Abstract
Both retinoid status and inflammation have been shown to control the level of expression of retinoid homeostatic genes. In the present study, DHRS3, previously shown to possess retinal reductase activity, was identified by microarray analysis of THP-1 monocytes as a possible gene target of all-trans-retinoic acid (RA). In these cells, DHRS3 mRNA increased 30- to 40-fold after treatment with ≤20 nM RA for 24 h, while DHRS3 protein also increased. Of several synthetic retinoids tested, only Am580, a RA receptor-α-selective retinoid, increased DHRS3 mRNA expression. The full-length DHRS3 cDNA was cloned from rat liver and subjected to in vitro transcription-translation. Two major ∼30- and 35-kDa proteins were detected. In adult rat tissues, DHRS3 mRNA was most abundant in the adrenal gland, liver, and ovary. In the liver, DHRS3 is expressed in hepatocytes and possibly in all liver cells. To evaluate whether DHRS3 is regulated in the liver by RA and/or inflammatory stimuli, we treated rats for 6 h with RA or LPS or both. DHRS3 mRNA was doubled by RA but reduced by >90% after treatment with LPS in the absence and presence of RA. On the basis of our results, DHRS3 mRNA expression is regulated by RA in a tissue- or cell-type specific manner; the RA-induced increase in DHRS3 may contribute to retinoid storage; and a reduction of DHRS3 expression in the liver during inflammation may contribute to the perturbation of whole body vitamin A metabolism that has previously been shown to occur in conditions of inflammatory stress.
Keywords: inflammation, β-carotene metabolism, liver, retinoid metabolism
the biochemical reactions and regulatory processes that maintain vitamin A (retinoid) homeostasis are incompletely understood. Retinal, the aldehyde form of vitamin A, is intrinsically reactive because of the ability of its carbonyl group to form Schiff bases with proteins, phospholipids, or retinal itself (38). Additionally, its biological oxidation product, all-trans-retinoic acid (RA), is a potent hormone capable of regulating gene transcription through binding to nuclear retinoid receptors (1, 13, 44). Retinal is metabolized intracellularly through reductive and oxidative reactions. All-trans-retinal is not only an intermediate in the oxidation process that converts retinol to RA, but it is also the first intermediate in the oxidative cleavage of β-carotene (2, 24, 25, 42), a nutritional precursor of vitamin A. Once retinal is reduced enzymatically to retinol, the retinol may be esterified with long-chain fatty acids and stored. In the retina, the light-catalyzed bleaching of 11-cis-retinal also produces all-trans-retinal, which is enzymatically reduced to all-trans-retinol in the visual process (43). Although high concentrations (3 mM) of retinal have been reported in the outer segment of photoreceptor cells (10), the concentration of retinal in the liver and most peripheral tissues is maintained at much lower levels.
The conversion of retinal to retinol has been reported to be mediated through several retinol dehydrogenases (29, 35, 36), of which the well-conserved microsomal protein DHRS3, a member of the classical short-chain dehydrogenase reductase (SDR) superfamily, has been previously identified (15). Although DHRS3 was originally cloned from the retina, it is expressed in several non-ocular human tissues, including the liver, pancreas, heart, kidney, and lung (3, 15). In cell extracts prepared from HEK-293T cells transfected with DHRS3 cDNA, retinal was reduced to retinol in an NADPH-dependent manner (15). In neuroblastoma cell lines transfected with DHRS3, the metabolism of retinal or retinol to retinyl ester was favored (3). During embryonic development, which is critically dependent on finely regulated concentrations of RA (30), DHRS3 was required in the embryo, primarily within the central nervous system (12), where it apparently functions as an RA-induced feedback inhibitor of RA synthesis and is highly active at the onset of gastrulation (17). The DHRS3 gene maps to a region of human chromosome 1 at band p36.1 (15), which is frequently lost in aggressive neuroblastoma tumors (3). DHRS3 expression was reported to be induced by retinoids in neuroblastoma cell lines (3) and in naïve CD4+ T cells (19) and by the retinoid X receptor (RXR) rexinoid ligand bexarotene in MMTV-erbB2 mice (23) and to be responsive to serum concentration in normal and transformed human buccal keratinocytes (39). Recent studies have shown that DHRS3 is regulated by p53 and p63 tumor suppressor proteins (20) and is able to promote lipid droplet storage in HepG2 cells, consistent with the localization of DHRS3 in the endoplasmic reticulum (8).
Little is known about the physiological regulation of DHRS3 expression in vivo. Several enzymes in the liver that have been implicated in the regulation of retinol metabolism have been shown to be regulated by RA, including lecithin:retinol acyltransferase (LRAT), and CYP26 family genes (33). Conversely, the expression of these genes was rapidly downregulated after the induction of inflammation in a rat model of LPS-induced acute inflammation (45), while inflammation also interfered with retinol transport in plasma (14). However, it is not known whether inflammation also alters the oxidative metabolism of retinol by DHRS3. Using two models, the human monocytic cell line THP-1, which has been shown to undergo differentiation in response to RA as well as acute response to inflammatory stimuli (4, 6), and rat liver, which is relevant to the regulation of whole body retinoid homeostasis and to the acute-phase response to infection (31), we have tested the hypothesis that DHRS3 expression is regulated by RA and inflammation.
MATERIALS AND METHODS
Reagents.
All-trans-RA, 9-cis-RA, oleic acid, actinomycin D, and cycloheximide were purchased from Sigma-Aldrich (St. Louis, MO). TNFα was obtained from R & D Systems (Minneapolis, MN). LPS from Pseudomonas aeruginosa was purchased from Biological Laboratory (Campbell, CA) and dissolved in PBS before use. Receptor-selective retinoids were kindly provided by Michael Klaus (Hoffmann-La Roche, Nutley, NJ). Stock solutions of retinoids were made in absolute ethanol, and oleic acid was dissolved directly in serum-free cell culture medium. RPMI 1640 medium, DMEM, heat-inactivated FBS, and TRIzol reagent were purchased from Invitrogen Life Technology (Carlsbad, CA).
