Abstract
The Deleted in AZoospermia (DAZ) genes encode potential RNA-binding proteins that are expressed exclusively in prenatal and postnatal germ cells and are strong candidates for human fertility factors. Here we report the identification of an additional member of the DAZ gene family, which we have called BOULE. With the identification of this gene, it is clear that the human DAZ gene family contains at least three members: DAZ, a Y-chromosome gene cluster that arose 30–40 million years ago and whose deletion is linked to infertility in men; DAZL, the “father” of DAZ, a gene that maps to human chromosome 3 and has homologs required for both female and male germ cell development in other organisms; and BOULE, a gene that we propose is the “grandfather” of DAZ and maps to human chromosome 2. Human and mouse BOULE resemble the invertebrate meiotic regulator Boule, the proposed ortholog of DAZ, in sequence and expression pattern and hence likely perform a similar meiotic function. In contrast, the previously identified human DAZ and DAZL are expressed much earlier than BOULE in prenatal germ stem cells and spermatogonia; DAZL also is expressed in female germ cells. These data suggest that homologs of the DAZ gene family can be grouped into two subfamilies (BOULE and DAZL) and that members of the DAZ family evolved from an ancestral meiotic regulator, Boule, to assume distinct, yet overlapping, functions in germ cell development.
Deletions encompassing the Y chromosomal DAZ genes are the most common molecularly defined cause of infertility in humans (1, 2). An array of four DAZ genes in two clusters is located on the Y chromosome and encodes RNA-binding proteins with a common RNA-recognition motif and a series of 8–18 DAZ repeats, consisting of 24 amino acids each that are rich in N, Y, and Q residues (1). An autosomal homolog of DAZ, DAZL (DAZ-Like) maps to chromosome 3 (3–7). The predicted protein product of DAZL is 95% identical to that of DAZ except that DAZL contains just one DAZ repeat.
Homologs of DAZ have been identified in diverse organisms (8–15). Homologs in these organisms are required for germ cell development but differ in null phenotypes and expression patterns (Table 1). In flies, disruption of the DAZ homolog, Boule, causes male meiotic arrest (8). In Caenorhabditis elegans, disruption of the homolog of DAZ causes meiotic arrest in oogenesis only (11), and in mice, disruption of the DAZ homolog, Dazl, interferes with germ cell development in both males and females (12). In zebrafish and frogs, DAZ homologs encode components of germplasm, a region of oocyte cytoplasm that allocates the germ lineage and contains clusters of RNAs, RNA-binding proteins, ribosomes, and mitochondria that segregate to give rise to germ cells (14–17). In frogs, Dazl is required for embryonic germ cell production and migration (16). Below we report the identification and characterization of an additional member of the human DAZ gene family, BOULE. With identification of BOULE, our phylogenetic analyses suggest that the DAZ gene family is composed of two subfamilies required for different stages of germ cell development: DAZL for early germ cell function and BOULE for meiotic function. BOULE is the ancestral gene that is conserved from flies to humans, whereas, DAZL arose in the early vertebrate lineage and DAZ arrived on Y chromosome during primate evolution (5).
Table 1.
Expression and null phenotypes associated with DAZ/BOL homologs
| Genes/organisms | Expression
|
Phenotypes | Ref. | ||
|---|---|---|---|---|---|
| Testis | Ovary | PGC/Germplasm | |||
| BOULE | |||||
| Drosophila | + | − | − | Meiotic arrest | 8 |
| Mouse | + | − | − | Unknown | * |
| Human | + | − | − | Unknown | * |
| Worm | − | + | − | Meiotic arrest | 11 |
| DAZL | |||||
| Zebrafish | + | + | + | Unknown | 17 |
| Xenopus | + | + | + | Early germ cell defect | 14–16 |
| Mouse | + | + | + | Early germ cell defect | 12 |
| Human | + | + | + | Unknown | 6, 18, * |
| DAZ | |||||
| Human | + | − | + | Azoospermia & oligospermia | 6, 18, * |
PGC, primordial germ cell. +, Protein or mRNA expression observed. −, No expression.
, This paper.
Materials and Methods
Two-Hybrid Screening of DAZ Interacting Proteins.
We used the yeast two-hybrid system to screen for proteins that interact with DAZ/DAZL protein expressed from a pAS2 vector (CLONTECH). The DAZ/DAZL was derived from a Y chromosome-encoded cDNA, pRR102, which encodes the N-terminal RNA-binding domain and a single DAZ repeat. A testis library of cDNAs fused to the GAL4 activation domain was transformed into yeast containing the DAZ/DAZL fusion according to the manufacturer's instructions (CLONTECH).
