Abstract
Background
Ongoing innovation in phylogenetics and evolutionary biology has been accompanied by a proliferation of software tools, data formats, analytical techniques and web servers. This brings with it the challenge of integrating phylogenetic and other related biological data found in a wide variety of formats, and underlines the need for reusable software that can read, manipulate and transform this information into the various forms required to build computational pipelines.
Results
We built a Python software library for working with phylogenetic data that is tightly integrated with Biopython, a broad-ranging toolkit for computational biology. Our library, Bio.Phylo, is highly interoperable with existing libraries, tools and standards, and is capable of parsing common file formats for phylogenetic trees, performing basic transformations and manipulations, attaching rich annotations, and visualizing trees. We unified the modules for working with the standard file formats Newick, NEXUS and phyloXML behind a consistent and simple API, providing a common set of functionality independent of the data source.
Conclusions
Bio.Phylo meets a growing need in bioinformatics for working with heterogeneous types of phylogenetic data. By supporting interoperability with multiple file formats and leveraging existing Biopython features, this library simplifies the construction of phylogenetic workflows. We also provide examples of the benefits of building a community around a shared open-source project. Bio.Phylo is included with Biopython, available through the Biopython website, http://biopython.org.
Background
Comparative methods in biology have been an active area of scientific software development since the wider availability of computing resources made such large-scale quantitative analyses feasible [1]. In recent years, the number and range of tools for working with phylogenetic information alone has expanded dramatically, creating new opportunities as well as challenges in data integration [2]. At the same time, we have seen new efforts to create standards for data exchange and storage [3-6]. There is thus a growing need for modular software toolkits that can integrate cleanly into workflows for bioinformatics analyses that include phylogenetic data, as well as facilitate tool development by providing a high-level application programming interface (API) independent of data storage formats.
Freely available, open-source software libraries now play a major role in integrating software components for bioinformatics analyses. Generalized toolkits for working with phylogenetic information are already available and in wide use, including the Ape package for the R statistical programming language [7], BioPerl [8] and Bio::Phylo [2] for Perl, and Mesquite [9] for Java. The Open Bioinformatics Foundation in particular supports an ecosystem of broadly aimed bioinformatics libraries, collectively referred to as the Bio* or “Bio-star” projects. These include BioPerl [8], Biopython [10], BioJava [11], BioRuby [12], BioLib [13] and BioSQL [14]. All of these libraries provide common functionality to each of their target programming languages or environments: read and write a variety of file formats used in bioinformatics, communicate with public web services such as NCBI Entrez [15], access widely used stand-alone programs such as BLAST [16,17], and define fundamental data types for storing biological information, such as annotated sequence records, protein structures and phylogenetic trees.
We chose Python as the implementation language for our library due to its increasing usage in scientific work [18]. It allows for particularly concise, easy-to-read code, has extensive library support, and enables the same code to be run on all major operating systems. In addition to Biopython, several other high-quality Python libraries are available for phylogenetics: PyCogent [19], DendroPy [20], ETE [21] and p4 [22] each serve specific problem domains well. However, they focus specificially on phylogenetics and are not intended to be general-purpose frameworks for processing biological data.
In this article we present a framework for phylogenetics in the Python programming language, fully integrated with the Biopython toolkit. We supplemented existing support for the NEXUS [23] and Newick [24] standards in Biopython with a full-featured phylogenetic module, Bio.Phylo, incorporating new input/output support for the phyloXML standard [5]. We then integrated the new I/O infrastructure with related code in Biopython and added more features that would be useful for researchers, with a focus on easy interoperability and rapid script development.
Our contributions include an I/O framework with parsers and serializers for the standard Newick, NEXUS and phyloXML formats for phylogenetic trees, integrated through a consistent and simple API that is familiar to users of Biopython and BioPerl, as well as a set of common functions to perform on trees independently of the source data format. This framework includes comprehensive class definitions for the rich annotation types which can be serialized in the standard phyloXML format. We also provide several options for visualizing trees, and convenient integration with popular third-party tools.
Implementation
Bio.Phylo is written as a sub-package within the Biopython code base, and is installed along with the Biopython distribution. It has been available as part of the Biopython library since version 1.54, released in May 2010.