Cell culture.
THP-1 cells were grown as previously described in RPMI 1640 medium (4) supplemented with 10% heat-inactivated FBS, which was reduced to 3% FBS for 24 h prior to treatment. Treatments included incubation with 20 nM RA in ethanol (final concentration 0.01%), 5 ng/ml of TNFα (5, 28), or 100 ng/ml of LPS (5, 28), alone or in combination, for 6 h. In separate experiments, the cells were also treated with receptor-selective retinoids (20 nM each), including Am580 [a RA receptor (RAR)-α (RARα) agonist], Ro19-0645 (a RARβ agonist), CD437 (a RARγ agonist), and Ro25-7386 (a RXR pan-agonist). DHRS3 gene expression in THP-1 cells was detected by microarray analysis, as described previously for galactose mutarotase (aldose 1-epimerase) (GALM) gene expression (28), using total RNA from cells treated with 20 nM RA, without or with 5 ng/ml of TNFα, under the conditions described above.
Human hepatoma HepG2 cells were grown in six-well plates in the presence of DMEM with 10% FBS. Upon reaching ∼80% confluence, the cells were treated with 1 μM RA for 0, 0.5, 1, 2, 4, and 6 h and then washed and collected for RNA isolation with TRIzol reagent.
Animal treatments.
Animal protocols were approved by the Animal Care and Use Committee of Pennsylvania State University. Briefly, vitamin A-adequate adult Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA) were fed a vitamin A-adequate diet (45) and treated with a single dose of 500 μg of RA, delivered orally in ∼30 μl of vegetable oil, which was also used as the placebo control, or with a single injection of P. aeruginosa LPS (50 μg/100 g body wt ip), or both, as previously reported (45). After 6 h, the rats were killed by CO2 inhalation, blood samples were drawn for preparation of plasma, and tissues were collected and immediately snap-frozen for RNA isolation (45). Liver samples from rats that had been fed the same diet lacking vitamin A to produce a state of vitamin A deficiency were also used; the rats were then treated with a single dose of 100 μg of RA in a time-course study, as described previously (32, 33). Rats were euthanized, and the liver was collected at time 0 (vehicle control) and 3, 6, 10, and 16 h after RA administration.
RNA isolation and analysis.
Total RNA was isolated from individual tissue samples with TRIzol Reagent. Poly(A)+ RNA was isolated from total RNA and subjected to agarose gel electrophoresis and transferred to Nytran membranes, which were hybridized with full-length rat 32P-labeled cDNA probes (45). The membranes were washed and then exposed to Kodak BioMax X-ray film (Eastman Kodak, Rochester, NY).
Cloning of rat liver DHRS3 cDNA.
A pair of primers was designed from the predicted sequence available in the GenBank database (http://www.ncbi.nlm.nih.gov/) and used to clone the 5′ and 3′ ends of the DHRS3 mRNA by rapid amplification of cDNA ends (RACE), following the strategy reported previously (47), using poly(A)+-enriched RNA of rat liver from RA-treated rats. The 5′- and 3′-RACE products were cloned by TA cloning (pGEM-T Easy vector, Promega, Madison, WI) and subjected to sequencing. Upon alignment, the sequence of the full-length DHRS3 cDNA clone was assembled and deposited to the GenBank database (accession no. EF125189).
In vitro transcription and translation of rat DHRS3.
On the basis of the sequence of the full-length rat DHRS3 cDNA (accession no. EF125189), a pair of primers, including 5′-atctcgagTGGCAATCAGATCGCGTTTAA-3′ (XhoI site in lowercase letters) as the sense and 5′-tatctagaATTCCTGCCGCGTGACCAGT-3′ (XbaI site in lowercase letters) as the antisense, were designed and used to amplify cDNA covering almost the entire region (2,042 bp) of DHRS3 cDNA by RT-PCR from rat liver total RNA, as described previously (47). The amplified cDNA was first cloned in pGEM-T Easy vector and then subcloned into the XhoI/XbaI site of the pcDNA3.1(+) vector (Invitrogen Life Technology). The pcDNA3.1(+) vector containing the rat DHRS3 clone was then used as a template in an in vitro transcription-translation assay using the Single Tube Protein System 3 kit (Novagen, EMD Chemicals, Gibbstown, NJ) in the presence of [35S]methionine (GE Healthcare Bio-Science, Piscataway, NJ) following the manufacturer's instructions exactly. A 10-μl aliquot of the reaction product was subjected to electrophoresis in an SDS-polyacrylamide gel (12%) under reducing conditions to size-fractionate the labeled protein products. The gel was dried under vacuum and then exposed to X-ray film (Eastman Kodak, Rochester, NY).
Plasmid DNA for preparation of RNA probes.