Sequence Analyses.
Human BOULE sequence was obtained by sequencing a clone we obtained from the two hybrid analysis that contained the entire ORF. Mouse Boule was assembled from an expressed sequence tag clone. A mouse testis cDNA was cloned from a mouse testis cDNA library (Genome Systems, St. Louis). CLUSTALW 1.4 alignment of MACVECTOR 6.5 was used for multiple sequence alignments; parameters were open gap penalty of 10, extend gap penalty of 0.1, delay divergence of 40%, and gap distance of 8.
Phylogenetic analyses were performed by using maximum parsimony (protpars) and distance-related (protdist and neighbor-joining) methods with the phylip analysis package (J. Felsenstein, University of Washington, Seattle). The reliability of the assignment of the branches on the phylogeny trees produced was estimated by bootstrapping (bootseq, J. Felsenstein). Trees were constructed by the programs, and a consensus tree was produced with the program consensus and plotted with the program TREEVIEW 1.5 (R. Page, University of Glasgow, Glasgow, U.K.).
Western Blotting and Immunocytochemistry.
Polyclonal antibodies were raised in rabbits by injection of a synthetic oligopeptide (ETQEDAQKILQEAEKLNYKDKKLN) coupled to a multiple-antigen peptide (Research Genetics, Huntsville, AL). Twelve to 20 weeks after initial injection of peptides, serum antibodies were purified as described (6, 18). Testes tissue extracts were used for Western blotting as described with anti-Boule antisera (1:1,000) (6, 18). Mouse testis sections were from 60-day adult mice. Human testis sections were from an adult male with complete spermatogenesis but physical obstruction of ducts.
Northern and Radiation Hybrid Mapping.
Northern hybridization was done on polyadenylated RNA blots (CLONTECH) using human and mouse cDNA clones. For radiation hybrid mapping, primers GAGGAGGTGGATGTGACCC and CTTATTGCTGGACCAATGTTCA were used to amplify a 1.5-kb fragment encompassing the second intron of BOULE. A second primer set (TTTTTCCATTCAGTCTTCCTGA and GCAGAGAAAATAAAAGACACCTCA) was used to confirm linkage. PCR of radiation hybrid panels (GENEBRIDGE 4.0; Research Genetics) was done in duplicate. For mouse mapping, the primers GCTCAGTTGCAGTGTGTTTTTC and TGCTCCATTCCTATATCTGCAA were used and the consensus of two duplicate experiments was used to map the position.
Results
Identification of Human BOULE.
To better understand the molecular function of the human DAZ gene family, we identified proteins that interact with DAZ/DAZL proteins. Using a DAZ/DAZL construct as bait in a two-hybrid screen, we identified a gene remarkably similar to DAZ and DAZL. To our surprise, the protein encoded by this gene is more similar to Drosophila Boule, the proposed fly ortholog of DAZ (19), than to DAZ or DAZL.
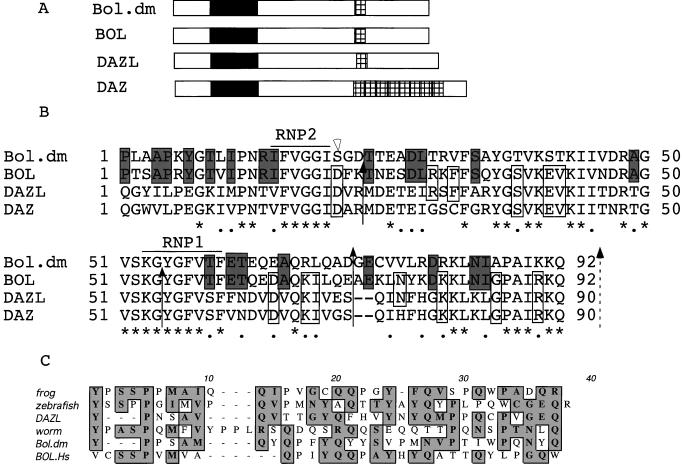
Protein sequence alignments of fly Boule, human DAZ, DAZL, and the product of this gene identified in our screen called BOULE show that BOULE is a closer homolog to Drosophila Boule than either human DAZ or DAZL. The overall identity and similarity between human BOULE and Drosophila Boule is greater (30% identity, 42% similarity) than that between DAZ or DAZL and Drosophila Boule (13% identity and 19% similarity or 22% identity and 32% similarity respectively). Like DAZL, BOULE encodes a potential RNA-binding protein that contains a single RNA-binding domain with signature RNP-1 and RNP-2 motifs (Fig. 1 A and B). The extensive similarity shared by the potential RNA-binding domains of fly Boule, human BOULE, DAZ and DAZL distinguishes these proteins as a unique family of ribonucleoprotein (RNP) proteins, the DAZ family. There is 80% similarity between the domains of BOULE and Boule, 59% between DAZ and Boule, and 61% between DAZL and Boule. We note that BOULE and Drosophila Boule share 21 aa between them that differ in DAZ whereas, in contrast, only 13 aa are shared between DAZ and Boule that differ in human BOULE (Fig. 1A). Furthermore, BOULE and Boule have identical RNP-1 and RNP-2 motifs whereas DAZ and DAZL differ slightly. A DAZ repeat is also present in BOULE, although the repeats are less conserved than the RNA-binding domains (Fig. 1C).
