Abstract
Cardiomyopathies are a major health problem, with inherited cardiomyopathies, many of which are caused by mutations in genes encoding sarcomeric proteins, constituting an ever-increasing fraction of cases. To begin to study the mechanisms by which these mutations cause disease, we have employed an integrative modelling approach to study the interactions between tropomyosin and actin. Starting from the existing blocked state model, we identified a specific zone on the actin surface which is highly favourable to support tropomyosin sliding from the blocked/closed states to the open state. We then analysed the predicted actin-tropomyosin interface regions for the three states. Each quasi-repeat of tropomyosin was studied for its interaction strength and evolutionary conservation to focus on smaller surface zones. Finally, we show that the distribution of the known cardiomyopathy mutations of α-tropomyosin is consistent with our model. This analysis provides structural insights into the possible mode of interactions between tropomyosin and actin in the open state for the first time.
Keywords: cardiomyopathy mutations, cardiac thin filament, modeling, tropomyosin, period 2
Video Abstract
Video Abstract Available from http://la-press.com/t.php?i=9798
Introduction
Cardiac muscle is striated like skeletal muscle. The muscle fibre consists of thin and thick filaments, composed of actin and myosin respectively, forming the basic unit of the sarcomere. F-actin, tropomyosin and the troponin complex together form the thin filament and the thick filament is made up of myosins, in which the coiled-coil tail domains are bundled and the head domains interact with actin, forming the cross-bridges.1 The double stranded F-actin filament is wrapped with an elongated tropomyosin dimer. The tropomyosin is a coiled-coil that can interact end-to-end with adjacent tropomyosins so that it extends throughout the actin filament. The thin filament also includes the troponin complex which is composed of three different subunits, troponin C (TnC), troponin I (TnI) and the troponin T (TnT). Troponin C is the calcium binding subunit, troponin I is the inhibitory subunit and troponin T is the tropomyosin-binding subunit. Upon an increase in intracellular free calcium level the binding of Ca2+ to TnC produces conformational changes to other proteins of the troponin complex. Such changes are responsible for the movement of tropomyosin relative to actin that modulates actin-myosin interaction.2
The acto-myosin ATPase activity and the contraction/relaxation process of muscle can be explained by a three-state muscle regulation model.3 The three states are blocked (B), closed (C) and open (M). In the B-state, the myosin binding sites on actin are blocked or not kept accessible by the tropomyosin coiled-coil dimer. The tropomyosin dimer interacts with specific residues on actin which are also required for interaction with the myosin head. Thus, tropomyosin sterically blocks myosin binding and cross-bridge formation. In the closed state, increases in intracellular Ca2+ levels increases binding of Ca2+ to TnC which through TnI and TnT induces conformational change and azimuthal movement of tropomyosin on the surface of the actin filament. This movement renders the myosin binding residues on actin to be more solvent-accessible and available for weak interactions with myosin. A further increase in myosin binding moves tropomyosin so that more myosin-binding sites on actin become accessible, allowing strong interaction of myosin and actin. This represents the open state, with maximum myosin binding and force production. Several studies based on Förster resonance energy transfer (FRET), electron microscopy (EM) and cryo-EM have been performed by other groups that are consistent with the three-state model of muscle regulation in detail.4–9
Although structures of actin, tropomyosin and troponin core domain are available, much uncertainty remains in understanding the conformational changes responsible for muscle regulation. The structural models of skeletal muscle thin filament are well studied by 3D-EM and FRET methods, the models are constructed for blocked and closed states or states with and without calcium.5,7–9 These models differ from each other in defining the precise orientation of the components and the interface formed between the structural units at different muscle regulation states. However, there is no structural model reported to date which describes the cardiac thin filament in its open state. This has led us to study the cTF in detail and understand the structural significance of interface regions of actin and tropomyosin.
These studies are clinically significant because hundreds of mutations in genes encoding cardiac muscle proteins cause inherited cardiomyopathies. The two major types of cardiomyopathies are dilated (DCM) and hypertrophic (HCM). In DCM, the heart muscle fibers are damaged, causing the weakness and reduction of thickness of walls of heart chambers. In HCM, the hypertrophy causes thickening of the walls with disordered growth of heart muscle fibres. This prevents efficient filling of the heart.10 Mutations in cardiac muscle proteins, including β-cardiac myosin heavy chain, cardiac myosin binding protein C, cardiac troponin T (cTnT), cardiac troponin I (cTnI), α-tropomyosin (Tm), cardiac α-actin, cardiac regulatory myosin light chain and cardiac essential myosin light chain have been shown to cause DCM and HCM phenotypes.11
Since high resolution structures of macromolecular assemblies like cTF are not available, we have employed an integrative approach to build the models, using data from multiple sources including X-ray crystallography, NMR, EM, footprinting, chemical cross-linking, FRET, SAXS and proteomics.12–15
In light of the known structural information of skeletal muscle thin filament, we have constructed an atomic model of cardiac thin filament in blocked and closed states by using an integrative template-based modelling approach. We also propose a path for movement of tropomyosin dimer on actin surface from blocked-closed states to open state and the actin-tropomyosin interface at the open state. We have also attempted to provide structural rationale for the effects of known cardiomyopathy mutations and anticipate important regions which will be vital for its structural states and functional consequences.
Results and Discussion
Integrative modelling of macromolecular cardiac thin filament
cTF consists of actin, tropomyosin (Tm) and troponin (Tn) complex in a ratio of 7:1:1, where seven actin subunits, one Tm and one Tn complex forms a single unit of cTF. Multiple units form the stretched thin filament macromolecular structure. In order to study the structural importance of cTF and the effect of associated disease mutations, it becomes essential to construct a three-dimensional model of cTF. Knowing the size of the system and the different proteins present, it becomes difficult to obtain the atomic model based only on high resolution X-ray structures. On the other hand, macromolecular assemblies are well captured using three-dimensional electron microscopy (3D-EM), cryo-EM and FRET studies. Thus, we have used the structural information from the available resources of EM data and also impose the 3D-atomic coordinates of high resolution X-ray structures along with them. Using this approach, we constructed the cTF-one unit atomic structure.
Available structural data were EM structure of F- actin,16,17 X-ray high resolution structures of partial Tm18–20 and low resolution structure of full-length Tm,21 and structures of partial Tn complex from skeletal and cardiac muscle.22,23 The overall macromolecular organization of thin filament is well-defined in EM studies.4,8,9
We have used an integrative approach to build the three-dimensional model of cTF, by this we mean that information from multiple resources like EM, X-ray and FRET has been used to construct the model and the model has been constructed for two states, the blocked and closed states.
The initial template of the model is an EM structure.9 The reconstructed structure of the EM map had data for both blocked and closed states, where the template has actin filament, partial tropomyosin structure wrapped throughout the filament and troponin core domain at low and high Ca2+ levels. The information from the EM template gives the idea about the azimuthal position of Tm at B- and C-state, but not the axial position or the interacting zones between Tm and actin. Li and coworkers were the first group to give details of tropomyosin position on F-actin at the Ca2+ saturated state in the absence of troponin complex using EM and computational chemistry. Thus, the interacting residue information between actin and Tm at closed state is taken into account to obtain the model for our study.8 Additionally, EM structure of actin-myosin interactions24 by Lorenz and coworkers agree with Li and coworkers on the predicted interacting residues and the mode of interactions between tropomyosin dimer and actin filament and the manner in which the myosin binding residues are kept inaccessible at the blocked and closed states. Based on FRET analysis, the interacting zone of troponin with Tm is identified: the Tn core domain interacts with Tm at residues 160–200.7 This zone is approximately found near to the fifth quasi-repeat of Tm, thus Tn might have interaction with Tm at P5 and actin3 (the numbering is based on specific periods of Tm and corresponding actin unit starting from the barbed end). For the present study, we have considered only the Tn core domain and not the TnT tail due to the lack of structural information for the latter. A recent work of Manning and coworkers has reported a model of TnT domain, along with other structures of cTF, but we have not used those structures as template since they were generated by a MD study.25 Instead, we have directly used structures from EM data. The Pirani et al model, which is considered to be the initial template for our integrative modelling, does not have full-length 284 residue long tropomyosin reconstructed, thus we have used the full-length Tm crystal structure21 to fit the Tm dimer of the model. All the information from different sources have thus been combined together to obtain the model.