The rat cDNA fragment covering nt 17–1440 of the rat DHRS3 cDNA was subcloned from pGEM-T Easy vector containing DHRS3 cDNA into the SmaI site of the pGEM4 vector. The pGEM4-rDHRS3 clone was then linearized with EcoRI or XbaI and used as a template to synthesize antisense or sense RNA probes with T7 or SP6 RNA polymerase, respectively. The rat retinol-binding protein 4 (RBP4) cDNA (548 bp) in the pGEM4 vector (32) was linearized with NarI or HindIII and used as the template to synthesize the antisense or sense RNA probe by T7 or SP6 RNA polymerase, respectively. On the basis of the sequence of rat chemokine (C-X-C motif) ligand 1 (melanoma growth-stimulating activity-α, CXCL1) mRNA (accession no. NM_030845), a pair of primers, including 5′-AAACCAGCTCCAGCACTCC-3′ as the sense and 5′TTTCATTTGTAACAGTCCTTTGAA-3′ as the antisense, were designed and used to amplify cDNA covering almost the entire region of rat CXCL1 cDNA (900 bp) by RT-PCR from total RNA of the rats treated with LPS as described. The amplified cDNA was first cloned in the pGEM-T Easy vector and then subcloned into the SmaI site of the pGEM4 vector. The pGEM4-rCXCL1 clone was then linearized with EcoRI or XbaI and used as a template to synthesize antisense or sense RNA probes, respectively, as described above. For each probe, a digoxigenin (DIG)-labeled sense or antisense RNA riboprobe was prepared using the DIG RNA labeling kit in a reaction with ribonucleotides including DIG-UTP (Roche Biotechnology). The labeled RNA was isolated, checked for size by ethidium bromide-stained agarose gel electrophoresis, and then treated in alkaline solution to prepare 100- to 150-base polynucleotide lengths as probe (32).
In situ hybridization.
Frozen liver sections (10 μm thick) were placed on glass slides, fixed with 4% paraformaldehyde, and hybridized in a 50% formamide buffer solution with DIG-labeled riboprobe at 42°C overnight, as previously described (32). After they were washed and blocked, the slides were incubated overnight with alkaline phosphatase (AP)-conjugated anti-DIG antibody or horseradish peroxidase-conjugated anti-DIG antibody (Roche Biotechnology, Indianapolis, IN) in blocking buffer overnight (32). After the slides treated with AP-conjugated antibody were washed, they were incubated with 5-bromo-4-chloro-3-indolyl phosphate-nitro blue tetrazolium as synthetic substrate for AP (Roche Biotechnology) and then evaluated under a digital color microscope in our laboratory. The slides treated with horseradish peroxidase-conjugated antibody were amplified (Tyramine Signal Amplification Plus Fluorescein System, Perkin Elmer and Life Analytical Sciences) following the protocol provided by the manufacturer, and the resulting fluorophores were detected by fluorescence visualization using the fluorescence microscope in our laboratory.
Quantification of DHRS3 mRNA.
The mRNA level of DHRS3 and other transcripts in cells and in individual tissue samples was determined by quantitative RT-PCR (45) using total RNA prepared as described above. The PCR cycling program was first set at 95°C for 10 min to activate the Taq polymerase and then at 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, with sense and antisense primers as follows: 5′-ACTTTCCACCTCCGGCTTCC-3′ and 5′-CGTTCTCCCGCGACAGGTC-3′ for human DHRS3 (GenBank accession no. NM_004753), 5′-TTGTCCACCGCCTCCTACCT-3′ and 5′-GTGCCCATCTGCCGAATCT-3′ for rat DHRS3 (GenBank accession no. EF125189), 5′-CACGGGGCTTTGTGTCTATT-3′ and 5′-CCTCGTCGTAATGAGGGTGT-3′ for rat β-carotene 15,15′-monooxygenase 1 (GenBank accession no. NM_053648), 5′-CGGACCCATTTTACCCACTA-3′ and 5′-CAGACTGCAGGAAGGGTCAT-3′ for rat LRAT (GenBank accession no. AF255060), and 5′-CGCGGTTCTATTTTGTTGGT-3′ and 5′-AGTCGGCATCGTTTATGGTC-3′ for 18S rRNA (GenBank accession no. X01117). Each RNA transcript was measured separately and calculated using 18S rRNA as an internal control. Data were normalized to the average value for the control group, set at 1.00, before statistical analysis.
Immunoblot analysis.
Cells washed in PBS and lysed in RIPA buffer were prepared, and equal amounts (30–60 μg) of lysate protein were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with anti-human DHRS3 monoclonal antibody (kindly provided by Dr. Krzysztof Palczewski, Case Western Reserve University School of Medicine, Cleveland, OH) and then with secondary antibody, as described previously (46). For loading controls, the membranes were stained for protein with Ponceau solution.
Statistical analysis.
Values are means ± SE. Data were analyzed by one-way ANOVA with Fisher's post hoc test.
RESULTS
DHRS3 mRNA and protein are rapidly increased by RA in THP-1 cells.
When THP-1 cells were treated with a concentration of RA typical of levels found in plasma (20 nM) (26), DHRS3 mRNA expression, determined by microarray analysis, was significantly upregulated after 2 h, increased further by 6 h, and then remained stable for up to 40 h (Fig. 1A), indicating a ≥16-fold increase in the level of DHRS3 mRNA. In a second study designed to determine the response of THP-1 cells to RA and TNFα, both of which have been shown to induce THP-1 cell differentiation toward a macrophage-like phenotype (4), RA (20 nM) was a significant factor for DHRS3 mRNA expression (Fig. 1B), whereas TNFα (5 ng/ml) alone had no effect. The presence of TNFα did not alter the response of THP-1 cells to RA (Fig. 1B).