The library can be used in Python versions 2.5 through 2.7 and 3.1 or later, without any external libraries required at the time of installation. Functions that rely on external libraries are written within a separate module of the code base, and import their dependencies at run-time. This design makes it possible to install Biopython and use the rest of Bio.Phylo without having installed the dependencies for these specific functions. Because Bio.Phylo is written entirely in Python, it also runs on alternative implementations of Python: Jython 2.5 and Pypy 1.6 through 1.9 in particular pass the module’s unit testing suite.
I/O functions for standard file formats
A unified API for input and output is provided for the Newick, NEXUS and phyloXML formats with the same underlying object structures. The simple API style is shared with Bio.SeqIO, AlignIO, Motif and other modules within Biopython; the API also resembles that of BioPerl. The read and write functions accept a filename or handle, so they also work with Unix pipes, network handles or other file-like objects. As in Biopython’s SeqIO and AlignIO, a convert function is available to convert between any two of the supported formats with a single call.
The I/O code is designed for simple addition of other file formats without disturbing the existing code for Newick, NEXUS and phyloXML. Parsing and serialization code is separated from the internal tree object representation. All parsers return a common object type, the tree, independently of the source data format, and the parsed tree objects all support a common set of operations.
Common tree representation
A phylogeny is represented by a Tree object which contains global information about the phylogeny, such as whether it is rooted, and a reference to the root Clade. Each Clade contains a reference to its child clades, a simple Python list of further Clade objects, nested recursively. The Clade object also contains information about the node occuring at the split or tip it represents, such as the length of the branch leading to it and the name of the node. There is no additional complex data structure operating “under the hood”, and trees are not required to be bifurcating, although the functions in Bio.Phylo currently assume that each Clade has a single parent, i.e., the topology of the Tree is indeed a tree and not a network. This straightforward design is conducive to implementing algorithms in a form that is easy to read and understand, with minimal need for management of the object’s internal data representation.
The basic Tree and Clade objects store the intersection of the information that can be represented in the Newick and phyloXML formats. To store additional format-specific attributes, we defined separate Newick and PhyloXML sub-classes which inherit from the basic Tree and Clade classes. Classes for each element type defined in the phyloXML specification have been implemented in the Bio.Phylo.PhyloXML module, allowing richer annotation types to be attached to Tree objects. For convenience in adding graphical cues, the Clade class also has attributes for the displayed color and width of a branch. The properties of the branch color and width attributes follow the phyloXML specification, and are available on the common Clade class due to their usefulness, particularly during interactive work. In accordance with the phyloXML specification, these attributes apply in a cascading manner down the clade: for example, if the root Clade object is assigned the color blue, the entire tree will be displayed as blue unless a child clade overrides this attribute.
Methods for tree inspection and manipulation
The Tree and Clade objects also implement common methods for tree manipulation and simple analyses that might be used routinely in bioinformatics workflows. These include methods for tree search and traversal, extracting basic information, and modifying or manipulating the tree. A listing of these methods and other functions available in Bio.Phylo is given in Table 1.
Table 1.
