Abstract
CD38, a membrane protein expressed in airway smooth muscle (ASM) cells, plays a role in cellular Ca2+ dynamics and ASM contractility. In human ASM (HASM) cells, TNF-α induces CD38 expression through activation of MAPKs, NF-κB, and AP-1, and its expression is differentially elevated in cells from asthmatic patients compared with cells from nonasthmatic subjects. The CD38 3′-untranslated region (UTR) has targets for miR-140-3p. We hypothesized that miR-140-3p regulates CD38 expression in HASM cells by altering CD38 mRNA stability. Basal and TNF-α-induced expression of miR-140-3p was determined in nonasthmatic ASM (NAASM) and asthmatic ASM (AASM) cells. NAASM and AASM cells were transfected with control, miR-140-3p mimic, or miR-140-3p antagomirs, and CD38 expression and CD38 mRNA stability were determined. Luciferase reporter assays were used to determine miR-140-3p binding to the CD38 3′-UTR. Activation of p38, ERK, and JNK MAPKs, NF-κB, and AP-1 was determined in miR-140-3p mimic-transfected NAASM. TNF-α attenuated miR-140-3p expression in NAASM and AASM cells, but at a greater magnitude in AASM cells. CD38 mRNA expression was attenuated by miR-140-3p mimic at comparable magnitude in NAASM and AASM cells. Mutated miR-140-3p target on the CD38 3′-UTR reversed the inhibition of luciferase activity by miR-140-3p mimic. CD38 mRNA stability was unaltered by miR-140-3p mimic in NAASM or AASM cells following arrest of transcription. TNF-α-induced activation of p38 MAPK and NF-κB was attenuated by miR-140-3p mimic. The findings indicate that miR-140-3p modulates CD38 expression in HASM cells through direct binding to the CD38 3′-UTR and indirect mechanisms involving activation of p38 MAPK and NF-κB. Furthermore, indirect mechanisms appear to play a major role in the regulation of CD38 expression.
Keywords: asthma, gene expression, microRNA
cd38 is a cell surface protein expressed in a variety of mammalian cells, including airway smooth muscle (ASM) cells (30). CD38 possesses multiple enzymatic activities, with ADP-ribosyl cyclase activity generating cyclic ADP-ribose (cADPR), a Ca2+-mobilizing agent (12, 18). Past studies in our laboratory established that the CD38/cADPR pathway plays an important role in cellular Ca2+ dynamics and ASM contractility (6, 7). In human ASM (HASM) cells, TNF-α induces CD38 expression through activation of the transcription factors NF-κB and AP-1 and MAPK kinases (27). Among the MAPKs, p38 and ERK MAPKs mediate TNF-α-induced CD38 expression through modulation of transcript stability (27). We recently reported that, in ASM cells from subjects with a history of asthma, TNF-α-induced CD38 expression was differentially elevated, although the mechanistic basis of this differential elevation was not clearly understood (15).
MicroRNAs (miRNAs) are noncoding small RNAs emerging as posttranscriptional regulators in various biological processes, including inflammation; they can regulate expression of their target genes by destabilizing the transcripts or by translational repression (2). Accumulating evidence has proven that miRNAs are associated with various pathological conditions in humans. Recent studies have shed light on the role of miRNAs in airway disorders such as asthma, chronic obstructive pulmonary disorder, and idiopathic pulmonary fibrosis (reviewed in Ref. 22). Recent studies report a role for miR-21 in determining the T helper (Th) type 1 (Th1)/Th2 immune response to antigen, thus playing a role in the pathogenesis of allergic asthma (19, 20). Other studies have attempted to determine the role of specific miRNAs in airway inflammation and allergic airway hyperresponsiveness (AHR). Studies of the roles of let-7 and miR-155 in IL-13 signaling and the potential role of miR-133 in RhoA expression were among those that highlight the roles of miRNA in the pathogenesis of airway inflammatory disorders (5, 19–21). In the present study, bioinformatic tools were used to determine potential miRNA response elements in the 3′-untranslated region (UTR) of the human CD38 gene. Expression of miR-140-3p, which came as a top hit in one of the target prediction algorithms, and its functional role in CD38 expression were determined in asthmatic ASM (AASM) and nonasthmatic ASM (NAASM) cells. We hypothesized that miR-140-3p downregulates CD38 expression in HASM cells through posttranscriptional mechanisms.
MATERIALS AND METHODS
Reagents.
Tris-base, glucose, HEPES, and other chemicals were purchased from Sigma Chemical (St. Louis, MO), unless otherwise noted. Human recombinant TNF-α (rhTNF-α) was purchased from R & D Systems (Minneapolis, MN); HBSS and DMEM from GIBCO-BRL (Grand Island, NY); TRIzol, SuperScript III reverse transcriptase, NCode miRNA first-strand synthesis kit, Platinum SYBR Green quantitative PCR (qPCR) mix, and Lipofectamine RNAiMax from Invitrogen Life Technologies (Carlsbad, CA); miRVana RNA isolation kit from Ambion Life Technologies (Carlsbad, CA); NE-PER nuclear/cytoplasmic extraction kit from Pierce (Rockford, IL); antibodies for MAPKs and NF-κB and lamin A/C, actin, α-tubulin, MAPK kinase 3 (MKK3), the dual-specificity phosphatase MKP-1, and nuclear receptor-interacting protein (NRIP-1) from Cell Signaling Technology; TransAM ELISA kits (NF-κB and AP-1) from Active Motif (Carlsbad, CA); QuikChange Lightning Multi Site-Directed mutagenesis kit from Agilent Technologies (Santa Clara, CA); control (C.ele-miR-67), miR-140-3p mimic, and antagomir (hsa-miR-140-3p mature sequence 5′-UACCACAGGGUAGAACCACGG-3′) oligonucleotides from Dharmacon (Lafayette, CO); and chemiluminescent substrate for horseradish peroxidase (HRP) from Millipore (Billerica, MA).
HASM cell cultures and treatments.