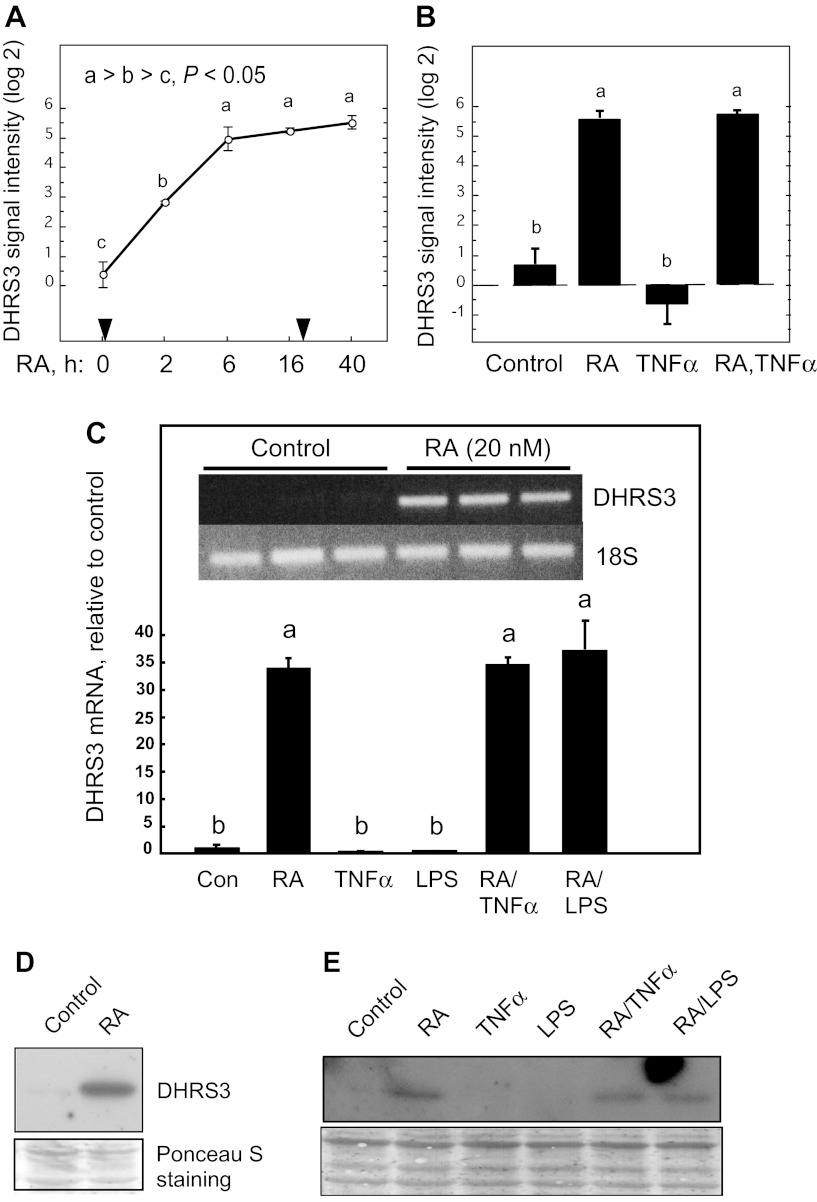
Fig. 1.
Expression and regulation of DHRS3 mRNA in THP-1 cells treated with retinoic acid (RA), TNFα, or LPS, and combinations of these agents. A: DHRS3 mRNA in THP-1 cells treated with 20 nM RA at time 0; an equal amount of RA was added after 24 h of incubation. B: DHRS3 mRNA after 40 h of treatment with RA, TNFα (5 ng/ml, added 16 h after time 0), and RA (added at time 0) followed by TNFα (added at 16 h for a total of 40 h). Results in A and B represent 2–6 replicates, determined by microarray analysis. For 2 different target sequences of DHRS3 mRNA, log2 difference (maximum − minimum) was 5.16 (A) and 4.16 (not shown), respectively. C: relative intensity of DHRS3 mRNA in THP-1 cells treated with vehicle or RA (20 nM), each in triplicate, for 6 h and then harvested for RNA analysis by semiquantitative RT-PCR. Inset: ethidium bromide-stained agarose gel. Treatments were as follows: vehicle only (Con), RA (20 nM), TNFα (5 ng/ml), LPS (100 ng/ml), RA + TNFα, and RA + LPS (n ≥ 3/group). D: DHRS3 protein detected by immunoblot on pooled cell extracts using human DHRS3 monoclonal antibody. THP-1 cells were cultured with vehicle only (control) and with RA (20 nM, 6 h). Ponceau S staining was used as a loading control. E: DHRS3 protein detected by immunoblot using human DHRS3 monoclonal antibody. Cell extracts were prepared from THP-1 cells treated as described in C. Ponceau S staining was used as a loading control. Differences were determined by 1-way ANOVA: different letters (a, b, c) indicate significant difference: P < 0.05.
To verify the microarray results of DHRS3 mRNA expression, we performed two separate experiments on THP-1 cells: we measured DHRS3 mRNA by quantitative PCR and protein by immunoblot analysis. In the first experiment, DHRS3 mRNA, which was barely detected in the control cells (Fig. 1C), was highly expressed after 6 h (Fig. 1C, inset) to 24 h (Fig. 1C) of treatment with 20 nM RA. Similarly, DHRS3 protein was not detectable in the control cells; however, a protein band that corresponded to the size of DHRS3 was significantly expressed in RA-treated THP-1 cells within 6 h (Fig. 1D). In a second experiment, THP-1 cells were treated with 20 nM RA in the presence or absence of TNFα (5 ng/ml) or LPS (100 ng/ml) for 24 h. Whereas DHRS3 mRNA was increased >30-fold by RA (Fig. 1C and inset), neither TNFα nor LPS had a significant effect on the level of DHRS3 mRNA in these cells, in the absence or presence of RA. Similar results were obtained for DHRS3 protein levels analyzed by immunoblot analysis (Fig. 1E).
Agonist-selective induction of DHRS3 mRNA.
RA is considered a pan-agonist ligand for all RAR subtypes, including RARα, RARβ, and RARγ. To determine which receptor subtype(s) is involved, directly or indirectly, in the induction of DHRS3 mRNA expression by RA, we treated THP-1 cells with various agonists that have been shown to be selective for binding to and activating members of the RAR family. Of several retinoids tested, only Am580, a retinoid with greatest affinity for RARα (9, 40), was as effective as RA in inducing DHRS3 mRNA expression in THP-1 cells (Fig. 2A). Retinoids selective for binding to RARβ (Ro19-0645) and RARγ (CD437) had no significant effect, although these receptors are expressed in this cell line (28); 9-cis-RA, a ligand capable of activating RAR and RXR subtypes (18), produced a small but significant effect, while a pan-RXR ligand (Ro25-7386) had no significant effect on expression of DHRS3 mRNA (Fig. 2A). Oleic acid, used as a lipophilic compound with physicochemical properties similar to RA, also was ineffective (Fig. 2A).