Built-in functions and tree methods
| Source | Function | Description |
|---|---|---|
|
Bio.Phylo |
read |
Parse a file in the given format and return a single tree. |
|
Bio.Phylo |
parse |
Iteratively parse a file and return each of the trees it contains. |
|
Bio.Phylo |
write |
Write a sequence of trees to file in the given format. |
|
Bio.Phylo |
convert |
Convert between two tree file formats. |
|
Bio.Phylo |
draw |
Plot the given tree using matplotlib (or pylab). |
|
Bio.Phylo |
draw_ascii |
Draw an ascii-art phylogram of the given tree. |
|
Bio.Phylo |
draw_graphviz |
Display a tree or clade as a graph, using the graphviz engine. |
|
Bio.Phylo |
to_networkx |
Convert a Tree object to a NetworkX graph object. |
|
Bio.Phylo.BaseTree | ||
|
TreeMixin |
find_elements |
Find all tree elements matching the given attributes. |
|
TreeMixin |
find_clades |
Find each clade containing a matching element. |
|
TreeMixin |
find_any |
Return the first matching element found by find_elements, if any. |
|
TreeMixin |
get_path |
List the clades directly between the current node and the target. |
|
TreeMixin |
get_nonterminals |
List of all of the tree or clade’s internal nodes. |
|
TreeMixin |
get_terminals |
List of all of the tree or clade’s “leaf” nodes. |
|
TreeMixin |
trace |
List of all clade object between two targets in the tree/clade. |
|
TreeMixin |
common_ancestor |
Most recent common ancestor (clade) of all the given targets. |
|
TreeMixin |
count_terminals |
Count the number of terminal nodes within the tree. |
|
TreeMixin |
depths |
Create a mapping of tree clades to depths (by branch length). |
|
TreeMixin |
distance |
Calculate the sum of the branch lengths between two targets. |
|
TreeMixin |
is_bifurcating |
Return True if tree downstream of node is strictly bifurcating. |
|
TreeMixin |
is_monophyletic |
If the given terminals comprise a complete subclade, return the MRCA. |
|
TreeMixin |
is_parent_of |
True if target is a descendent of the tree. |
|
TreeMixin |
is_preterminal |
True if all direct descendents are terminal. |
|
TreeMixin |
total_branch_length |
Calculate the sum of all the branch lengths in the tree. |
|
TreeMixin |
collapse |
Deletes target from the tree, relinking its children to its parent. |
|
TreeMixin |
collapse_all |
Collapse all the descendents of the tree, leaving only terminals. |
|
TreeMixin |
ladderize |
Sort clades in-place according to the number of terminal nodes. |
|
TreeMixin |
prune |
Prunes a terminal clade from the tree. |
|
TreeMixin |
split |
Generate n (default 2) new descendants. |
|
Tree, Clade |
is_terminal |
True if the node has no descendents. |
|
Tree |
root_with_outgroup |
Reroot the tree with the specified outgroup clade. |
|
Tree |
root_at_midpoint |
Reroot the tree at the midpoint between the two most distant terminals. |
|
Tree |
format |
Serialize the tree as a string in the specified file format. |
|
Tree |
as_phyloxml |
Convert the tree to its PhyloXML subclass equivalent. |
|
Tree |
from_clade |
Create a new Tree object given a clade. |
|
Tree |
randomized |
Create a randomized bifurcating tree, given a list of taxa. |
|
Bio.Phylo.PhyloXML | ||
|
Phylogeny |
to_alignment |
Construct an alignment from the aligned sequences in this tree. |
|
Bio.Phylo.PAML | ||
|
chi2 |
cdf_chi2 |
χ2 cumulative distribution function, for log-likelihood ratio tests. |
|
baseml |
read |
Parse a BASEML results file. |
|
codeml |
read |
Parse a CODEML results file. |
|
yn00 |
read |
Parse a yn00 results file. |
|
_paml.Paml |
write_ctl_file |
Dynamically build a program-specific control file. |
|
_paml.Paml |
read_ctl_file |
Parse a control file to create a program-specific class instance. |
|
_paml.Paml |
print_options |
Print all of the program options and their current settings. |
|
_paml.Paml |
set_options |
Set the value of a program option. |
|
_paml.Paml |
get_option |
Return the value of a program option. |
|
_paml.Paml |
get_all_options |
Return the current values of all the program options. |
| _paml.Paml | run | Run a PAML program and parse the results. |
Public methods and functions provided in the Bio.Phylo module and sub-modules. An up-to-date version of this information is available at http://biopython.org/DIST/docs/api/.
Both the Tree and Clade classes inherit from a third class, TreeMixin, which implements these common tree methods. Therefore, most of the same methods are available on both the Tree object and any of the Clade objects it contains; in practice, one can usually ignore the distinction between the global Tree object and the root Clade.