Procedures for isolation and culture of HASM cells are described elsewhere (6, 15). Briefly, HASM cells were obtained from the laboratory of R. A. Panettieri, Jr. The cells were from deidentified healthy donors (NAASM cells) and donors who died due to severe asthma (AASM cells). All experiments were conducted in HASM cells in passage 4 or 5. In all the experiments, prior to exposure to TNF-α, the cells were growth-arrested for 48 h in arresting medium without serum, but in the presence of transferrin and insulin. In experiments to determine the expression of CD38 and miR-140-3p, the cells were exposed to vehicle (0.1% BSA in PBS) or 10 ng/ml rhTNF-α for 0–24 h. To determine CD38 mRNA stability, the cells were exposed to TNF-α for 12 h and then washed to remove TNF-α before addition of actinomycin D (5 μg/ml) to arrest transcription. Total RNA was collected at 0, 6, 12, and 24 h after transcriptional arrest. Data are reported for only 0- and 24-h time points for brevity. In experiments to determine the activation of NF-κB and MAPKs, cells were exposed to rhTNF-α (10 ng/ml) for 1 h and 15 min, respectively.
Extraction of total RNA and cDNA synthesis.
Total RNA was extracted from HASM cells following the manufacturer's instructions (miRVana, Ambion). Briefly, HASM cells (5 × 104–2 × 105) were collected in sterile PBS and centrifuged, and the pellet was homogenized in a buffer provided in the kit. Column-eluted total RNA was quantified in a bioanalyzer (Agilent NanoDrop). To synthesize cDNA from small RNA, 200 ng of total RNA were polyadenylated, and cDNA was synthesized using NCode first-strand synthesis kit according to the manufacturer's instructions. In parallel, 200 ng of total RNA were used to synthesize cDNA from larger RNA using the SuperScript III reverse transcription kit.
Extraction of whole cell/nuclear lysates.
Whole cell lysates were obtained by sonication of HASM cells (2–5 × 105) in lysis buffer (50 mM Tris, 100 mM NaCl, 50 mM NaF, 40 mM β-glycerol phosphate, 2 mM EDTA, 0.2 mM Na3VO4, 1% Triton X-100, and protease inhibitor cocktail, pH 7.4). Nuclear extracts were collected from HASM cells (5–6 × 105) using the NE-PER cytoplasmic and nuclear extraction kit, according to the manufacturer's protocols.
Transient transfection of HASM cells.
HASM cells were plated into appropriate formats and cell number (2 × 105 cells/well in a 6-well plate or 5 × 105 cells/plate in a 100-mm culture plate) 24 h prior to transfection. Transfection was performed using Lipofectamine RNAimax according to the manufacturer's instructions. Briefly, control (C.ele-miR-67), miR-140-3p mimic, or miR-140-3p antagomir oligonucleotides were transfected at 5–100 nM. Control oligonucleotide was transfected at 50 or 100 nM.
qRT-PCR.
To determine miR-140-3p expression, a forward primer specific to miR-140-3p and a universal primer targeting the poly(T) of the cDNA were used according to the manufacturer's instructions. Mammalian small nuclear RNA U6 was used for normalization of miR-140-3p expression. To determine CD38 expression, qRT-PCR was performed using Brilliant SYBR Green Master Mix, as described elsewhere (27). Cyclophilin was amplified as the housekeeping control.
Site-directed mutagenesis.
Site-directed mutagenesis was performed in luciferase-CD38 3′-UTR reporter plasmids. CD38–3′-UTR (481 bases long; UGC genome browser) has a predicted miR-140-3p target site at 8 bases after the stop codon in the CD38 mRNA. The first miR-140-3p target had a 7-base complementarity to the miR-140-3p. A second target site with a 6-base complementarity to miR-140-3p was found 21 bases from the first target site. The first target site (mutated 1) or both target sites (mutated 1+2) were mutated to determine the selectivity of miR-140-3p binding to these sites. The QuikChange Lightning Multi Site-Directed mutagenesis kit was used to mutate 4 bases (taGaGGa) at the first target site (CTGTGGT) or 3 bases (aGaGGa) at the second target site (TGTGGT) (Fig. 2A, top). Twenty bases flanking the target site with mutation were designed as primers for mutation. The primers for each mutagenesis were as follows: first target site mutation primer (TCTGAGATCTGAGCCAGTCGtaGaGGaTGTTTTAGCTCCTTGACTCC) and second target site mutation primer (TTTAGCTCCTTGACTCCTaGaGGaTTATGTCATCATACATGACTCAGC). Wild-type and mutant plasmids were expanded in Escherichia coli and sequenced to confirm mutations.
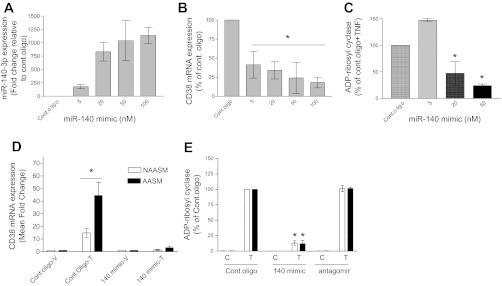
Fig. 2.
Inhibition of TNF-α-induced CD38 expression in HASM cells by miR-140-3p mimic. Mimics of miR-140-3p (140 mimic) or antagomirs for the microRNA (miRNA) were transiently transfected into NAASM and AASM cells, and CD38 mRNA expression and ADP-ribosyl cyclase activity were determined. A: transient transfection of NAASM cells with miR-140-3p mimic oligonucleotide resulted in a concentration-dependent increase in miR-140-3p expression (n = 3). B: transfection of miR-140-3p mimics in NAASM cells resulted in a concentration-dependent attenuation of TNF-α-induced CD38 mRNA expression, although the relationship between mimic concentration and CD38 expression was not linear (n = 3). *P < 0.05, all concentrations vs. control oligonucleotide (Cont oligo). C: in NAASM cells transiently transfected with miR-140-3p mimic oligonucleotides, ADP-riboysl cyclase activity was significantly attenuated at higher concentrations of the mimic (n = 3). *P < 0.05 vs. Cont oligo. D: when NAASM and AASM cells were transiently transfected with 50 nM control oligonucleotide or miR-140-3p mimic oligonucleotide, CD38 mRNA expression was comparably attenuated in both groups of cells (n = 2). *P < 0.05. E: TNF-α-induced ADP-ribosyl cyclase activity was attenuated to a comparable magnitude in NAASM and AASM cells in the presence of miR-140-3p mimic oligonucleotides (n = 3). Basal [control (C)] and TNF-α-induced (T) ADP-ribosyl cyclase activities were unaltered in NAASM or AASM cells transiently transfected with miR-140-3p antagomir oligonucleotides (n = 3). *P < 0.05 vs. Cont oligo(T).