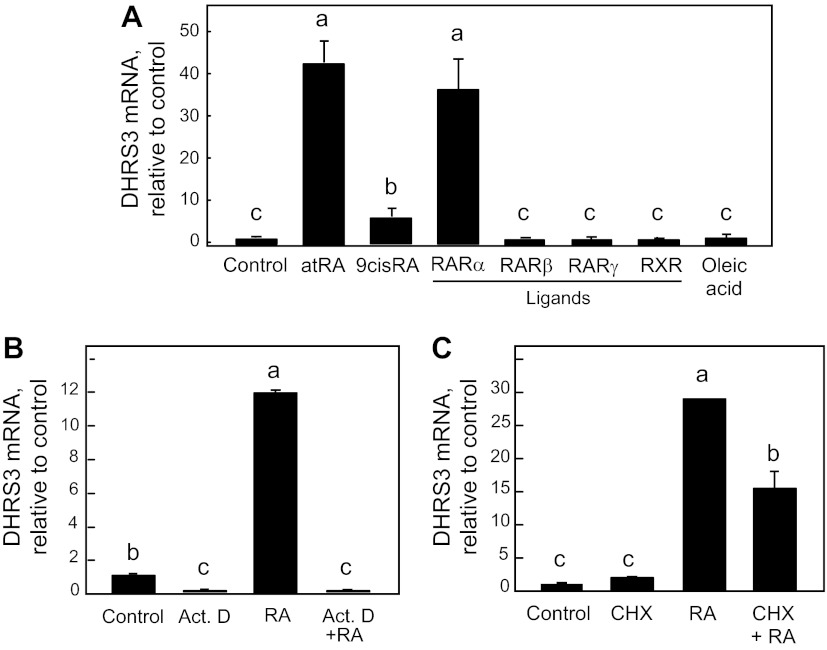
Fig. 2.
DHRS3 mRNA expression is regulated by RA receptor (RAR)-specific retinoids, actinomycin D, and cycloheximide in THP-1 cells. A: cells were treated in triplicate with various RAR subtype-specific and retinoid X receptor (RXR)-specific retinoids (20 nM) for 6 h. Receptor ligands were as follows: Am580 for RARα, Ro19-0645 for RARβ, CD437 for RARγ, and Ro25-7386 as a pan-agonist for RXR. atRA, all-trans-RA. B: cells were treated with actinomycin D (ActD, 5 μg/ml) for 1 h before addition of RA (20 nM) for another 6 h. C: cells were treated with cycloheximide (CHX, 5 μg/ml) for 1 h before addition of RA (20 nM) for another 6 h. After each experiment, RNA was analyzed by quantitative RT-PCR for DHRS3 mRNA and 18S RNA, and results were normalized. Different letters indicate significant difference (P < 0.05).
The ability of RA to increase the expression of DHRS3 mRNA is likely to be at the transcriptional level, as the increase in DHRS3 mRNA after RA treatment was completely prevented in cells pretreated with actinomycin D (5 μg/ml) for 6 h (Fig. 2B). On the other hand, cycloheximide, an inhibitor of protein synthesis, blocked the increased expression after RA treatment significantly, but not completely (Fig. 2C). However, cycloheximide has previously been shown to attenuate the induction by RA of a particular form of DHRS3 transcript in neuroblastoma cells (3).
DHRS3 is expressed in various tissues of the rat.
Next, we examined whether such regulation occurs in the whole organism. We selected the rat because of the numerous similarities in the absorption, transport, and metabolism of retinol between rats and humans. After cloning, alignment, and assembly of the rat DHRS3 cDNA (see materials and methods), a cDNA clone with 2,111 bases was obtained (GenBank accession no. EF125189). Unlike the human DHRS3 mRNA previously reported (15), the rat DHRS3 mRNA contains an ATG located 680 bases upstream of the predicted ATG, but not in the same reading frame. Therefore, to verify the size of the predicted rat DHRS3 protein, we subcloned the nearly full-length cDNA sequence into the pcDNA3.1 vector and used an in vitro transcription-translation assay with [35S]methionine incorporation for detection of the newly translated protein. After the 35S-labeled protein product was subjected to SDS-PAGE, two major 30- to 35-kDa bands were observed on the film (Fig. 3A), indicating that the downstream ATG at position 746 may be considered the initiation codon for an open reading frame similar to that of human DHRS3 (15) and ruling out the upstream ATG as an effective initiation codon in this assay. On this basis, the rat DHRS3 mRNA contains an open reading frame of 909 bases with 5′- and 3′-untranslated regions of 745 and 457 bases, respectively. Rat DHRS3 mRNA is significantly longer than human DHRS3 mRNA, yet the number of amino acid residues is similar to that of human and mouse DHRS3 protein, with 95% and 99% amino acid identity, respectively (http://www.ncbi.nlm.nih.gov/homologene; Fig. 3B).
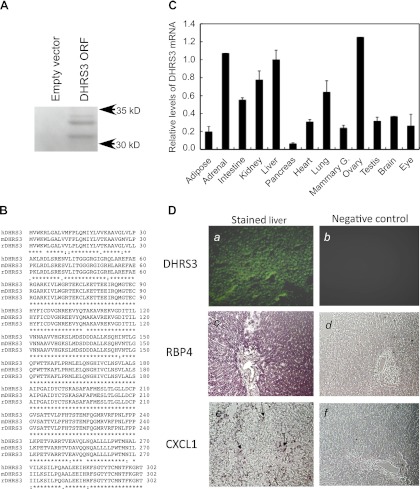
Fig. 3.
