Abstract
Coenzyme Q10 (CoQ10, ubiquinone), a potent antioxidative dietary supplement, was produced by submerged fermentation using Agrobacterium tumefaciens instead of chemical synthesis or solvent extraction. Agrobacterium tumefaciens 1.2554 was subjected to mutagenesis using a series of treatments including high hydrostatic pressure (HHP) treatment, UV irradiation, and diethyl sulfate (DES) treatment to obtain mutant strains showing higher CoQ10 production than wild-type strains. A mutant strain PK38 with four genetic markers was isolated: the specific CoQ10 content of the mutant strain increased by 52.83% compared with the original strain. Effects of carbon and nitrogen sources on CoQ10 production with PK38 were studied. Sucrose at concentration of 30 g/l was tested as the best carbon source, and yeast extract at concentration of 30 g/l supplemented with 10 g/l of ammonium sulfate was identified to be the most favorable for CoQ10 production using PK38. Fed-batch culture strategy was then used for increasing production of CoQ10 in 5-l fermentor. Using the exponential feeding fed-batch culture of sucrose, cell growth and CoQ10 formation were significantly improved. With this strategy, the final cell biomass, CoQ10 production, and specific CoQ10 production increased by 126.11, 173.12, and 22.76%, respectively, compared to those of batch culture.
1. Introduction
Coenzyme Q10 (CoQ10), also known as ubiquinone or ubiquinone-10, occurs widely in animals, plants, and the cells of microorganisms. It plays a crucial role in generation of cellular energy and in free radical scavenging in the human body [1]. Accordingly, it has been used in therapeutic applications for several diseases such as heart disease [2], breast cancer [3], and Alzheimer's and Parkinson's diseases [4]. Moreover, CoQ10 has been used as a food supplement and cosmetic ingredient because of its various physiological functions. Extensive attempts have been made to increase CoQ10 production to meet growing demands for this product. To date, production of CoQ10 is produced by one of three methods: extraction from biological tissues [5], chemical synthesis [6], and microbial fermentation [7]. In the wake of recent environmental awareness, the first two methods became least desirable because of the inherent uses of solvents and chemicals in the process. Microbial fermentation, conversely, offers an environmentally benign option based on the enzymatic catalysis at the cellular level for CoQ10 assembly. Also, this approach is attractive to the industry because the process is easy to control and has a relatively low production cost [8, 9]. Among all strains investigated so far, A. tumefaciens has been shown to be an excellent producer of CoQ10 [7, 10–13]. However, the yield of CoQ10 in liquid cultivation using the wild-type strain of A. tumefaciens remains limited because of its low specific CoQ10 content.
To increase the specific CoQ10 content of A. tumefaciens, further strain development by physical and chemical mutagenesis has been used to obtain high-CoQ10-producing mutants. Well-known mutagens such as ultraviolet radiation and diethyl sulfate (DES) have been tested. However, the chemicals used for these mutagenesis procedures are harmful to human health. It is desirable to find new mutagenic treatments to increase CoQ10 yield.
In recent years, there were reports about high hydrostatic pressure (HHP) treatment that influenced the structure of genes and proteins in microorganisms [14–16]. There were also reports that HHP treatment in laboratory could cause beneficial mutagenesis to E. coli [17, 18], R. glutinis [19, 20], and G. xylinus [21]. Compared with traditional physical and chemical mutagenesis methods, HHP treatment as a mutagen could offer some advantages such as easier handling, savings in time and money, and negligible effects to the environment [19, 20]. However, little is known related to the effectiveness of HHP treatment on improving the production of CoQ10 in microorganisms.
Carbon and nitrogen sources significantly affected cell growth and CoQ10 production [12]. Aside from the optimization of carbon and nitrogen sources, the use of fed-batch culture by controlling the nutrient feeding is one of the most popular methods to achieve high cell density of E. coli [22–24] and enhance the production of CoQ10 [11, 12]. Exponential feeding is a simple method that was widely employed for E. coli cultivation [23]. So far, there seem to be no published reports related to the CoQ10 production under exponential feeding fed-batch control.
In this study, HHP treatment was investigated as a new mutagenic treatment to increase the specific CoQ10 content of A. tumefaciens. A mutant strain PK38 was isolated using selection markers and the optimal carbon and nitrogen sources for CoQ10 production with PK38 were selected. A new exponential feeding strategy was proposed for improving CoQ10 production using PK38.
2. Materials and Methods
2.1. Microorganism and Culture Medium
The parental strain A. tumefaciens 1.2554 was purchased from China General Microbiological Culture Collection Center (CGMCC, Beijing, China). This strain was inoculated on mannitol agar slants, incubated for 2 days at 28°C, and then stored at 4°C.
All media were sterilized by autoclaving at 121°C for 20 min and were adjusted to pH 7.2 before sterilization. The complete medium contained 5 g/L glucose, 3 g/L beef extract, 3 g/L yeast extract, 10 g/L peptone, and 0.2 g/L MgSO4·7H2O. The selective medium was made by adding a certain amount of one of the following four substances: sodium azide, ethionine, daunomycin, or vitamin K3. The seed medium consisted of 10 g/L glucose, 5 g/L peptone, 5 g/L yeast extract, and 5 g/L NaCl. The basal fermentation medium contained 20 g/L glucose, 10 g/L peptone, 10 g/L yeast extract, 0.5 g/L K2HPO4, 0.5 g/L KH2PO4 0.5 g/L, and 0.5 g/L MgSO4·7H2O.