SDS-PAGE and Western blotting.
Total protein (10 μg) was resolved in a 4–20% Tris·HCl SDS gel and electrophoretically transferred onto a polyvinylidene difluoride membrane. The blot was blocked in 5% skim milk solution in PBS containing 0.05% Tween 20 for ≥4 h. The blot was probed with relevant primary antibodies and then incubated with HRP-conjugated secondary antibodies. After washes in PBS containing 0.05% Tween 20, the blots were treated with the chemiluminescent substrate for HRP and exposed to X-ray film for visualization of bands.
ELISA.
For determination of NF-κB or AP-1 activation, ELISA was performed according to the manufacturer's instructions (Activ Motif) and as previously described (15). Briefly, 3 μg of nuclear extracts from HASM cells were incubated in a multiwell plate coated with oligonucleotides carrying consensus NF-κB or AP-1 sequences. Competitor oligonucleotide (20 pmol, 20× excess) was added to some reactions to determine the specificity of the binding.
ADP-ribosyl cyclase assay.
The ADP-ribosyl cyclase activity of HASM cell lysates was quantified by measurement of the reverse cyclase activity of CD38. HASM whole cell lysates containing 5 μg of total protein were incubated for 1 h at 37°C with or without 10 mM nicotinamide in the presence of 0.45 mM cADPR. The reverse cyclase reaction was stopped by addition of 25 μl of 1 M HCl, vacuum-filtered through a 0.45-μm protein-binding membrane (Immobilon, Millipore), and neutralized with 15 μl of 2 M Tris-base. The filtrate was incubated with reagent mixture containing 2 μM rezasurin, 0.76% (vol/vol) ethanol, 4 μM flavin mononucleotide, 40 μg/ml alcohol dehydrogenase, and 0.04 U/ml diaphorase in NaH2PO4/Na2HPO4 buffer, pH 6.8, at room temperature. Fluorescence was quantified (excitation at 544 nm and emission at 590 nm) in a fluorometer (FLUO star Galaxy, BMG Biotechnologies), and the rate of fluorescence emission was calculated. The quantity of NAD generated in the reaction was calculated from a standard curve generated from known NAD.
Data analysis.
Each experiment was performed three to six times (NAASM or AASM cells obtained from 3–6 donors were used). Values are means ± SE. Data were statistically analyzed by Student's t-test or one-way ANOVA (depending on the number of experimental groups analyzed) using GraphPad Prism software. Differences were considered significant at P ≤ 0.05.
RESULTS
Expression of miR-140-3p in HASM cells.
HASM cells were exposed to vehicle (0.1% BSA in sterile PBS) or TNF-α (10 ng/ml) for 24 h, and expression of miR-140-3p and CD38 mRNA was determined. In TNF-α-treated NAASM cells, miR-140-3p expression was marginally reduced compared with vehicle-treated cells (Fig. 1A). In TNF-α-treated AASM cells, miR-140-3p expression was significantly reduced compared with vehicle-treated cells (Fig. 1B). The basal miR-140-3p expression levels were comparable in NAASM and AASM cells (Fig. 1C). Exposure to TNF-α for 24 h resulted in downregulation of miR-140-3p expression in NAASM and AASM cells, but to a greater magnitude in AASM cells (Fig. 1C). Exposure to IL-13, a Th2 cytokine with a critical role in asthma pathogenesis, did not alter miR-140-3p expression in either group of HASM cells (data not shown).
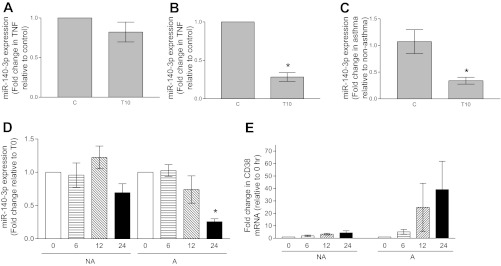
Fig. 1.
Expression of miR-140-3p in vehicle-treated (C) and TNF-α-treated (T10) nonasthmatic airway smooth muscle (NAASM) and asthmatic airway smooth muscle (AASM) cells. A: in the presence of TNF-α, miR-140-3p expression was marginally attenuated in NAASM cells compared with cells treated with vehicle (n = 5). B: in AASM cells, TNF-α significantly attenuated miR-140-3p expression (n = 6). *P < 0.05. C: basal miR-140-3p expression levels were comparable between NAASM and AASM cells. In the presence of TNF-α, miR-140-3p expression was significantly attenuated in AASM cells compared with NAASM cells. *P < 0.05. (Data in A–C are from the same experiments.) D: when NAASM (NA) and AASM (A) cells were exposed to TNF-α for 0–24 h, both showed attenuated miR-140-3p expression at 24 h, although reduction was statistically significant only in AASM cells (n = 3). *P < 0.05. E: TNF-α caused a time-dependent increase in CD38 mRNA expression in NAASM and AASM cells. Magnitude of CD38 induction in AASM cells was higher in AASM cells, although differential increase was not statistically significant because of larger standard error in AASM cells.
In another set of experiments, miR-140-3p expression was determined at different times following exposure to TNF-α. Significant attenuation of miR-140-3p expression was noted at 24 h of TNF-α exposure in AASM cells compared with NAASM cells (Fig. 1D). TNF-α induced CD38 mRNA expression in a time-dependent manner in NAASM and AASM cells, with larger magnitudes of CD38 expression in AASM cells, confirming our earlier findings (15) (Fig. 1E).
miR-140-3p mimic inhibits TNF-α-induced CD38 upregulation in HASM cells.