In vitro transcription of cloned rat DHRS3 and expression of DHRS3 mRNA in various tissues of adult rats. A: in vitro transcription and translation of rat liver DHRS3. Full-length DHRS3 cDNA was isolated from rat liver and cloned into pcDNA3.1 vector for in vitro transcription-translation using rabbit reticulocyte lysate in the presence of [35S]methionine. PAGE was conducted under denaturing conditions for separation of the 35S-labeled protein products. Gel was dried under vacuum at 80°C and exposed to X-ray film, as described elsewhere (47). ORF, open reading frame. B: alignment of protein sequence of rat DHRS3 (rDHRS3) with human and mouse DHRS3 (hDHRS3 and mDHRS3). Protein sequence of rat DHRS3 (accession no. NP_001032276) was aligned with human DHRS3 (accession no. NP_004744) and mouse DHRS3 (accession no. NP_035433) sequences using http://www.ebi.ac.uk/Tools/msa/clustalw2/. Rat DHRS3 protein is 95% identical to human and 99% identical to mouse DHRS3 protein (http://www.ncbi.nlm.nih.gov/homologene). C: relative expression level of DHRS3 mRNA in different tissues of rats. Total RNA from individual tissues of adult rats was subjected to quantitative RT-PCR for DHRS3 mRNA analysis. Values are means ± SE of 4 different rats relative to value for liver, which is considered 1.00. Values for adrenal gland and ovary are from pooled RNA samples. D: localization of DHRS3 mRNA in liver of rats by in situ hybridization. Liver sections (10 μm) in glass slides were hybridized to digoxigenin-labeled antisense (images a, c, and e) or sense (images b, d, and f) riboprobes for DHRS3 (images a and b), retinol-binding protein 4 (RBP4; images c and d), and CXCL1 (images e and f) and then analyzed by fluorescence for DHRS3 or color detection system for RBP4 and CXCL1. Arrows in image e indicate site of expression of CXCL1, which appeared as isolated mononuclear cells in liver sections of rats treated with LPS.
To investigate the tissue distribution of DHRS3 mRNA, total RNA samples from each tissue of four different adult rats were individually subjected to reverse transcription followed by quantitative PCR analysis (see materials and methods). DHRS3 is expressed in most tissues (Fig. 3C), with the highest relative abundance of DHRS3 mRNA transcripts in the adrenal gland, liver, and ovary; intermediary level of expression in the small intestine, kidney, and lung; and lower level of expression in the adipose tissue, heart, mammary tissue, testis, brain, and eye. Rat DHRS3 mRNA is apparently absent from or expressed at only very low levels in the pancreas.
In an in situ hybridization assay, rat liver sections in glass slides were hybridized to DIG-labeled antisense or sense riboprobes for DHRS3, RBP4, and CXCL1 and then analyzed by fluorescence (for DHRS3) or color detection system (for RBP4 and CXCL1). Whereas significant signals were detected in the liver sections by the antisense riboprobes (Fig. 3D, images a, c, and e), no detectable signals were observed with the sense riboprobes (Fig. 3D, images b, d, and f). DHRS3 mRNA is apparently expressed in hepatocytes (Fig. 3D, image a), although not as exclusively as RBP4 mRNA, the hepatocyte-specific gene (Fig. 3D, image c), which has been reported by us (32) and others (37) to have a strong expression and hepatocyte-exclusive distribution. As a nonparenchymal cell-specific gene, we used CXCL1, which has been reported to be expressed in neutrophils and Kupffer cells in response to liver injury (16). We found no signal for CXCL1 in the section of the liver of normal rats (data not shown), but in the liver of rats treated with LPS, CXCL1 was detected in isolated cells or small clusters appearing as mononuclear cells (Fig. 3D, image e).
DHRS3 expression has been reported in human hepatoma HepG2 cells (8), a well-differentiated cell model for hepatocytes (21). In cell culture experiments, we measured DHRS3 mRNA levels in HepG2 cells treated without or with RA for up to 6 h. DHRS3 mRNA is comparatively expressed in HepG2 cells (average cycle number for detection was ∼17.3 ± 0.2 in HepG2 cells vs. 29.2 ± 0.6 in THP-1 cells), and its expression is induced after 4 h of treatment with RA by about two- to threefold (data not shown), which is much lower than that in THP-1 cells.
DHRS3 expression in rat liver is correlated with that of LRAT.
Having observed that DHRS3 mRNA is highly expressed in the liver, the major site of vitamin A storage (34), we wished to determine if DHRS3 is regulated by vitamin A status or RA treatment in this organ. Thus we analyzed rat liver RNA samples (45) from adult vitamin A-adequate rats and similar rats treated with RA or a low dose of LPS to induce inflammation (31) or both. Blood and tissues were collected after 6 h. These short-term treatments did not alter the concentration of retinol in plasma or liver (45). Total RNA extracted from individual liver samples was subjected to quantitative RT-PCR analysis for expression of DHRS3, as well as β-carotene monooxygenase (BCMO1) and LRAT, the genes that putatively may act upstream and downstream of DHRS3, respectively (Fig. 4A). In rats treated with RA alone, DHRS3 mRNA increased to about twice the control level (Fig. 4B; P < 0.05). Although this increase was significant, it was much less than the 30-fold increase in RA-treated THP-1 cells. However, in contrast to the results for THP-1 cells in which LPS treatment had no effect on DHRS3 expression, in vivo treatment with LPS reduced the expression of DHRS3 mRNA in the liver by >90% (P < 0.0001) in the absence and presence of RA (Fig. 4B). Although the induction effect of RA was low in the expression of DHRS3 in the liver, it occurs apparently through a transcriptional process similar to that observed in THP-1 cells (Fig. 2B), since treatment of the individual rats with actinomycin D, alone or with RA, for 6 h suppressed the expression of DHRS3 mRNA in the liver by >90% (P < 0.0001; Fig. 4C). This suggests that the steady-state level and RA-induced increase in DHRS3 mRNA are mainly due to the transcriptional process.