2.2. Preparation of Cell Suspensions
One loop of bacterial cells grown overnight on a mannitol agar slant was inoculated to 250 mL Erlenmeyer flask containing 50 mL of a seed medium and incubated on a rotary shaker (200 rpm) at 28°C for 24 h. After the culture entered the log phase (about 18–24 h in this study), 20 mL culture broth was centrifuged under 4°C at 10,000 rpm (21,000 ×g) for 10 min (PM180R, ALC International, Milan, Italy), washed twice with sterile physiological saline, and suspended in sterile 0.1 M potassium phosphate buffer of pH 7.2 (cells density 106–107 cell/mL) for HHP treatment.
2.3. HHP Mutation
The cell suspension in potassium phosphate buffer was transferred aseptically into sterile polyethylene pouches and heat-sealed following the expulsion of air. The prepared pouches were placed into the HHP equipment, containing a 2 l working pressure chamber (UHPF-750MP, Baotou Kefa New Type Hi-Tech Food Machine Limited Company, Baotou, China). HHP treatments at constant pressures from 100 to 400 MPa with holding time (10 to 30 min) were carried out at room temperature (about 25°C) using castor oil as the pressure medium. Control samples were maintained at atmospheric pressure within the constant temperature housing during the experiment.
To assess loss of viability caused by the HHP treatment, untreated and treated cell suspensions were serially diluted in PBS and plated on the basal agar plates. Agar plates were incubated at 28°C for 2 days for colony counting. Inactivation was expressed as a logarithmic viability reduction log(N0/N), with N0 and N representing the colony counts before and after treatment, respectively [25].
2.4. UV + DES Mutation
The cell suspension was spread onto a presterilized plate (9 cm diameter), and a UV lamp (254 nm, 30 W) was used for the mutation by irradiating cells at a distance of 30 cm from the plate for 60 s. After UV radiation treatment, the cell suspension was treated with 1% (v/v) DES for 20 min. The results show that the frequency of positive mutant generation is 12%.
2.5. Screening for High-CoQ10-Producing Mutant
After each treatment, cell suspensions were spread on the selective medium and incubated at 28°C for 48 h. The selective medium contained L-ethionine (500 mg/L), daunomycin (20 mg/L), vitamin K3 (160 mg/L), or sodium azide (20 mg/L). All these chemicals were purchased from Ding Guo Biological Technology Co., LTD (Beijing, China). The fast-growing, large, and single colony was transferred onto the selective medium plate and then incubated for 48 h at 28°C. The slant was preserved at 4°C.
For screening, each mutant was taken and used in fermentation, and the content of CoQ10 from each mutant was determined.
2.6. Fermentation
The seed culture was transferred into a stirred tank fermentor (BioFlo 110, New Brunswick, NJ, USA) with a working volume of 2 l production medium. The temperature, agitation speed, and air flow rate during the culture were 28°C, 300 rpm, and 0.6 l/min, respectively. The pH was controlled at 7.2 ± 0.1 by addition of 3 M NaOH or 2 M HCl.
2.7. Analytical Methods
The cell mass concentration was determined using a calibration curve constructed by optical density at 620 nm and dry cell weight (DCW). The optical density at 620 nm was measured with a spectrophotometer (UV-2500, Shimadzu, Tokyo, Japan). The DCW was determined after the culture broth was centrifuged at 10,000 rpm (21,000 ×g) for 10 min under 4°C using an ultracentrifuge. Cell lysis and CoQ10 extraction conducted in this study were similar to that described by Tian et al. [26]. The extracted CoQ10 was dissolved in ethanol and applied to a high-performance liquid chromatography (HPLC) system (LC-2010A, Shimadzu, Tokyo, Japan) with a Hypersil ODS C18 (5 μm, 4.6 mm × 250 mm, Germany) coupled with a UV detector (Waters 486). The column was eluted with ethanol and methanol (9 : 1, v/v) at a flow rate of 1.0 mL/min, and a chromatogram was obtained by monitoring the absorbance at 275 nm. With an authentic CoQ10 standard (Sigma Co., Shanghai, China), CoQ10 was identified according to retention time and quantified by using a calibration curve. The residual sugar was quantified by Fehiling's reaction.
2.8. Statistical Analysis
All analyses were performed in triplicate. The experimental results obtained were expressed as means ± SD. Statistical analysis was performed using the SPSS package (version 11.5, SPSS Inc., Chicago, IL). Data were analyzed by analysis of variance (P < 0.05), and the means were separated by Duncan's multiple range test.
3. Results and Discussion
3.1. Effect of HHP Treatment on Lethal Rate and Mutation of A. tumefaciens
To determine the optimum mutagenic conditions by HHP treatment, cell suspensions of A. tumefaciens were subjected to different HHP treatments combining pressure with holding time at 25°C. Figure 1 shows the death curve of A. tumefaciens with HHP treatment. The data revealed that HHP treatment had a significant effect on A. tumefaciens, a Gram-negative bacterium. In general, Gram-negative bacteria are less resistant than the Gram-positive bacteria to HHP treatments [25]. In this study, the cell viability during HHP treatment decreased with the increase of processing pressure and holding time. Based on the curve (Figure 1), deactivation of cells occurred between 4.0 and 4.5 log units at 200 MPa for 30 min, 250 MPa for 20 min, or 400 MPa for 10 min. Under these three levels, the mortality of A. tumefaciens was close to 100% (data not shown). Therefore, these three levels were chosen for subsequent mutagenesis [21].