To determine the functional role of miR-140-3p in CD38 expression in HASM cells, we transfected NAASM and AASM cells with miR-140-3p mimic or control oligonucleotides and determined the effects on CD38 mRNA expression and ADP-riboysl cyclase activity. Transfection of NAASM cells with a range of miR-140-3p mimic oligonucleotides resulted in a concentration-dependent increase in miR-140-3p expression (Fig. 2A). In cells transfected with miR-140-3p mimics, upregulation of TNF-α-induced CD38 mRNA was significantly attenuated in a concentration-dependent manner, although without a linear relationship to the mimic concentration (Fig. 2B). At higher concentrations of miR-140-3p mimic (20 and 50 nM), TNF-α-induced ADP-ribosyl cyclase activity was significantly reduced compared with the cells transfected with 50 nM control oligonucleotide (Fig. 2C). At the optimal concentration (50 nM), miR-140-3p mimic transfection attenuated TNF-α-induced CD38 mRNA expression in NAASM and AASM cells to a similar magnitude (Fig. 2D). Transfection of NAASM and AASM cells with miR-140-3p mimic oligonucleotides also resulted in comparable inhibitory effects on TNF-α-induced ADP-ribosyl cyclase activity (Fig. 2E). Transfection of antagomirs for miR-140-3p did not alter basal or TNF-α-induced ADP-ribosyl cyclase activity in either group of HASM cells (Fig. 2E).
miR-140-3p targets the CD38 3′-UTR.
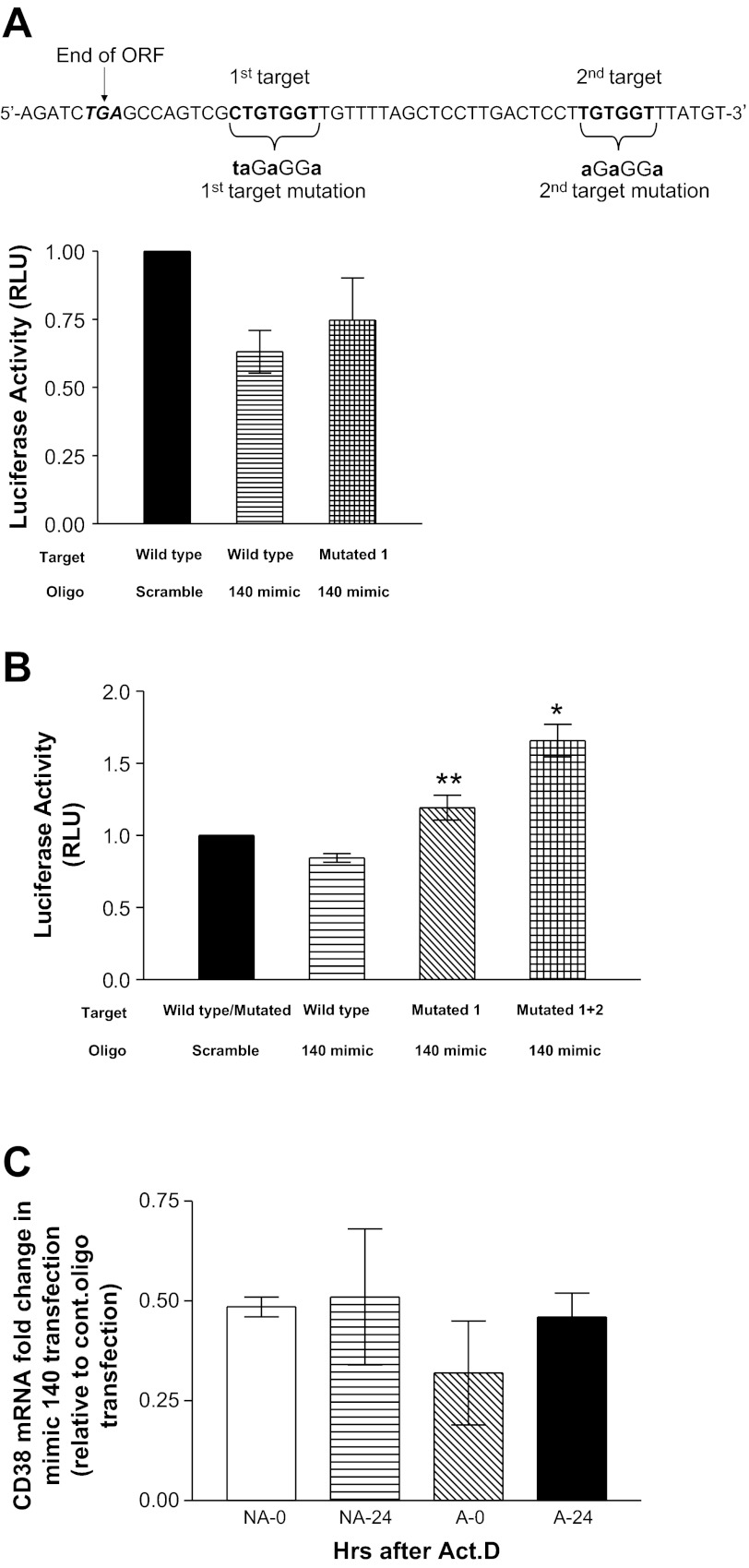
To determine whether miR-140-3p brings about its effects through direct binding to the 3′-UTR of CD38, dual luciferase reporter assays were performed in human embryonic kidney (HEK)-293 or NIH 3T3 cells. Cotransfection of cells with miR-140-3p mimic oligonucleotides and the wild-type Luc-CD38 3′-UTR resulted in a marginal 10–20% inhibition of luciferase activity in HEK-293 or NIH 3T3 cells. Site-directed mutation of the first miR-140-3p target on the CD38 3′-UTR partially reversed the inhibition by miR-140-3p mimic oligonucleotide in HEK-293 cells (Fig. 3A). When the luciferase reporter assay studies were repeated in NIH 3T3 cells, site-directed mutation of the first miR-140-3p target completely reversed the luciferase inhibition by miR-140-3p mimic (Fig. 3B). Mutation of both miR-140-3p targets on the CD38 3′-UTR resulted in elevated luciferase activity in the presence of miR-140-3p mimic oligonucleotides (Fig. 3B). To determine the effect of miR-140-3p mimic transfection on CD38 mRNA stability, NAASM and AASM cells were transfected with 20 nM control oligonucleotide or miR-140-3p mimic. At 12 h following TNF-α exposure (0 h), mimic-transfected NAASM and AASM cells showed attenuated CD38 mRNA levels compared with control oligonucleotide-transfected cells (Fig. 3C). There were no further reductions in CD38 mRNA levels in mimic-transfected cells at 6, 12, and 24 h in either group of HASM cells (Fig. 3C; 6- and 12-h data not shown).