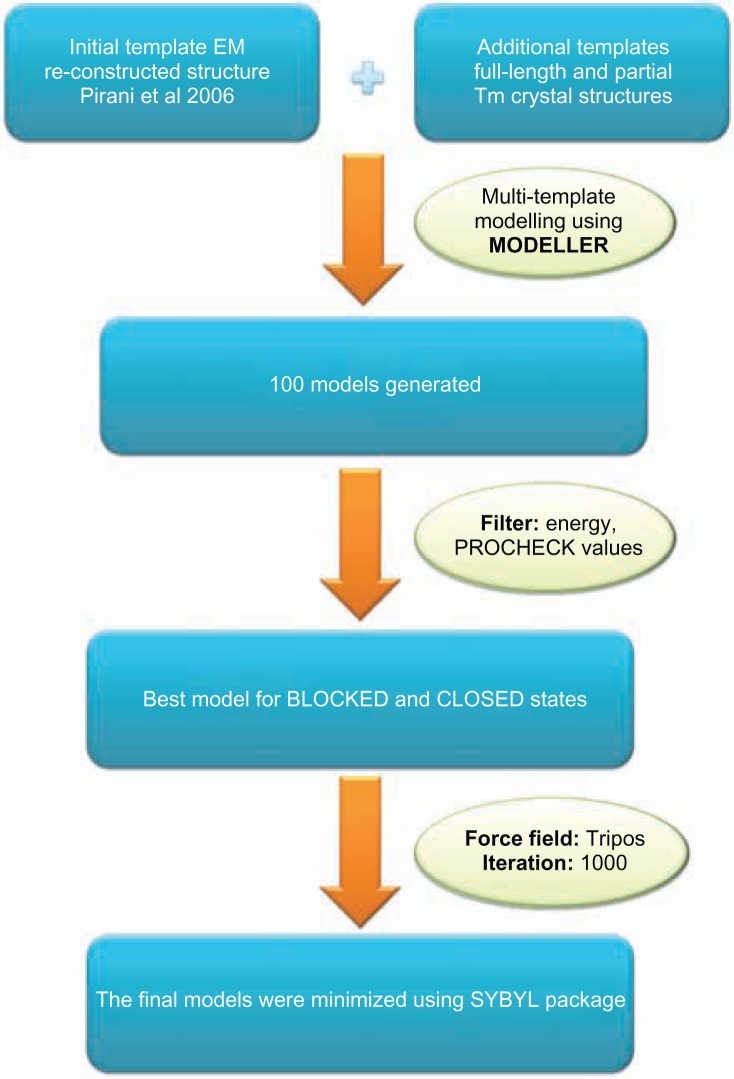
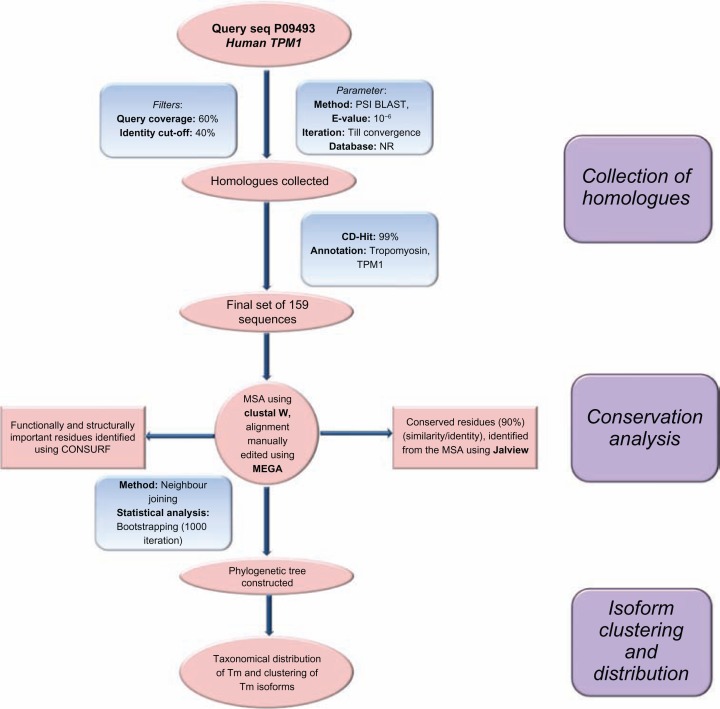
Steps followed to build the 3D model of cTF were as follows: the template was a reconstructed EM structure from Pirani et al model which has a resolution of 25 Å. The model had template for both blocked and closed states. The cTF model was built using the MODELLER software (version 9.8),26 a multi-template modelling approach was used so that good resolution structures of Tm dimer (PDB ID: 1IC2, 2B9C, 1KQL) and the full-length Tm structure (PDB id: 1C1G) was used simultaneously. A total of 100 models were generated and the best model was chosen based on their energy and PROCHECK27 values. From the pool of generated models, the lowest-energy model was chosen as the best one for the blocked and closed states. The final model was minimized using SYBYL package (SYBYL7.1, Tripos Inc) with the following parameters: Tripos force field, 1000 iterations using Powell’s gradient and was terminated at a convergence of 0.05 Kcal mol Å−1. Finally, the atomic model of cTF was built for the B- and C-states and the minimized models were used for the rest of the study. The protocol followed to build the model is shown as a flowchart in Figure 1 and the structure of the model is shown in Figure 2. The quality of the final models was assessed using structure analysis program PROSA.28 All the independent chains of actin, Tm and Tn complex of the both blocked and closed models have Z-scores within the acceptable range (please see Supplementary File). Other structure validation tools became difficult to apply due to the huge macromoleular size of cTF.
Figure 1.
Complete protocol used to build the cardiac thin filament model in blocked (low calcium) and closed (high calcium) states.
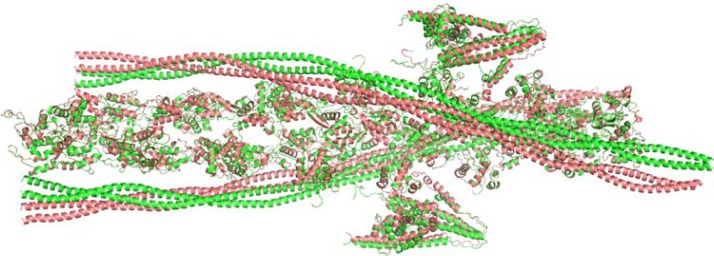
Figure 2.
Systematic representation of the models (green-blocked state, salmon-closed state).
Note: The cTF one unit models of blocked and closed states consist of F-actin, Tm dimer and the partial Tn complex [2(7:1:1)].
We have constructed the models with the available structural information present and integrated the known structural knowledge of cardiac thin filament. However, we have not included the TnT tail domain of cTnT due to the lack of structural template. This troponin T tail plays a crucial role in cTF, retains interaction with Tm overlap complex and most of the cardiomyopathy mutation in cTnT gene resides at this domain. Second, we have constructed a one unit cTF model which means the model does not have information about the tropomyosin overlap region. Thirdly, the models were constructed using coalescence of the high resolution crystal structures, still, lack of high resolution full-length thin filament structure creates difficulty in building the model. Such a derived model cannot precisely reflect the exact orientation of the side chains and their interacting residues prominently. However, this integrated model can be used to study the interface regions between the components of cTF and the change of spatial relationship between these components from blocked to closed states.
Actin-tropomyosin interface in the open state could be predicted
The actin-Tm interaction plays an important role in the cTF structural stability and the azimuthal rotation of Tm on actin outer domains is essential for myosin activation and function of muscle. Li and coworkers had proposed a list of interacting residues between actin and Tm in the closed state using EM analysis,8 independently Miki and coworkers had proposed actin-Tm interface in B- and C-states,5 but the results on interacting residues are not in agreement. In our models, we have used the results from recent EM data of Li and coworkers and Lorenz and coworkers to build the cTF structure. Both the models agree on the concept that relieving myosin binding sites on actin will facilitate TF activation and cross-bridge formation.
We have used the models of blocked and closed states to analyse the energetically more stable state. Tm interaction with actin in the blocked state is the native position in cTF and its interaction at the closed and open states requires an external force to move it to that interaction zone. Logically, we anticipate that the blocked model interactions will be energetically more stable than that of the closed and open states. Using our models of blocked and closed states; we calculated the energy contributed by the electrostatic interactions between actin and Tm dimer. The energy was calculated using COILCHECK+29 (please see supplementary file for the details of energy calculation). The results show that the model of blocked state is found to be more energetically stable than the closed state (Energies: B-state: −60.286504 kJ/mol and C-state: −46.375797 kJ/mol). Since the blocked state is energetically more favourable the interface formed at this state is very crucial for the cTF function. Thus, the interface formed between actin and Tm in the blocked state shares specific residues from both the interacting partners which are highly important to maintain the stability of the cTF.
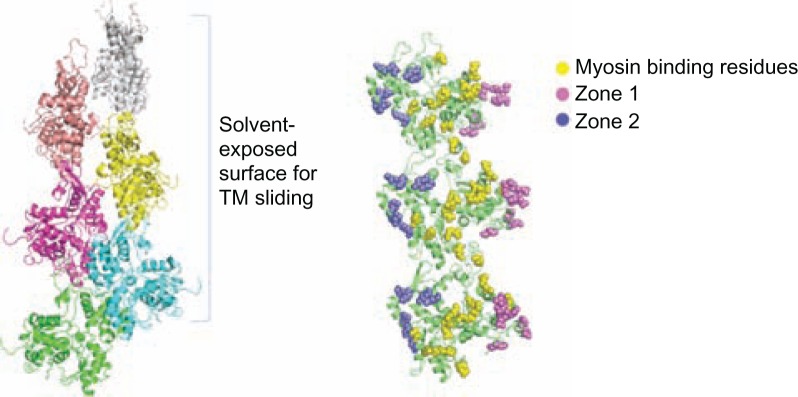
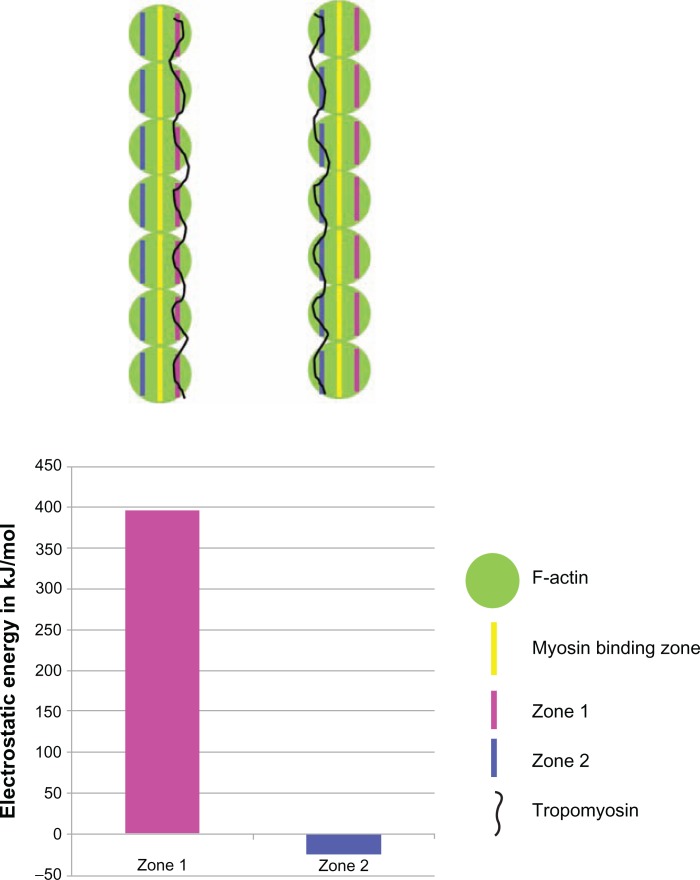
From the F-actin structure, there could be two zones (zone 1 and zone 2) of charged residues on the actin surface on either side of myosin binding residues where Tm could slide (Figure 3). These two zones were identified based on their surface accessibility using JOY program.30 We have employed computational tools to identify which of the two zones will be energetically more favourable for the Tm to move in its closed and open state. Tm has been modelled at both zone 1 and zone 2 on the outer surface of actin and the interaction energy at the site of actin-Tm interface was calculated using our in-house program COILCHECK+, using standard Coulomb’s law, where inter-atomic distance was considered as 15 angstrom and distant dependent dielectric was employed. As shown in Figure 4, zone 1 has high positive energy values and it is highly unfavourable for the Tm interaction. This is due to the presence of clustered negatively charged residues at that region which imposes more unfavourable electrostatic interactions with the Tm dimer. On the other hand, zone 2 is much favourable in its energetic contribution. Thus, it becomes more convincing that the direction of Tm movement from blocked to closed and then to open states on the surface of actin will be towards zone 2. (please see details on the Tm dimer modelling at the two zones in Supplementary data). Supplementary Figure 1 shows the electrostatic energy at the different interfaces.