Figure 1.
Human BOULE (BOULE) encodes a protein that is homologous to fly Boule and human DAZ/DAZL. (A) Diagram of BOULE, DAZL, and DAZ proteins. Black boxes represent the RNA-binding domains, hatched boxes represent the DAZ repeats. (B) Alignment of RNA-binding domains of fly Boule, human BOULE, human DAZL, and human DAZ proteins. * indicates conserved residues, and • indicates similar residues among all four proteins. Shadowed boxes outline amino acids shared between Boule and BOULE but different from DAZ or DAZL. Open boxes indicate amino acids shared between DAZ or DAZL and BOULE but not with fly Boule. Solid arrows indicate shared splice sites and open arrows indicate unique splice sites in Boule and/or BOULE. Dashed arrow indicates a shared splice site if we consider the two amino acids (positions 73 and 74) as a single addition/deletion. (C) Conservation of the DAZ repeats in DAZ and BOULE homologs. Shade indicates identical or similar amino acids.
Human BOULE Maps to Chromosome 2.
To determine whether human BOULE is simply a variant copy from the Y chromosome DAZ gene cluster, we mapped the BOULE gene by radiation hybrid mapping and placed BOULE on chromosome 2 at a position 0.4 cR (centiRads) from the marker D2S348 (logarithm of odds = 21). This mapping places BOULE in human 2q33, a position verified by mapping of two genomic bacterial artificial chromosome clones by fluorescence in situ hybridization (data not shown). We further verified these findings by isolating a cDNA fragment from mouse testes and mapping mouse Boule to its chromosomal position. The predicted protein of a mouse cDNA clone is 86% identical to human BOULE and its position on chromosome 1 is syntenic to human 2q33. A male sterile mutation, jsd (juvenile spermatogonial degeneration), maps to the same region.
Expression of Human and Mouse DAZ, DAZL, and BOULE Proteins.
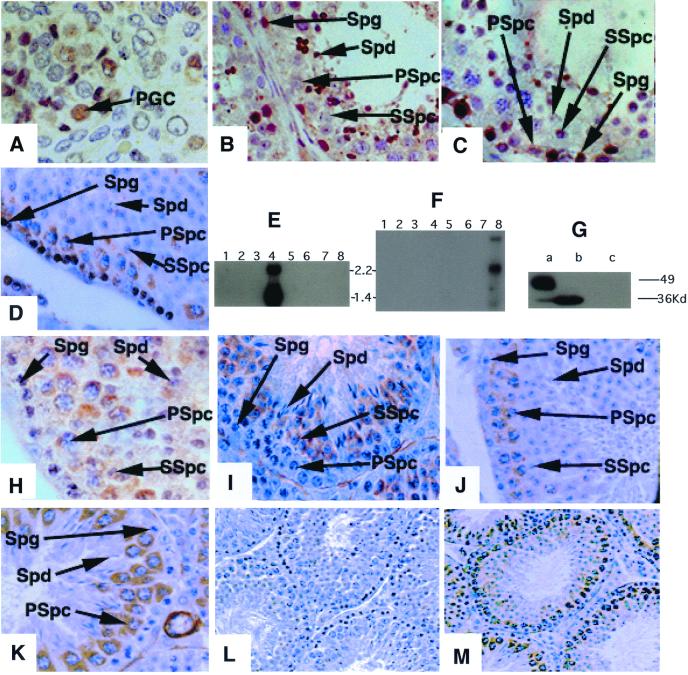
We next compared the expression patterns of the different members of the DAZ family by using antibodies specific to DAZ, DAZL and BOULE. Expression of the DAZ and DAZL genes is restricted to germ cells (5, 6, 12–15, 18, 20–23). As shown, DAZ and DAZL are expressed early in development in both the human male and female (Fig. 2A; refs. 6 and 18). Staining of the nucleus and cytoplasm of primordial germ cells is abundant in both sexes. In the adult, both proteins are expressed only in germ cells with expression most abundant in the nucleus and cytoplasm of spermatogonia and in the cytoplasm of the meiotic spermatocytes (Fig. 2 B and C). DAZL is also abundant in the cytoplasm of oocytes (6). The cellular and subcellular expression pattern of mouse Dazl is identical to that of the human (Fig. 2D). Thus, as expected (6, 12, 18), the expression of proteins encoded by the DAZL and DAZ genes begins early in development in the germ stem cell populations and continues through the meiotic divisions of gametogenesis.