Figure 1.
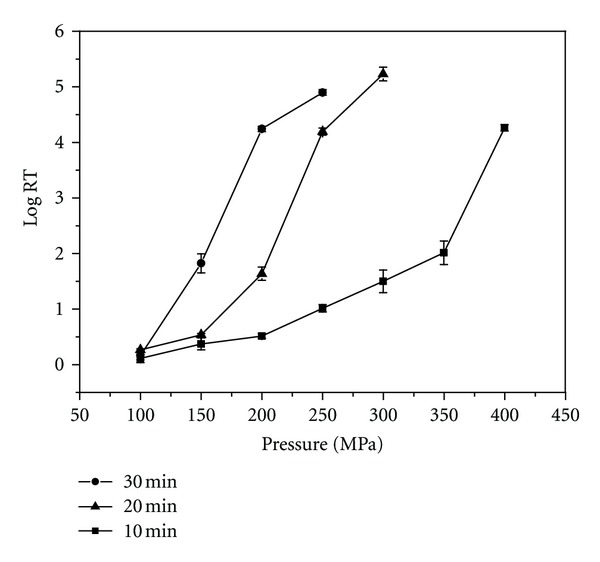
Death curve of A. tumefaciens caused by HHP treatment. Process temperature 25°C. logRF = log(N0/N). Data are shown as mean ± SD (n = 3).
Table 1 shows the effect of HHP treatments on mutation of A. tumefaciens. It is evident that the HHP treatment, at 200 MPa for 30 min, is better than the other two treatments: 250 MPa for 20 min and 400 MPa for 10 min. Positive mutations accounted for 16% of these mutants, and 12 strains had a specific CoQ10 content higher than that from the wild-type strain. Among those 12 strains, maximum specific CoQ10 content was achieved from the mutant PN07 with an approximate 17.5% increase compared with the wild-type strain. So the HHP treatment at 200 MPa for 30 min was selected as the optimal condition for inducing mutation.
Table 1.
HHP treatments on mutation of A. tumefaciens.
| Mutation | No. of strains (N) | Positive mutant (r %) | Specific CoQ10 content (mg/g DCW) | Improvement (r %) |
|---|---|---|---|---|
| 400 Mpa/10 min | 56 | 5 | 1.645 | 8.4 |
| 250 Mpa/20 min | 80 | 10 | 1.690 | 11.3 |
| 200 Mpa/30 min | 78 | 16 | 1.784 | 17.5 |
CoQ10 biosynthesis is typically composed of three parts: synthesis of a quinonoid ring, synthesis of decaprenyl diphosphate, and quinonoid ring modification [9]. The formation of each part is catalyzed by several enzymes. For example, decaprenyl diphosphate synthase (DPPS) can catalyze the synthesis of decaprenyl diphosphate, which appears to be a rate-limiting step and critical in CoQ10 production [9, 27]. The present study found that HHP treatment can have beneficial mutagenic effects on A. tumefaciens for CoQ10 production, which agrees with the finding of Wu et al. [21] who found that the bacterial cellulose mutant M438 was obtained after HHP treatment at 250 MPa for 15 min. Wang et al. [19, 20] obtained the barotolerant mutant PR68 after five repeated cycles of HHP treatment at 300 MPa for 12 min. They found that the DNA segments of mutant PR68 were different from the original strain. Lauro et al. [28] reported that pressure triggered a stress response which activated distinct chaperones and DNA repair proteins. So the change of specific CoQ10 content of A. tumefaciens might occur at the gene level, and HHP treatment might have effects on the three enzymatic steps of CoQ10 biosynthesis. Study of these effects should be carried out in future research.
3.2. Screening A. tumefaciens
Random mutagenesis is an easy tool to use in achieving genetic and functional modifications of an organism. Using progressive stepwise mutagenesis-selection protocols and various mutagens with differing modes of action has been proven effective in increasing product yield [29]. To obtain a high-CoQ10-producing strain from the wild type, mutagenesis was carried out by the HHP treatment (200 MPa, 30 min) and UV+DES mutation. After each mutagenic treatment, the mutant strain was selected and assessed in shake flask cultures. The most promising strain was subjected to the next mutagenic treatment.
According to the general pathway of CoQ10 synthesis, two means of additional CoQ10 production are possible: the mutant could overcome growth inhibition during CoQ10 biosynthesis or its related metabolisms might overproduce CoQ10 [9]. These chemicals included L-ethionine (an analogue of L-methionine, which is a precursor for the methoxy moiety of CoQ10), daunomycin, and vitamin K3 (structural analogs of CoQ10). CoQ10 is an electron carrier in the respiration chain with antioxidant activity [30]. An electron flux inhibitor, such as sodium azide, can be used for screening the mutant, which can be resistant to this inhibitor because of high intracellular CoQ10 [9]. Moreover, tyrosine is located at a final branch point in the pathway of CoQ10 biosynthesis. So a tyrosine auxotrophic mutant might enhance CoQ10 production.
As Figure 2 shows, the mutant strains isolated with respective chemicals at each mutation step were tested for their CoQ10 production by flask culturing, and the best strain was used as the parent for successive mutations. Figure 2 also shows the methods and the maximal specific CoQ10 content for such mutants. Among those chemicals used for selecting a high producer, daunomycin had no significant effect on the CoQ10 content. Finally, the strain with the highest specific CoQ10 content, named PK38, was obtained, containing about 52.83% higher specific CoQ10 content when compared with the original strain.
Figure 2.