Figure 3.
F-actin structure from the cardiac thin filament model and the predicted charged zones on the surface of actin filament.
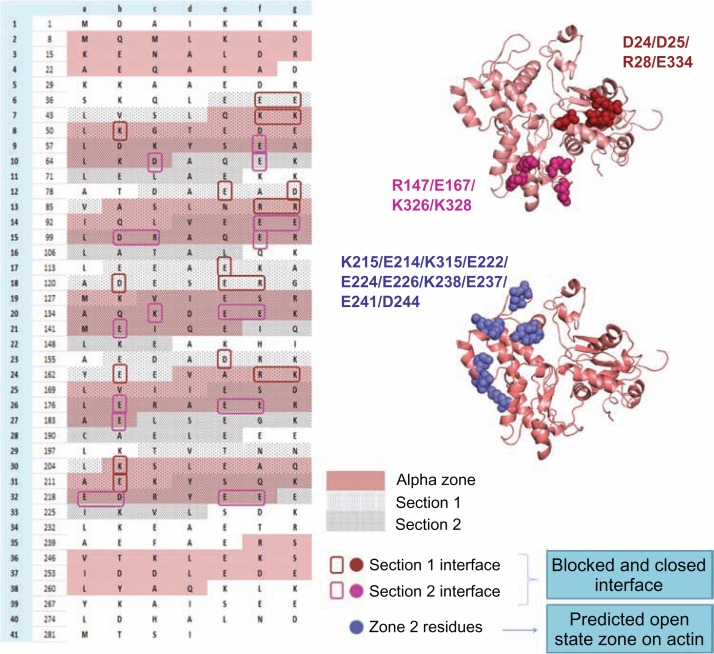
Notes: The two zones are present on either side of myosin binding residues. [Zone 1: D1, D2, E3, E4, R95, E100, E363, E364, K373; Zone 2: K215, E214, D222, E224, E226, K238, E237, E241, D244, K315]. The view of the actin filament on the right is rotated 90 degrees from the view on the left, and the actin monomers are recolored green.
Figure 4.
The tropomyosin dimer was placed on both of the predicted charged zones and the electrostatic energy was calculated using COILCHECK+ program.
Notes: The energies were very highly unfavourable for zone 1 interface when compared to zone 2. Cartoon representation of the thin filament is shown for better understanding.
The interface at blocked and closed states could be identified from the models, and based on the expectation that all myosin binding sites will be accessible to the myosin head in the open state (M-state), we find that zone 2 will be more favourable for Tm interaction. Thus, a list of possible interfaces between actin and tropomyosin in the three states (B-, C- and M-states) could be derived. Since the models were built from low resolution data, it is difficult to provide exact residue interactions and therefore probable interacting regions could been identified. Moreover, to our knowledge, there is no specific report documenting the potential Tm binding residues on actin in the open state (M-state). Thus, this work is the first to predict the interaction zone on actin in the open state.
From the tropomyosin perspective, quasi-repeats of Tm (please see below for quasi-repeats) are the seven equivalent parts of Tm that interact with actin in the cTF named as periods (P1–P7). From the zones of interactions between actin and Tm in the three states, it is noticed that each quasi-repeat of Tm can be divided into two sections; let us denote them as the first section containing the N-terminal part of a quasi-repeat and second containing the C-terminal part. In the blocked and closed states, interactions of the second section of the period are conserved. Tm residues that interact with actin residues R147, E167, K326, and K328 retain their interactions whereas interactions in the first section of the period are quite weak. Just as zone 2 in actin (described above) is the most favourable region for Tm to shift in its open state, it can be anticipated that the first section of Tm could form new interactions with specific residues on zone 2 in its open state. The details of predicted interacting regions of the three states are systematically given in Figure 5. Different sections of different quasi-repeats interact with actin in the three different states.
Figure 5.
The probable actin-tropomyosin interface in the three states (B-, C-, M-states) is shown systematically.
Notes: In the left side the Tm sequence is represented based on their heptad pattern and the pink shaded region is the α zone of each quasi-repeat. Each quasi-repeat is divided into two sections (section 1 and 2-dotted regions) and the probable interface residues which may interact with actin at the blocked and closed states are boxed with different colours. In the right side F-actin structure with mapped interface residues at the three states are displayed. Actin interface residues at blocked and closed states are found to be common but the strength of interaction with the Tm residues differs in both the states. In the blocked state section 1 and section 2 forms strong interactions. But in the closed state section 1 becomes weaker but section 2 is still stronger in its interaction with actin. The predicted open state interface of actin consists of the zone 2 residues, and the charged residues from section 1 of each quasi-repeat contribute for the Tm interface. The periods 1 and 7 are not considered for the study.
Quasi-repeats of tropomyosin interacts differently
The stability and the structural organization of cTF is maintained by the actin-tropomyosin interactions throughout the thin filament. The G- to F-actin conversion is due to a relative rotation of two major domains which give rise to the F-actin flat conformation.13 The tropomyosin, which is an elon-gated long coiled-coil structure, interacts with actin units throughout the TF. Tm is a 284-residue protein which is completely made up of coiled-coil domain; it is one of the longest coiled-coil proteins which is predicted to have complete heptad pattern. As is well-known, coiled-coils are structural domains which follow a seven residue heptad pattern ‘abcdefg’, in which ‘a’ and ‘d’ are hydrophobic residues which are in the interior, ‘e’ and ‘g’ are charged residues which help in maintain the stability of the dimer by forming intrachain salt-bridges and ‘b’, ‘c’ and ‘f’ positions are polar residues which may have interactions with other proteins.31,32 Full-length Tm would interact with seven actin subunits forming the filament, and hence it could be divided into seven quasi-repeats based on their interaction with corresponding actins. Though Tm consists of seven quasi-repeats, not all of them are similar in function; each quasi-repeat has specific properties and function.33,34
The interface between actin and Tm is dominated by charged residues: both actin and Tm are made up of large number of positive and negatively charged residues at the surface. Though the F-actin structure has same charged zones exposed on the surface for Tm binding, Tm interacts differently with actin at different quasi-repeats. The composition of residues in different quasi-repeats is slightly different which makes variation in interaction and stability between actin and Tm. The difference in residue specificity within quasi-repeat and the presence of destabilising interactions in the Tm coiled-coil dimer is an in-built property of Tm and is required for its function.35
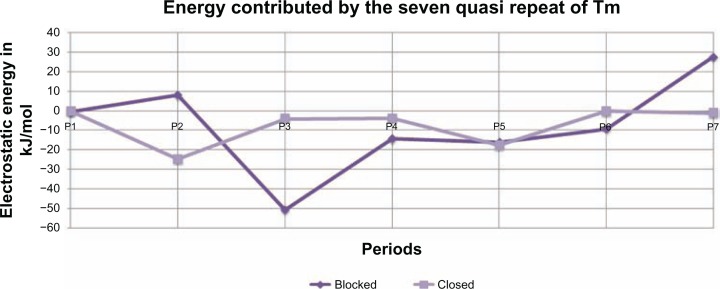
Since the interactions between actin and tropomyosin will be different at the seven quasi-repeats of Tm, the electrostatic energy contributed by different quasi-repeats was calculated. Both the blocked and closed state models were divided into seven parts, where each part would contain an actin and its corresponding Tm quasi-repeat alone and electrostatic energy was calculated for the different parts as stated above. The results are shown in Figure 6, where it is clearly evident that different zones energetically interact differently with actin. P2 and P7 are unstable in the blocked state and becomes more stable in the closed state, and P3 is found to be highly stable in the blocked state and loses its stability considerably in the closed state. P1 and P5 do not exhibit much difference in energy in both the states. P4 and P6 do not show very high deviation in energy. These observations do have a meaningful explanation. Studies on cryo-EM data of insect flight muscle have shown that there is a ‘target zone’ on the actin filament and it is located in the region between the two troponin globular core domains, and this is the region where myoS1 binds more frequently.36 This suggests to us that the actin-Tm interaction in these target zones plays a crucial role in calcium-mediated change in interface which is functionally required. The target zone seen between the troponin core domains corresponds to specific tropomyosin quasi-repeats. The energy calculations at different quasi-repeats of different states also suggest that these regions play a vital role in cTF organization. P1 and P7 are already strained by the adjacent end-to-end overlap which makes them very important for cTF stability. The next is the important regions of P2 and P3, the energy contribution by these quasi-repeats is quite interesting. It can be seen from the graph that P2 is unstable in the blocked state whereas P3 is highly stable. This indicates that these two repeats could be sensitive to Ca2+ mediated change in interface that falls in the target zone of myosin interaction. This clearly shows us a way in which the individual quasi-repeat of Tm contributes differently to the interaction and stability of cTF and has a specific role in the actomyosin cross-bridge and muscle contraction.