Figure 2.
BOULE expression is distinct from that of DAZ and DAZL. DAZ is expressed in prenatal primordial germ cells, spermatogonial stem cells, and spermatocytes. DAZL is expressed in both the male and female germ line. BOULE is expressed in the cytoplasm of pachytene spermatocytes, persists through meiosis, and decreases in early spermatids. (A) DAZ staining of fetal primordial germ cells of the human testes; similar staining is observed in female primordial germ cells (6, 18). (B) Human testes staining with antisera that recognize DAZL only; DAZL is expressed in spermatogonia, early and late spermatocytes, and postmeiotic cells. Staining of human ovary with this antisera indicate cytoplasmic staining of oocytes (6, 18). (C) Human testis section stained using antisera that recognize DAZ only. DAZ is expressed in spermatogonia and early spermatocytes, but is absent from late spermatocytes or postmeiotic cells. (D) Mouse Dazl is present in spermatogonia and early and late spermatocytes as in human testes. As in humans, female mice also express Dazl in the germ cells (6, 12). (E) A Northern blot with polyadenylated RNA from different human tissues. Blot was hybridized with human BOULE cDNA that detects two testes specific transcripts. Lanes: 1, spleen; 2, thymus; 3, prostate; 4, testis; 5, ovary; 6, small intestine; 7, colon; and 8, leukocyte. (F) A Northern blot with polyadenylated RNA from mouse tissues. Blot was hybridized with human BOULE cDNA that detects three testes-specific transcripts. Lanes: 1, heart; 2, brain; 3, spleen; 4, lung; 5, liver; 6, muscle; 7, kidney; and 8, testis. (G) Anti-BOULE antisera detects a single 32-kDa protein in mouse testes (b) and a similar size protein in human testes (a) but not in other human or mouse tissues (data not shown), nor does it recognize DAZ protein expressed in yeast strain, RRY618 (c). The 50-kDa band in human testes is nonspecific as it is detected by preimmune also. (H) BOULE staining in human testis section is also restricted to cytoplasm of spermatocytes; no staining of spermatogonial stem cells is observed. (I) Stage III seminiferous tubules. BOULE is expressed in round spermatids (Spd) and secondary spermatocytes but not in spermatogonia (Spg) or primary spermatocytes (Spc). (J) Stage VII seminiferous tubule. BOULE is expressed in the cytoplasm of pachytene spermatocytes. There is no staining in spermatogonia and spermatids. (K) Stage X–XI seminiferous tubules. BOULE expression peaks in late pachytene stage spermatocytes. (L and M) Lower-magnification view of staining with preimmune and anti-BOULE antisera. (L) Preimmune of BOULE antisera (×200). (M) BOULE antisera (×200). Spg, spermatogonial cells; Spc, spermatocytes; SSpc, secondary spermatocytes; Spd, spermatids; Pgc, primordial germ cell. Unless noted, all pictures were taken at the same magnification (×630). DAZ and DAZL antisera are described (6, 18).
Considering that the human and mouse BOULE genes are members of the DAZ gene family, we investigated whether the BOULE genes are transcribed and whether BOULE proteins are found in the same cellular and subcellular compartments as DAZ and DAZL. We found that in both mice and men, BOULE transcripts are restricted to the testes (Fig. 2 E and F). Western blotting with antisera specific to BOULE also shows that a 32-kDa BOULE protein is confined to testes in both species (Fig. 2G). Thus, superficially, the expression of BOULE resembles that of human DAZ and DAZL.