Fig. 3.
miR-140-3p targets the CD38 3′-untranslated region (UTR). Luciferase reporter assays were performed in Luc-CD38-3′-UTR constructs containing wild-type or mutated (Mutated) target. A: 2 miR-140-3p targets on CD38 3′-UTR and mutations on each target (top). ORF, open reading frame. Bottom: in human embryonic kidney (HEK)-293 cells cotransfected with wild-type Luc-CD38-3′-UTR and miR-140-3p mimic oligonucleotides, luciferase activity [relative light units (RLU)] was marginally reduced compared with cells cotransfected with scrambled-sequence oligonucleotides; in HEK-293 cells cotransfected with target-mutated Luc-CD38-3′-UTR and miR-140-3p mimic oligonucleotides, inhibition of luciferase activity was partially reversed (n = 3). B: in NIH 3T3 cells cotransfected with wild-type Luc-CD38-3′-UTR and miR-140-3p mimic oligonucleotides, luciferase activity was marginally reduced compared with cells cotransfected with scrambled-sequence oligonucleotides. When the first miR-140-3p target on Luc-CD38-3′-UTR was mutated (mutated 1), inhibition of luciferase activity was completely reversed. When both miR-140-3p targets on Luc-CD38-3′-UTR were mutated (mutated 1+2), luciferase activity was significantly elevated in the presence of miR-140-3p mimic oligonucleotides (n = 4). C: in NAASM and AASM cells transfected with miR-140-3p mimic (mimic 140), CD38 mRNA levels were comparably attenuated at 0 h (NA-0 and A-0; following TNF-α removal and transcriptional arrest) compared with cells transfected with control oligonucleotides. There were no further reductions in CD38 mRNA levels at 6, 12, or 24 h (NA-24 and A-24) following transcriptional arrest in mimic-transfected cells of either group. *P < 0.05 vs. wild-type or mutated vector cotransfected with scramble oligonucleotide. **P < 0.05 vs. wild-type vector cotransfected with miR-140-3p mimic oligonucleotide. ActD, actinomycin D.
miR-140-3p mimic attenuates activation of p38 MAPK in HASM cells.
Results of the 3′-UTR luciferase reporter assay indicated that direct binding of miR-140-3p mimic on the 3′-UTR of CD38 only partially accounted for the inhibition of CD38 expression. We previously reported that the MAPKs mediate TNF-α-induced CD38 expression in HASM cells (27). Therefore, to determine whether changes in MAPK activation were involved in the miR-140-3p effect on CD38 expression, activation of p38, ERK, and JNK MAPKs was determined in HASM cells following transfection with miR-140-3p mimic oligonucleotides. Transfection with miR-140-3p mimic reduced the TNF-α-induced p38 phosphorylation, with no significant changes in the expression of total p38 (Fig. 4, A and B). TNF-α-induced activation of ERK or JNK activation was not altered by transfection with miR-140-3p mimic (Fig. 4, C and D). Expression of MKK3, a Ser/Thr protein kinase upstream of p38 MAPK, was not altered in the presence of miR-140-3p mimic oligonucleotides (Fig. 4E). We also determined the expression of MKP-1, which is known to inactivate p38, ERK, and JNK MAPKs. Transfection of HASM cells with miR-140-3p antagomir or mimic oligonucleotides did not alter MKP-1 expression (Fig. 4F).
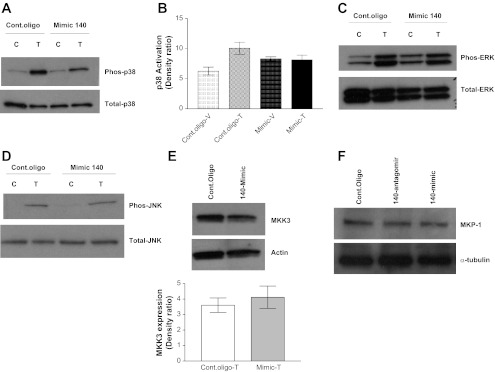
Fig. 4.
miR-140-3p mimic attenuates activation of p38 MAPK in NAASM cells. TNF-α-induced activation of p38, ERK, and JNK MAPKs was determined in NAASM cells transiently transfected with 50 nM control oligonucleotides or miR-140-3p mimic oligonucleotide. A: representative Western blot showing marginally reduced TNF-α-induced p38 MAPK activation in miR-140-3p mimic transfection. Phos, phosphorylated. B: average relative densitometry measurement (n = 5) of Western blot showing marginally reduced p38 MAPK activation in miR-140-3p mimic-transfected NAASM cells. C and D: basal and TNF-α-induced activation of ERK and JNK MAPKs was not altered in the presence of miR-140-3p mimics. Blots are representative of 5 independent experiments. E: expression of MAPK kinase 3 (MKK3), a kinase upstream of p38 MAPK, was not altered by transient transfection with miR-140-3p mimic oligonucleotides. Blot is representative of 3 independent experiments. F: representative blot showing unaltered expression of MKP-1, a dual-specificity phosphatase, in HASM cells transfected with miR-140-3p antagomir or miR-140-3p mimic oligonucleotides (n = 3).
miR-140-3p mimic attenuates activation of NF-κB in HASM cells.
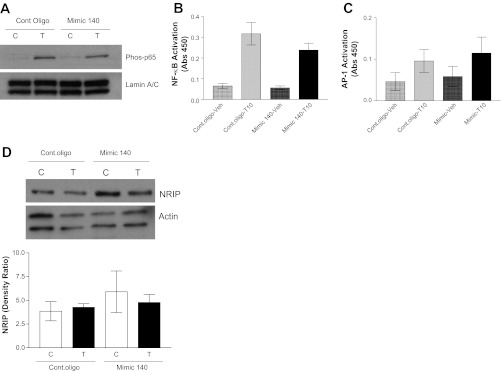
We previously reported that the transcription factors NF-κB and AP-1 mediate TNF-α-induced CD38 expression in HASM cells (16, 27). Transient transfection of HASM cells with miR-140-3p mimic oligonucleotides marginally attenuated activation of the transcription factor NF-κB (Fig. 5, A and B). Transfection with miR-140-3p did not have a significant effect on TNF-α-induced AP-1 activation (Fig. 5C). Expression of NRIP was not altered by miR-140-3p mimic transfection in HASM cells (Fig. 5D).