Visualization
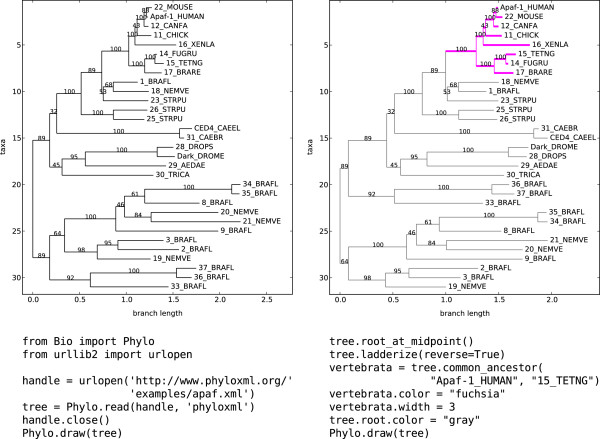
We implemented several mechanisms for displaying trees. The draw function displays a rooted phylogram (Figure 1), in the style of Phylip’s drawgram program, while draw_graphviz displays an unrooted cladogram using the Graphviz programs [25] for layout and the NetworkX library [26] as an intermediate graph representation.
Figure 1.
Tree visualization. An example tree with the code to generate each plot shown below each plot. First, a phyloXML tree of the Apaf-1 protein family [27] is downloaded, read by Bio.Phylo, and plotted with default settings. The tree is then rerooted at the midpoint of its two most divergent tips and ladderized such that sibling clades with a larger number of descendents are listed first. The clade of genes belonging to vertebrate species is identified as the common ancestor of the human and zebrafish, after inspection of the original tree. The vertebrate clade is highlighted with the color fuchsia and an increased branch width, and the rest of the tree is colored gray. Finally, the tree is plotted again
The generated plots can be further modified using the built-in functions of matplotlib or PyLab [28]. In case no visualization libraries are available on a system, plain-text representations of a tree are also possible. The function draw_ascii draws an “ASCII-art” tree to standard output or a given file handle. Using the built-in Python print statement on a Tree object shows the nested object hierarchy, including class names and the values of attributes such as branch length.
Since the complete phyloXML specification is implemented, files can be saved by Bio.Phylo with graphical annotations and then rendered with popular visualization tools such as Archaeopteryx [29]. The cascading behavior of clade colors and branch widths is shared by Archaeopteryx and other tree visualization software that implements phyloXML.
Wrappers for third-party programs
Biopython includes a common framework for building wrappers for external programs. This framework allows us to leverage the functionality of widely used third-party programs from within Biopython, rather than reimplement those functions. For example, Biopython supports the Phylip suite of programs via EMBOSS [30], with wrappers implemented in the Bio.Emboss.Applications module. We follow this rationale in Bio.Phylo.
Within the sub-module Bio.Phylo.Applications, we currently provide wrappers for the tree inference programs PhyML [31] and RAxML [32]. The code scaffolding is in place to add more, using the same common Bio.Application framework.
PAML integration
Bio.Phylo also includes wrappers for the PAML suite of programs [33], specifically the analysis programs yn00, baseml, and codeml. Additionally, we created a pure-Python re-implementation of the program chi2 as a simple means to perform likelihood ratio tests. Since the analysis programs are operated through the use of configuration files rather than command-line arguments, a different approach was required than what Bio.Application enables. We therefore provide wrappers for these programs in a separate sub-module, Bio.Phylo.PAML.
For each of the three analysis programs, the wrapper defines a corresponding class that the user instantiates to store the configuration state. The programs’ standard configuration options are set through the set_options method and are automatically written to a configuration file when the run method is invoked or through the write_ctl_file method. Existing configuration files may be parsed via the method read_ctl_file. Finally, each module of Bio.Phylo.PAML provides a read method, which parses the output files of the respective programs. The results of an analysis are stored hierarchically in a set of nested dictionary objects, allowing quick access by keywords.
Biopython integration
Bio.Phylo integrates cleanly with other parts of the Biopython toolkit. We reuse existing Biopython API conventions, including classes for exceptions and warnings, as well as the packaging and installation mechanisms and the testing framework. As mentioned above, the wrappers for running external programs use a common Biopython framework, Bio.Application. NEXUS and Newick support were obtained through a refactoring of the Bio.Nexus module, originally written by Frank Kauff and Cymon J. Cox [34].