Figure 6.
Electrostatic contribution of different quasi-repeats at the blocked and closed states of the model.
Conservation and phylogenetic analysis of tropomyosin
The study of interacting residues between actin and Tm prompted us to identify the conserved and variable regions in tropomyosin. This would guide us to understand the importance of different zones and would allow us to predict the most vital residues and region of Tm that would have significance in cTF function. To study the conservation of Tm, we collected homologues of tropomyosin by using human-α tropomyosin TPM1 (Uniprot id: P09493) as query using PSI-BLAST.37 The homologues were aligned using CLUSTALW protein sequence alignment program38 and manually edited using MEGA ver.539 to obtain a proper multiple sequence alignment (MSA). CONSURF40 was used to identify the conservation patterns and to determine the structurally and functionally important residues. Further, to understand the taxonomic distribution and clustering of collected homologues, a phylogenetic analysis was carried out. Neighbour-joining bootstrapped tree was constructed using MEGA software. The tree was analysed in terms of taxonomic clustering, iso-form distribution and conservation pattern. The steps followed for the analysis are shown in Figure 7. The homologues collected cover a broad range of taxonomic groups. Tm sequences from mammals, birds, fishes, insects, worms were included in the analysis. The distribution of the collected homologues in various levels of classification is shown in Supplementary file.
Figure 7.
Complete protocol for homologue collection, clustering based on isoforms and analysis of the conservation pattern.
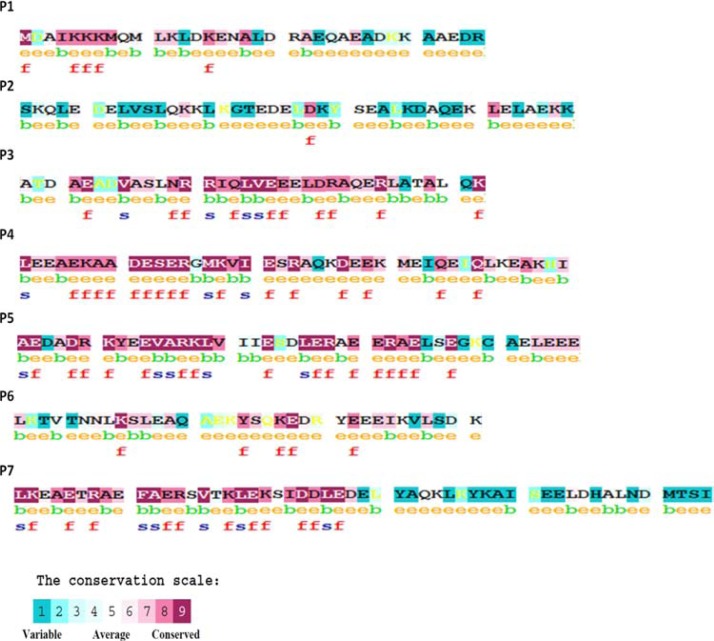
From the CONSURF output, it can be seen that the first 12 residues of the N-terminus of P1 were highly conserved, it also supports the fact these residues are highly important for the Tm head-to-tail overlap with its adjacent Tm chain.41 P3, P4 and P5 are found to be highly conserved, but P2 and the extreme C-ter of P7 are found to be variable (Fig. 8). We further analysed the alignment and phylogenetic tree to note if this conservation pattern may be class-specific amongst the homologues chosen for analysis.
Figure 8.
Conservation pattern of the seven quasi-repeat (results from CONSURF).
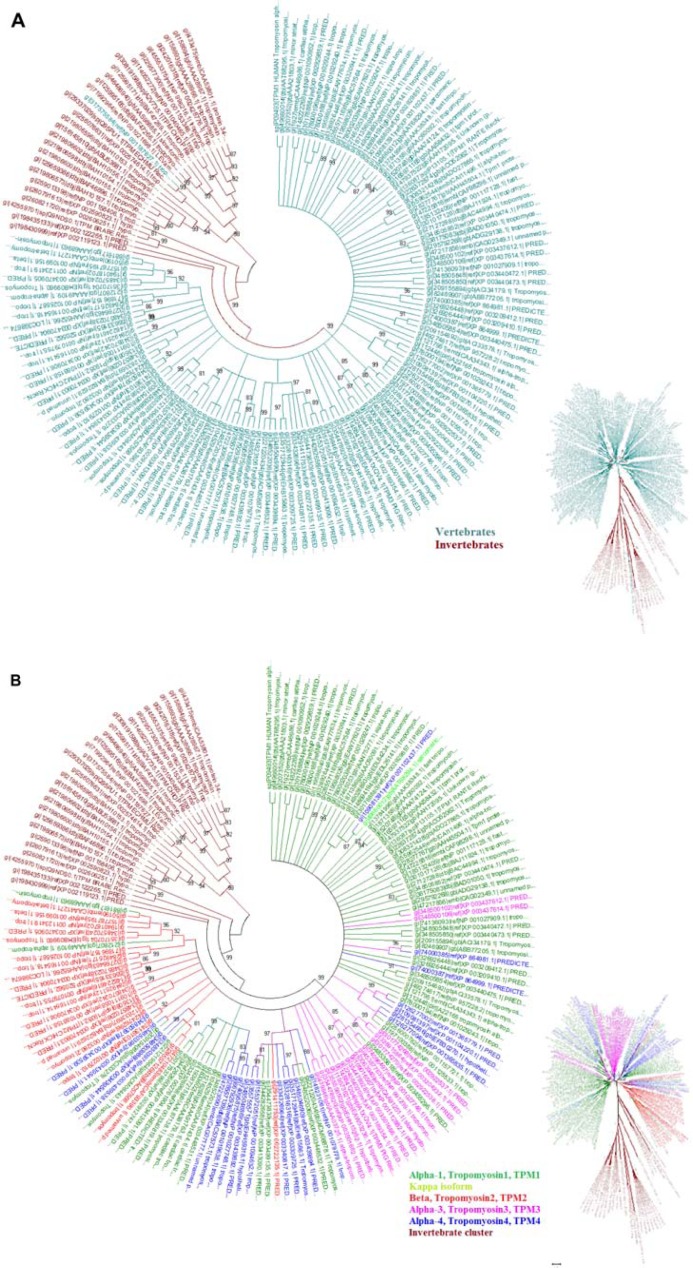
The tree displayed two distinct clusters of homologues. The first cluster retains homologues from vertebrates and other from invertebrates (Fig. 9A). This shows that tropomyosin sequence differs considerably between major taxonomic groups. This suggests that the mechanism of action of Tm in invertebrates could be different from vertebrates. Vertebrate Tm can be classified into two groups based on molecular weight; they are high molecular weight (HMW) group and low molecular weight (LMW) group. Four Tm genes, identified in mammalian species, form 20 different isoforms which are expressed by alternative RNA processing events.42 The four genes, identified in the human genome, are named as TPM1 (α), TPM2 (β), TPM3 (γ) and TPM4 (δ). The clustering of the major four isoforms is shown in Figure 9B. The cardiac muscle is dominated by the presence of α-isoform and few β-isoforms, where both the isoforms could function efficiently.43 Recently, a new specific isoform has been identified in heart muscles, during the onset of cardiomyopathies, which is named the kappa-isoform.44,45 Human α- and β-isoforms share 85.6% identity. Amino acid differences are distributed throughout the length of the Tm sequence, but the residue exchanges seem to be permitted (with similar chemical groups) suggesting that the function of Tm may not be affected. Whereas human α- and κ-isoforms are 90% identical, the amino acid differences are exactly targeted at Tm residues 41–80 which falls in the P2 quasi-repeat. Therefore, the amino acid conservation patterns at P2 in different isoforms were examined in detail.
Figure 9.
Clustering of the collected homologues. (A) Two different clusters separating the vertebrates and invertebrate Tm sequence. (B) clustering of the four isoforms of Tm is shown, isoforms were classified based on their annotation.
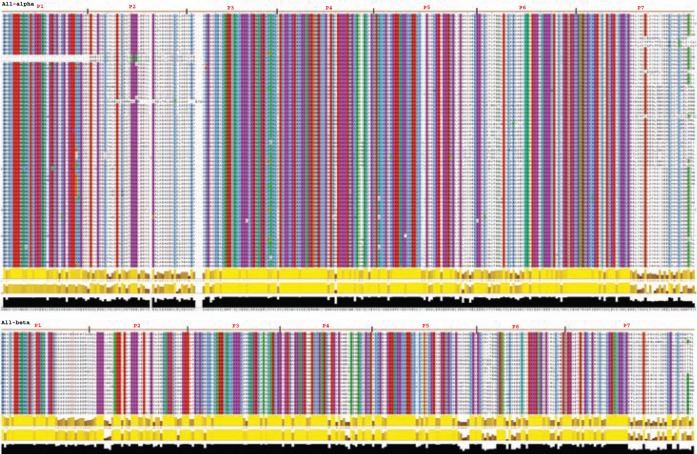
We collected all-α and all-β homologues based on their annotation and aligned them to check the conservation pattern. In all-α set, the quasi-repeat P2 and C-ter of P7 is not conserved. Whereas in all-β set, the C-ter of P7 is less conserved, and P2 does not show much deviation (Figure 10A). Cardiac muscle is a hybrid system, which shares the property of striated muscle, but it is involuntary like the smooth muscle. Thus, from the all-α and all-β sets, the smooth and striated muscle sequences were divided into two subsets. The classification of sequences to smooth and striated category was done by checking for the tissue type from which the protein was purified and the annotation given in NCBI. In both the α-smooth and α-striated, the P2 region was less conserved, but the same trend was not seen in β-smooth and β-striated subset (Figure 10B).