Fig. 5.
miR-140-3p mimic attenuates activation of transcription factor NF-κB in NAASM cells. TNF-α-induced activation of NF-κB was determined in NAASM cells transiently transfected with 50 nM control oligonucleotide or miR-140-3p mimic oligonucleotides. A: TNF-α-induced nuclear translocation of NF-κB (phosphorylated p65 subunit) was marginally attenuated in NAASM cells transfected with miR-140-3p mimic oligonucleotides. B: TNF-α-induced activation of NF-κB, measured as binding of p65 subunit to a consensus DNA motif [absorbance at 450 nm (Abs 450)], was marginally reduced in NAASM cells transfected with miR-140-3p mimic oligonucleotides (n = 5). C: TNF-α-induced activation of AP-1 (measured as binding of phosphorylated c-Jun to consensus DNA sequence) was unaltered in the presence of miR-140-3p mimic oligonucleotides (n = 4). D: expression of nuclear receptor-interacting protein (NRIP), which is associated with NF-κB activation in other cell systems, was not altered in HASM cells transfected with mimic 140 (n = 3). Blots (A and D) are representative of results from 3 independent experiments. C, vehicle; T, TNF-α.
DISCUSSION
This is the first report of the regulatory role of miR-140-3p in CD38 expression in HASM cells. Our findings show that miR-140-3p expression is lower in HASM cells from donors with a history of severe asthma, in the presence of TNF-α. Our findings also indicate that the effects of miR-140-3p on CD38 expression are mediated through direct binding of miRNA to the CD38 transcript and indirect mechanisms involving activation of p38 MAPK and NF-κB. The net result of the modest inhibition of activation of p38 and NF-κB by miR-140-3p transfection is the robust attenuation of CD38 expression in HASM cells.
Previous studies conducted in our laboratory established that TNF-α-induced CD38 expression in HASM cells is mediated at the transcriptional and posttranscriptional levels (16, 27). The MAPKs ERK1/2 and p38 play a role in posttranscriptional regulation, whereas p38 and JNK MAPKs have a role in transcriptional regulation of the CD38 gene (27). One of the objectives of the current study was to determine the role of miR-140-3p in the regulation of CD38 expression. Studies by other investigators revealed that specific miRNAs, such as miR-21 and miR-133a, contribute to the pathogenesis of airway inflammatory disorders (5, 20). Chiba et al. (5) showed that IL-13, a Th2 cytokine with a prominent role in allergic asthma, downregulates the expression of miR-133a. It is suggested that the downregulation of miR-133a leads to elevated RhoA, a procontractile protein in the ASM cells. CD38 contributes to the development of AHR in mouse models of asthma (10, 11). The CD38-null mouse developed significantly lower levels of airway responsiveness than the wild-type mouse in response to the contractile agonist methacholine (7). The CD38-null mouse also developed reduced AHR compared with the wild-type mouse following brief exposure to TNF-α or IL-13 (10, 11). Therefore, investigating the miRNAs that target CD38 gene expression in HASM cells may lead to an understanding of the signaling pathways involved in the pathogenesis of AHR and asthma.
Multiple web-based target prediction algorithms [Target Scan (www.targetscan.org), miRWalk (www.ma.uni-heidelberg.de), and miRbase (www.mirbase.org)] were used to determine potential miRNA targets in the CD38 3′-UTR. From the list of predicted miRNAs, miR-140-3p was chosen for further investigation, because it has been reported as one of the highly expressed miRNAs in HASM cells (32). Furthermore, in HASM cells exposed to a mixture of inflammatory cytokines including TNF-α, there is significant downregulation of several miRNAs, including miR-140 and some miRNAs involved in the regulation of the smooth muscle phenotype (17). In earlier publications by other investigators, miR-140 has been reported as a cartilage-specific miRNA in mouse and zebrafish (29, 31). However, these reports were largely referring to miR-140-5p, one of the two mature miRNAs originating from the precursor miR-140. In the present study, we focused on miR-140-3p and its role in regulation of CD38 expression.
An altered miRNA expression profile has been reported in T cells obtained from patients with severe asthma, indicating that miRNAs are among the mechanisms involved in the cellular phenotypic changes observed in asthma (28). Attenuation of miR-140-3p expression in AASM cells by TNF-α suggests that this miRNA may have a role in the asthmatic phenotype in ASM cells. The mechanisms involved in miR-140-3p downregulation by TNF-α in HASM cells are yet to be determined. Recent studies in human chondrocytes showed that the proximal upstream region of pri-miR-140 has functional response elements for chondrogenic transcription factors Sox5/Sox6/Sox9, indicating transcriptional regulation of miR-140-3p and miR-140-5p expressions (33). Different transcriptional regulators and epigenetic mechanisms such as DNA methylation may be involved in the altered miR-140-3p expression in AASM cells. The finding that miR-140-3p expression is attenuated to a larger magnitude in AASM cells in the presence of TNF-α suggests an anti-inflammatory role for this miRNA. The potential anti-inflammatory role for miR-140-3p is supported by previously reported findings of downregulated miR-140 expression in whole lung lysate from rats exposed to cigarette smoke extract (14). We also found that IL-13, a Th2 cytokine with a major role in asthma, did not alter the expression levels of miR-140-3p in NAASM or AASM cells (data not shown). These observations suggest that miR-140-3p may have a functional role selective to TNF-α signaling. TNF-α is a cytokine with a major role in the pathogenesis of asthma (1, 26). TNF-α expression is elevated in the airways of asthmatic patients, and some recent therapeutics for asthma target TNF-α and its receptors in lungs (3, 4, 13, 34). Therefore, defining the role of miRNAs involved in the regulation of TNF-α-induced genes in ASM may have a therapeutic potential. Although TNF-α attenuates miR-140-3p expression to a greater magnitude in AASM cells, it does not appear to solely contribute to the differential induction of CD38 expression by TNF-α that we reported in a recent study (15).