Where appropriate, classes in the PhyloXML module support methods for conversion to and from instances of the general-purpose Biopython classes for molecular sequences, annotated sequence regions and multiple sequence alignments. For example, phyloXML defines a ProteinDomain element type which represents a functional domain within a protein sequence that appears in the tree; our implementation of the ProteinDomain class includes a to_seqfeature method to convert a ProteinDomain instance to an instance of the Biopython class SeqFeature, a generalized representation of an annotated sequence region. These SeqFeature objects could then be used with the Biopython’s GenomeDiagram module [35] to create a diagram of the protein domain architecture of each sequence appearing in the phyloXML tree. The ProteinDomain class also provides a complementary method, from_seqfeature, which could be used to add domain architecture annotations to the sequences in a phyloXML record, given the corresponding GenBank files.
Validation
A complete suite of unit tests to verify correct functioning of each module is included with the Biopython distribution. This includes the round-trip parsing and serialization of example files in each supported file format; conversion between formats; proper construction, behavior and serialization of all phyloXML element types; methods for tree inspection, traversal and manipulation; and succesful loading of optional third-party libraries, if available.
Results
Use of Bio.Phylo in published studies
Since the Bio.Phylo module has been made available to the community throughout its development, researchers have had the opportunity to use it in studies that have since been published.
A recent study of microbial phylogenomics used Bio.Phylo for large-scale processing of microbial gene trees, generating permutations of tree topologies by rerooting over 100,000 gene trees at each internal and external node [36]. Additional file 1 shows a simplified example of how such rerooting could be performed.
In another study of the eukaryotic protein kinase superfamily in the protozoan phylum Apicomplexa, Bio.Phylo was used to identify putative lineage-specific ortholog groups by selecting clades with significant bootstrap support from gene trees [37]. The corresponding sequences were then automatically extracted from a matching FASTA file, using Biopython’s SeqIO module, for further analysis in a computational pipeline.
Performance
Despite the stark simplification of the underlying data structures, the majority of the built-in tree methods have a run time that is theoretically linear or constant in proportion to the number of nodes in the tree. The tree traversal methods find_clades and find_elements are Python generator functions which evaluate and emit values incrementally, as needed by the caller; this “lazy evaluation” approach uses computer memory more efficiently and avoids performing more computation than necessary; for example, the method find_any uses this property to stop traversal after finding the first element matching the user’s query and thus avoid traversing the remainder of the tree structure or evaluating any further comparisons.
We timed several benchmark operations on large tree files to evaluate the speed of Bio.Phylo on several different tasks (Table 2; Additional file 2). On more modestly sized trees and input files, these operations typically complete in a small fraction of a second.
Table 2.
Performance
| Task | Input tree | Python 2.7 | Python 3.2 | PyPy 1.9 |
|---|---|---|---|---|
| Read a very large Newick tree |
Smith 2011 angiosperm supertree |
|||
| |
(55473 terminal nodes) [38] |
17.45 |
16.85 |
1.214 |
| Read the same large tree in phyloXML |
Smith 2011, converted to phyloXML with |
|||
| |
phylo_converter (http://phylosoft.org/) |
3.805 |
4.318 |
3.937 |
| Write the same large tree as Newick |
Smith 2011 |
0.5238 |
0.7704 |
0.4378 |
| Write the same large tree as phyloXML |
Smith 2011 |
10.39 |
10.85 |
24.17 |
| Read a medium-sized Newick tree |
Davies 2004 angiosperm supertree |
|||
| |
(440 terminal nodes) [39] |
0.1097 |
0.1087 |
0.007312 |
| Parse many Newick trees |
Davies 2004, copies rerooted at |
|||
| |
each node (816 trees) |
84.91 |
84.29 |
6.812 |
| Reroot at each node |
Davies 2004 |
1.347 |
1.167 |
0.3450 |
| Collapse all splits with bootstrap values less than 50 |
Davies 2004 |
2.266 |
2.312 |
2.411 |
| Total branch length |
Davies 2004 |
0.01322 |
0.01310 |
0.01448 |
| Ladderize the tree |
Davies 2004 |
0.1274 |
0.1190 |
0.1127 |
| Count terminal nodes | Davies 2004 | 0.006838 | 0.006323 | 0.005914 |
Performance of Bio.Phylo functions and tree methods under different Python versions on several benchmark tasks. Reported execution times are the median of 101 replications of each task, in seconds (Additional file 2). Benchmarks were evaluated with Python versions 2.7.3 and 3.2.3 and PyPy version 1.9 on an Intel Xeon E5405 2.00 GHz processor with 8 GB memory, running under 64-bit Ubuntu Linux 12.04 with Biopython 1.60 installed.