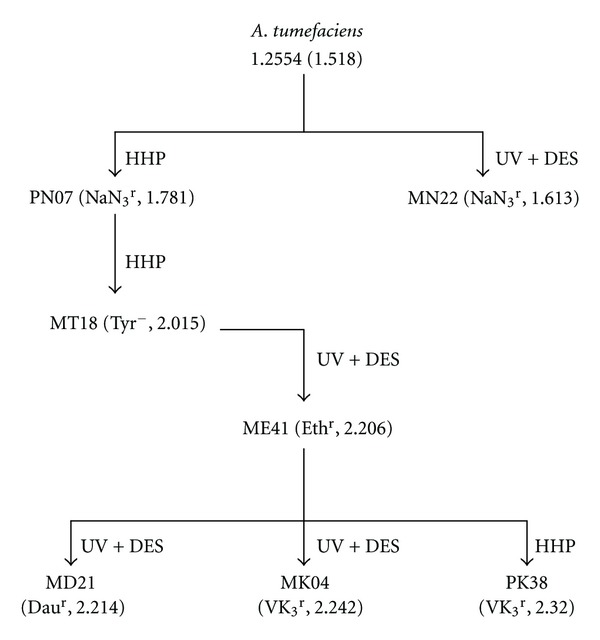
Genealogy of mutant strains derived from AT 1.2554. Mutation markers: NaN3r, sodium azide (NaN3) resistance; Tyr−, tyrosine auxotroph; Ethr, ethionine resistance; Daur daunomycin resistance; VK3r, vitamin K3 resistance. All the mutants were induced by HHP or UV + DES treatment. The specific CoQ10 content (mg/g dry cell weight) was obtained in flask tests.
3.3. Stability of PK38 for CoQ10 Production
The mutant PK38 was selected, and its stability of producing specific CoQ10 content was investigated (Table 2). There was little fluctuation in specific CoQ10 content among 10 generations. Also, the genetic markers were stable. This illustrated PK38 strain has a good genetic stability and has the potential to be used for CoQ10 production.
Table 2.
The experiment testing genetic stability and specific CoQ10 content of PK38.
| Generations | 2 | 4 | 6 | 8 | 10 |
|---|---|---|---|---|---|
| Genetic markersa | + | + | + | + | + |
| Specific CoQ10 content (mg/g) | 2.321 | 2.320 | 2.318 | 2.319 | 2.318 |
aGenetic markers included NaN3r, Tyr−, Ethr, and Vk3r.
3.4. Optimization of Carbon and Nitrogen Sources
To determine the optimal carbon and nitrogen source for CoQ10 production, cultures were prepared in 250 mL flasks containing 100 mL of fermentation medium with various carbon and nitrogen sources and incubated for 72 h on a rotary shaker (at 180 rpm) at 28°C.
Figure 3 shows the effect of different carbon sources on PK38 growth and CoQ10 production. Among the various carbon sources (glucose, fructose, lactose, maltose, sucrose, and xylose), sucrose proved to be the best carbon source for the growth of PK38 with biomass reaching 5.28 g/L after 72 hours of cultivation. Also, CoQ10 production was the highest when sucrose was used as a carbon source, reaching 12.27 mg/L. These results are in agreement with the work conducted by Ha et al. [12]. To evaluate the effect of the initial concentrations of sucrose on PK38 growth and CoQ10 production, different concentrations of sucrose (10, 20, 30, 40, and 50 g/L) were used. As Table 3 shows, CoQ10 production was highest at 30 g/L of sucrose, reaching 14.88 mg/L. So sucrose, at a concentration of 30 g/L, was selected as carbon source for CoQ10 production with PK38.
Figure 3.
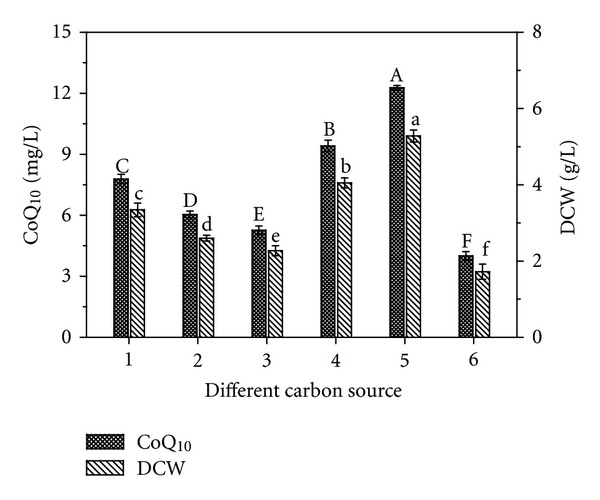
Effect of various carbon sources on the cell growth and CoQ10 production. Cells were cultivated in the test tube at 28°C for 72 h. The carbon sources included glucose (1), fructose (2), lactose (3), maltose (4), sucrose (5), and xylose (6). Data are shown as mean ± SD (n = 3). Means with the same letter are not significantly different (P < 0.05).
Table 3.
Effect of sucrose on DCW and CoQ10 production by PK38 with batch culture.
| Sucrose (g/L) | DCW (g/L) | CoQ10 concentration (mg/L) | Specific CoQ10 content (mg/g DCW) |
|---|---|---|---|
| 10 | 4.11 + 0.065d | 9.55 + 0.196d | 2.325 + 0.037a |
| 20 | 5.28 + 0.157b | 12.27 + 0.110b | 2.322 + 0.029a |
| 30 | 6.40 + 0.161a | 14.88 + 0.172a | 2.326 + 0.069a |
| 40 | 4.90 + 0.181c | 11.40 + 0.284c | 2.324 + 0.101a |
| 50 | 4.12 + 0.148d | 9.56 + 0.056d | 2.320 + 0.019a |
Data are shown as mean ± SD (n = 3). Means with the same letter are not significantly different as indicated by Duncan's multiple range test (P < 0.05).