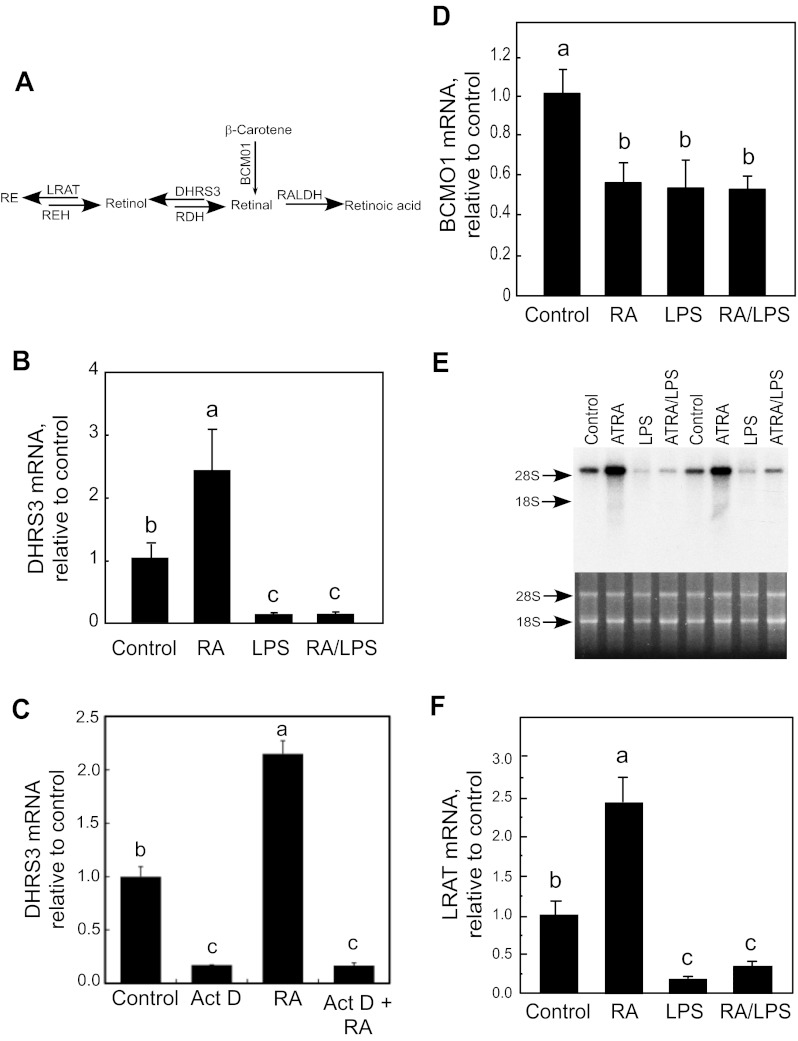
Fig. 4.
Regulation of DHRS3, β-carotene 15,15′-monooxygenase 1 (BCMO1), and lecithin:retinol acyltransferase (LRAT) mRNA expression in liver of adult rats treated in vivo with RA, LPS, or both. A: schema of pathway involving genes metabolizing retinal. RE, retinyl ester; REH, RE hydrolase; RDH, retinol dehydrogenase; RALDH, retinaldehyde dehydrogenase. B, D, and F: total RNA was extracted from individual liver samples (n = 6/group) and then measured by real-time quantitative RT-PCR for DHRS3, BCMO1, or LRAT, as well as for 18S RNA as the control. B: DHRS3 expression in liver of adult rats fed vitamin A-adequate diet and treated with RA (500 μg/kg body wt), LPS (50 μg/kg body wt) (45), or both for 6 h. C: effect of actinomycin D (Act D) on expression of DHRS3 mRNA in liver of adult rats. Rats were treated with actinomycin D alone or with RA for 6 h, and liver tissues were collected for total RNA extraction and analysis. Values are means ± SE (n = 3/group). D: BCMO1 mRNA from samples used in B. E: Northern blot analysis of LRAT mRNA from liver of rats treated as described in B. ATRA, all-trans-RA. F: quantitative RT-PCR for LRAT mRNA from samples used in B. Different letters indicate significant difference: P < 0.05 for B, D, and F; P < 0.0001 for C.
We also measured BCMO1 and LRAT, as genes involved 1) in production of retinal from β-carotene and 2) in removal of the putative product of the DHRS3 reaction, retinol. After vitamin A-adequate rats were treated with RA or LPS for 6 h, the level of BCMO1 mRNA in the liver was reduced by ∼50% (P < 0.05; Fig. 4D). However, administration of RA and LPS together did not cause a further reduction. We also analyzed the expression of LRAT, which catalyzes the esterification of retinol, the putative product of the enzymatic reaction of DHRS3. Treatment of vitamin A-adequate rats with RA increased LRAT mRNA in the liver by ∼2.5-fold (P < 0.05; Fig. 4, E and F). In contrast to treatment with RA, treatment with LPS, alone or in combination with RA, attenuated the expression of LRAT mRNA significantly (P < 0.05) by ∼80% (Fig. 4, E and F). As expression levels of DHRS3 and LRAT mRNA are upregulated by RA and downregulated by LPS at almost the same magnitude in the liver of normal rats, we found a strong correlation between DHRS3 mRNA levels and those of LRAT across several experiments (R2 = 0.83, P < 0.0001, n = 24).