Cookbook and additional documentation
In keeping the Bio.Phylo module general-purpose and simple to begin using, we have chosen not to include niche functions, or approaches that are still the topic of active research, in the Biopython distribution. However, we nonetheless anticipate that other users of the Bio.Phylo module will want to use these features, and there is a benefit to sharing this code. We resolve this by maintaining an online “cookbook” on the Biopython wiki (http://biopython.org/wiki/Phylo_cookbook). This cookbook contains working code samples for common usage patterns, as well as exporting to object types of other libraries. In particular, we provide functions to convert a Bio.Phylo tree object to a distance or adjacency matrix using the NumPy module for Python, and to export a Bio.Phylo tree to the native tree objects used by the R package Ape [7], via the Rpy2 module, and the Python package PyCogent [19].
The main Biopython tutorial, included with the Biopython distribution and available online at http://biopython.org/, contains a chapter on the Bio.Phylo module describing its use in detail.
Discussion
Bio.Phylo organizes phylogenetic trees as a primary data type, filling a previously underserved area of data handling within Biopython. The module directly implements tree parsing and serialization in three standard formats, as well as navigation, visualization and manipulation of phylogenetic trees, and conversion of tree data to other data types. Other important aspects of phylogenetics, including phylogenetic tree reconstruction and analysis of rate variation and ancestral character states, exist as optimized stand-alone programs by other authors; RAxML and PAML are examples of such programs that are already well accepted by the phylogenetics community. Rather than re-implement the functionality of these established third-party programs ourselves, we have opted to provide wrappers for these, and focus on providing “glue” utilities to ease the process of assembling computational workflows that involve phylogenetic data.
By building our work into an existing, popular library, we were able to take advantage of both the software infrastructure and the community of developers and users associated with Biopython. By reusing core objects and maintaining common API conventions for file parsing, the resulting software package has a familiar feel to new users who have prior experience with Biopython or BioPerl. To ensure code correctness and minimize errors introduced during additional development, we reuse the existing test framework and packaging mechanisms in Biopython. A continuous integration server http://testing.open-bio.org/biopython/ runs an automated test suite nightly on all supported Python versions, operating systems and implementations, including the Jython Java-based port of Python.
Our work also provides an example of the short- and long-term benefits of building a community of developers and users around a shared open-source project. By integrating with Biopython from the beginning, we gained access to an existing community of developers and users who have continually tested the software under a variety of environments and use cases, reported bugs, requested new features, and provided new code. A notable example of a contribution from the Biopython user community is Bio.Phylo.PAML, which began as an independent project, pypaml. We successfully integrated the pypaml source code into Biopython, with further enhancements based on feedback from Biopython developers and users.
The permissive open-source license that governs both Biopython and Bio.Phylo allows this code to be reused freely in other software, which could help overcome the pervasive problem of incompatible software support for widely used file formats.
Future development
In future releases we intend to provide support for another recently standardized XML-based format, NeXML, an XML-based successor to NEXUS [6]. Since the core classes for phylogenies have already been defined and implemented in Bio.Phylo, the implementation of NeXML I/O is expected to be straightforward.
Another area for future growth is the addition of wrappers for other third-party applications in Bio.Phylo.Applications. Using the existing Bio.Application framework, it is straightforward to add other tools that can be run with a standard command-line interface. Other widely used programs that require specialized input, notably MrBayes and PhyloBayes, may be implemented with an approach similar to Bio.Phylo.PAML, providing a module containing both the functions to generate the configuration file and to run the program itself with the generated configuration.
Smaller features may appear first on the Cookbook wiki page, implemented as stand-alone functions, and may later be added to the official Biopython distribution.
Conclusions
Bio.Phylo meets a growing need in bioinformatics for working with heterogeneous types of biological and phylogenetic data. By supporting interoperability with multiple file formats and leveraging existing Biopython features, this library simplifies the construction of phylogenetic workflows and computational pipelines, addressing practical issues of data integration and exchange in the bioinformatics community.