Although the inhibitory effect of miR-140-3p mimic on luciferase activity was modest, mutation of both miR-140-3p target sites reversed the inhibition, indicating binding of the miRNA to the 3′-UTR of CD38. Since the 3′-UTR of the CD38 transcript possesses targets for other miRNAs as well, a significant effect on stability or translatability of the CD38 transcript may require multiple miRNAs. However, the significant inhibition of CD38 mRNA and protein expression by miR-140-3p mimic oligonucleotides suggests that additional mechanisms are involved in miR-140-3p regulation of CD38 expression in HASM cells. In this context, we found that miR-140-3p mimic attenuated p38 MAPK activation with no apparent effects on ERK or JNK MAPKs, demonstrating the selectivity of miR-140-3p for specific targets on cell signaling pathways in HASM cells. The marginal inhibitory effect of miR-140-3p mimic on p38 MAPK activation does not appear to be related to the upstream kinase MKK3 or the MAPK dual-specificity phosphatase. However, it remains to be determined whether other MAPK phosphatases with higher substrate specificity to p38 MAPK mediate the miR-140-3p-mediated attenuation of p38 activation.
The significant attenuation of TNF-α-induced CD38 mRNA expression by miR-140-3p mimic suggests that the regulation by miRNA may be through transcription of the CD38 gene in HASM cells. The transcription factors NF-κB and AP-1 mediate TNF-α-induced CD38 expression in HASM cells (16). The role of NF-κB signaling in airway inflammatory disorders has been demonstrated through in vitro studies and animal models (8, 24). For this reason and because of the availability of numerous synthetic inhibitors of NF-κB signaling, this pathway remains an attractive therapeutic target in airway inflammatory disorders such as asthma and chronic obstructive pulmonary disorder (reviewed in Ref. 9). Although the DNA binding activity of NF-κB was only marginally reduced by miR-140-3p mimic transfection, the reduction was a consistent finding in five independent experiments. Furthermore, a recent study reported attenuation of NF-κB activation by miR-140-3p in hepatocytes, primarily by targeting NRIP-1, a coactivator of NF-κB (23). This is an unlikely mechanism in HASM cells, since NRIP-1 expression was unaltered in cells transfected with miR-140-3p. The results of CD38 mRNA stability also showed that, following transcription arrest, CD38 mRNA levels were maintained for up to 24 h. The expression levels of miR-140-3p remain high for up to 72 h following transfection. If the effects of miR-140-3p on CD38 transcript levels were due to binding to 3′-UTR target sites, we would have seen a significant decline in transcript levels following transcription arrest. A lack of correlation between miR-140-3p and CD38 expression following TNF-α treatment strengthens the argument that miR-140-3p effects are largely indirect and time-dependent. Furthermore, overexpression of miR-140-3p causes a significant decline in CD38 transcript levels before arrest of transcription, with no further decline in transcript levels for up to 24 h, favoring a mechanism that involves indirect transcriptional regulation of CD38 expression in HASM cells.
In summary, miR-140-3p regulates TNF-α-induced CD38 expression in HASM cells through direct interaction with the 3′-UTR of CD38 mRNA and indirect mechanisms involving activation of p38 MAPK and the transcription factor NF-κB. However, we cannot rule out other indirect mechanisms, such as competing endogenous RNAs (25), in the regulation of CD38 expression in HASM cells. Understanding the signaling pathways involved in miR-140-3p regulation of CD38 expression in HASM cells may reveal novel therapeutic targets for AHR and asthma.
GRANTS
This work was supported by National Institutes of Health Grants HL-057498 (to M. S. Kannan) and ES-013508 and HL-097796 (to R. A. Panettieri, Jr.), a Seed Grant from the Academic Health Center at the University of Minnesota, and a grant from the Comparative Medicine Signature Program of the College of Veterinary Medicine, University of Minnesota (to M. S. Kannan).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.A.J., M.D., and T.F.W. performed the experiments; J.A.J., M.D., and T.F.W. analyzed the data; J.A.J., M.D., S.S., J.S., R.A.P., T.F.W., and M.S.K. interpreted the results of the experiments; J.A.J. and M.D. prepared the figures; J.A.J., M.D., T.F.W., and M.S.K. drafted the manuscript; J.A.J., M.D., S.S., J.S., R.A.P., T.F.W., and M.S.K. edited and revised the manuscript; J.A.J., S.S., J.S., R.A.P., T.F.W., and M.S.K. approved the final version of the manuscript; S.S. and M.S.K. are responsible for conception and design of the research.
ACKNOWLEDGMENTS
We thank S. Aggarwal and Dr. Lihua Li for technical support and assistance.