Figure 4 shows the effects of different nitrogen sources on biomass and CoQ10. The results demonstrate that crop steep powder (CSP) was more desirable than other nitrogen sources. The highest CoQ10 production (17.65 mg/L) was obtained with 20 g/L of CSP, with the highest biomass at 7.16 g/L. CSP comprises the water soluble components of the crop, the composition of which is primarily amino acids, peptides, carbohydrates, and salts. CSPs are rich in nitrogen, vitamins, and other growth stimulating compounds. Therefore, CSP was used as an ingredient in media for cultivation of microorganisms. Different concentrations of CSP (10, 20, 30, 40, and 50 g/L) were used to evaluate the effect of the initial concentrations of CSP on PK38 growth and CoQ10 production. As Table 4 shows, CoQ10 production was the highest with 30 g/L of sucrose, reaching 18.92 mg/L. Ammonium may also have effects on CoQ10 synthesis. Knowles and Redfearn [31] found that the cells of Azotobacter vinelandii grown on ammonium medium produced comparatively high concentrations of CoQ10. Obviously, the complex nitrogen source (CSP + ammonium sulfate) was more desirable than single nitrogen sources (Table 5). CoQ10 production reached 20.62 mg/L with 30 g/L CSP and 10 g/L ammonium sulfate in the production medium.
Figure 4.
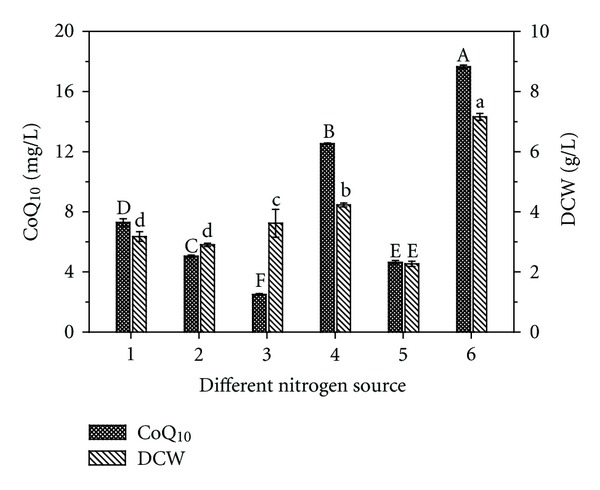
Effect of various nitrogen sources on the cell growth and CoQ10 production. Cells were cultivated in the test tube at 28°C for 72 h. The nitrogen sources included yeast extract (1), peptone (2), tryptone (3), soytone (4), ammonium sulfate (5), and CSP (6). Data are shown as mean ± SD (n = 3). Means with the same letter are not significantly different (P < 0.05).
Table 4.
Effect of CSP on DCW and CoQ10 production by PK38 with batch culture.
| CSP (g/L) | DCW (g/L) | CoQ10 production (mg/L) | Specific CoQ10 content (mg/g-DCW) |
|---|---|---|---|
| 10 | 6.67 + 0.061c | 15.67 + 0.080d | 2.351 + 0.016c |
| 20 | 7.16 + 0.116b | 17.65 + 0.120b | 2.466 + 0.017b |
| 30 | 7.46 + 0.095a | 18.92 + 0.146a | 2.536 + 0.047a |
| 40 | 7.07 + 0.080b | 16.60 + 0.062c | 2.347 + 0.035c |
| 50 | 7.62 + 0.038a | 16.81 + 0.182c | 2.207 + 0.022d |
Data are shown as mean ± SD (n = 3). Means with the same letter are not significantly different as indicated by Duncan's multiple range test (P < 0.05).
Table 5.
Effect of complex nitrogen sources on DCW and CoQ10 production by PK38 with batch culture.
| CSP + (NH4)2SO4 (g/L) | DCW (g/L) | CoQ10 production (mg/L) | Specific CoQ10 content (mg/g-DCW) |
|---|---|---|---|
| 20 + 0 | 7.16 + 0.116d | 17.65 + 0.098d | 2.466 + 0.017c |
| 20 + 5 | 6.73 + 0.139f | 16.17 + 0.091f | 2.403 + 0.010c |
| 20 + 10 | 6.92 + 0.097e | 17.08 + 0.082e | 2.467 + 0.027c |
| 20 + 15 | 7.13 + 0.067d | 17.60 + 0.115d | 2.469 + 0.031c |
| 30 + 0 | 7.46 + 0.095c | 18.92 + 0.146c | 2.536 + 0.047b |
| 30 + 5 | 7.65 + 0.107b | 19.40 + 0.284b | 2.537 + 0.068b |
| 30 + 10 | 7.86 + 0.090a | 20.62 + 0.121a | 2.624 + 0.032a |
| 30 + 15 | 7.60 + 0.512b | 18.54 + 0.085c | 2.439 + 0.162c |
Data are shown as mean ± SD (n = 3). Means with the same letter are not significantly different as indicated by Duncan's multiple range test (P < 0.05).