Availability and requirements
Project name: BiopythonProject home page:http://biopython.org/Operating system: Platform independentProgramming language: PythonOther requirements: Python 2.5 or higher; optional libraries matplotlib, Graphviz, NetworkXLicense: Biopython LicenseAny restrictions to use by non-academics: None
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ET wrote the code for Bio.Phylo. BI wrote the code for Bio.Phylo.PAML. BAC and PJAC mentored and supervised the development of Bio.Phylo and contributed enhancements and bug fixes. ET, BI, PJAC and BAC wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Tree rerooting. Example code to read a tree from a file and write copies of the tree rerooted at every internal and external node. The method find_clades produces an iterator over the tree rather than a new list, in order to make more efficient use of memory in general. In this case, because the method root_with_outgroup modifies the tree in-place, which could change the ordering of nodes during traversal, list is used to create an unchanging copy of references to the original nodes in the tree.
Performance benchmark script. Python script to time the execution of the benchmark tasks shown in Table 2.
Contributor Information
Eric Talevich, Email: etal@uga.edu.
Brandon M Invergo, Email: brandon.invergo@upf.edu.
Peter JA Cock, Email: peter.cock@hutton.ac.uk.
Brad A Chapman, Email: bchapman@hsph.harvard.edu.
Acknowledgements
We acknowledge the role of Google Summer of Code 2009 and the National Evolutionary Synthesis Center (NESCent) Phyloinformatics program in sponsoring and initiating this project. We also thank the Biopython community and Open Bioinformatics Foundation for ongoing advice and support. Several individuals made direct contributions to Bio.Phylo: Aaron Gallagher, Connor McCoy, Joel Berendzen, Robert Beiko and Tanya Golubchik provided bug fixes, test cases, feature requests and example code; Frank Kauff wrote most of the tree parsing code in the Bio.Nexus module, which Bio.Phylo builds upon, and gave helpful comments on a draft of the manuscript; Cymon J. Cox gave advice on the port of Bio.Nexus code; and Christian M. Zmasek co-mentored the original Google Summer of Code project with Brad A. Chapman, guiding the implementation of phyloXML. We also thank the peer reviewers for helpful suggestions on this manuscript.
BI is supported by a FI-DGR from AGAUR (Catalonia, Spain). Harvard Bioinformatics Core provided for publication costs and moral support.
References
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/284325. [DOI] [PubMed] [Google Scholar]
- Vos RA, Caravas J, Hartmann K, Jensen MA, Miller C. Bio:Phylo - phyloinformatic analysis using Perl. BMC Bioinformatics. 2011;12:63. doi: 10.1186/1471-2105-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack J, Vision T, Brenner E, Bowers JE, Cannon S, Clement MJ, Cunningham CW, DePamphilis C, DeSalle R, Doyle JJ, Eisen Ja, Gu X, Harshman J, Jansen RK, Kellogg Ea, Koonin EV, Mishler BD, Philippe H, Pires JC, Qiu YL, Rhee SY, Sjölander K, Soltis DE, Soltis PS, Stevenson DW, Wall K, Warnow T, Zmasek C. Taking the first steps towards a standard for reporting on phylogenies: Minimum Information About a Phylogenetic Analysis (MIAPA) Omics: J Integr Biol. 2006;10(2):231–237. doi: 10.1089/omi.2006.10.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosdocimi F, Chisham B, Pontelli E, Thompson JD, Stoltzfus A. Initial implementation of a comparative data analysis ontology. Evolutionary Bioinf. 2009;5:47–66. doi: 10.4137/ebo.s2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos Ra, Balhoff JP, Caravas Ja, Holder MT, Lapp H, Maddison WP, Midford PE, Priyam A, Sukumaran J, Xia X, Stoltzfus A. NeXML: rich, extensible, and verifiable representation of comparative data and metadata. Syst Biol. 2012;61(4):675–689. doi: 10.1093/sysbio/sys025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Bolker B, Claude J, Cuong HS, Desper R, Dutheil J, Gascuel O, Heibl C, Lawson D, Lefort V, Lemon J, Noel Y, Nylander J, Opgen-rhein R, Vienne DD. Package ‘ape’. 2010. [ http://ape.mpl.ird.fr/]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz Sa, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, Lehväslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12(10):1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W, Maddison D. Mesquite: A modular system for evolutionary analysis. 2011. [ http://mesquiteproject.org]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland RCG, Down TA, Pocock M, Prlić A, Huen D, James K, Foisy S, Dräger A, Yates A, Heuer M, Schreiber MJ. BioJava: an open-source framework for bioinformatics. Bioinformatics. 2008;24(18):2096–2097. doi: 10.1093/bioinformatics/btn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N, Prins P, Nakao M, Bonnal R, Aerts J, Katayama T. BioRuby: bioinformatics software for the Ruby programming language. Bioinformatics. 2010;26(20):2617–2619. doi: 10.1093/bioinformatics/btq475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins P, Contributors. BioLib. 2008. [ http://biolib.open-bio.org/]
