excellent agreement with predictions based on crystal structures, indicating minimal effects of native biomolecular structure.33–38
Fig. 1.
(a) Modification of protein or peptide amino groups by the thioimidate reagents described in the text. (b) The structural features each reagent and the mass increase per modified amino group.
Brome Mosaic Virus (BMV) was chosen to demonstrate the application of SMTA labeling to a dynamic macromolecular complex because of its simplicity (180 copies of a single capsid protein), the availability of a high-resolution crystal structure of the virion,39 and its high sequence homology (70% sequence identity) to Cowpea Chlorotic Mottle Virus (CCMV). High sequence homology to CCMV allows us to use crystallographic and electron microscopic structural models of CCMV virions to interpret our results from BMV.40 Both viruses undergo a pH-dependent swelling transition that results in a 10% increase in the virion’s 28 nm diameter as the solution pH increases from 5.0 to 8.0, with a transition midpoint at pH 6.5.15,39–41 The swelling transition was first characterized by analytical ultracentrifugation.42 Subsequent research described the sensitivity of the virion’s RNA and protein components to nuclease and protease digestion as a function of pH,42–45 and the role of divalent cation binding to virions.42,46 More recently, the CCMV swelling transition was the subject of a molecular modeling study that compared in silico structural parameters to previous in vitro results.47 The generally accepted mechanism of bromovirus swelling involves the loss of divalent metal ions ligated by clusters of glutamic acid residues and consequent structural expansion driven by charge-charge repulsion.39,41,46 Tama and Brooks’ molecular dynamics study indicated that the pKa of CCMV residue E81 (homologous to BMV E80) was elevated in the virion’s compact form relative to more expanded swelling intermediates (pKa = 7.2–8.5 in the compact virion vs. pKa = 4.0–4.5 in the most expanded intermediate), implicating this residue as a keystone in the structural transition.47 Assuming a typical pKa of 10.5,48 the BMV CP’s 12 lysines are unlikely to be directly involved in the transition. However their distribution on the exterior and interior surfaces of the virion and at the interfaces between capsid protein monomers makes their SMTA reactivity a useful readout of virion quaternary structure as a function of pH. The present report demonstrates that the chemical reactivity of lysine residues at the interface of capsid subunits is an accurate monitor of the pH-dependent structural transition of BMV virions. To further illustrate the utility of SMTA modification, we applied our labeling procedure to mutant virion that has an altered pH dependent swelling transition. The results demonstrate that a residue at a protein-protein interface in a virion’s structure can influence the conformations of distal residues or the global flexibility of the entire virion.
Results
BMV virions demonstrate pH-dependent SMTA reactivity
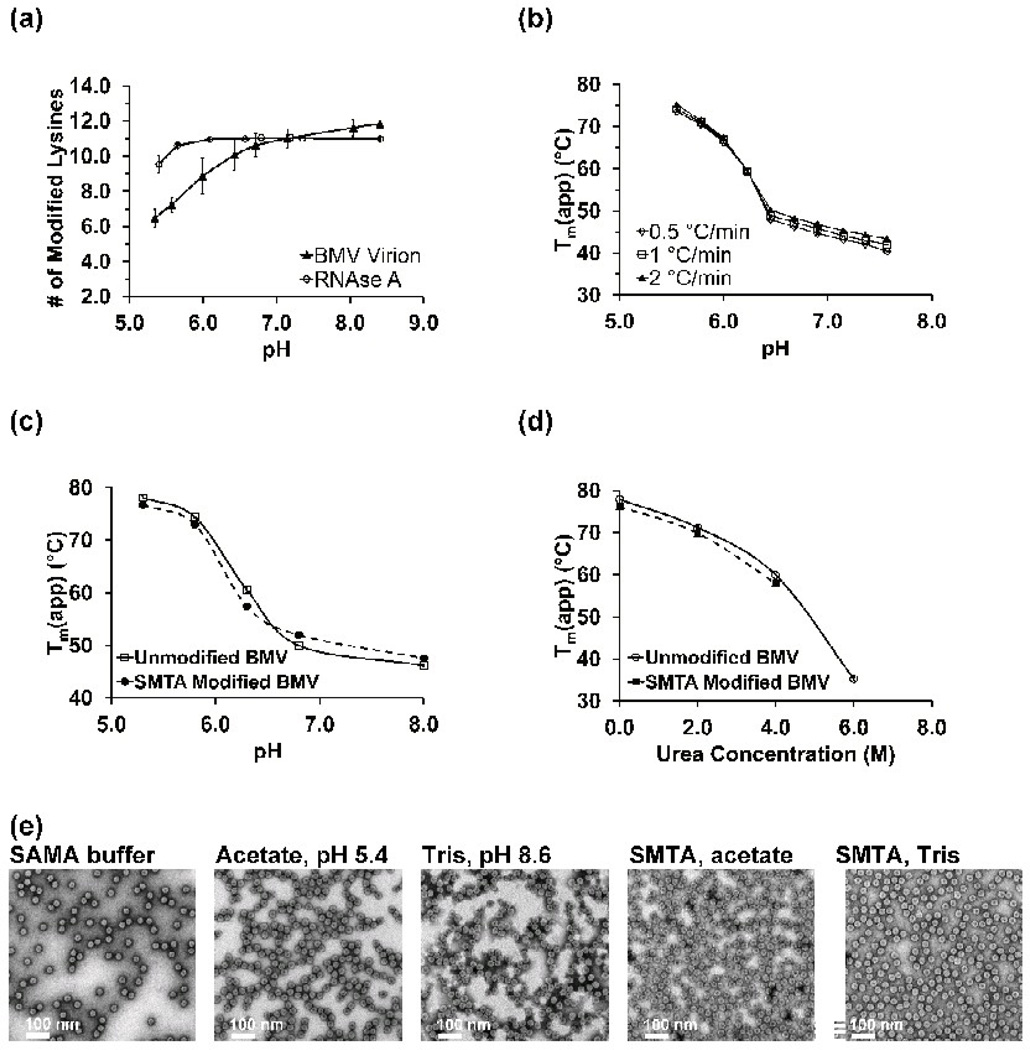
Thioimidate reagents have a lower tendency to engage in unproductive side reactions compared to oxygen-containing analogs like O-methyacetimidate.49–51 However, thioimidates can still be hydrolyzed to unreactive amide and thiol products. Protonation of the imine (pKa ~ 7.0) increases the rate of hydrolysis,52–54 which would compete directly with the rate of lysine modification and decrease the SMTA concentration. Consequently, the observed pH-dependence of the extent of lysine modification would contain contributions from both protein structure and SMTA hydrolysis. In order to control for the effect of hydrolysis of SMTA, conditions that allowed complete modification of protein amino groups across the pH range from 5.4 to 8.5 were developed. The ten lysines and amino terminus of bovine pancreatic RNase A all appear to be surface accessible54 and the protein exists predominately as a monomer in solution at the concentrations used for our experiments (2.5 µM, 0.03 g/L).55–56 When RNase A is subjected to 10 cycles of modification between pH 5.7 and 8.4, the result is nearly complete modification of all eleven of the protein’s amino groups across the entire pH range(Fig. S1 and Fig. 2a, open circles).The deconvoluted spectrum of RNase A reacted with SMTA at pH 5.4 (Fig. S1A) shows a distribution of labeled forms of the protein ranging from 7 to 11 amidinated lysine residues. At pH 5.7, the distribution has converged to either 10 or 11 amidinated lysines (Fig. S1B). At pH values higher than 5.7, complete modification was observed (Fig S1C and S1D). After 10 cycles of modification, the only indication of pH dependence for RNase A’s SMTA reactivity is at low pH values: as the pH decreases from 5.6 to 5.4, the average extent of modification decreases by 1 lysine residue (Fig 2a, open circles). This decrease is most likely the effect completion between hydrolysis of the SMTA and lysine modification.
Fig. 2.
(a) The pH dependence of the extent of amidination of RNase A and the capsid protein of the BMV virion. The values on the ordinate axis are intensity weighted averages of the extent of modification. Error bars are the standard deviations of three determinations. (b) Apparent melting points (Tm(app)) of wild-type BMV virions as a function of pH for three heating rates. Each point is the average of three determinations. (c) Melting points of unmodified (open circles) and SMTA-modified (closed squares) BMV virions as a function of pH. (d) Melting points of unmodified (open circles) and SMTA-modified (closed squares) BMV virions as a function of urea concentration. (e) Electron micrographs of unmodified and SMTA-modified virions. Virions labeled “acetate, pH 5.4” and “Tris, pH 8.6” were subjected to 10 cycles of buffer exchange with no SMTA. Virions labeled “SMTA, acetate” or “SMTA, Tris” were subjected to 10 cycles of SMTA modification as described in the text.
The same reaction conditions developed for RNase A were applied to BMV virions, whose capsid protein contains twelve lysine residues and a post-translationally acetylated N-terminus, 57 the average number of modified lysines in the BMV capsid protein increases from six to twelve (Fig. S2 and Fig. 2a, closed triangles). Significantly, as the pH increases from 5.4 to 8.5 an inflection point appears in the data at around pH 6.5. These changes in the SMTA reactivity of capsid protein lysines as a function of pH suggests that lysine modification by SMTA can be used to probe of the relative solvent accessibility of the capsid protein structure during the pH-dependent swelling transition as long as chemical modification of the lysines does not significantly alter fundamental properties of the capsid protein.