3.5. Fed-Batch CoQ10 Fermentation in 5-l Fermentor
Figure 5(a) shows the batch profile of DCW and CoQ10 production with the mutant PK38 in the 5-l fermentor under the optimal carbon and nitrogen sources. During the batch fermentation of PK38, cell aggregation was observed after incubation, and after 42 h, the cells were not grown further. The final yield of CoQ10 reached 43.94 mg/L, which was significantly higher than that reached in the flask culture (20.62 mg/L), indicating that the low constant agitation (300 rpm) provided in the 5-l fermentor might have enhanced cell-substrate contact [32], leading to the increase in CoQ10 productivity. The final biomass of PK reached 13.75 g/L, which was almost 2 times that obtained in the flask culture (7.86 g/mL). This indicates that the specific CoQ10 content was 3.196 mg/g-DCW, which is much higher than that achieved in the flask culture (2.624 mg/g-DCW).
Figure 5.
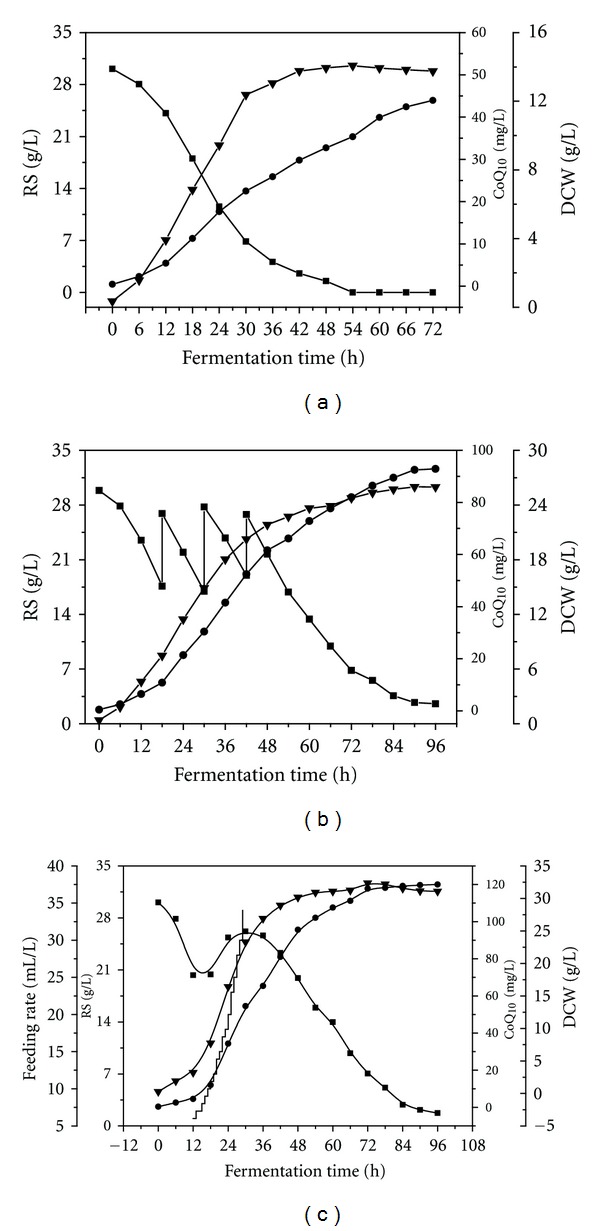
Profiles of DCW and CoQ10 production in batch (a), fed-batch (b), and exponential fed-batch (c) culture with PK38 in the 5-L fermentor at 28°C; residual sugar (RS, filled square), CoQ10 (filled circle), DCW (filled inverted triangle).
As shown in the batch fermentation, CoQ10 production is mainly dependent on cell growth. A fed-batch fermentation of PK38 was carried out to get a high cell density and to increase more CoQ10 accumulation. When the residual sugar concentration of the broth dropped to about 17.5 g/L, 100 l of a sucrose solution (30%) was fed to the 5 l fermentor. As Figure 5(b) shows, sucrose feed was performed at 18, 30, and 40 h, respectively. The feed strategy was designed so the residual sugar was kept at a concentration of 17–28 g/L. The feeding profile was found to be optimum, by maintaining a higher concentration of the residual sugar in the broth during the log phase of PK38. As a result, the cell density increased to 25.99 g/L, which was almost twice the amount obtained in the batch fermentation. Specifically, the final CoQ10 production reached 92.87 mg/L in 96 h of incubation, which was significantly higher than the batch operation. This demonstrates that the specific CoQ10 content achieved 3.575 mg/g-DCW, which is much higher than that achieved in the batch fermentation.
In general, microorganisms will grow exponentially under the optimal condition. If the feed rate of the substrate is increased in proportion to the exponential growth rate, it is possible to maintain a high rate of cell growth for a long time [33]. Also, exponential feeding can limit the negative effect on the cell growth exerted by the sudden increase of sucrose. Figure 5(c) shows the profiles of DCW and CoQ10 production with the mutant PK38 in the 5-l fermentor under the exponential feeding fed-batch control. The sucrose feed started from 12 h when the residual sugar concentration in the broth dropped to 21 g/L. The sucrose solution (30%) flow rate profile was established in the equation described by Martínez et al. [34]. CoQ10 production increased quickly after 18 h of incubation, and the final yield reached 120.01 mg/L. The production of CoQ10 was proportional to cell growth, and the final biomass obtained was 31.09 g/L. Also, the exponential feeding fed-batch culture resulted in the highest specific CoQ10 content within the cell biomass, reaching 3.860 mg/g, superior to that of the aforementioned two cultures.