Figure 10.
Conservation at different isoform level is shown, the quasi-repeats are marked above the alignment and the P2 which was found to be highly variable in α sequences are highlighted.
Note: The conserved region is brightly coloured whereas the variable region is not coloured (Jalview colouring: clustalX + 90% conservation).
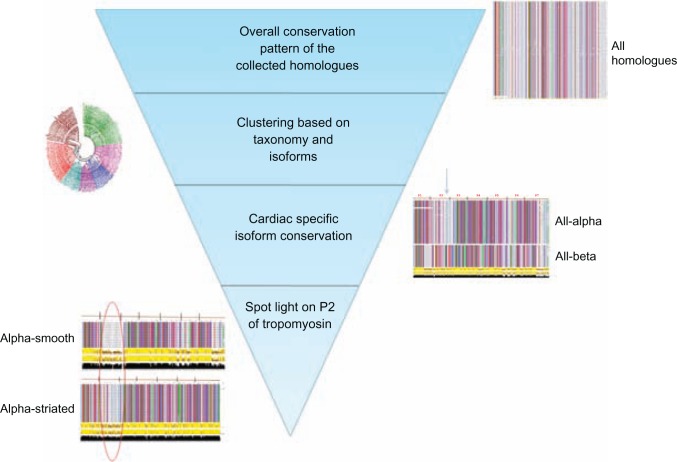
The conservation study was started with a broader data with all the homologues, where a number of residues of Tm sequences were conserved. Hence, a definite zone of conserved residues was not identified. Next, the sequences were clustered based on the major taxonomic classification based on the phylogeny and cardiac specific isoforms were analysed. Isoform specific clustering and conservation showed that regions near the second quasi-repeat and extreme C-ter of P7 were less conserved. Different isoforms of Tm from different organisms have a distinct end-to-end overlap pattern between adjacent Tm chains.46 Next, since it is crucial to understand the importance of P2 conservation, the study was narrowed down to tissue-specific sequences which belong to the cardiac muscle isoforms. It was seen that α-smooth and α-striated sequences still retain the property of the less conserved P2 region. At this stage, it was clear that variable P2 is a characteristic feature of tropomyosin sequences at all levels of our study. The conservation patterns were examined from a broad range of sequences to close families within a specific zone. This particular area which composes the second period (P2) could have a very crucial role in cTF function. An overview of the entire conservation study and the identification of the hot spot region is shown in Figure 11.
Figure 11.
Overview of the conservation study; starting from a broad range of collected homologues the pattern of variable P2 region is preserved.
Note: narrowed down analysis on specific sequences in turn shows the importance of period 2 of tropomyosin.
Forecast on spotted trend in known cardiomyopathy mutations
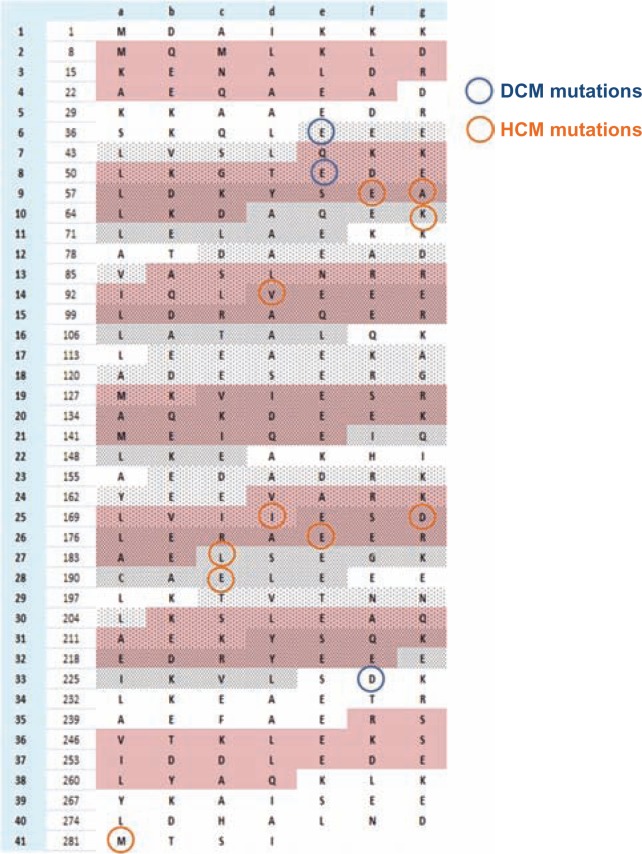
Cardiomyopathy is characterised by large number of mutations in the sarcomeric proteins. A total of 459 mutations in eight genes are documented in a cardiomyopathy mutation database.11 Among them, a set of 13 mutations are recorded for the α-tropomyosin gene. The mutations that cause DCM are E40K, E54K and D230N and HCM are E62Q, A63V, K70T, V95A, I172T, D175N, E180G, E180V, L185R, E192K and M281T.47–51 These mutations are mapped on the Tm sequences shown in Figure 12. Even though the number of documented mutation of Tm is too few to make a statistical validation, most of the cardiomyopathy mutations are clustered in P2 and P5 of tropomyosin. From our energetic analysis and conservation study, it was noted that P2 is susceptible to interaction energy fluctuations. In the interface energy analysis, the change from blocked to closed states had imposed increased stability to P2 and it was also found to be highly variable. P5 was very stable energetically in both blocked and closed states and was also found to be in the most conserved region of Tm. P2 falls within the target zone of myosin interaction and P5 is the area where troponin complex interacts. These two pivotal points play a crucial role in the function of cardiac muscle.
Figure 12.
Spatial distribution of the known DCM and HCM mutations in cardiac α-tropomyosin.
At this juncture, it is crucial to consider the point that increasing in myosin concentration increases actin-Tm interaction.52,53 This means that apart from myosin interacting with actin it also interacts with tropomyosin. Though a target zone has been defined between two Tn complexes, where the myosin head binds frequently on actin, the important region of Tm where myosin could interact is not well understood. Tropomyosin is present in most of the actin-based architecture and performs critical functions in different environments.
Since tropomyosin interacts with the actin filament throughout its length with the seven quasi-repeats, it is important that all the quasi-repeats are equally conserved to maintain its interaction. But it is not the case because P2 is less conserved when compared to the other repeats which are shown from our analysis. The less conserved nature of P2 could be required for its interaction with multiple forms of myosin motors. This leads to a conclusion that the myosin head has interacting partners on tropomyosin P2 region that are required for cross-bridge formation. From our energetic analysis, it is evident that the P2 region becomes more stable from blocked to closed states in terms of actin interaction. Upon calcium activation, P2 moves to a much more favourable and stable zone on actin and exposes residues that may interact with myosin. This would facilitate myosin binding at the P2 region and its corresponding actin which in turn initiates acto-myosin activity. Thus, any residue mutation in the P2 region that will affect the stable binding of Tm-actin at the closed and open state or residues or that may perturb its interaction with the myosin head domain could lead to a disease phenotype.
Next, while concentrating on the P5 region of Tm, we could anticipate that this region is highly conserved to maintain stable interaction with actin and the troponin globular complex. Non-muscle and smooth muscle actin filament is not regulated by troponin,54 thus in the striated muscle thin filament, where the Tn complex mediates the regulation, it is highly important for the binding region to be conserved. Since the Tn complex is identified to bind at a region near P5 and its corresponding actin, it is crucial for this region to be highly stable. From our analysis of the conservation pattern and interface energies of P5, it is evident that this region is highly protected to fulfill its function. Thus, any residue mutation that affects Tn binding or actin binding may lead to a diseased phenotype.
From HCM and DCM studies, it is proposed that these phenotypes are linked with high calcium sensitivity/high force production and low calcium sensitivity/low force production respectively.55,56 In the HCM phenotype, the force production is high, which gives us a sense that the number of times myosin binds to actin to form a cross-bridge is high. This, in turn, correlates with the fact that myosin binding sites on the actin surface should be vacant for an extended time period so that myosin binds easily leading to high force production. In the DCM phenotype, acto-myosin cross-bridge formation is lowered which is coupled with less force production. Out of the 14 analysed mutations, 10 involves charged residues and of these charged mutations all the HCM type mutations fall in section 2 (Fig. 5) and all DCM mutations fall in section 1 of the Tm quasi-repeat. From our analysis, it was stated that section 2 of each quasi-repeat could be important to maintain stable interactions between the actin and Tm at blocked and closed states. Thus, the HCM charge mutations are related to its function to keep Tm stably on actin at both the states (B- and C-) and the reversal in charges in these residues in disease condition may destabilize the interface. This would, in turn, move Tm out from its blocking position and keep the myosin binding sites open for a longer duration. An opposite effect could be related to the DCM phenotype, where mutations seen in section 1 of the quasi-repeat are essential to hold the Tm stably in the open state. So residues in DCM conditions may have an effect in not moving the Tm out of its blocking region. This suggests that HCM phenotype is connected with specific residues at the C-ter of each quasi-repeat and DCM phenotype with the N-ter of each quasi-repeat.