Virion structure is preserved after SMTA modification
Differential scanning fluorimetry (DSF), a method to examine the thermal denaturation profile of proteins,58 and electron microscopy results demonstrate that the structure of BMV virions is essentially unchanged by amidination. Wild-type virions exhibit a sharp decrease in stability as a function of increasing pH, with an inflection point at pH ~6.5 (Fig. 2b). When the Tm(app) curve of unmodified virions (Fig. 2c, open circles) is compared to SMTA-modified virions (Fig. 2c, closed squares), differences between the two curves are minimal. First derivative plots from raw melting point data for unmodified and SMTA-derivatized virions are presented in Supplemental Figures S3 and S4, respectively. Modified virions exhibit a 2 °C decrease in Tm(app) at low pH that is likely due to a general loosening of the virion structure, a phenomenon that has been observed in other chemically modified viral capsids.59–60 A modest 2 °C increase in the melting point for SMTA-modified virions at high pH values as also observed, likely due to the more intense positive charge conferred by the amidinated lysine’s elevated pKa (estimated to be between 12.5– 1350–51), which could increase virion stability by enhancing either protein-protein61 or protein-RNA62 interactions. To compare RNA binding by the WT and SMTA-modified BMV capsid proteins, a fluorescence titration of a well characterized ligand, fluoresce in-labeled B-Box stem loop RNA6–7 was performed. Unmodified or amidinated capsid protein give nearly identical protein-RNA dissociation constants (0.13 µM for unmodified vs. 0.11 µM for amidinated CP), as shown in Figure S5. These results suggest that the stabilizing effect of amidination is due to enhanced protein-protein interactions. Previous fluorescence anisotropy titration experiments determined a KD value for B Box binding to WT capsid protein of 449 nM.6 However, our Kd determination of the unmodified and modified was made in parallel and the small difference suggest that the stabilizing effect of amidination is due to enhanced protein-protein interactions. To further demonstrate the minimal structural effect of SMTA modification, the Tm(app) of virions was determined as a function of urea concentration. The nearly identical decreases of unmodified and SMTA-derivatized virions’ Tm(app) (Fig. 2d, open circles and close squares, respectively) to increasing urea concentrations again demonstrates that the structural effects of lysine amidination are minimal. To put the changes observed in these analyses in context, virions containing capsid protein mutants with single amino acid substitutions of key residues or empty virus like particles result in 5 to 15°C decreases in BMV virion Tm(app) values (Fig. S6 and Ref. 63). The series of electron micrographs in Figure 2(e) demonstrate that there are no gross structural differences between virions taken directly from SAMA storage buffer, virions that were repeatedly diluted and concentrated in either acetate or Tris buffer, or virions reacted with SMTA at pH 5.4 or 8.6.
SMTA reactivity of lysine residues in the BMV virion
In order to map the locations of SMTA modified lysine residues in BMV virions, modified virions were proteolytically digested and the resulting peptide mixtures were analyzed by LC-MS/MS. Extracted ion chromatograms for identified peptides were integrated to yield intensities for each peptide in a digest sample, and the resulting data were used to determine the extent of labeling of each lysine within the capsid protein as a function of pH. Data on the extent of amidination of BMV capsid protein lysines as a function of pH are shown in Figure S8. Amidination of lysine residues eliminates tryptic digest sites in a protein50, resulting in digest mixtures containing progressively larger peptides. Manual integration of the TIC data is a partial remedy, as discussed below, but inevitably information on some portions of the protein is lost. Digestion of modified BMV virions with chymotrypsin is a better solution, and these data are discussed below. Results from the analysis of tryptic and chymotryptic digests are presented in Table 1. Peptides from all enzymatic digestions were propionamidinated to maintain uniform ionization efficiency. Propionamidinated lysines have a 14 Da higher mass than acetamidinated lysines, allowing unambiguous identification of residues protected from SMTA modification in the native virion (Fig. 1b). Although the peptide Acetyl-STSGTGKMTR with an amidination at K8 was detected irregularly, quantitatively reliable labeling data could not be obtained for K8, and results of this residue’s reactivity are not included in this work. Figures S9 and S10 in the supplemental material illustrate the distribution of tryptic and chymotryptic cleavage sites in the BMV capsid protein sequence, respectively. Partial lists of tryptic and chymotryptic peptide sequences are tabulated in Tables S4 and S5, respectively, along with the peptides predicted unmodified and modified mass-to-charge ratios.
Table 1.
Extent of amidination of lysines of the BMV capsid protein. The extent of SMTA modification is quantified as a percentage of peptides containing the indicated lysine with an amidination out of all identified peptides containing that lysine. Mean values and standard deviations (in parenthesis) are the averages of three to five determinations.
| Tryptic Data | % amidinated lysine (standard deviation) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lysine | Classificationb | pH 5.4 | pH 5.6 | pH 6.0 | pH 6.5 | pH 6.7 | pH 7.2 | pH 8.0 | pH 8.5 |
| 8a | Internal Surface | - | - | - | - | - | - | - | - |
| 41 | Internal Surface | 58.9 (11.2) | 63.3 (16.3) | 80.4 (6.8) | 91.1 (5.2) | 93.8 (4.4) | 94.0 (3.0) | 85.2 (5.0) | 88.1 (1.8) |
| 44 | Internal Surface | 50.2 (3.8) | 49.3 (14.1) | 75.6 (8.1) | 65.4 (24.8) | 85.0 (9.9) | 76.5 (7.8) | 82.4 (0.6) | 72.6 (8.4) |
| 53 | Interface | 6.9 (12.4) | 3.3 (5.4) | 2.7 (1.8) | 10.9 (15.1) | 0.1 (0.2) | 1.8 (2.5) | 36.7 (14.3) | 34.2 (17.4) |
| 64 | External Surface | 24.9 (29.0) | 34.4 (33.7) | 86.5 (3.0) | 93.9 (10.0) | 93.4 (1.2) | 98.7 (1.2) | 74.9 (7.1) | 84.1 (12.6) |
| 81 | Interface | 8.4 (4.3) | 21.7 (12.9) | 39.1 (4.3) | 44.3 (50.4) | 91.8 (10.9) | 97.8 (1.4) | 99.1 (1.7) | 98.0 (3.2) |
| 83 | External Surface | 13.3 (8.7) | 25.3 (21.9) | 83.1 (14.2) | 82.7 (24.3) | 94.2 (6.4) | 98.9 (0.7) | 100.0 (0.0) | 99.3 (1.1) |
| 86 | Internal Surface | 21.2 (20.8) | 22.1 (11.1) | 80.8 (11.1) | 100.0 (0.0) | 96.1 (7.7) | 100.0 (0.0) | 100.0 (0.0) | 100.0 (0.0) |
| 105 | External Surface | 99.9 (0.1) | 99.7 (0.4) | 92.6 (10.4) | 98.5 (2.7) | 98.4 (2.3) | 98.0 (2.4) | 96.4 (3.9) | 100.0 (0.0) |
| 111 | External Surface | 100.0 (0.0) | 80.4 (26.2) | 71.5 (11.0) | 97.7 (3.3) | 91.3 (12.3) | 65.4 (6.7) | 92.6 (4.2) | 84.2 (10.2) |
| 130 | Interface | 7.7 (10.4) | 9.0 (8.5) | 28.2 (19.7) | 23.3 (33.0) | 11.3 (17.2) | 33.1 (18.5) | 39.0 (21.3) | 23.9 (33.2) |
| 165a | External Surface | - | - | - | - | - | - | - | - |
| Chymotryptic Data |
% amidinated lysine (standard deviation) |
||||||||
| Lysine | Classificationb | pH 5.4 | pH 5.6 | pH 6.0 | pH 6.5 | pH 6.7 | pH 7.2 | pH 8.0 | pH 8.5 |
| 8a | Internal Surface | - | - | - | - | - | - | - | - |
| 41 | Internal Surface | 91.7 (5.5) | 88.1 (6.1) | 96.4 (5.4) | 99.4 (0.8) | 99.7 (0.6) | 99.9 (0.3) | 99.7 (0.5) | 99.9 (0.1) |
| 44 | Internal Surface | 49.7 (5.2) | 58.3 (4.8) | 92.3 (8.6) | 91.4 (12.9) | 99.5 (0.7) | 92.6 (0.3) | 98.7 (1.2) | 97.9 (4.4) |
| 53 | Interface | 24.1 (19.7) | 10.6 (9.8) | 18.3 (7.9) | 21.7 (3.2) | 44.8 (20.7) | 51.1 (21.2) | 63.6 (8.1) | 86.7 (17.6) |
| 64 | External Surface | 43.4 (12.6) | 60.3 (6.3) | 89.6 (8.5) | 90.9 (6.3) | 99.1 (1.5) | 97.7 (3.5) | 98.8 (1.5) | 98.1 (3.9) |
| 81 | Interface | 5.3 (3.0) | 33.6 (23.1) | 56.8 (10.2) | 95.3 (5.4) | 98.2 (2.5) | 99.9 (0.1) | 99.3 (1.4) | 96.5 (6.0) |
| 83 | External Surface | 28.1 (9.5) | 34.5 (8.6) | 86.7 (8.4) | 94.1 (3.3) | 97.6 (3.0) | 95.5 (7.8) | 97.8 (3.0) | 97.2 (3.3) |
| 86 | Internal Surface | 63.8 (29.5) | 43.4 (38.1) | 21.0 (34.2) | 65.4 (20.6) | 34.7 (28.1) | 95.5 (7.7) | 99.4 (1.1) | 94.0 (10.3) |
| 105 | External Surface | 98.8 (0.4) | 88.1 (15.8) | 100.0 (0.0) | 99.5 (0.5) | 96.9 (5.2) | 99.9 (0.2) | 99.7 (0.4) | 97.4 (44.0) |
| 111 | External Surface | 99.0 (1.0) | 81.6 (15.2) | 96.9 (3.6) | 97.3 (3.2) | 96.8 (6.3) | 94.9 (9.4) | 98.7 (0.8) | 98.4 (2.8) |
| 130 | Interface | 52.2 (13.0) | 56.6 (17.9) | 66.8 (9.5) | 58.6 (15.5) | 78.8 (4.0) | 86.9 (1.4) | 98.2 (3.0) | 96.8 (3.1) |
| 165 | External Surface | 90.5 (8.1) | 90.5 (11.9) | 86.8 (22.9) | 98.9 (1.5) | 100.0 (0.0) | 98.0 (4.4) | 100.0 (0.0) | 97.3 (5.9) |
Notes:
Peptides containing lysine 8 were not observed reliably in either tryptic or chymotryptic digests. Peptides containing K165 were not observed in tryptic digests. See text for discussion.
Classification as “Surface” or “Interface” residues was accomplished using ViPERdb. Surface residues were further classified as “External” or “Internal” by inspection of a reconstructed capsid structure.
We identify four classes of lysines, based on inspection of the crystal structure and patterns of SMTA labeling: highly reactive surface residues, residues that are exposed on either the exterior or interior surface of the virions, and residues that are sequestered at the interfaces of tertiary or quaternary structural features.