However, when we examined the cellular and subcellular distribution of BOULE in mouse and human testes, results indicated that BOULE, unlike DAZ, is not detected in early embryos and there is no BOULE expression in primordial germ cells (data not shown). Furthermore, BOULE is not present in spermatogonial cells but instead is first detectable in the cytoplasm of spermatocytes and then persists through meiosis in both species (Fig. 2 H–K). In mice, which are amenable to staging of germ cell types, BOULE expression begins in stage III spermatocytes and peaks in late pachytene or diplotene stage spermatocytes (Fig. 2 I–K; low-power magnification is shown in Fig. 2 L and M). BOULE is detectable in secondary spermatocytes and early spermatids, then decreases until it is undetectable in spermatids. Thus, the expression of BOULE differs from that of DAZ and DAZL and instead is identical to the meiotic expression pattern of fly Boule (19, 24).
The DAZ Family Contains Two Subgroups.
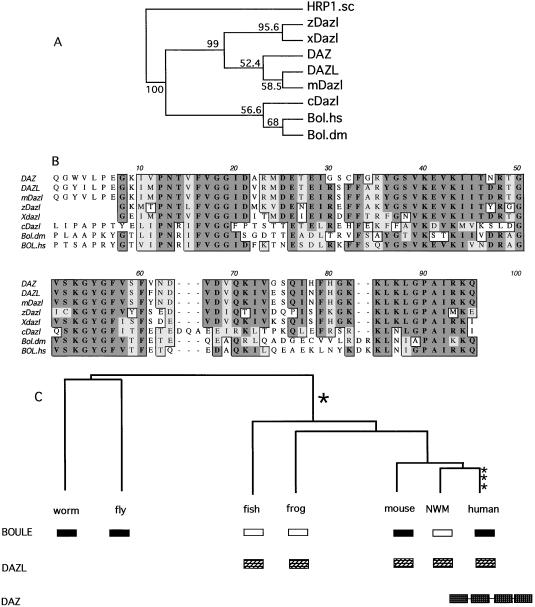
Phylogenetic analyses and alignments of DAZ homologs from major groups of vertebrates and invertebrates suggest that the DAZ family can be divided into two subgroups—DAZ and BOULE. The phylogenetic analyses of RNA-binding domains in homologous DAZ proteins by maximum parsimony and distance methods group DAZ homologs into two separate clades (Fig. 3A). As an outgroup for the phylogenetic tree construction, we use yeast HRP1 protein (U38535), a member of the same RNA-binding protein family to which DAZ and BOULE belong. The DAZ clade contains mammalian, zebrafish, and frog Dazl homologs whereas the BOULE clade includes mammalian BOULE, fly Boule, and worm Dazl. Indeed alignment of functional motifs indicates homologs in zebrafish and frog are closer to DAZL in overall sequence and that worm Daz-1 is almost equally divergent from BOULE and DAZ although slightly closer to the BOULE group. This suggests that the homologs in zebrafish and frog are indeed DAZL homologs whereas worm Daz-l is more likely a BOULE homolog, a suggestion that is corroborated by distinct features in the RNA binding domains unique to each subgroup of DAZ family (Fig. 3B). First, there is a 2-aa deletion shared by human, mouse, frog, and zebrafish DAZ homologs that is not present in human BOULE (BOULE.hs) and fly Boule (BOULE.dm). Second, in the most conserved RNP-1 and RNP-2 motifs, all Boule homologs encode amino acids IFVGG whereas DAZ homologs encode V/LFVGG. RNP-1 in Boule encodes KGYGFV/IT, but KGYGFV/IS/Y in DAZ. The sequence between RNP-1 and the C terminus of the RNA-binding domain yields the same grouping.
Figure 3.
DAZ/DAZL evolved from Boule. (A) Phylogenetic tree based on the RNA-binding domains of human BOULE (BOULE.hs), fly Boule (BOULE.dm), human DAZL (DAZL), and known homologs from mouse (mDazl), worm (cDazl), zebrafish (zDazl), and frog (Xdazl). Yeast HRP1 (HRP1.sc), which belongs to the same RNP family as DAZ/BOULE, was used as an outgroup. Both maximum parsimony and distance-based methods using PHYLIP 3.5 (J. Felsenstein) produce trees of similar topology. The consensus tree presented here is built with maximum parsimony method (PROTPARS). Numbers indicate the percentage of the bootstrapping trials in which an identical node was produced. The number of bootstrap replicates was 100. The position of DAZ was placed outside its ancestor mouse Dazl due to greater divergence in DAZ than DAZL. (B) Multiple sequence alignment of RNP domains in BOULE and DAZL homologs. Conserved residues are boxed (dark shadow is identical and light shadow is similar). (C) Model of evolutionary history of BOULE/DAZ family. Dark box represents BOULE homologs and hatched box represents DAZL homologs. Open box indicates the inferred presence of a BOULE homolog that has yet to be identified. The DAZ gene cluster on the Y chromosome is represented by four hatched boxes linked together to indicate the presence of at least four genes in tandem (3, 25). Vertical hatch box indicates the probable period of gene duplication. NWM, New World monkey.