Materials and Methods
Integrative template modelling
The integrative template modelling of cTF was carried out by using the MODELLER26 software. The initial template which represents the full cTF macromolecular assemble was obtained from a reconstructed EM data.9 Additionally, good resolution crystal structures of actin, tropomyosin and troponin core domain are used as templates to best-fit the cTF models. A multi template modelling mode of the MODELLER software was used to build the model. In general MODELLER models the three-dimensional structure of target sequences using a known template structure, it uses the spatial restraints of the template structure and builds the target sequence. In the integrative structural modelling approach, the spatial restrains of the thin filament macromolecular assembly was obtained from the re-constructed electron microscopy data and independent component of the cTF was provided with additional restraints from high resolution crystal structures. The region on actin filament where the Tn core domain had to fit was obtained from FRET analysis.7 The alignment between the target and the multiple templates were manually edited to obtain the best optimal alignment. Based on these known structural details the cTF, one unit was modelled in its blocked and closed states. 100 models were generated and the best models for both B- and C- were selected based on the molecular probability density function (molpdf) which gives the current energy of each model.
Energy minimization
The best models of both B- and C-states were energy minimized using the SYBYL package (SYBYL 7.1, Tripos Inc). The models were built using fitted EM structure as a template, even though high resolution crystal structures were also used simultaneously the interfaces between the different components of the cTF assembly would have unfavourable short contacts. The cTF macromolecular assembly in B- and C-states consist of 7154 amino acids independently, taking into account the size of the system and the expected deviation from nativity and in order to minimize the number short contacts and to increase the quality of the models an extensive energy minimization was carried out. The structures were minimized using Tripos force field for 1000 iterations using Powell’s gradient and was terminated at a convergence of 0.05 Kcal mol Å−1. The distant dependent cutoff was kept as 1 and the non-bonded interaction cutoff was kept as 8 for all rounds of minimization. At every step of energy minimization, the structures were monitored manually to avoid distortion of the model structure due to over optimization, it was also noticed for the removal of maximum number of short contacts within the system.
Sequence and phylogenetic analysis
The homologues of human-α tropomyosin (TMP1) were collected using NCBI/BLAST/blastp suite. PSI-BLAST37 was used with an E-value of 10−6 the homologous sequences were collected till the round of convergence where TMP1 sequence was queried against the NR database dated 09/MAR/2012. Default parameters of word size 3 and BLOSUM62 matrix were used. The collected homologues from each iterations were refined with a query coverage filter of 60% and identity filter with 40% cutoff. The set of homologues were further filtered using CD-hit (99%) in order to remove the redundant entries. The final set of homologues were aligned using CLUSTALW38 program using BLOSUM matrix and the default parameters. The alignment quality was improved by manually editing the MSA using the MEGA package.39 The phylogenetic tree was constructed using the neighbour-joining method using the poisson model for protein sequences with 1000 iterations of bootstrapping. The bootstrapped consensus tree was accepted as the final tree.
Conclusions
In this study, we have used the available structural data from various resources to construct a three-dimensional atomic model of the cardiac thin filament (one unit: 7 + 7 actin, 1 + 1 Tm and 1 + 1 Tn globular complex) in the blocked and closed states. Numerous studies have been concentrating on various aspects of thin filament organisation, its associated protein structures and their conformation changes during the process of muscle regulation. A recent model of a complex, derived from three-dimensional FRET analysis, defines the orientation and positioning of the troponin core domain and tropomyosin on the actin filament with and without calcium. That model proposes a view that with Ca2+ the Tn core domain and the interacting Tm quasi repeat is located near actin subdomain 3 and 4, and on the release of Ca2+ they move towards the actin outer domain.57 The model also supports our view on actin subdomain position where Tm and Tn could be present in both B- and C-states. However, these models do not contain the myosin head domain in their system, which raises a question of what change in interactions could occur when the myosin is present. Though there are different such models present to decipher the thin filament structure, the overall atomic model of the thin filament and their subsequent changes in the three states of blocked, closed and open states are not available at high resolution. Hence, we used an integrative modelling approach to construct the cardiac thin filament utilizing the available information from other studies which were based on various methods like X-ray structures, EM models and FRET data. Keeping the models as starting points, we studied the actin-tropomyosin interface at different states and identified the most stable state to be the blocked state. From the surface of the actin filament, it was predicted that zone 2 was the most favourable area where the tropomyosin dimer could slide. A very recent work on actin-myosin has revealed the important interaction between the N-ter segment of actin and the activation loop of myosin; this interaction is required for muscle contraction.58 That the N-ter segment which is shown to bind with myosin in their work falls in zone 1 of our analysis. That the N-ter negatively charged patch of actin is required for myosin binding and its function supports our hypothesis that zone 1 is an unfavourable region for tropomyosin sliding from its blocked to closed and open states. Thus, based on our model and the predicted direction of actin-tropomyosin interactions at open state, probable interface regions at the three states were compiled. From our proposed interface regions, each of the tropomyosin quasi-repeats could be divided into two sections, where each of the sections plays an important role in maintaining the stability of the interface in the three states. Similarly, the actin surface could have two major interfaces, one for the blocked/closed states and the other for the open state. The interface formed between actin and tropomyosin in blocked and closed states share similar regions, but the strength of interacting zones differ considerably. In both the states, section 2 interface is preserved, showing that these residues are functionally important to maintain stability between actin-tropomyosin and sterically blocks acto-myosin interaction. Calcium-regulated conformation changes of the troponin complex followed by the azimuthal sliding of tropomyosin dimer on the actin surface will lead to the open state interface which is contributed by the zone 2 residues. Based on the interface energy at blocked and closed states, it was also identified that different quasi-repeats interact differently with actin. This shows the importance of the specific function carried out by each of the tropomyosin periods. To understand the vitality of tropomyosin quasi-repeats, a conservation study was carried out. From the conservation analysis, it was identified that period 2 of tropomyosin is highly variable. The less conserved nature of period 2 could be related to its function of myosin binding during cross-bridge formation. This suggests that tropomyosin P2 and its corresponding actin unit could be the hotspot in the target zone for myosin S1 binding. This also shows that for the muscle contraction/relaxation process and cross-bridge formation, interaction of myosin and tropomyosin is highly required. From the actin-tropomyosin interface energy calculations, it is evident that the blocked state is highly stable compared to the closed state and the predicted zone 2 interface is very unstable when compared to the other two states. This shows that the tropomyosin position on the actin surface in the open state is stabilized by other factors suggesting that although troponin-mediated conformational change triggers the movement of the tropomyosin dimer from blocked to closed states, interaction between myosin and tropomyosin is essential for holding the tropomyosin dimer in the open state. Our results concerning the importance of tropomyosin P2 region was observed from the actin-Tm interaction and interface energy of the B- and C-state models. In addition, the skeletal muscle tropomyosin shows high degree of sequence variation at P2 and C-terminal region across different organisms. It is been shown recently that the less conserved nature of P2 of smooth muscle tropomyosin does not affect the structural curvature or interaction with its actin unit and the region could have a gate-keeping role in determining the interaction of actin with its binding partners.59 This finding, along with our studies of cardiac tropomyosin, suggests that P2 likely has a crucial role in binding the myosin head domain.
The three-dimensional atomic model of the cTF has permitted us to visualize the structural effects of the deleterious cardiomyopathy mutations. The energetic analysis of the B- and C-state models has shown that tropomyosin quasi-repeat plays an important role in its function. Most of the cardiomyopathy mutations in α-tropomyosin involve changes in local charges; the change in charge of the solvent accessible disease-causing mutations means reduced local interaction with the corresponding actin residues. This might result in unstable actintropomyosin interface that eventually leads to the distortion in assembled components of the thin filament.
Our study started with a broad goal of examining the cardiac thin filament and then narrowed down to the actin-tropomyosin interface and the effect of specific interacting regions. The energetics and the conservation pattern, using our COILCHECK+ program, enabled the recognition of a particular variable zone which is the tropomyosin period 2. From our study, we were able to show that P2 of tropomyosin could nevertheless be an important region which is essential for some crucial function of the system. We have used the recent known data while constructing the models, but high resolution data with the entire myosin-actin-tropomyosin-troponin complex are required to obtain precise biological information. In our study, we have not included the TnT domain of troponin T due to the lack of structural information, but this region plays a crucial role in regulating the thin filament function. Hence, in the future, we will try to construct a two-unit model of cardiac thin filament where the TnT domain details could be fitted and analysed. The result of our study has also thrown light in the direction of known cardiomyopathy mutations. This gives us an insight to understand the structural basis and study the effects of the known mutations in detail and to identify if there could be any specific residue mutations that have not been identified as disease causing ones but may be predicted to lead to the disease state.
Acknowledgments
We would like to thank the National Centre for Biological Sciences (NCBS) for infrastructure. MS and JAS are supported by the Human Frontier Science Program (HFSP).
Footnotes
Author Contributions
Conceived and designed the experiments: RS, MS, JM, JS. Analysed the data: MS. Wrote the first draft of the manuscript: MS. Contributed to the writing of the manuscript: RS. Agree with manuscript results and conclusions: RS, MS, JM, JS. Jointly developed the structure and arguments for the paper: RS, MS. Made critical revisions and approved final version: RS. All authors reviewed and approved of the final manuscript.
Funding Sources
This work was supported by Human Frontier Science Program (HFSP) Grant (Code: RGP0054/2009-C/).
Competing Interests
JAM’s institution received consulting fees or honorarium from inStem in consideration for work as a visiting professor. Other authors disclose no competing interests.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Supplementary Data
Fitting the Tm dimer on zone 1 and zone 2
Using a rotational matrix the coordinates of F-actin was rotated keeping the Tm dimer constantly in place. For every 5 degree of rotation of the F-actin the structure was minimized to get rid of the short contacts. The minimization process was carried out using SYBYL package with the parameters mentioned earlier. At every rotation the electrostatic energy of the interface was calculated using COILCHECK+. Additionally it was checked if all the myosin binding residues on actin are open up and relieved from Tm blocking. It was observed that interface of Tm and actin in the direction of zone 1 was very unfavourable where as the direction of zone 2 was favourable. Supplementary Figure 1 shows the electrostatic energy at the Tm-actin interface for every 5 degree rotation on either side of myosin binding residues.