Highly reactive surface exposed lysines of the BMV virion
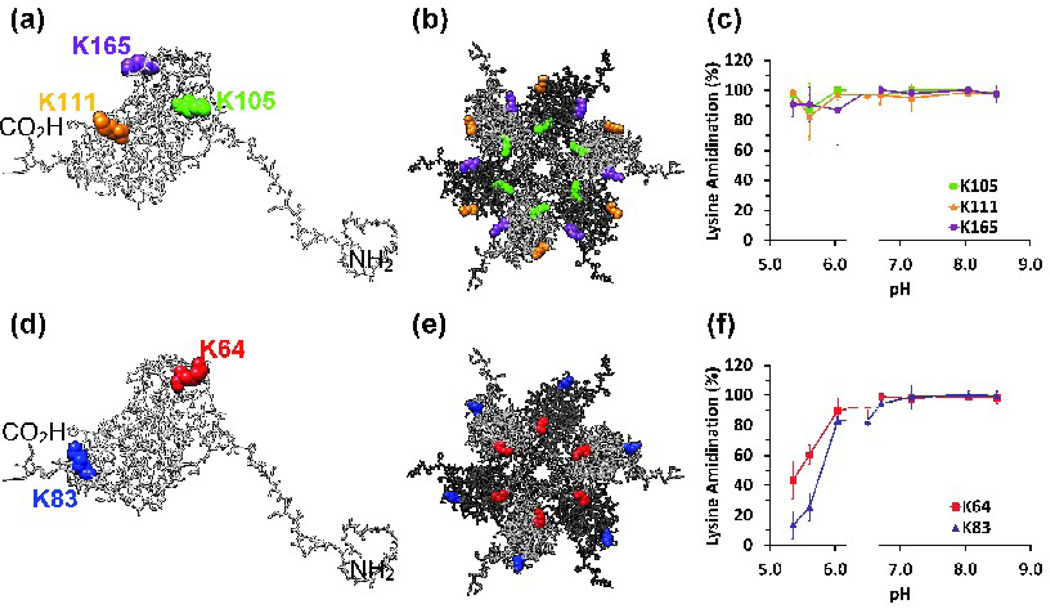
Data for K105, K111, and K165 introduce the format used in all subsequent figures (Fig.3). The locations of these three residues are depicted in an isolated capsid protein monomer and on the external surface of a hexamer of capsid proteins (Fig. 3a and Fig. 3b, respectively). The color used to depict each residue in the structure is also used to plot that residue’s extent of labeling data (Fig. 3c). The extent of modification of these three lysine residues is 80% or greater at all pH values, demonstrating that our procedure compensates adequately for SMTA hydrolysis at low pH. The 10% decrease in the reactivity of these residues at the lowest pH values serves as an estimate of the contribution of SMTA hydrolysis to any observed decreases in lysine reactivity at low pH.
Fig. 3.
(a) The location of external, surface exposed residues that show pH-independent reactivity in a capsid protein monomer: K105 (green), K111 (orange), K165 (purple). The N- and C-termini are also indicated. (b) K105, K111, and K165 displayed in the structural context of the external, solvent exposed face of a hexamer of capsid proteins. Adjacent monomers are displayed in contrasting shades of gray. (c) Chymotryptic digest data for K105, K111, and K165. (d) External, surface exposed lysine residues in an isolated capsid protein monomer: K64 (red) and K83 (blue). (e) Lysine residues K64 and K83 displayed on the solvent exposed face of a hexamer. (f) Chymotryptic digest data for K64 and K83. Error bars represent the standard deviations of at least three determinations.
There are few tryptic peptides that contain residue K165. Even in unmodified capsid protein the smallest tryptic peptide containing K165 has a mass of 2420.8 Da (G145 - K165); amidination of K165 increases the size of the smallest possible tryptic peptide to 5772.8 Da (G145 – R189). Chymotryptic digestion produces more easily separated and identified peptides such as A158 – L171 (1389.8 Da). Chymotryptic digest data in Table 1 indicate that the modification of K165 is nearly complete at all pH values. As a result, K165 is included in this class of lysine residues. The reactivities of K105, K111, and K165 imply that the capsid protein’s other nine lysines are responsible for the pH dependence of SMTA reactivity.
Surface exposed, exterior lysines of the BMV virion
K64 and K83 are on the exterior of the BMV capsid but are located near the interface of capsid subunits (Fig.3d and 3e). At pH values below 6.0, chymotryptic digest data show that less than 40% of the K64 and K83 residues were modified by SMTA (Fig. 3f). At pH 6.0, however, approximately 80% of the K64 and K83 residues were modified. These results demonstrate that some residues on the exterior of the BMV particle are partially protected at low pH, but become more solvent accessible as the pH increases. The observation that the ~30% and ~50% increases in SMTA reactivity for K64 and K83 occur before the pH transition midpoint of pH 6.5 suggests that the associated structural changes are sequential, and affect the lysines exposed on the external surface of the virion before either the interfacial or interior surface lysine residues.
Surface exposed, interior lysines of the BMV virion
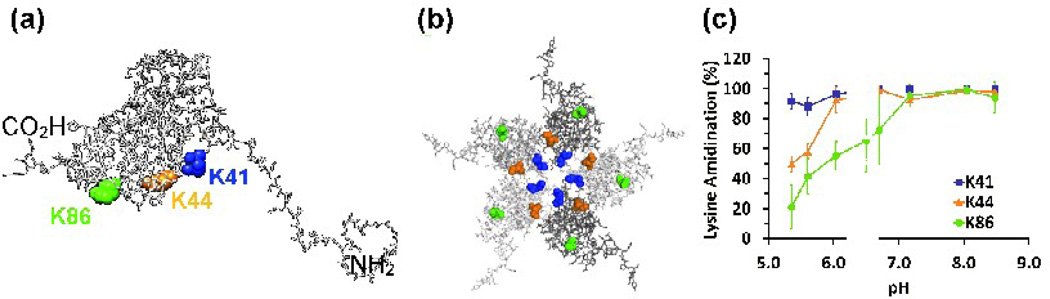
Lysines K41, K44, and K86 are found on the interior surface of the virion (Fig.4a and 4b). In the crystal structure of BMV, these lysines exist in a region of low electron density above the shell of encapsulated RNA.39 Chymotryptic digest data demonstrate that K41 is easily modified even at pH 5.4, indicating that SMTA can access the virion’s interior even in its low pH compact form (Fig. 4c). Approximately 50% of K44 positions and nearly 80% of K86 are less reactive at pH values below the major structural transition midpoint of 6.5, indicating that these lysines are partially protected by structural features of the compact virion (Fig. 4c and Table 1). Residue K44’s SMTA reactivity shows a 40% increase below the swelling transition midpoint of pH 6.5, suggesting that this position, adjacent to a pore on the pentamer axis, becomes more accessible before the main expansion of the virion. The reactivity of K41 and K44 is consistent with the currently accepted model of viral particles as dynamic structures whose multiple “breathing modes” allow solvent and small molecules access to the particle core without disrupting its structure.3–4,14,59 Partial tryptic digest experiments using BMV demonstrated that the N-terminal region containing K41 and K44 is flexible and can access bulk solution even in intact virions.43–44 The reactivity of K86 increases more slowly, consistent with its position close to the interface between subunits, and only reaches 90% reactivity at pH 7.0. The large standard error in the extent of modification for K86 at intermediate pH values is due to digest-to-digest variability in chymotrypsin activity.64 Cleavage at M69 or L73 and L85 provides peptides that report reliably on the labeling states of K81 and K83, resulting in the low standard deviations seen for these residues (Fig. 5c and Fig. 3c, respectively). However, the peptides containing K86 are formed by cleavage at L85 and one of six succeeding sites (L91, L92, W93, L94, L96 or L97), effectively diluting the signals that report on K86’s extent of labeling among a collection of seven peptides.
Fig. 4.
(a) The locations of internal, surface exposed lysine residues in a capsid protein monomer. The three lysine residues are displayed as space-filling models: K41 (blue), K44 (orange) and K86 (green). The molecule is in the same orientation shown in Fig. 3(a). (b) Lysine residues displayed in situ on the lumenal face of a pentamer structure. (c) Chymotryptic digest data for the same three lysine residues. Error bars represent the standard deviations of at least three determinations.
Fig. 5.
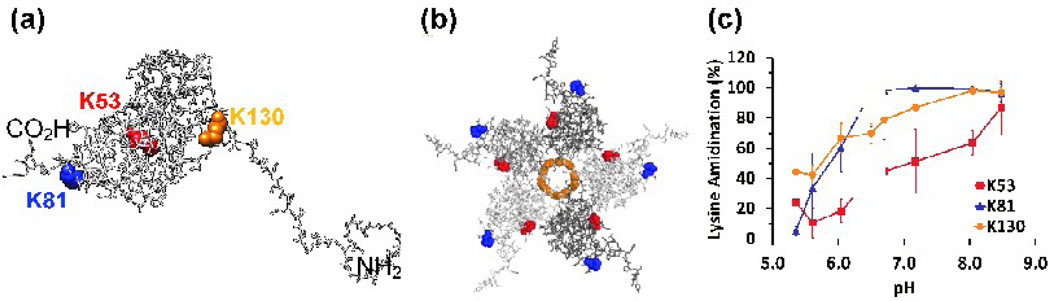
(a) Interfacial lysine residues K53 (red), K81 (blue) and K130 (orange), displayed in a capsid protein monomer. (b) Lysine residues displayed in situ in a pentamer structure viewed from its lumenal face. (c) Chymotryptic digest data for the same three lysine residues. Error bars represent the standard deviations of at least three determinations.
SMTA labeling of interfacial lysines of the BMV virion
K53, K81 and K130 occupy positions on capsid protein surfaces that are buried by quaternary protein-protein interactions (Fig.5a and 5b). K81 shows a dramatic increase in reactivity, starting at less than 10% modified below the transition midpoint pH and increasing to almost complete modification around pH 6.5, tracking the pH dependence of the overall virion swelling almost exactly.
The interpretation of the data for residues K53 and K130 is more complex. SMTA modification of K53 increases steadily through the midpoint for the main structural transition but never reaches 100% at pH 8.5 (Fig 5c), suggesting that it is exposed to solution late in the swelling transition and still somewhat protected from SMTA even in the expanded form of the virion. K130’s 40% reactivity at pH 5.4 and 5.6 shows it to be the most accessible interfacial residue in the compact virion. Modification of K130 shows a constant increase to complete reactivity over the pH range from 5.4 to 8.5. These results demonstrate that the interfacial residues make the most dramatic transition from a sequestered environment at low pH to full solvent exposure at high pH.