REFERENCES
- 1. Albuquerque RV, Hayden CM, Palmer LJ, Laing IA, Rye PJ, Gibson NA, Burton PR, Goldblatt J, Lesouef PN. Association of polymorphisms within the tumour necrosis factor (TNF) genes and childhood asthma. Clin Exp Allergy 28: 578–584, 1998 [DOI] [PubMed] [Google Scholar]
- 2. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, function. Cell 116: 281–297, 2004 [DOI] [PubMed] [Google Scholar]
- 3. Berry MA, Hargadon B, Shelley M, Parker D, Shaw DE, Green RH, Bradding P, Brightling CE, Wardlaw AJ, Pavord ID. Evidence of a role of tumor necrosis factor-α in refractory asthma. N Engl J Med 354: 697–708, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Bradding P, Roberts JA, Britten KM, Montefort S, Djukanovic R, Mueller R, Heusser CH, Howarth PH, Holgate ST. Interleukin-4, -5, and -6 and tumor necrosis factor-α in normal and asthmatic airways: evidence for the human mast cell as a source of these cytokines. Am J Respir Cell Mol Biol 10: 471–480, 1994 [DOI] [PubMed] [Google Scholar]
- 5. Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of RhoA in bronchial smooth muscle cells. Am J Respir Crit Care Med 180: 713–719, 2009 [DOI] [PubMed] [Google Scholar]
- 6. Deshpande DA, Walseth TF, Panettieri RA, Kannan MS. CD38/cyclic ADP-ribose-mediated Ca2+ signaling contributes to airway smooth muscle hyper-responsiveness. FASEB J 17: 452–454, 2003 [DOI] [PubMed] [Google Scholar]
- 7. Deshpande DA, White TA, Guedes AG, Milla C, Walseth TF, Lund FE, Kannan MS. Altered airway responsiveness in CD38-deficient mice. Am J Respir Cell Mol Biol 32: 149–156, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Desmet C, Gosset P, Pajak B, Cataldo D, Bentires-Alj M, Lekeux P, Bureau F. Selective blockade of NF-κB activity in airway immune cells inhibits the effector phase of experimental asthma. J Immunol 173: 5766–5775, 2004 [DOI] [PubMed] [Google Scholar]
- 9. Edwards MR, Bartlett NW, Clarke D, Birrell M, Belvisi M, Johnston SL. Targeting the NF-κB pathway in asthma and chronic obstructive pulmonary disease. Pharmacol Ther 121: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guedes AG, Jude JA, Paulin J, Kita H, Lund FE, Kannan MS. Role of CD38 in TNF-α-induced airway hyperresponsiveness. Am J Physiol Lung Cell Mol Physiol 294: L290–L299, 2008 [DOI] [PubMed] [Google Scholar]
- 11. Guedes AG, Paulin J, Rivero-Nava L, Kita H, Lund FE, Kannan MS. CD38-deficient mice have reduced airway hyperresponsiveness following IL-13 challenge. Am J Physiol Lung Cell Mol Physiol 291: L1286–L1293, 2006 [DOI] [PubMed] [Google Scholar]
- 12. Howard M, Grimaldi JC, Bazan JF, Lund FE, Santos-Argumedo L, Parkhouse RM, Walseth TF, Lee HC. Formation and hydrolysis of cyclic ADP-ribose catalyzed by lymphocyte antigen CD38. Science 262: 1056–1059, 1993 [DOI] [PubMed] [Google Scholar]
- 13. Howarth PH, Babu KS, Arshad HS, Lau L, Buckley M, McConnell W, Beckett P, Al Ali M, Chauhan A, Wilson SJ, Reynolds A, Davies DE, Holgate ST. Tumour necrosis factor (TNFα) as a novel therapeutic target in symptomatic corticosteroid dependent asthma. Thorax 60: 1012–1018, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J 23: 806–812, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jude JA, Solway J, Panettieri RA, Jr, Walseth TF, Kannan MS. Differential induction of CD38 expression by TNF-α in asthmatic airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 299: L879–L890, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang BN, Tirumurugaan KG, Deshpande DA, Amrani Y, Panettieri RA, Walseth TF, Kannan MS. Transcriptional regulation of CD38 expression by tumor necrosis factor-α in human airway smooth muscle cells: role of NF-κB and sensitivity to glucocorticoids. FASEB J 20: 1000–1002, 2006 [DOI] [PubMed] [Google Scholar]
- 17. Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol 42: 506–513, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee HC, Aarhus R. ADP-ribosyl cyclase: an enzyme that cyclizes NAD+ into a calcium-mobilizing metabolite. Cell Regul 2: 203–209, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, Orkin SH, Aronow BJ, Rothenberg ME. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-γ pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol 187: 3362–3373, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol 182: 4994–5002, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor α1 (IL13Rα1). J Biol Chem 286: 1786–1794, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pagdin T, Lavender P. MicroRNAs in lung diseases. Thorax 67: 183–184, 2011 [DOI] [PubMed] [Google Scholar]
- 23. Takata A, Otsuka M, Kojima K, Yoshikawa T, Kishikawa T, Yoshida H, Koike K. MicroRNA-22 and microRNA-140 suppress NF-κB activity by regulating the expression of NF-κB coactivators. Biochem Biophys Res Commun 411: 826–831, 2011 [DOI] [PubMed] [Google Scholar]
- 24. Tao Y, Williams-Skipp C, Scheinman RI. Mapping of glucocorticoid receptor DNA binding domain surfaces contributing to transrepression of NF-κB and induction of apoptosis. J Biol Chem 276: 2329–2332, 2001 [DOI] [PubMed] [Google Scholar]
- 25. Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, Lieberman J, Rigoutsos I, Pandolfi PP. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147: 344–357, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomas PS, Yates DH, Barnes PJ. Tumor necrosis factor-α increases airway responsiveness and sputum neutrophilia in normal human subjects. Am J Respir Crit Care Med 152: 76–80, 1995 [DOI] [PubMed] [Google Scholar]
- 27. Tirumurugaan KG, Jude JA, Kang BN, Panettieri RA, Walseth TF, Kannan MS. TNF-α induced CD38 expression in human airway smooth muscle cells: role of MAP kinases and transcription factors NF-κB and AP-1. Am J Physiol Lung Cell Mol Physiol 292: L1385–L1395, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, Adams OD, Macedo P, Booton R, Gibeon D, Chung KF, Lindsay MA. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol 129: 95–103, 2012 [DOI] [PubMed] [Google Scholar]
- 29. Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 580: 4214–4217, 2006 [DOI] [PubMed] [Google Scholar]
- 30. White TA, Johnson S, Walseth TF, Lee HC, Graeff RM, Munshi CB, Prakash YS, Sieck GC, Kannan MS. Subcellular localization of cyclic ADP-ribosyl cyclase and cyclic ADP-ribose hydrolase activities in porcine airway smooth muscle. Biochim Biophys Acta 1498: 64–71, 2000 [DOI] [PubMed] [Google Scholar]
- 31. Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Williams AE, Larner-Svensson H, Perry MM, Campbell GA, Herrick SE, Adcock IM, Erjefalt JS, Chung KF, Lindsay MA. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLos One 4: e5889, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamashita S, Miyaki S, Kato Y, Yokoyama S, Sato T, Barrionuevo F, Akiyama H, Scherer G, Takada S, Asahara H. L-Sox5 and Sox6 enhance chondrogenic miR-140 expression by strengthening dimeric Sox9 activity. J Biol Chem 287: 22206–22215, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ying S, Robinson DS, Varney V, Meng Q, Tsicopoulos A, Moqbel R, Durham SR, Kay AB, Hamid Q. TNF-α mRNA expression in allergic inflammation. Clin Exp Allergy 21: 745–750, 1991 [DOI] [PubMed] [Google Scholar]