4. Conclusion
This work demonstrates that the use of HHP could be a promising approach for mutagenesis to a CoQ10 producer A. tumefaciens strain. A mutant strain PK38 was obtained by HHP treatment: the intracellular CoQ10 content of this strain increased by 52.83% when compared to the wild-type strain. Even though the mechanism of HHP-induced mutagenesis is not clear, HHP treatment might affect the three enzymatic steps of CoQ10 biosynthesis. Sucrose was the best carbon source, and CSP and ammonium sulfate were the optimal nitrogen sources for CoQ10 production with PK38. Fed-batch culture could significantly improve cell growth and CoQ10 production. By the exponential feeding strategy, the cell biomass and CoQ10 production increased 126.11 and 173.12%, much higher than those obtained in the batch culture. There may be opportunities for further enhancement of the CoQ10 production in this species.
Authors' Contribution
Y. Yuan and Y. Tian contributed equally to this work.
Acknowledgments
The research was supported by China State ‘‘12th Five-Year Plan” scientific and technological support schemes (2012BAK17B06, 2012BAD31B01); National Natural Science Foundation of China (31071550, 31171721); Special Major Science and Technology Project in Shaanxi Province (2006 KZ09-G1); New Century Talent Plan of the Ministry of Education (2005); Beyond Plan of the Ministry of Agriculture (2005-4).
References
- 1.Ernster L, Dallner G. Biochemical, physiological and medical aspects of ubiquinone function. Biochimica et Biophysica Acta. 1995;1271(1):195–204. doi: 10.1016/0925-4439(95)00028-3. [DOI] [PubMed] [Google Scholar]
- 2.Singh RB, Niaz MA, Rastogi SS, Verma SP. Coenzyme Q10 and its role in heart disease. Journal of Clinical Biochemistry and Nutrition. 1999;26(2):109–118. [Google Scholar]
- 3.Portakal O, Ozkaya O, Inal ME, Bozan B, Koşan M, Sayek I. Coenzyme Q10 concentrations and antioxidant status in tissues of breast cancer patients. Clinical Biochemistry. 2000;33(4):279–284. doi: 10.1016/s0009-9120(00)00067-9. [DOI] [PubMed] [Google Scholar]
- 4.Beal MF. Mitochondrial dysfunction and oxidative damage in Alzheimer’s and Parkinson’s diseases and coenzyme Q10 as a potential treatment. Journal of Bioenergetics and Biomembranes. 2004;36(4):381–386. doi: 10.1023/B:JOBB.0000041772.74810.92. [DOI] [PubMed] [Google Scholar]
- 5.Laplante S, Souchet N, Bryl P. Comparison of low-temperature processes for oil and coenzyme Q10 extraction from mackerel and herring. European Journal of Lipid Science and Technology. 2009;111(2):135–141. [Google Scholar]
- 6.Ehud I, Doron E. Total synthesis of polyprenoid natural products via Pd(O)-catalyzed oligomerizations. Pure and Applied Chemistry. 1988;60:89–98. [Google Scholar]
- 7.Yoshida H, Kotani Y, Ochiai K, Araki K. Production of ubiquinone-10 using bacteria. Journal of General and Applied Microbiology. 1998;44(1):19–26. doi: 10.2323/jgam.44.19. [DOI] [PubMed] [Google Scholar]
- 8.Cluis CP, Burja AM, Martin VJJ. Current prospects for the production of coenzyme Q10 in microbes. Trends in Biotechnology. 2007;25(11):514–521. doi: 10.1016/j.tibtech.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Choi JH, Ryu YW, Seo JH. Biotechnological production and applications of coenzyme Q10. Applied Microbiology and Biotechnology. 2005;68(1):9–15. doi: 10.1007/s00253-005-1946-x. [DOI] [PubMed] [Google Scholar]
- 10.Choi GS, Kim YS, Seo JH, Ryu YW. Restricted electron flux increases coenzyme Q10 production in Agrobacterium tumefaciens ATCC4452. Process Biochemistry. 2005;40(10):3225–3229. [Google Scholar]
- 11.Gu SB, Yao JM, Yuan QP, Xue PJ, Zheng ZM, Yu ZL. Kinetics of Agrobacterium tumefaciens ubiquinone-10 batch production. Process Biochemistry. 2006;41(8):1908–1912. [Google Scholar]
- 12.Ha SJ, Kim SY, Seo JH, Oh DK, Lee JK. Optimization of culture conditions and scale-up to pilot and plant scales for coenzyme Q10 production by Agrobacterium tumefaciens. Applied Microbiology and Biotechnology. 2007;74(5):974–980. doi: 10.1007/s00253-006-0744-4. [DOI] [PubMed] [Google Scholar]
- 13.Ha SJ, Kim SY, Seo JH, Sim WI, Moon HJ, Lee JK. Lactate increases coenzyme Q10 production by Agrobacterium tumefaciens. World Journal of Microbiology and Biotechnology. 2008;24(6):887–890. [Google Scholar]
- 14.Bartlett DH, Kato C, Horikoshi K. High pressure influences on gene and protein expression. Research in Microbiology. 1995;146(8):697–706. doi: 10.1016/0923-2508(96)81066-7. [DOI] [PubMed] [Google Scholar]