Characterization of mutant virions
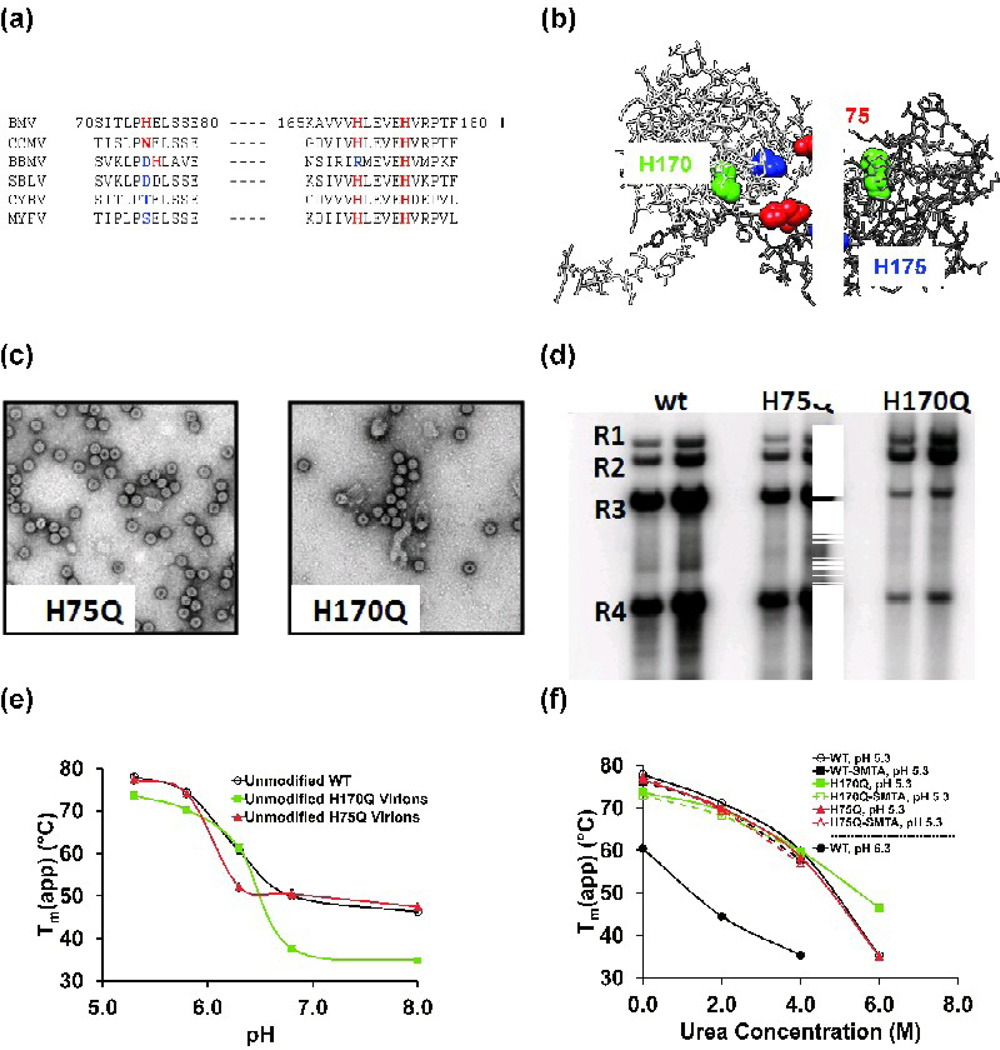
Lysine modifications report on the responses of residues in three structural regions of the BMV virion. However, the high pKa of lysine side chains (ε-amino pKa of 10.5)48 suggests that they probably do not participate directly in the BMV virions’ swelling transition. Histidine residues, with pKa values of ~6.448, are better candidates for sensing solution pH or triggering the pH-dependent conformational change of BMV virions. The BMV capsid protein contains three histidines, at positions 75, 170 and 175 (Fig 6a). Perhaps relevant to the swelling transitions, all three histidines are located at the interface of dimers of the capsid subunits (Fig. 6b). Analysis of the conservation of the three residues revealed that, among members of the Bromoviridae, H175 is absolutely conserved, H170 is conserved in all but one member, and H75 is the least conserved (Fig. 6a). To investigate the structural role of these residues, glutamine substitutions of each histidine were produced to generate viruses H75Q, H170Q and H175Q.
Fig. 6.
(a) Partial sequence alignment of histidine residues in the capsid protein sequences from members of the Bromoviridae. Conservative sequence substitutions are highlighted in red, others are highlighted in blue. Sequence numbering is for the BMV capsid protein. Additional abbreviations are BBMV: broad bean mottle virus, SBLV: Spring beauty latent virus, CYBV: Cassia yellow blotch virus, MYFV: Melandrium yellow fleck virus. (b) The location of residues H75, H170 and H175 at the interface of a capsid protein dimer. The two capsid protein subunits are colored white and gray. (c) Transmission electron micrographs of virions isolated from tobacco leaves transfected with the H75Q or H170Q mutants. (d) Northern blot analysis of the RNAs extracted from WT and mutant virions. The RNAs were detected with a riboprobe that recognizes the nearly identical 3’ untranslated regions of the BMV RNAs as described by Hema, et al.63 Each sample was from an independent preparation of virion from N. benthamiana plants. R1-R3: BMV genomic RNAs 1–3; R4: subgenomic RNA 4. (e) Virion melting points determined by DSF as a function of pH for wild-type BMV, H75Q, and H170Q containing mutant virions. Each value represents the mean Tm(app) of three determinations. Error bars for the standard deviation of each measurement are smaller than the symbols used for each data point. (f) Wild-type and mutant virion melting points as a function of urea concentration. Solution pH was as indicated in the figure legend. Each value is the mean of three measurements, and standard deviations are smaller than the size of the symbols used for each data point.
Virion production was quantified by Bradford protein assays of the final virion isolates using Bovine serum albumin as a standard. Mutants H75Q, H170Q, and H175Q produced virions at approximately 50%, 10%, and less than 1% of wild type, respectively. Isolated virions were also examined by electron microscopy. The yields of mutant H175Q precluded further investigation. Mutants H75Q and H170Q produced viral particles whose diameter and morphology is similar to that of WT (Fig. 6c). The RNAs packaged in wild-type and the two remaining mutants were extracted from CsCl-purified virions and electrophoresed on an agarose gel (Fig. 6d). Mutant H75Q has a distribution of genomic RNA comparable to wild type, while the H170Q virions show decreased packaging of BMV RNA3 and RNA4. This aberrant RNA packaging pattern suggests that H170 exerts a direct or indirect influence on interactions between the BMV capsid and RNA3 or RNA4.
The DSF assay was used to characterize the stability H75Q and H170Q virions as a function of pH (Fig. 6e). Mutant H170Q exhibits an inflection point at ~ pH 6.5 similar to that of the WT, with a dramatic 30 °C decrease in the Tm(app) above the transition midpoint. Mutant H75Q exhibits stability comparable to the WT both above and below the swelling transition. However, H75Q’s transition midpoint occurs at pH 6.2, suggesting that the presence imidazole of H75Q is a significant factor in the virions’ response to changing pH. Wild-type and H75Q virions show nearly identical decreases in their denaturation temperatures as a function of increasing urea concentration at pH 5.3 (Fig. 6f). The H170Q mutant has a marginally lower Tm(app) at low urea concentrations and significantly higher Tm(app) at high urea concentrations. These changes should be considered in the context of the 15 to 20 °C changes in the Tm(app) for the WT virions when the pH in increased from 5.3 to 6.3 (Figure 6f, closed black squares vs. closed black circles). Furthermore, the Tm(app) values for SMTA-modified H75Q and H170Q virions are nearly identical to the wild-type and their unmodified counterparts, lending further support to the conclusion that SMTA modification does not alter or destabilize virion structure. Denaturation of the compact, low pH form of the virions is less likely to reflect contributions from molecular interactions that affect the swelling transition, but would be expected to contain contributions from intramolecular interactions that destabilize the capsid protein’s structure.
The three histidines at the interface of BMV capsid subunits have a range of effects on virion production, RNA encapsidation and pH-dependent conformational changes. The H75Q mutant’s structural integrity and the perturbation of its response to pH prompted us to examine the SMTA labeling of its lysines as a function of pH. To decrease virion heterogeneity, we made H75Q in a form that packaged only RNA3 and RNA4 (referred to as R3/4H75Q). Virions composed of wild-type capsid protein and containing only RNA3 and RNA4 (referred to as R3/4) were prepared for comparison to the R3/4H75Q particles. Both R3/4 and R3/4H75Q virions were purified at pH 5.5 and were indistinguishable from each other in electron micrographs (data not shown).
SMTA labeling of mutant virions
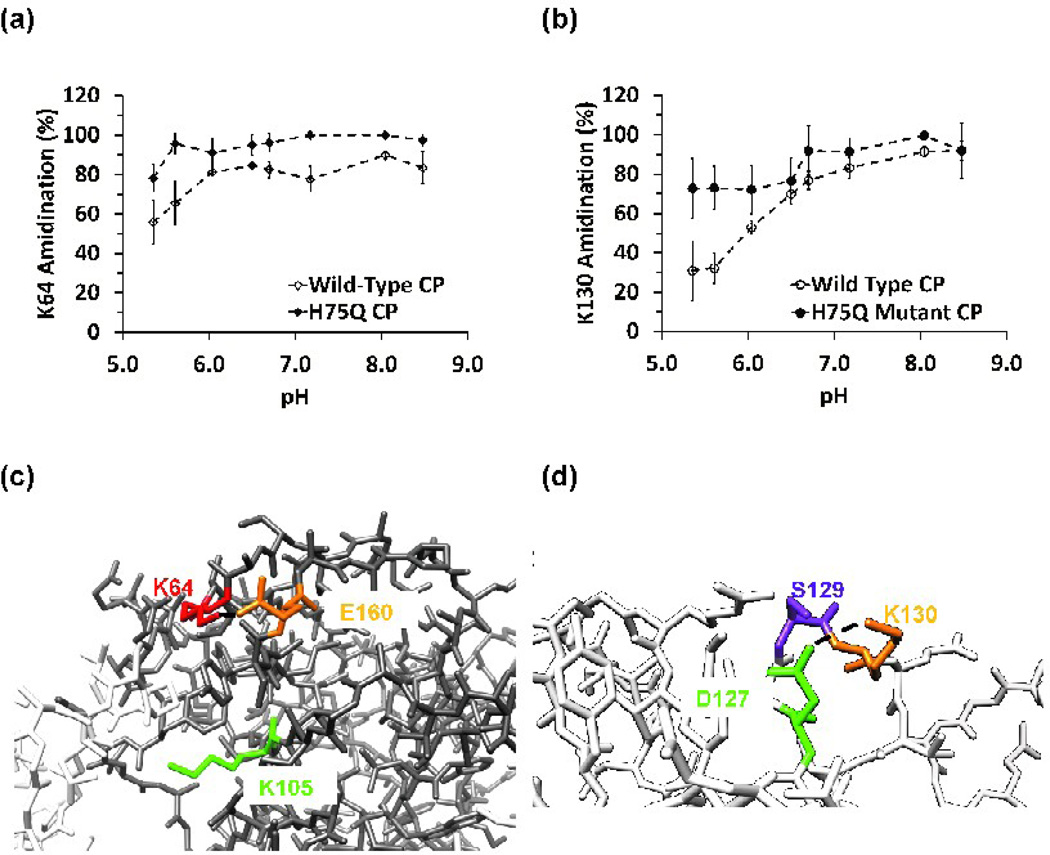
Both R3/4 and R3/4H75Q virions were subjected to the SMTA modification procedure, enzymatic digestion, and LC-MS/MS analysis. The WT R3/4 virions give results that are nearly identical to virions containing a natural distribution of all three BMV genomic RNAs (Fig. S10a–f and Table S6). The major differences are that R3/4 exhibited lower average standard deviations (5.6% and 5.8% for the tryptic and the chymotryptic digest data), compared to natural, heterogeneous virions (9.5% and 9.2% for tryptic and chymotryptic data) (Tables S6 and 1, respectively). In contrast, R3/4H75Q revealed some significant changes in the pH-dependent reactivity of lysine residues (Fig. 7a–b). Although K64 was inaccessible to modification until the solution pH was above 6.0 in R3/4 virions, it was nearly completely modified even at pH values below 6.0 in R3/4H75Q particles (Fig. 7a). The two-fold increase in interfacial residue K130’s SMTA reactivity at pH values below the transition midpoint (Fig 7b) indicates that effect of the H75Q mutation can be detected using SMTA modification, and that this mutation has a detectable structural influence on residues distal from the dimer interface occupied by H75.