Boule Is the Ancestor of the DAZ Family.
We next explored the evolutionary history of the DAZ family. It is certain that only one BOULE homolog exists in flies and worms and that there are no DAZ homologs in these organisms because both genomes have been completely sequenced. Yet both BOULE and DAZL exist in mammals. Thus, DAZL was either lost in the invertebrate lineage or arose during vertebrate evolution. Comparison of genomic structure among human DAZL genes, human BOULE, and fly Boule supports a rise of Dazl from BOULE during early vertebrate lineage. We determined the genomic structure of BOULE and found that, like DAZL and DAZ, BOULE has 11 exons and shares six splicing positions with DAZ/DAZL (Fig. 1B). Eight splice positions are shared if we consider junctions with a single deletion/addition difference. The observations that there are more exon-intron splicing sites shared between human BOULE and DAZL than between BOULE and fly Boule, and that there is an identical number of exons in BOULE and DAZL, suggest that the historical relationship between BOULE and DAZL is closer than that between BOULE and fly Boule. Such observations argue against loss of DAZL in invertebrates. Our phylogenetic analyses also support the scenario that the DAZL genes were derived from Boule via duplication after separation of protostomes and deuterostomes (Fig. 3A). This finding is not in conflict with the greater similarity of protein sequence between human BOULE and fly Boule, which reveals either similar functional constraint on these proteins or selection for optimization of a new function for DAZL or both. Hence, we propose that the single copy gene, Boule, is the DAZ ancestor of the vertebrate and invertebrate lineages and that DAZL arose in the vertebrate lineage through gene duplication. Based on sequence alignments and expression patterns of DAZL homologs in frog and zebrafish (14–17), DAZL arose through a duplication of Boule that occurred before the divergence of modern vertebrates, but after splitting of invertebrates and vertebrates (Fig. 3C). Later during early primate evolution, after divergence of New World monkey and Old World monkeys, another duplication took place from Dazl, resulting in a new Y-linked DAZ. Y-linked DAZ went through two more duplications as recently as 55,000 years ago, giving rise to a cluster of four DAZ genes (9, 10, 25) (Fig. 3C).
Discussion
Here we report identification of human BOULE and compare its expression to that of DAZ and DAZL in humans and mice. Our results suggest that human BOULE and mouse Boule are members of the DAZ family, a gene family that encodes proteins that are distinct from other RNA-binding proteins based on three features: First, DAZ, DAZL, and BOL share a degree of similarity in the RNA-binding domain that is not shared with other RNA-binding proteins (26). Second, the proteins of the DAZ family have a DAZ repeat. The function of this repeat is not known, but may be involved in protein–protein interactions (27). Finally, members of the DAZ family are germ cell-specific in their expression.
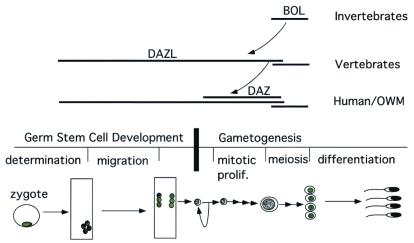
The phenotypes associated with disruption of genes in the DAZ family are diverse as are the expression profiles (Table 1). Such differences in expression and function among DAZ family members suggest that in humans, BOULE, not DAZL, may be the ortholog of Drosophila boule and that homologs of DAZL and BOULE perform functions at different stages of germ cell development (Fig. 4). Although definitive functional relationships will be determined when mutations in human and mouse BOULE are identified and phenotypes can be compared, our data clearly indicate that a component of meiotic regulation involving BOULE has been conserved from flies, to worms to humans.
Figure 4.