The process of rotation of actin is systematically shown for clear understanding. The graph shows the electrostatic energy at different interface position between actin and tropomyosin.
Calculation of electrostatic energies using COILCHECK+
The energy of electrostatic interaction energies are calculated according to the Coulomb’s law. All inter-protomer charged atomic pairs 15 Å or less apart have been considered for energy calculation.
| Amino acid | Atomic charge | Atomic charge |
|---|---|---|
| ASP | OD1 = −0.76 | OD2 = −0.76 |
| GLU | OE1 = −0.76 | OE2 = −0.76 |
| ARG | NH1 = 0.80 | NH1 = 0.80 |
| LYS | NZ = 0.30 | |
| HIS | ND1 = 0.38 | NE2 = 0.57 |
where q1 and q2 are the partial atomic charges, r is the distance between the atoms and D is the dielectric constant of the medium, distance dependent diletric (DDD, where D = 2r) is been used for the energy calculation. The values for the different parameters are as in CHARMM (Brooks et al, 1983 and Brooks et al, 2009).
Validation of the models using Prosa method
The final models of B- and C-states were validated using Prosa—protein structure analysis tool. Individual structures of actin, tropomyosin, troponin C, troponin I, troponin T which frames the of cTF macromolecular assembly are validated. The Z-scores of these components fall in the acceptable range of known crystal and NMR structures of their corresponding lengths.
Multiple sequence alignment of α-smooth and α-striated and β-smooth and β-striated tropomyosin sequences.
Note: Quasi-repeats are indicated as P1, P2 until P7.
Prosa—web server results for the components of blocked and closed state cTF models.
Supplementary Table 1
A video abstract by the authors of this paper is available.
video-abstract9798.mov
Table S1.
Distribution of the collected homologues at the taxonomic level of phylum and class.
| No. of homologues | |
|---|---|
| Vertebrates | |
| Phylum—chordata | |
| Class—mammalia | |
| Homo sapiens | 7 |
| Rattus norvegicus | 7 |
| Sus scrofa | 4 |
| Ailuropoda melanoleuca | 6 |
| Mus musculus | 5 |
| Macaca mulatta | 5 |
| Cervus elaphus | 1 |
| Canis lupus familiaris | 2 |
| Pongo abelii | 1 |
| Macaca fascicularis | 1 |
| Bos taurus | 1 |
| Loxodonta africana | 2 |
| Oryctolagus cuniculus | 2 |
| Pan troglodytes | 2 |
| Heterocephalus glaber | 2 |
| Cavia porcellus | 4 |
| Monodelphis domestica | 4 |
| Cricetulus griseus | 3 |
| Callithrix jacchus | 1 |
| Mesocricetus auratus | 1 |
| Class—aves | |
| Meleagris gallopavo | 4 |
| Gallus gallus | 3 |
| Coturnix coturnix | 2 |
| Taeniopygia guttata | 1 |
| Class—actinopterygii | |
| Takifugu rubripes | 7 |
| Ictalurus furcatus | 1 |
| Danio rerio | 5 |
| Salmo salar | 6 |
| Thunnus thynnus | 2 |
| Gadus chalcogrammus | 1 |
| Oreochromis niloticus | 15 |
| Epinephelus coioides | 1 |
| Salmo trutta | 2 |
| Ictalurus punctatus | 1 |
| Tetraodon nigroviridis | 4 |
| Class—chondrichthyes | |
| Scyliorhinus retifer | 1 |
| Class—amphibia | |
| Ambystoma mexicanum | 4 |
| Xenopus (Silurana) tropicalis | 2 |
| Xenopus laevis | 7 |
| Rana temporaria | 1 |
| Rana catesbeiana | 1 |
| Branchiostoma floridae | 2 |
| Branchiostoma belcheri | 1 |
| Ciona intestinalis | 2 |
| Phylum—arthropoda | |
| Pediculus humanus corporis | 1 |
| Bombyx mori | 1 |
| Drosophila melanogaster | 4 |
| Nasonia vitripennis | 1 |
| Homarus americanus | 1 |
| Erimacrus isenbeckii | 1 |
| Chionoecetes opilio | 1 |
| Paralithodes camtschaticus | 1 |
| Balanus rostratus | 1 |
| Phylum—mollusca | |
| Fulvia mutica | 1 |
| Ruditapes philippinarum | 1 |
| Solen strictus | 1 |
| Tresus keenae | 1 |
| Sinonovacula constricta | 1 |
| Pseudocardium sachalinensis | 1 |
| Phylum—nematoda | |
| Caenorhabditis elegans | 1 |
| Phylum—hemichordata | |
| Saccoglossus kowalevskii | 1 |
| Phylum—platyhelminthes | |
| Echinococcus multilocularis | 1 |
| Schistosoma mansoni | 1 |
References
- 1.Squire JM. Muscle filament structure and muscle contraction. Annu Rev Biophys Bioeng. 1975;4(00):137–63. doi: 10.1146/annurev.bb.04.060175.001033. [DOI] [PubMed] [Google Scholar]
- 2.Zot AS, Potter JD. Structural aspects of troponin-tropomyosin regulation of skeletal muscle contraction. Annu Rev Biophys Biophys Chem. 1987;16:535–9. doi: 10.1146/annurev.bb.16.060187.002535. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T, Solaro RJ. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu Rev Physiol. 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 4.Poole KJ, Lorenz M, Evans G, et al. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155(2):273–84. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Miki M, Makimura S, Saitoh T, et al. A three-dimensional FRET analysis to construct an atomic model of the actin-tropomyosin complex on a reconstituted thin filament. J Mol Biol. 2011;414(5):765–82. doi: 10.1016/j.jmb.2011.10.033. [DOI] [PubMed] [Google Scholar]
- 6.Ueda K, Kimura-Sakiyama C, Aihara T, Miki M, Arata T. Interaction sites of tropomyosin in muscle thin filament as identified by site-directed spin-labeling. Biophys J. 2011;100(10):2432–9. doi: 10.1016/j.bpj.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura-Sakiyama C, Ueno Y, Wakabayashi K, Miki M. Fluorescence resonance energy transfer between residues on troponin and tropomyosin and in the reconstituted thin filament: modeling the troponin-tropomyosin complex. J Mol Biol. 2008;376(1):80–91. doi: 10.1016/j.jmb.2007.10.078. Epub Nov 4, 2007. [DOI] [PubMed] [Google Scholar]
- 8.Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys J. 2011;100(4):1005–13. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pirani A, Vinogradova MV, Curmi PM, et al. An atomic model of the thin filament in the relaxed and Ca2+-activated states. J Mol Biol. 2006;357(3):707–17. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 10.Lee FA. Yale university school of medicine heart book-Heart muscle disease. 15:185–94. [Google Scholar]
- 11.Genomics of Cardiovascular Development, Adaptation, and Remodeling. NHLBI Program for Genomic Applications, Harvard Medical School. URL: http://www.cardiogenomics.org. [February 15, 2012]
- 12.Webb B, Lasker K, Schneidman-Duhovny D, et al. Modeling of proteins and their assemblies with the integrative modeling platform. Methods Mol Biol. 2011;781:377–97. doi: 10.1007/978-1-61779-276-2_19. [DOI] [PubMed] [Google Scholar]
- 13.Russel D, Lasker K, Webb B, et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10(1):e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasker K, Phillips JL, Russel D, et al. Integrative structure modeling of macromolecular assemblies from proteomics data. Mol Cell Proteomics. 2010;9(8):1689–702. doi: 10.1074/mcp.R110.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lasker K, Förster F, Bohn S, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109(5):1380–7. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467(7316):724–8. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 17.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457(7228):441–5. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 18.Brown JH, Kim KH, Jun G, et al. Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci U S A. 2001;98(15):8496–501. doi: 10.1073/pnas.131219198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown JH, Zhou Z, Reshetnikova L, et al. Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc Natl Acad Sci U S A. 2005;102(52):18878–83. doi: 10.1073/pnas.0509269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Mui S, Brown JH, et al. The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc Natl Acad Sci U S A. 2002;99(11):7378–83. doi: 10.1073/pnas.102179999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitby FG, Phillips GN., Jr Crystal structure of tropomyosin at 7 Angstroms resolution. Proteins. 2000;38(1):49–59. [PubMed] [Google Scholar]
- 22.Takeda S, Yamashita A, Maeda K, Maéda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424(6944):35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 23.Vinogradova MV, Stone DB, Malanina GG, et al. Ca(2+)-regulated structural changes in troponin. Proc Natl Acad Sci U S A. 2005;102(14):5038–43. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz M, Holmes KC. The actin-myosin interface. Proc Natl Acad Sci U S A. 2010;107(28):12529–34. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning EP, Tardiff JC, Schwartz SD. A model of calcium activation of the cardiac thin filament. Biochemistry. 2011;50(34):7405–13. doi: 10.1021/bi200506k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, Mac Arthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereo chemical quality of protein structures. J Appl Crystallogr. 1993;26:283–91. [Google Scholar]