Fig. 7.
(a) Summary of SMTA modification of K64 as a function of pH in R3/4 virions or H75Q mutant virions. Data were derived from chymotryptic digests. (b) Labeling of K130 in R3/4 virions containing wild type CP or H75Q mutant CP as a function of pH. Plot of chymotryptic digest data. (c) A potential interaction between K64 with E160. Disruption of this interaction by greater conformational flexibility in the H75Q mutant could explain the observed labeling behavior. The position of K64 in the capsid protein and the virion’s quaternary structure can be seen in Fig. 3(d)–(e). (d) Close up of potential hydrogen bonding or salt bridge interactions between K130 and D127 or S129. The position of K130 in the capsid protein monomer and in a pentamer can be seen in Fig. 5(a)–(b).
Discussion
Chemical labeling of lysines followed by mass spectrometric analysis of peptides was previously used to probe protein-nucleic acid interactions in bacterial ribosomes.34–38 In order to extend this technique to different solution conditions, a procedure for the amidination of lysine residues that mitigated the effect of changes in solution pH on labeling efficiency was optimized using RNase A. When subjected to 10 cycles of SMTA modification, the SMTA reactivity of monomeric RNase A is nearly pH independent (Fig. S1 and Fig. 2a). In contrast, only half of the BMV capsid protein’s lysine residues are modified at low pH, and as the reaction pH increases the broad distribution of modified states converges to a single mass representing 12 completely amidinated lysines (Fig. S2 and Fig. 2a). These results demonstrate that protein-protein interactions in the BMV capsid can preclude the SMTA modification of the twelve lysines that are on the external surface, the internal surface and the interfacial regions of the virion. Furthermore, the degree of modification of the lysines a function of pH will provide insights to the conformational dynamics of the BMV virion. The details of the extent of modification were examined using proteolytic digestion and LC-MS/MS analysis.
Lysine modification and pH-dependent conformational changes of the BMV virion
There are four interesting exceptions to expected labeling behavior among the surface residues. At pH values above the pH 6.5 midpoint for the swelling transition, residues K44, K64, K83 and K86 are uniformly modified. As the pH decreases to values below 6.5, their reactivity is significantly attenuated, suggesting that the environments occupied by K44, K64, K83 and K86 become more sterically restricted in the compact, low pH virion structure. These observations are consistent with Wang et al.’s15 experiments on HDX of backbone amides in BMV capsid protein. They found that the backbone amide protons of 6 residues in the capsid protein sequence containing residues 1–47 and 4 residues in the range from 70–90 decreased their rates of hydrogen-deuterium exchange when the solution pH was lowered from 7.30 to 5.43, indicating a decrease in solvent exposure or conformational flexibility in the regions of the molecule containing K44, K83 and K86. A single backbone amide in a peptide containing residues 62–69 also decreased its rate of exchange, but the HDX exchange data did not provide residue-level resolution, preventing this change from being correlated directly to K64. Formaldehyde cross-linking of BMV RNA and capsid protein recovered peptides containing residue K44,7 indicating that this residue is capable of interacting with the viral RNA. Crystal structures of the closely related CCMV virion showed distinct bulges in the RNA-associated electron density directly below the lysine homologous to BMV K86, but the structure of the RNA could not be resolved due to crystallographic disorder.40 The combination of observed proximity and pH-dependent differential protection from SMTA modification suggests that the interaction of K44 and K86 with the phosphodiester backbone of the virion’s RNA changes as the virus swells.
Figures 7a and 7b show the effect of the H75Q mutation on the SMTA labeling of K64 and K130. The H75Q mutant is of special interest since it could encapsidate the RNA similarly to WT BMV and has a urea denaturation profile comparable to the WT virions at pH 5.3. The fact that the swelling transition of the H75Q mutant was altered suggests that the imidazole side chain of H75 either contributes to or is affected by the swelling transition. Capsid protein dimers such as the one depicted in Figure 6b have contact surface areas between 1244 and 1589 Å2,39 and are thought to be the basic assembly unit of the virion,39,65–66 leading to the expectation that any disruption of the interfacial region would have measurable effects on the stability of the virion. Comparing the locations of H75 in Figure 6b with the location of K64 or K130 in Figures 3b or 5b, it seems unlikely that direct interactions take place between theses residues. When the low and high pH forms of the CCMV virion were compared, the largest radial displacement of capsid protein backbone α-carbons occurred at the dimer interface, centered on residue N76. This asparagine occupies a position in CCMV’s capsid protein sequence homologous to BMV’s H75 (Fig. 6a). We hypothesize that the 30–40% differences in the low pH extent of labeling of K64 and K130 (Fig. 7a and 7b) indicate greater conformational flexibility in the virion structure due to the loss of a hydrogen bond donated by the H75 δ-N in the protonated imidazole. Interestingly, asparagine or aspartate residues are found in 4 of the 6 Bromovirus sequences compared in Figure 6a. The localized molecular interactions responsible for the differential reactivity of K64 and K130 are probably hydrogen bonds or salt bridges, as observed in previous experiments involving SMTA modification of bacterial ribosomal proteins.34–36,38 Inspection of the BMV capsid protein crystal structure (PDB:1JS9) reveals that the ε-amino group of K64 (red) is within 4.4 Å of one of the carboxyl oxygens of E160 (orange). If allowance is made for conformational flexibility of lysine and glutamate side chains (Fig. 7c), the distance can be decreased to 1.7 Å. Figure 7c also contrasts the relatively more exposed position of K105 (green), explaining this residue’s paradoxically higher reactivity compared to K64. Although K64 is located further toward the particle exterior than K105, its interaction with E160 apparently protects it from SMTA modification. On the other hand, K105 hangs over anopen volume on the hexamer six-fold axis, surrounded by hydrophobic and aromatic residues such as L123, Y157, and P98 from the adjacent monomer. K130 is apparently involved in hydrogen bonds or salt bridges with D127 and S129 (Fig. 7d). The proposed salt bridge between K130 and D127 would be less flexible due to the short aspartyl side chain, and would be disrupted more easily by virion expansion, consistent with this residue’s reactivity (Fig. 5c).
Characteristics of the SMTA modification procedure
In addition to the information on the swelling transition of the BMV virion, this study should provide guidance to analysis of other proteins whose activities are affected by pH. These include endosome-resident proteins such as mannose-binding proteins and Toll-like receptors.67–68 With regard to the activities of the BMV protein, we observed that SMTA modification had only minimal effects on the quaternary structures of the BMV virions or RNA binding by the capsid protein. These observations are consistent with SMTA modifications having only minimal effects on the structure of prokaryotic ribosomes.34–36
An issue with the analysis of amidinated samples is trypsininability to cleave at SMTA-modified lysines. Elimination of tryptic digest sites creates errors and inconsistencies in the data for three reasons. First, large peptides generated from samples modified at high pH do not always elute effectively from our C18 columns, resulting in the outright loss of information for some lysine residues. A dramatic example can be seen by comparing the K81 labeling data from R3/4 virions generated from tryptic and chymotryptic digests (Fig. S5c and S5f, respectively). As the pH increases in the tryptic digest data, K81’s extent of modification approaches 80% then drops suddenly to ~40% (Fig. S5c). The chymotryptic data for K81 show a smooth increase to ~90% modification at pH 8.5 (Fig. S5f). This difference occurs because the number of tryptic peptides that report on the labeling of K81 decreases as K53, K64, K83 and K86 are more completely amidinated at higher pH, eliminating these lysines as tryptic cleavage sites. Conversely, the chymotryptic data are based on a uniform collection of peptides at every pH value. Second, the progressively larger tryptic peptides that do elute into the mass spectrometer support higher charge states. Although the Orbitrap mass analyzer can accurately measure the precursor masses of such peptides, the MS/MS spectra produced in the linear ion trap contain +3 and +4 fragment ions that are not recognized by automated database searches. Moreover, if more than one lysine in such a peptide is differentially modified, assignment of the position of the amidination will be ambiguous, increasing the errors in the extent of modification data. Finally, the current version of the ProtParser software was designed for use in bottom up proteomics experiments and does not parse peptides with charge states >+4. The +5, +6 or +7 charge states of the large peptides generated in tryptic digests of extensively modified BMV display higher intensities than the +3 or +4 charge states. A remedy for this problem is inspection of the Mascot output to identify large peptides, followed by manual extraction and integration of XICs for their higher charge states. Tryptic digest data presented in Table 1 and Table S6, and in Figures S4a–c and Figures S5a–c have been corrected this way.
Summary and conclusions
Our data provide a dynamic view of the BMV virion structure in solution to augment static crystallographic or cryoelectron microscopic data. The reactivity of some “surface exposed” lysine residues was essentially pH-independent, as expected from their classification. However, other surface lysine residues showed pH-dependent changes in SMTA reactivity, specifically protection at lower pH. These results demonstrate the ability of SMTA modification to provide experimental evidence of interactions between amino acid residues that otherwise must be inferred from structural data, or that might be overlooked.