The ancient meiotic regulator, Boule, is conserved in all metazoans and gave rise to a gene family required for novel vertebrate germ cell functions. Expression pattern of each member is indicated by extent of each horizontal line. Meiotic expression of BOULE is seen in fly, mice and humans (primary spermatocytes) and in female oocytes before meiosis (8, 11). This meiotic function is likely conserved throughout metazoans. DAZL evolved a novel function required for germ stem cell proliferation or differentiation that is unique to vertebrates. DAZL is expressed in multiple compartments from germ stem cells to mature spermatocytes. DAZ arose from DAZL recently in the primate lineage and is expressed in multiple compartments from germ stem cells to meiotic cells. OWM, Old World monkey. Major stages of germ cell development are diagramed in the middle to provide a timeline for DAZ family gene expression. The curved arrow indicates a duplication.
Our phylogenetic analyses, taken together with previous studies on DAZL homologs in the frog and mouse, suggest DAZL has evolved a novel premeiotic function unique to vertebrates. Whereas BOULE homologs appear to perform a meiotic function, the derived DAZL homologs acquired an essential function in early germ cell development. This is suggested by premeiotic expression of DAZL in germ cells of males and females and an essential requirement in germ stem cell development in frog and mouse (12, 16). In frogs and mice, disruption of DAZL homologs leads to loss of premeiotic stem cell populations early in development. In contrast, in flies and worms, disruption of the BOULE homologs leads to meiotic defects late in germ cell development. Yet, remnants of the traditional role for DAZL being maintained in vertebrates is evident by the observation that Xenopus Dazl can rescue a fly boule meiotic mutation (14). This indicates that although the DAZL genes may have acquired a new function they preserve an RNA-binding domain that can bind the same RNA as Boule and can substitute for Boule when expressed in the right place at the right time. The novel function of DAZL in premeiotic germ cell development has not come at the expense of the functional unit of Boule. Instead, we suggest that the RNA-binding domain of the ancient meiotic regulator, Boule, has been used as a module for innovation to evolve a family of genes with different functions in germ-line development.
Finally, in contrast to both DAZL and BOULE, DAZ is not essential for completion of spermatogenesis as evidenced by two facts. First, deletions encompassing the DAZ genes cause phenotypes that include low numbers of sperm. Second, analysis of human DAZ exons and introns suggests that they are evolving at the same rate, an observation consistent with neutral genetic drift or positive selection on the DAZ proteins (1, 28–30). Thus, we speculate that perhaps DAZ has yet to evolve a function essential for completion of spermatogenesis. The distinct expression patterns of the human DAZ family suggests that through evolution, members of the family have already or may gradually acquire new functions in the human reproductive pathway (Fig. 4). Perhaps the DAZ genes, which recently duplicated twice (25), are probing the evolutionary possibilities of nature. Whether the emergence of DAZ on the human Y chromosome is a reinforcement of the ancient meiotic function of Boule or a novel function in the making remains to be seen as DAZ genes evolve.
Acknowledgments
We thank Ira Herskowitz, Holly Ingraham, Synthia Mellon, Robert Taylor, Paul Turek, Richard Weiner, Pauline Yen, and members of the Reijo lab, as well as two anonymous reviewers for comments on the manuscript, Jingly F. Weier for fluorescence in situ hybridization mapping, and Jadwiga Jaruzelska for assistance in the two-hybrid screen. This work is supported by a National Research Service Award (to E.Y.X.), a Ford Foundation and Woodrow Wilson Foundation fellowship (to F.L.M.), and grants from the National Institutes of Health, the Searle Foundation, and the Sandler Family Foundation (to R.A.R.P.).