- 28.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–10. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alva V, Syamala Devi DP, Sowdhamini R. COILCHECK: An interactive server for the analysis of interface regions in coiled coils. Protein and Peptide Letters. 2008;15:33–8. doi: 10.2174/092986608783330314. [DOI] [PubMed] [Google Scholar]
- 30.Mizuguchi K, Deane CM, Blundell TL, Johnson MS, Overington JP. JOY: protein sequence-structure representation and analysis. Bioinformatics. 1998;14(7):617–23. doi: 10.1093/bioinformatics/14.7.617. [DOI] [PubMed] [Google Scholar]
- 31.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11(2):82–8. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 32.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21(10):375–82. [PubMed] [Google Scholar]
- 33.Hitchcock-DeGregori SE, Song Y, Greenfield NJ. Functions of tropomyosin’s periodic repeats. Biochemistry. 2002;41(50):15036–44. doi: 10.1021/bi026519k. [DOI] [PubMed] [Google Scholar]
- 34.Singh A, Hitchcock-DeGregori SE. Tropomyosin’s periods are quasi-equivalent for actin binding but have specific regulatory functions. Biochemistry. 2007;46(51):14917–27. doi: 10.1021/bi701570b. [DOI] [PubMed] [Google Scholar]
- 35.Hitchcock-DeGregori SE, Singh A. What makes tropomyosin an actin binding protein? A perspective. J Struct Biol. 2010;170(2):319–24. doi: 10.1016/j.jsb.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S, Liu J, Reedy MC, et al. Electron tomography of cryofixed, isometrically contracting insect flight muscle reveals novel actin-myosin interactions. PLoS One. 2010;5(9):piie12643. doi: 10.1371/journal.pone.0012643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami K, Stewart M, Nozawa K, et al. Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T. Proc Natl Acad Sci U S A. 2008;105(20):7200–5. doi: 10.1073/pnas.0801950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22(1):5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 43.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15(6):333–41. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Rajan S, Jagatheesan G, Karam CN, et al. Molecular and functional characterization of a novel cardiac-specific human tropomyosin isoform. Circulation. 2010;121(3):410–8. doi: 10.1161/CIRCULATIONAHA.109.889725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karam CN, Warren CM, Rajan S, de Tombe PP, Wieczorek DF, Solaro RJ. Expression of tropomyosin-κ induces dilated cardiomyopathy and depresses cardiac myofilament tension by mechanisms involving cross-bridge dependent activation and altered tropomyosin phosphorylation. J Muscle Res Cell Motil. 2011;31(5–6):315–22. doi: 10.1007/s10974-010-9237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang CL, Coluccio LM. New insights into the regulation of the actin cytoskeleton by tropomyosin. Int Rev Cell Mol Biol. 2010;281:91–128. doi: 10.1016/S1937-6448(10)81003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borovikov YS, Karpicheva OE, Chudakova GA, Robinson P, Redwood CS. Dilated cardiomyopathy mutations in alpha-tropomyosin inhibit its movement during the ATPase cycle. Biochem Biophys Res Commun. 2009;381(3):403–6. doi: 10.1016/j.bbrc.2009.02.054. [DOI] [PubMed] [Google Scholar]
- 48.Borovikov YS, Karpicheva OE, Avrova SV, Robinson P, Redwood CS. The effect of the dilated cardiomyopathy-causing mutation Glu54 Lys of alpha-tropomyosin on actin-myosin interactions during the ATPase cycle. Arch Biochem Biophys. 2009;489(1–2):20–4. doi: 10.1016/j.abb.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 49.Chang AN, Harada K, Ackerman MJ, Potter JD. Functional consequences of hypertrophic and dilated cardiomyopathy-causing mutations in alpha-tropomyosin. J Biol Chem. 2005;280(40):34343–9. doi: 10.1074/jbc.M505014200. [DOI] [PubMed] [Google Scholar]
- 50.Hilario E, da Silva SL, Ramos CH, Bertolini MC. Effects of cardiomyopathic mutations on the biochemical and biophysical properties of the human alpha-tropomyosin. Eur J Biochem. 2004;271(20):4132–40. doi: 10.1111/j.1432-1033.2004.04351.x. [DOI] [PubMed] [Google Scholar]
- 51.Michele DE, Albayya FP, Metzger JM. Direct, convergent hypersensitivity of calcium-activated force generation produced by hypertrophic cardiomyopathy mutant alpha-tropomyosins in adult cardiac myocytes. Nat Med. 1999;5(12):1413–7. doi: 10.1038/70990. [DOI] [PubMed] [Google Scholar]
- 52.Ali LF, Cohen JM, Tobacman LS. Push and pull of tropomyosin’s opposite effects on myosin attachment to actin. A chimeric tropomyosin host-guest study. Biochemistry. 2010;49(51):10873–80. doi: 10.1021/bi101632f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tobacman LS, Butters CA. A new model of cooperative myosin-thin filament binding. J Biol Chem. 2000;275(36):27587–93. doi: 10.1074/jbc.M003648200. [DOI] [PubMed] [Google Scholar]
- 54.Ansari S, Alahyan M, Marston SB, El-Mezgueldi M. Role of caldesmon in the Ca2+ regulation of smooth muscle thin filaments: evidence for a cooperative switching mechanism. J Biol Chem. 2008;283(1):47–56. doi: 10.1074/jbc.M706771200. [DOI] [PubMed] [Google Scholar]
- 55.Olson TM, Kishimoto NY, Whitby FG, Michels VV. Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J Mol Cell Cardiol. 2001;33(4):723–32. doi: 10.1006/jmcc.2000.1339. [DOI] [PubMed] [Google Scholar]
- 56.Mirza M, Robinson P, Kremneva E, et al. The effect of mutations in alpha-tropomyosin (E40K and E54K) that cause familial dilated cardiomyopathy on the regulatory mechanism of cardiac muscle thin filaments. J Biol Chem. 2007;282(18):13487–97. doi: 10.1074/jbc.M701071200. [DOI] [PubMed] [Google Scholar]
- 57.Miki M, Makimura S, Sugahara Y, et al. A Three-Dimensional FRET Analysis to Construct an Atomic Model of the Actin-Tropomyosin-Troponin Core Domain Complex on a Muscle Thin Filament. J Mol Biol. 2012;420(1–2):40–55. doi: 10.1016/j.jmb.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 58.Várkuti BH, Yang Z, Kintses B, et al. A novel actin binding site of myosin required for effective muscle contraction. Nat Struct Mol Biol. 2012;19(3):299–306. doi: 10.1038/nsmb.2216. [DOI] [PubMed] [Google Scholar]
- 59.Rao JN, Rivera-Santiago R, Li XE, Lehman W, Dominguez R. Structural analysis of smooth muscle tropomyosin α and β isoforms. J Biol Chem. 2012;287(5):3165–74. doi: 10.1074/jbc.M111.307330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The process of rotation of actin is systematically shown for clear understanding. The graph shows the electrostatic energy at different interface position between actin and tropomyosin.
Multiple sequence alignment of α-smooth and α-striated and β-smooth and β-striated tropomyosin sequences.
Note: Quasi-repeats are indicated as P1, P2 until P7.
Prosa—web server results for the components of blocked and closed state cTF models.
Table S1.
Distribution of the collected homologues at the taxonomic level of phylum and class.
| No. of homologues | |
|---|---|
| Vertebrates | |
| Phylum—chordata | |
| Class—mammalia | |
| Homo sapiens | 7 |
| Rattus norvegicus | 7 |
| Sus scrofa | 4 |
| Ailuropoda melanoleuca | 6 |
| Mus musculus | 5 |
| Macaca mulatta | 5 |
| Cervus elaphus | 1 |
| Canis lupus familiaris | 2 |
| Pongo abelii | 1 |
| Macaca fascicularis | 1 |
| Bos taurus | 1 |
| Loxodonta africana | 2 |
| Oryctolagus cuniculus | 2 |
| Pan troglodytes | 2 |
| Heterocephalus glaber | 2 |
| Cavia porcellus | 4 |
| Monodelphis domestica | 4 |
| Cricetulus griseus | 3 |
| Callithrix jacchus | 1 |
| Mesocricetus auratus | 1 |
| Class—aves | |
| Meleagris gallopavo | 4 |
| Gallus gallus | 3 |
| Coturnix coturnix | 2 |
| Taeniopygia guttata | 1 |
| Class—actinopterygii | |
| Takifugu rubripes | 7 |
| Ictalurus furcatus | 1 |
| Danio rerio | 5 |
| Salmo salar | 6 |
| Thunnus thynnus | 2 |
| Gadus chalcogrammus | 1 |
| Oreochromis niloticus | 15 |
| Epinephelus coioides | 1 |
| Salmo trutta | 2 |
| Ictalurus punctatus | 1 |
| Tetraodon nigroviridis | 4 |
| Class—chondrichthyes | |
| Scyliorhinus retifer | 1 |
| Class—amphibia | |
| Ambystoma mexicanum | 4 |
| Xenopus (Silurana) tropicalis | 2 |
| Xenopus laevis | 7 |
| Rana temporaria | 1 |
| Rana catesbeiana | 1 |
| Branchiostoma floridae | 2 |
| Branchiostoma belcheri | 1 |
| Ciona intestinalis | 2 |
| Phylum—arthropoda | |
| Pediculus humanus corporis | 1 |
| Bombyx mori | 1 |
| Drosophila melanogaster | 4 |
| Nasonia vitripennis | 1 |
| Homarus americanus | 1 |
| Erimacrus isenbeckii | 1 |
| Chionoecetes opilio | 1 |
| Paralithodes camtschaticus | 1 |
| Balanus rostratus | 1 |
| Phylum—mollusca | |
| Fulvia mutica | 1 |
| Ruditapes philippinarum | 1 |
| Solen strictus | 1 |
| Tresus keenae | 1 |
| Sinonovacula constricta | 1 |
| Pseudocardium sachalinensis | 1 |
| Phylum—nematoda | |
| Caenorhabditis elegans | 1 |
| Phylum—hemichordata | |
| Saccoglossus kowalevskii | 1 |
| Phylum—platyhelminthes | |
| Echinococcus multilocularis | 1 |
| Schistosoma mansoni | 1 |