Materials and Methods
Chemicals
Water was purified by a Barnstead Thermolyne Nanopure system. Ammonium chloride, magnesium chloride hexahydrate, and methylamine as a 40% (v/v) solution in water were supplied by Aldrich. HPLC grade acetonitrile, 2-propanol, methanol, trifluoroacetic acid and formic acid were products of J. T. Baker. To determine the approximate pH of reaction mixtures, EMD ColorpHast pH strips with a pH range of 0–14 were used. Bovine chymotrypsin (EN-160) was a product of Princeton Separations. Reagent grade (ethylenedinitrilo)-tetraacetic acid was obtained from Mallinckrodt. 2-Amino-2-(hydroxymethyl)-1,3-propanediol (Tris free base), dithiothreitol, horse heart myoglobin, 4-(2-Hydroxyethyl)-piperazine-1-ethanesulfonic acid (HEPES, free acid), leucine enkephalin, 2-(N-Morpholino)-ethanesulfonic acid (MES, free acid), and proteomics-grade alkylated porcine trypsin were from Sigma. The response of all pH electrodes used was standardized with VWR pH 4, 7, and 10standard solutions.
Preparation of thioimidate reagents
SMTA was synthesized according to Thumm’s method as described by Beardsley and Reilly.49,69 The product of this synthesis is the hydroiodide salt of the compound, and when dissolved in a 250 mM solution of Tris-free base, the solution pH decreases from 10.6 to 8.5. SMTP was synthesized according to Matsuda’s method as described by Beardsley and Reilly.69–70 The product form of this reagent is the hydroiodide salt of SMTP.
S-ethylthiopropioimidate (SETP) was synthesized using an adaptation of the Pinner synthesis of Diethylsuberothioimidate (DEST) reported recently by Lauber and Reilly.71 Propionitrile (0.02 moles, 1.4 ml) was dissolved in methylene chloride (4.3 ml, dried over molecular sieves) in a round bottom flask. A 10-fold molar excess of ethanethiol (0.20 moles, 14.8 ml) was added and the solution was immersed in an ice bath and stirred while bubbling HCl gas into the solution for 1 hour. During this interval, the reaction flask was vented into a solution of 1 M sodium hydroxide. After charging the reaction flask with HCl, the flask was stoppered and placed in an airtight bag (to minimize release of ethanethiol) and placed in a cold room at 4°C. After incubation overnight, the hydrochloride salt of the product was precipitated by adding diethyl ether and placing the reaction mixture into a −20°C freezer. After 1 h at −20°C, 2.26 g of white solid were collected by filtration through Whatman’s #1 paper, a 73% yield of the clear and colorless S-ethylthioproionimidate hydrochloride.
The final products of all syntheses were stored in vacuo over Drierite pellets until used.
Production of BMV virions and capsid protein
Agrobacterium tumefaciens was used to launch BMV infection in the geranium tobacco, Nicotiana benthamiana, as described previously.72 To produce BMV virions containing RNA3 and its subgenomic copy RNA4 encapsidated by wild-type capsid protein (referred to as R3/4 BMV), Nicotiana benthamiana plants were infiltrated with a mixture of Agrobacterium cells engineered to express the BMV 1a and 2a replication proteins and the replication-competent RNA3 that also directs transcription of RNA4. Three separate cDNAs for BMV RNA3 containing mutations of histidines 75, 170, or 175 glutamines were made by PCR-mediated site-directed mutagenesis using the Quick change kit and protocols recommended by the manufacturer (Stratagene, Torrey Pines, CA). Primers for each mutant are available upon request. Wild-type virions and virions expressing each of the three histidine mutants were all purified from N. benthamiana leaves according to previously published procedures.6 Virion yields were quantified by Bradford dye binding assays (Quick Start Bradford Dye Reagent, Bio-Rad, Hercules, CA) using Bovine serum albumin as a standard.
Dissociated capsid protein samples were prepared from wild type virions by dialysis in Slide-A-Lyzer dialysis cassettes (Thermo-Pierce, Rockford, IL) against a buffer containing 50 mM Tris, pH 7.5, 500 mM CaCl2, and 1 mM DTT for 24 h at 4 °C. Following centrifugation at 97,000 × g for 1 h to remove the precipitated RNAs, the supernatant was dialyzed against 50 mM Tris, pH 7.5, 300 mM NaCl, and 1 mM DTT for 24 h. The final dialyzate had an A260/A280 ratio < 0.65, and the protein concentration was determined by Bradford assay using BSA as a standard.
Modification of viral particles with SMTA
The buffers used were 500 mM stock solutions of acetic acid (pH 5.4 and 5.8), the free acid forms of MES (pH 6.0 and 6.5) and HEPES (free pH 6.7 and 7.2), and Tris free base (pH 8.0 and 8.5), adjusted with concentrated ammonium hydroxide. Stock solution pH was determined with a Corning Scholar 425 pH meter equipped with a glass pH electrode. The buffer stocks were used to prepare solutions of SMTA, and to prepare more dilute 100 mM reaction buffer solutions. When modifying BMV virions at varying pH values, an IQ400 ion selective field effect transistor (ISFET, Hach Company, Loveland, CO) electrode interfaced to a Handspring Visor handheld computer (Palm Inc., Sunnyvale, CA) was used to measure the initial pH of the dilute buffer solution, the pH of the dilute buffer solution after addition of an aliquot of SMTA-containing solution, and the pH of the reaction solution after 1 h. Addition of the SMTA solution to the dilute buffer solution typically caused a 0.1 – 0.2 unit decrease in pH, but the pH stayed constant after 1 h. Tris-buffered solutions showed large decreases in pH after SMTA addition (from 8.7 to 8.0 and from 10.6 to 8.5). The reported reaction pH values were measured after the initial addition of SMTA to the dilute reaction buffer solutions.
The SMTA protein modification procedure was developed using Bovine Pancreatic RNase A before application to virions. When BMV virions were modified, 160 pmoles of BMV protein were diluted into a total volume of 100 µL in an Amicon Microcon centrifugal concentrator with a 10,000 Da molecular weight cutoff filter (protein concentration: 1.6 µM or 0.03 g/L). All SMTA reactions and subsequent sample clean-up were performed in the same filter cartridge at room temperature (22–24 °C). Reaction solutions containeda 100 mM concentration of one of the buffers described above and 20 mM MgCl2. The solution was centrifuged for 17 min., diluted with 80 µL of the reaction buffer, and then a 20 µL aliquot of 200 mM SMTA in concentrated buffer solution was added. The BMV capsid protein contains 12 lysines and an acetylated N-terminus, thus the SMTA:protein mole ratio was 25370:1 and the SMTA:amino group mole ratio was 2100:1. This process was repeated 10 times to ensure that any observed protection of lysine amino groups from SMTA reaction was due only to tertiary and quaternary structure rather than competing hydrolytic degradation of the SMTA. After 10 cycles of modification, residual reaction products were removed by diluting the reaction to 500 µL with 100 mM ammonium bicarbonate containing 50 mM DTT to reduce any mixed disulfide bonds formed by reaction with methanethiol produced by amidination of lysines in proteins.31 Residual DTT and ammonium bicarbonate were removed by diluting the protein twice to 500 µL with 0.1% (v/v) formic acid in water and concentrating it to a final volume of 50 µL. Samples were then transferred to septum-capped vials for whole protein LC-MS analysis.
Whole protein LC-MS
The average extent of SMTA modification of RNase A or BMV capsid protein was determined with a Micromass QTOF Micro spectrometer. Sample handling and gradient development were provided by a Waters Alliance 2795 chromatograph using the gradient of Table S1. Protein samples in 0.1% (v/v) formic acid in water from the previous section’s modification procedure were concentrated and desalted by injecting 30 to 40 pmoles of protein onto a reversed phase capillary column (Upchurch Scientific Blue PEEK, 10 cm × 0.0254 cm, packed with Phenomenex Jupiter C4 material). Raw profile mode spectra were summed and extracted from TICs manually, and the masses of SMTA modified forms of the proteins were determined by deconvolution of the raw spectra using MaxEnt 1 (Micromass). The resulting whole protein spectra were centroided and exported as text files containing mass/intensity pairs for calculation of the intensity weighted average extent of modification using Microsoft Excel 2007.
Differential scanning fluorimetry
The effect of SMTA modification on BMV structural integrity was examined using Differential Scanning Fluorimetry (DSF). After 10 cycles of modification with SMTA in Tris buffer at pH 8.0, aliquots containing 5 µg of of BMV capsid protein were diluted into 30 µL of reaction buffer solutions and then transferred to a 96-well PCR plate. A 1 µL aliquot of a 200X stock solution of SYPRO Orange dye in DMSO (Sigma) was added to each well.58 Samples of virions containing unmodified protein were diluted and loaded into a second set of wells at the same concentrations of BMV virions and SYPRO Orange as above. The temperature gradient and fluorescence were monitored with an Agilent Strategene Mx3005P (Agilent, Santa Clara, CA). The samples were equilibrated at 25°C for 5 min, and then their temperature was increased to a final value of 95°C in 0.5°C increments with a ramp time of 30 s. Fluorescence intensity was monitored at 570 nm. Raw data were processed with Microsoft Excel 2007.
To further characterize the effects of the H75Q and H170Q mutations on capsid protein and virion structure, wild-type mutant virions were diluted into buffers containing 0, 2, 4, and 6 M urea. Quantities of protein, SYPRO Orange, and the temperature gradient program controlling the Agilent Strategene were identical to the details above.
Fluorescence polarization titration
Samples contained 0.25 µM fluorescein-labeled B Box stem loop (Integrated DNA Technologies, Coralville, IA) in 50 mM Tris, pH 7.5, and 50 mM NaCl.6–7 Concentrations of unmodified or SMTA-modified BMV capsid protein ranged from 2 pM to 7 µM. Fluorescence polarization measurements were made using a PanVera Beacon 2000 fluorescence polarization system (PanVera Corp. Madison, WI) equipped with a 488 nm excitation and 535 nm emission filters. Prior to data collection, samples were incubated at 37 °C for 15 minutes, and the 37 °C temperature was maintained by the instrument. Dissociation constants for the RNA-protein complex were extracted by fitting the raw data against a non-cooperative saturation binding curve with OriginPro 8.6 (OriginLab, Northampton, MA).