- 15.Bartlett DH. Pressure effects on in vivo microbial processes. Biochimica et Biophysica Acta. 2002;1595(1-2):367–381. doi: 10.1016/s0167-4838(01)00357-0. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes PMB. How does yeast respond to pressure? Brazilian Journal of Medical and Biological Research. 2005;38(8):1239–1245. doi: 10.1590/s0100-879x2005000800012. [DOI] [PubMed] [Google Scholar]
- 17.Hauben KJA, Bartlett DH, Soontjens CCF, Cornelis K, Wuytack EY, Michiels CW. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Applied and Environmental Microbiology. 1997;63(3):945–950. doi: 10.1128/aem.63.3.945-950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Li J, Ruan KC. Barotolerant E. coli induced by high hydrostatic pressure. Acta Biochimica et Biophysica Sinica. 2001;33(1):77–81. [PubMed] [Google Scholar]
- 19.Wang SL, Sun JS, Han BZ, Wu XZ. Optimization of β-carotene production by Rhodotorula glutinis using high hydrostatic pressure and response surface methodology. Journal of Food Science. 2007;72(8):M325–M329. doi: 10.1111/j.1750-3841.2007.00495.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang SL, Chen DJ, Deng BW, Wu XZ. Effects of high hydrostatic pressure on the growth and β-carotene production of Rhodotorula glutinis. Yeast. 2008;25(4):251–257. doi: 10.1002/yea.1583. [DOI] [PubMed] [Google Scholar]
- 21.Wu RQ, Li ZX, Yang JP, Xing XH, Shao DY, Xing KL. Mutagenesis induced by high hydrostatic pressure treatment: a useful method to improve the bacterial cellulose yield of a Gluconoacetobacter xylinus strain. Cellulose. 2010;17(2):399–405. [Google Scholar]
- 22.Wang Y, Wu SL, Hancock WS, et al. Proteomic profiling of Escherichia coli proteins under high cell density fed-batch cultivation with overexpression of phosphogluconolactonase. Biotechnology Progress. 2005;21(5):1401–1411. doi: 10.1021/bp050048m. [DOI] [PubMed] [Google Scholar]
- 23.Kim BS, Lee SC, Lee SY, Chang YK, Chang HN. High cell density fed-batch cultivation of Escherichia coli using exponential feeding combined with pH-stat. Bioprocess and Biosystems Engineering. 2004;26(3):147–150. doi: 10.1007/s00449-003-0347-8. [DOI] [PubMed] [Google Scholar]
- 24.Korz DJ, Rinas U, Hellmuth K, Sanders EA, Deckwer WD. Simple fed-batch technique for high cell density cultivation of Escherichia coli. Journal of Biotechnology. 1995;39(1):59–65. doi: 10.1016/0168-1656(94)00143-z. [DOI] [PubMed] [Google Scholar]
- 25.Wuytack EY, Diels AMJ, Michiels CW. Bacterial inactivation by high-pressure homogenisation and high hydrostatic pressure. International Journal of Food Microbiology. 2002;77(3):205–212. doi: 10.1016/s0168-1605(02)00054-5. [DOI] [PubMed] [Google Scholar]
- 26.Tian Y, Yue T, Pei J, Yuan Y, Li J, Martin Lo Y. Effects of cell lysis treatments on the yield of coenzyme Q10 following Agrobacterium tumefaciens fermentation. Food Science and Technology International. 2010;16(2):195–203. doi: 10.1177/1082013210366788. [DOI] [PubMed] [Google Scholar]
- 27.Zahiri HS, Noghabi KA, Shin YC. Biochemical characterization of the decaprenyl diphosphate synthase of Rhodobacter sphaeroides for coenzyme Q10 production. Applied Microbiology and Biotechnology. 2006;73(4):796–806. doi: 10.1007/s00253-006-0524-1. [DOI] [PubMed] [Google Scholar]
- 28.Lauro FM, Tran K, Vezzi A, Vitulo N, Valle G, Bartlett DH. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. Journal of Bacteriology. 2008;190(5):1699–1709. doi: 10.1128/JB.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandra M, Kalra A, Sangwan NS, Gaurav SS, Darokar MP, Sangwan RS. Development of a mutant of Trichoderma citrinoviride for enhanced production of cellulases. Bioresource Technology. 2009;100(4):1659–1662. doi: 10.1016/j.biortech.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Kawamukai M. Biosynthesis, bioproduction and novel roles of ubiquinone. Journal of Bioscience and Bioengineering. 2002;94(6):511–517. doi: 10.1016/s1389-1723(02)80188-8. [DOI] [PubMed] [Google Scholar]
- 31.Knowles CJ, Redfearn ER. The effect of combined-nitrogen sources on the synthesis and function of the electron transport system of Azotobacter vinelandii. Biochimica et Biophysica Acta. 1968;162(3):348–355. doi: 10.1016/0005-2728(68)90121-7. [DOI] [PubMed] [Google Scholar]
- 32.Lo YM, Hsu CH, Yang ST, Min DB. Oxygen transfer characteristics of a centrifugal, packed-bed reactor during viscous xanthan fermentation. Bioprocess and Biosystems Engineering. 2001;24(3):187–193. [Google Scholar]
- 33.Yamanè T, Shimizu S. Fed-Batch Techniques in Microbial Processes. Berlin, Germany: Springer; 1984. [Google Scholar]
- 34.Martínez A, Ramírez OT, Valle F. Effect of growth rate on the production of β-galactosidase from Escherichia coli in Bacillus subtilis using glucose-limited exponentially fedbatch cultures. Enzyme and Microbial Technology. 1998;22(6):520–526. [Google Scholar]