- Lapp H, Contributors. BioSQL. 2003. [ http://www.biosql.org]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33(Database issue):D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer aa, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant TE. Python for Scientific Computing. Comput Sci Eng. 2007;9(3):10–20. [Google Scholar]
- Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC, Eaton M, Hamady M, Lindsay H, Liu Z, Lozupone C, McDonald D, Robeson M, Sammut R, Smit S, Wakefield MJ, Widmann J, Wikman S, Wilson S, Ying H, Huttley GA. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8(8):R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26(12):1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldón T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PG. p4: A Python package for phylogenetics. 2003. [ http://code.google.com/p/p4-phylogenetics/]
- Maddison DR, Swofford DL, Maddison WP. NEXUS: an extensible file format for systematic information. Syst Biol. 1997;46(4):590–621. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- Archie J, Day WH, Maddison W, Meacham C, Rohlf FJ, Swofford D, Felsenstein J. The Newick tree format. 1986. [ http://evolution.genetics.washington.edu/phylip/newicktree.html]
- Gansner E. An open graph visualization system and its applications to software engineering. Software Pract Experience. 2000;00(S1):1–5. [Google Scholar]
- Hagberg AA, Schult DA, Swart PJ. In: Proceedings of the 7th Python in Science Conference (SciPy2008), SciPy. Varoquaux G, Vaught T, Millman J, editor. 2008. Exploring network structure, dynamics, and function using NetworkX; 11 pp. Pasadena, CA USA–15. [Google Scholar]
- Zmasek CM, Zhang Q, Ye Y, Godzik A. Surprising complexity of the ancestral apoptosis network. Genome Biol. 2007;8(10):R226. doi: 10.1186/gb-2007-8-10-r226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD. Matplotlib: A 2D Graphics Environment. Comput Sci Eng. 2007;9(3):90–95. [Google Scholar]
- Zmasek CM, Eddy SR. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17(4):383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [ http://www.atgc-montpellier.fr/phyml/] [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Kauff F, Cox C, Lutzoni F. WASABI: an automated sequence processing system for multigene phylogenies. Systematic Biol. 2007;56(3):523. doi: 10.1080/10635150701395340. [DOI] [PubMed] [Google Scholar]
- Pritchard L, White JA, Birch PR, Toth IK. GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics. 2006;22(5):616–617. doi: 10.1093/bioinformatics/btk021. [DOI] [PubMed] [Google Scholar]
- Beiko RG. Telling the Whole Story in a 10,000-Genome World. Biol Direct. 2011;6:34. doi: 10.1186/1745-6150-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talevich E, Mirza A, Kannan N. Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evolutionary Biol. 2011;11:321. doi: 10.1186/1471-2148-11-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Beaulieu JM, Stamatakis A, Donoghue MJ. Understanding angiosperm diversification using small and large phylogenetic trees. Am J Bot. 2011;98(3):404–414. doi: 10.3732/ajb.1000481. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Darwin’s abominable mystery: Insights from a supertree of the angiosperms. Proc Natl Acad Sci USA. 2004;101(7):1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tree rerooting. Example code to read a tree from a file and write copies of the tree rerooted at every internal and external node. The method find_clades produces an iterator over the tree rather than a new list, in order to make more efficient use of memory in general. In this case, because the method root_with_outgroup modifies the tree in-place, which could change the ordering of nodes during traversal, list is used to create an unchanging copy of references to the original nodes in the tree.
Performance benchmark script. Python script to time the execution of the benchmark tasks shown in Table 2.