EM sample preparation and image collection
BMV virions were added to glow-discharged copper-coated grids at a final concentration of 0.01 mg/ml and stained with 1% (w/v) depleted uranyl acetate. Electron micrographs with a 60,000X magnification were recorded using a JEOL 1010 transmission electron microscope operating at 80 kV.73
Enzymatic digests and peptide LC-MS/MS
Labeled BMV capsid proteins were exchanged into 20 mM ammonium bicarbonate or 20 mM Tris for trypsin or chymotrypsin digestion, respectively. The protease was added to a final concentration of 4% or 3.3% (w/w) of the capsid protein, and enzymatic digestion was allowed to proceed under the conditions described in Table S2. Peptides produced by enzymatic digest were reacted with SMTP or SETP to ensure uniform ionization efficiency and to mass tag lysine residues that were protected from SMTA modification in the native virions. Because thioimidate modification of free amino groups proceeds rapidly at pH values above 7.5, enzymatic digestions were terminated by the addition of an aliquot of 200 mM SMTP or SETP stock solution in 250 mM Tris-free base to a final concentration of 100 mM reagent. The propionamidination reaction was incubated at 25°C for 1 h and terminated by the addition of 10 µL of 10% trifluoroacetic acid in water. Doubly-labeled peptide samples were purified using C18 spin columns (Pierce Biotechnology). Eluted peptides were concentrated in vacuo then resuspended in 25 µL of 0.1% aqueous formic acid. Peptide samples were analyzed by LC-MS/MS on a 75 µm i. d. fused silica column packed with C18 material (Magic C18, Michrom Bioresources, Auburn, CA), using a Dionex Ultimate 3000 to supply the gradient shown in Table S3. Eluting peptides were detected with a Thermo LTQ-Orbitrap hybrid mass spectrometer. Peptide masses were measured to a specified resolution of 30,000 in the Orbitrap cell while the instrument’s linear quadrupole ion trap was used to produce and measure CID MS/MS spectra of the 5 most intense peptides detected in a given precursor spectrum.
Bioinformatics and quantitation of labeling results
The extent of SMTA modification of each BMV capsid protein lysine was quantified by integration of total ion chromatogram (TIC) data using programs from the NCGG ProteinQuant Suite.74 Lists of precursor ions and associated fragment ions were extracted from raw TIC data files into Mascot Generic Format using the program TurboRAW2MGF. Peptides were identified by Mascot searches against either the SWISSPROT database filtered for “Viruses”, or a database containing only BMV protein sequences. Modifications used as search parameters included acetylation of the protein’s N-terminus, amidination of lysine, propionamidination of lysine or a peptide’s N-terminus, oxidation of methionine, and deamidation of asparagine or glutamine. Mascot results were parsed with ProtParser allowing for peptide charge of +1 to +4, the minimum MOWSE and maximum expectation scores for all peptides of 1 and 400, respectively, a minimum intensity for all peptides being 1, and with up to 5 allowed missed cleavages and no exclusion of doubled tryptic cleavage sites. Redundant appearances by the same peptide sequence were combined under the appearance with the highest MOWSE score.
Peptides were quantified using ProteinQuant 2.6. Parameters for peptide signal integration include the initial retention time for a peptide from the ProtParser results, a maximum chromatographic peak width of 1.0 min., a search window for peak reassignment of 1.5 min., beginning and ending edge definitions based on intensity alone, a baseline intensity calculated from the first 10 min. of the chromatogram, and normalization based only on the total intensity of the identified peptides. ProteinQuant output files were converted to Microsoft Excel 2007 spread sheets to tabulate each appearance by the 12 lysines from the BMV capsid protein.
Crystal structures of BMV and RNase A were obtained from the Protein Databank (ww.pdb.org), and visualized using PyMOL v. 0.99 (DeLano Scientific, www.pymol.org) or UCSF Chimera.75 The classification of lysine residues in the BMV virion as surface or interfacial used the Amino Acid Information utility available at VIPERdb (viperdb.scripps.edu).76
Supplementary Material
Highlights.
The pH dependence of virion structure is difficult to study crystallograpically
Chemical modification of lysine residues can be quantified using mass spectrometry
Extent of lysine modification depends on virion structure and its pH dependence
Capsid protein mutants show quantitatively different labeling patterns
MS-detected chemical modification reliably monitors virion conformational changes
Acknowledgments
WER would like to thank Dr. Stella Aniagyei and Professor Bogdan Dragnea for an introduction to BMV preparation and handling. This work was supported by NIH grant 1R01AI090280 to C. K. and NSF grants CHE-1012855 and CHE-0832651 to JPR
Abbreviations
- BMV
Brome Mosaic Virus
- CCMV
Cowpea Chlorotic Mottle Virus
- DSF
Differential Scanning Fluorimetry
- HDX
Hydrogen-deuterium exchange
- LC-MS
Liquid chromatography-mass spectrometry
- LC-MS/MS
Liquid chromatography – tandem mass spectrometry
- SETP
S-ethylpropionimidate
- SMTA
S-methylacetimidate
- SMTP
S-methylthiopropionimidate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levy JA, Fraenkel-Conrat H, Owens RA. Virology. 3rd ed. Englewood Cliffs, NJ: 1994. [Google Scholar]
- 2.Forterre P, Prangishvili D. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann. N.Y. Acad. Sci. 2009;1178:65–77. doi: 10.1111/j.1749-6632.2009.04993.x. [DOI] [PubMed] [Google Scholar]
- 3.Morton VL, Stockley PG, Stonehouse NJ, Ashcroft AE. Insights in virus capsid assembly from non-covalent mass spectrometry. Mass Spectrom. Review. 2008;27:575–595. doi: 10.1002/mas.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bothner B, Hilmer JK. Probing viral capsids in solution. RSC Biomol. Sci. 2011;21:41–61. [Google Scholar]
- 5.Kao CC, Ni P, Hema M, Huang X, Dragnea B. The coat protein leads the way: an update on basic and applied studies with the brome mosaic virus coat protein. Mol. Plant Pathol. 2011;12:403–412. doi: 10.1111/j.1364-3703.2010.00678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi G, Letteney E, Kim C-H, Kao CC. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA. 2009;15:615–626. doi: 10.1261/rna.1375509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi G, Vaughan RC, Yarbrough I, Dharmaiah S, Kao CC. RNA binding by the brome mosaic virus capsid protein and the regulation of viral RNA accumulation. J. Mol. Biol. 2009;391:314–326. doi: 10.1016/j.jmb.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor DJ, Wang Q, Bothner B, Natarajan P, Finn MG, Johnson JE. Correlation of chemical reactivity of Nudaurelia capensis ω virus with a pH-induced conformational change. Chem Commun. 2003:2770–2771. doi: 10.1039/b310533d. [DOI] [PubMed] [Google Scholar]
- 9.Strable E, Finn MG. Chemical modification of viruses and virus-like particles. Curr. Topics Microbiol. Immunol. 2009;327:1–21. doi: 10.1007/978-3-540-69379-6_1. [DOI] [PubMed] [Google Scholar]
- 10.Douglas T, Young M. Viruses: making friends of old foes. Science. 2006;312:873–875. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- 11.Douglas T, Young M. Host-guest encapsulation of materials by assembled virus protein cages. Natur. 1998;393:152–155. [Google Scholar]
- 12.Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. Natural supramolecular building blocks: wild-type cowpea chlorotic mottle virus. Chemistry & Biology. 2002;9:805–811. doi: 10.1016/s1074-5521(02)00165-5. [DOI] [PubMed] [Google Scholar]
- 13.Barnhill HN, Reuther R, Ferguson PL, Dreher T, Wang Q. Turnip yellow mosaic virus as a chemoaddressable bionanoparticle. Bioconjugate Chem. 2007;18:852–859. doi: 10.1021/bc060391s. [DOI] [PubMed] [Google Scholar]
- 14.Lewis JK, Bothner B, Smith TJ, Siuzdak G. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA. 1998;95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Lane LC, Smith DL. Detecting structural changes in viral capsids by hydrogen exchange and mass spectrometry. Protein Sci. 2001;10:1234–1243. doi: 10.1110/ps.100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanman J, Prevelige PE., Jr High-sensitivity mass spectrometry for imaging subunit interactions: hydrogen/deuterium exchange. Curr. Opinion Struct. Biol. 2004;14:181–188. doi: 10.1016/j.sbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Englander SW, Mayne L, Sosnick TR. Hydrogen exchange: the modern legacy of Linderstrøm-Lang. Protein Sci. 1997;6:1101–1109. doi: 10.1002/pro.5560060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehring H. Hydrogen exchange/electrospray ionization mass spectrometry studies of structural features of proteins and protein/protein interactions. Anal. Biochem. 1999;267:252–259. doi: 10.1006/abio.1998.3000. [DOI] [PubMed] [Google Scholar]
- 19.Smith DL, Deng Y, Zhang Z. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J. Mass. Spectrom. 1997;32:135–146. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 20.Neurath H. Limited proteolysis, protein folding and physiological regulation. In: Jaenicke R, editor. Protein Folding. NY: Elsevier; 1980. pp. 501–523. [Google Scholar]
- 21.Hubbard SJ. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta. 1998;1382:191–206. doi: 10.1016/s0167-4838(97)00175-1. [DOI] [PubMed] [Google Scholar]
- 22.Suh M-J, Pourshahian S, Limbach PA. Developing limited proteolysis and mass spectrometry for the characterization of ribosome topography. J. Am. Soc. Mass Spectrom. 2007;18:1304–1317. doi: 10.1016/j.jasms.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendoza VL, Vachet RM. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glocker MO, Borchers C, Fiedler W, Suckau D, Przybylski M. Molecular characterization of surface topology in protein tertiary structures by amino-acylation and mass spectrometric peptide mapping. Bioconjugate Chem. 1994;5:583–590. doi: 10.1021/bc00030a014. [DOI] [PubMed] [Google Scholar]
- 25.Glocker MO, Nock S, Sprinzl M, Przybylski M. Characterization of surface topology and binding area in complexes of the elongation factor proteins EF-Ts and EF-Tu from Thermus thermophilus: a study by protein chemical modification and ,ass spectrometry. Chem. Eur. J. 1998;4:707–715. [Google Scholar]