Abbreviation
- RNP
ribonucleoprotein
Footnotes
References
- 1.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 2.Cooke H J, Elliot D J. Trends Genet. 1997;13:87–92. doi: 10.1016/s0168-9525(97)01066-4. [DOI] [PubMed] [Google Scholar]
- 3.Saxena R, Brown L G, Hawkins T, Alagappan R K, Skaletsky H, Reeve M P, Reijo R, Rozen S, Dinulos M B, Disteche C M, Page D C. Nat Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 4.Seboun E, Barbaux S, Bourgeron T, Nishi S, Algonik A, Egashira M, Nikkawa N, Bishop C, Fellow M, McElreavey K, Kasahara M. Genomics. 1997;41:227–235. doi: 10.1006/geno.1997.4635. [DOI] [PubMed] [Google Scholar]
- 5.Yen P H, Chai N N, Salido E C. Hum Mol Genet. 1996;5:2013–2017. doi: 10.1093/hmg/5.12.2013. [DOI] [PubMed] [Google Scholar]
- 6.Dorfman D M, Genest D R, Pera R A R. Hum Reprod. 1999;14:2531–2536. doi: 10.1093/humrep/14.10.2531. [DOI] [PubMed] [Google Scholar]
- 7.Chai N N, Phillips A, Fernandez A, Yen P. Mol Hum Reprod. 1997;3:705–708. doi: 10.1093/molehr/3.8.705. [DOI] [PubMed] [Google Scholar]
- 8.Eberhart C G, Maines J Z, Wasserman S A. Nature (London) 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 9.Carani C, Gromoll J, Brinkworth M H, Simoni M, Weinbauer G F, Nieschlag E. Mol Hum Reprod. 1997;3:479–483. doi: 10.1093/molehr/3.6.479. [DOI] [PubMed] [Google Scholar]
- 10.Gromoll J, Weinbauer G F, Skaletsky H, Schlatt S, Rocchietti-March M, Page D C, Nieschlag E. Hum Mol Genet. 1999;8:2017–2024. doi: 10.1093/hmg/8.11.2017. [DOI] [PubMed] [Google Scholar]
- 11.Karashima T, Sugimoto A, Yamamoto M. Development (Cambridge, UK) 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- 12.Ruggiu M, Speed R, Taggart M, McKay S J, Kilanowski F, Saunders P, Dorin J, Cooke H. Nature (London) 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 13.Shan Z, Hirschmann P, Seebacher T, Edelmann A, Jauch A, Morell J, Urbitsch P, Vogt P H. Hum Mol Genet. 1996;5:2005–2011. doi: 10.1093/hmg/5.12.2005. [DOI] [PubMed] [Google Scholar]
- 14.Houston D W, Zhang J, Maines J Z, Wasserman S A, King M L. Development (Cambridge, UK) 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- 15.Mita K, Yamashita M. Mech Dev. 2000;94:251–255. doi: 10.1016/s0925-4773(00)00295-1. [DOI] [PubMed] [Google Scholar]
- 16.Houston D W, King M L. Development (Cambridge, UK) 2000;127:447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 17.Maegawa S, Yasuda K, Inoue K. Mech Dev. 1999;81:223–226. doi: 10.1016/s0925-4773(98)00242-1. [DOI] [PubMed] [Google Scholar]
- 18.Reijo R A, Dorfman D M, Slee R, Renshaw A A, Loughlin K R, Cooke H, Page D C. Biol Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- 19.Maines J Z, Wasserman S A. Nat Cell Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- 20.Cooke H J, Lee M, Kerr S, Ruggiu M. Hum Mol Genet. 1996;5:513–516. doi: 10.1093/hmg/5.4.513. [DOI] [PubMed] [Google Scholar]
- 21.Maiwald R, Luche R M, Epstein C J. Mamm Genome. 1996;7:628. doi: 10.1007/s003359900292. [DOI] [PubMed] [Google Scholar]
- 22.Menke D B, Mutter G L, Page D C. Am J Hum Genet. 1997;60:237–241. [PMC free article] [PubMed] [Google Scholar]
- 23.Reijo R, Seligman J, Dinulos M B, Jaffe T, Brown L G, Disteche C M, Page D C. Genomics. 1996;35:346–352. doi: 10.1006/geno.1996.0366. [DOI] [PubMed] [Google Scholar]
- 24.Cheng M H, Maines J Z, Wasserman S A. Dev Biol. 1998;204:567–576. doi: 10.1006/dbio.1998.9098. [DOI] [PubMed] [Google Scholar]
- 25.Saxena R, de Vries J W A, Repping S, Alagappan R K, Skaletsky H, Brown L G, Ma P, Chen E, Hoovers J M N, Page D C. Genomics. 2000;67:256–267. doi: 10.1006/geno.2000.6260. [DOI] [PubMed] [Google Scholar]
- 26.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 27.Tsui S, Dai T, Roettger S, Schempp W, Salido E C, Yen P H. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 28.Reijo R, Alagappan R K, Patrizio P, Page D C. Lancet. 1996;347:1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- 29.Agulnik A I, Zharkikh A, Tong H B, Bourgeron T, McElreavey K, Bishop C E. Hum Mol Genet. 1998;7:1371–1377. doi: 10.1093/hmg/7.9.1371. [DOI] [PubMed] [Google Scholar]
- 30.Bielawski J P, Yang Z. Mol Biol Evol. 2001;18:523–529. doi: 10.1093/oxfordjournals.molbev.a003831. [DOI] [PubMed] [Google Scholar]