- 26.Fiedler W, Borchers C, Macht M, Deininger S-O, Przybylski M. Molecular characterization of a conformational epitope of hen egg white lysozyme by differential chemical modification of immune complexes and mass spectrometric peptide mapping. Bioconjugate Chem. 1998;9:236–241. doi: 10.1021/bc970148g. [DOI] [PubMed] [Google Scholar]
- 27.Guan J-Q, Chance MR. Structural proteomics of macromolecular assemblies using oxidative foot printing and mass spectrometry. Trends Biochem. Sci. 2005;30:583–592. doi: 10.1016/j.tibs.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Sharp JS, Becker JM, Hettich RL. Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal. Chem. 2004;76:672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 29.Sharp JS, Guo J-T, Uchiki T, Xu Y, Dealwis C, Hettich RL. Photochemical surface mapping of C14S-Sml1p for constrained computational modeling of protein structure. Anal. Biochem. 2005;340:201–212. doi: 10.1016/j.ab.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Tong X, Wren JC, Konerman L. Effects of protein concentration on the extent of γ-ray-mediated oxidative labeling studied by electrospray mass spectrometry. Anal. Chem. 2007;79:6376–6382. doi: 10.1021/ac070724u. [DOI] [PubMed] [Google Scholar]
- 31.Lundblad RL. Chemical Reagents for Protein Modification. 3rd ed. Boca Raton, FL: CRC Press; 2005. [Google Scholar]
- 32.Janecki DJ, Beardsley RL, Reilly JP. Probing protein tertiary structure with amidination. Anal. Chem. 2005;77:7274–7281. doi: 10.1021/ac050891z. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Broshears WC, Reilly JP. Probing the structure and activity of trypsin with amidination. Anal. Biochem. 2007;367:13–19. doi: 10.1016/j.ab.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Reilly JP. Correlating the chemical modification of Escherichia coli ribosomal proteins with crystal structure data. J. Proteome Res. 2009;8:4466–4478. doi: 10.1021/pr9002382. [DOI] [PubMed] [Google Scholar]
- 35.Beardsley RL, Running WE, Reilly JP. Probing the structure of the Caulobacter crescentus ribosome with chemical labeling and mass spectrometry. J. Proteome Res. 2006;5:2935–2946. doi: 10.1021/pr060170w. [DOI] [PubMed] [Google Scholar]
- 36.Running WE, Reilly JP. Ribosomal proteins of Deinococcus radiodurans: their solvent accessibility and reactivity. J. Proteome Res. 2009;8:1228–1246. doi: 10.1021/pr800544y. [DOI] [PubMed] [Google Scholar]
- 37.Lauber MA, Running WE, Reilly JP. B. subtilis ribosomal proteins: structural homology and post-translational modifications. J. Proteome Res. 2009;8:4193–4206. doi: 10.1021/pr801114k. [DOI] [PubMed] [Google Scholar]
- 38.Running WE, Reilly JP. Variation of the chemical reactivity of Thermus thermophilus HB8 ribosomal proteins as a function of pH. Proteomics. 2010;10:3669–3687. doi: 10.1002/pmic.201000342. [DOI] [PubMed] [Google Scholar]
- 39.Lucas RW, Larson SB, McPherson A. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 2002;317:95–108. doi: 10.1006/jmbi.2001.5389. [DOI] [PubMed] [Google Scholar]
- 40.Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bancroft JB. The self assembly of spherical plant viruses. Advan. Virus Res. 1970;16:99–134. doi: 10.1016/s0065-3527(08)60022-6. [DOI] [PubMed] [Google Scholar]
- 42.Incardona NL, Kaesberg P. A pH-induced structural change in bromegrass mosaic virus. Biophys. J. 1964;4:11–21. doi: 10.1016/s0006-3495(64)86766-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeiffer P, Hirth L. The effect of conformational changes in brome mosaic virus upon its sensitivity to trypsin, chymotrypsin and ribonuclease. FEBS Lett. 1975;56:144–148. doi: 10.1016/0014-5793(75)80129-3. [DOI] [PubMed] [Google Scholar]
- 44.Tremaine JH, Ronald WP, Agrawal HO. Some tryptic peptides of bromovirus proteins. Virology. 1977;83:404–412. doi: 10.1016/0042-6822(77)90185-4. [DOI] [PubMed] [Google Scholar]
- 45.Pfeiffer P. Changes in the organization of bromegrass mosaic virus in response to cation binding as probed by changes in susceptibility to degradative enzymes. Virology. 1980;102:54–61. doi: 10.1016/0042-6822(80)90069-0. [DOI] [PubMed] [Google Scholar]
- 46.Pfeiffer P, Durham ACH. The cation binding associated with structural transitions in bromegrass mosaic virus. Virology. 1977;81:419–432. doi: 10.1016/0042-6822(77)90157-x. [DOI] [PubMed] [Google Scholar]
- 47.Tama F, Brooks CL., III The mechanism and pathway of pH induced swelling in cowpea chlorotic mottle virus. J. Mol. Biol. 2002;318:733–747. doi: 10.1016/S0022-2836(02)00135-3. [DOI] [PubMed] [Google Scholar]
- 48.Nelson DL, Cox MM, editors. Lehninger Principles of Biochemistry. 3rd. ed. NY: Worth Publishers; 2000. [Google Scholar]
- 49.Thumm M, Hoenes J, Pfleiderer G. S-methylthioacetimidate is a new reagent for the amidination of proteins at low pH. Biochim. Biophys. Acta. 1987;923:263–267. [Google Scholar]
- 50.Inman JK, Perham RN, DuBois GC, Apella E. Amidination. Methods Enzymol. 1983;91:559–569. doi: 10.1016/s0076-6879(83)91051-0. [DOI] [PubMed] [Google Scholar]
- 51.Hunter MJ, Ludwig ML. The reaction of imidoesters with proteins and related small molecules. J. Am. Chem. Soc. 1962;84:3491–3504. [Google Scholar]
- 52.Lienhard GE, Jencks WP. The reaction of carbanions with N,S-diacetylcysteamine. A model for enzymatic carbon-carbon condensation. J. Am. Chem. Soc. 87:3863–3874. doi: 10.1021/ja01095a013. (196). [DOI] [PubMed] [Google Scholar]
- 53.Chaturvedi RK, MacMahon AE, Schmir GL. The hydrolysis of thioimidate esters. Intermediates and general catalysis. J. Am. Chem. Soc. 1967;89:6984–6993. [Google Scholar]
- 54.Chatani E, Hayashi R, Moriyama H, Ueki T. Conformational strictness required for maximum activity and stability of bovine pancreatic ribonuclease A as revealed by crystallographic study of three Phe120 mutants at 1.4 Å resolution. Protein Sci. 2002;11:72–81. doi: 10.1110/ps.31102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park C, Raines RT. Dimer formation by a “monomeric” protein. Protein Sci. 2000;9:2026–2033. doi: 10.1110/ps.9.10.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nenci A, Gotte G, Bertoldi M, Libonati M. Structural properties of trimers and tetramers of ribonuclease A. Protein Sci. 2001;10:2017–2027. doi: 10.1110/ps.14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.She Y-M, Haber S, Seifers DL, Loboda A, Chernushevich I, Perreault H, Ens W, Standing KG. Determination of the complete amino acid sequence for the coat protein of brome mosaic virus by time-of-flight mass spectrometry. J. Biol. Chem. 2001;276:20039–20047. doi: 10.1074/jbc.M100189200. [DOI] [PubMed] [Google Scholar]
- 58.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature Protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 59.Broo K, Wei J, Marshall D, Brown F, Smith TJ, Johnson JE, Schneemann A, Siuzdak G. Viral capsid mobility: a dynamic conduit for inactivation. Proc. Natl. Acad. Sci. USA. 2001;98:2274–2277. doi: 10.1073/pnas.051598298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillitzer E, Willits D, Young M, Douglas T. Chemical modification of a viral cages for multivalent presentation. Chem. Commun. 2002:2390–2391. doi: 10.1039/b207853h. [DOI] [PubMed] [Google Scholar]
- 61.Reynolds JH. Amidination of bovine pancreatic ribonuclease A. Biochemistry. 1968;7:3131–3135. doi: 10.1021/bi00849a016. [DOI] [PubMed] [Google Scholar]
- 62.Speir JA, Bothner B, Qu C, Willits DA, Young MJ, Johnson JE. Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics. J. Virol. 2006;80:3582–3591. doi: 10.1128/JVI.80.7.3582-3591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hema M, Murali A, Ni P, Vaughan RC, Fujisaki K, Tsvetkova I, Dragnea B, Kao CC. Effects of amino-acid substitutions in the brome mosaic virus capsid protein on RNA encapsidation. MPMI. 2010;23:1433–1447. doi: 10.1094/MPMI-05-10-0118. [DOI] [PubMed] [Google Scholar]
- 64.Liebler DC. Introduction to Proteomics: Tools for the New Biology. Totowa, NJ: 2002. [Google Scholar]
- 65.Cuillel M, Berthet-Colominas C, Timmins PA, Zulauf M. Reassembly of Brome Mosaic Virus from dissociated virus. Eur. J. Biophys. 1987;15:169–176. [Google Scholar]
- 66.Zhao X, Fox JM, Olson NH, Baker TS, Young MJ. In vitro assembly of Cowpea Chlorotic Mottle virus from coat protein expressed in Escherichia coli and in vitro transcribed viral cDNA. Virology. 1995;207:486–494. doi: 10.1006/viro.1995.1108. [DOI] [PubMed] [Google Scholar]
- 67.Ranjith-Kumar CT, Miller W, Santos J, Cleveland L, Park M, Duffy KE, Lamb R, Sarisky RT, Mbow L, Kao CC. Biochemical and Functional Analyses of the Human Toll-like Receptor 3 Ectodomain. J. Biol. Chem. 2007;282:7668–7678. doi: 10.1074/jbc.M610946200. [DOI] [PubMed] [Google Scholar]
- 68.Wileman T, Boshans R, Stahl P. Uptake and transport of mannosylated ligands by alveolar macrophages. J. Biol. Chem. 1985;260:7387–7393. [PubMed] [Google Scholar]
- 69.Beardsley RL, Reilly JP. Quantitation using enhanced signal tags: a technique for comparative proteomics. J. Proteome Res. 2003;2:15–21. doi: 10.1021/pr0255506. [DOI] [PubMed] [Google Scholar]
- 70.Matsuda K, Yanagisawa I, Isomura Y, Mase T, Shibanuma T. Alternative synthesis of dibenzo- and pyrido-[1,3]diazepines from thioamidates and o,o’diaminobiaryls. Synthetic Commun. 1997;27:2393–2402. [Google Scholar]
- 71.Lauber MA, Reilly JP. Novel amidinating cross-linker for facilitating analyses of protein structures and interactions. Anal Chem. 2010;82:7736–7743. doi: 10.1021/ac101586z. [DOI] [PubMed] [Google Scholar]
- 72.Gopinath K, Dragnea B, Kao CC. Interactions between brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J. Virol. 2005;79:14222–14234. doi: 10.1128/JVI.79.22.14222-14234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wild P. Electron microscopy of viruses and virus-cell interactions. Methods Cell Biol. 2008;88:497–524. doi: 10.1016/S0091-679X(08)00425-1. [DOI] [PubMed] [Google Scholar]
- 74.Mann B, Madera M, Sheng Q, Tang H, Mechref Y, Novotny MV. ProteinQuant suite: a bundle of automated software tools for label-free quantitative proteomics. Rapid Commun. Mass Spectrom. 2008;22:3823–3834. doi: 10.1002/rcm.3781. [DOI] [PubMed] [Google Scholar]
- 75.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 76.Shepherd CM, Borelli IA, Lander G, Natarajen P, Siddanvanhalli V, Bajaj C, Johnson JE, Brooks CL, III, Reddy V. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 2006;34:D386–D389. doi: 10.1093/nar/gkj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.