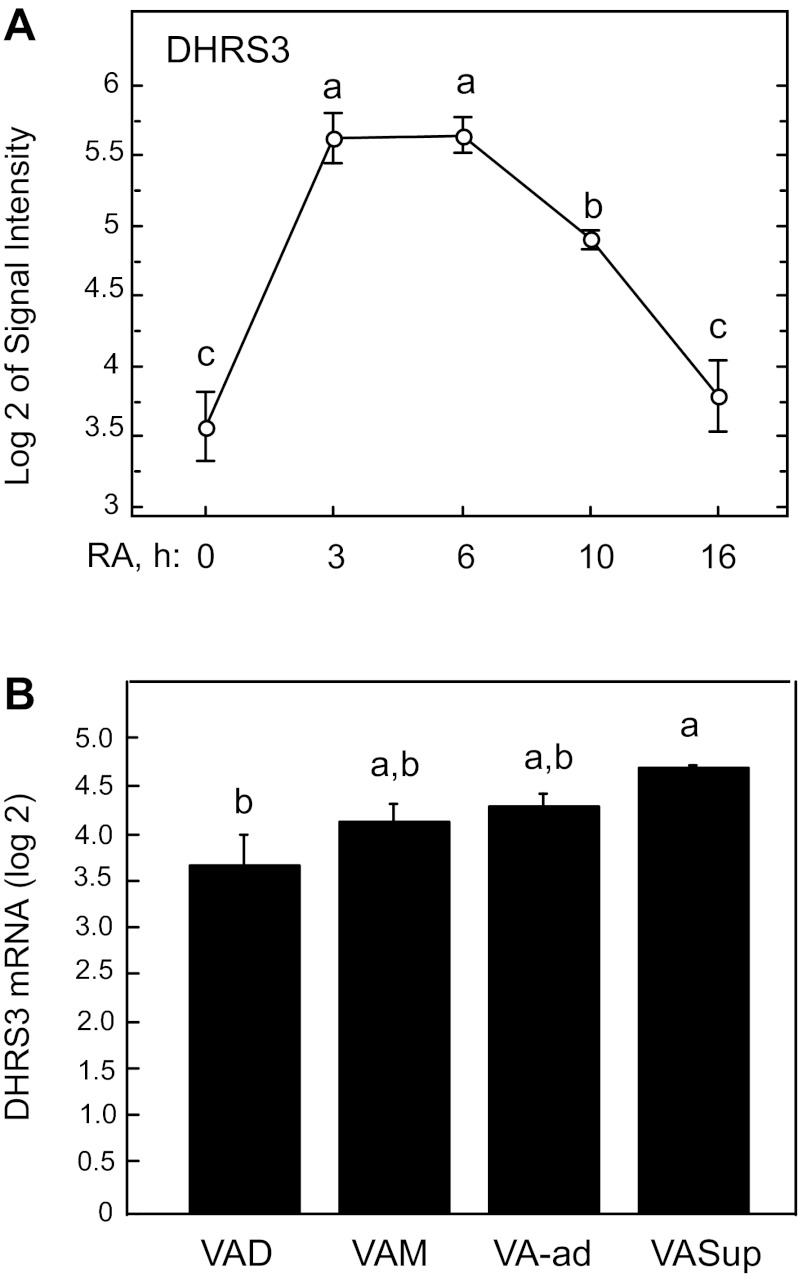
Recently, we reported the kinetic response to in vivo treatment with RA for several retinoid homeostatic genes, including LRAT, known to be responsive to RA (47), in the liver of rats first made deficient in vitamin A and then given a single dose of RA (32). Gene expression was measured at baseline (deficient state) and 3, 6, 10, and 16 h after administration of RA. LRAT mRNA increased significantly in the first 3 h, reached a peak at 6–10 h, and then declined. In the current study, we used the same liver samples to examine the response of DHRS3 mRNA levels and the correlation of its expression to the response of LRAT (32). DHRS3 mRNA (Fig. 5A) followed nearly the same kinetics as those of LRAT, with a more than fourfold increase within the first 3 h, a plateau to 6 h, and then a decline. DHRS3 mRNA was significantly and strongly correlated with LRAT (R2 = 0.509, P = 0.0006).
Fig. 5.
Relative mRNA levels of DHRS3 in liver of adult rats. A: kinetics of regulation of DHRS3 expression in liver of vitamin A-deficient rats after a single bolus dose of RA. Livers of vitamin A-deficient rats were analyzed 0, 3, 6, 10, and 16 h following injection of a single bolus dose of 100 μg RA, as previously reported (32). B: relative mRNA levels of DHRS3 mRNA in liver of rats fed vitamin A-deficient (VAD), -marginal (VAM), -adequate (VA-ad), and -supplemented (VASup) diets from weaning to 8 wk of age. Values are means ± SE; n = 5, 7, 6, and 2 rats for VAD, VAM, VA-ad, VASup, respectively. Different letters indicate significant difference: P < 0.05.
To evaluate gene expression profile in the liver under different dietary regimens, we established conditions that produce a wide range of vitamin A status from deficient to marginal, adequate, and supplemented vitamin A status in growing healthy rats (32). As an indicator of vitamin A status, the average liver total retinol in vitamin A-deficient, -marginal, and -supplemented groups was 0.0035, 0.035, and 40 times that in the vitamin A-adequate group, respectively (32). Although DHRS3 mRNA expression correlated well with the total retinol levels in the liver, the mRNA levels were not significantly lower in deficient and marginal animals or not significantly higher in supplemented rats than in vitamin A-adequate control rats. However, the average expression levels of DHRS3 mRNA were lower in vitamin A-deficient than vitamin A-supplemented rats (P < 0.05; Fig. 5B). DHRS3 mRNA expression levels were correlated with those of LRAT (R2 = 0.572, P < 0.0001), as previously reported (32).
Comparison with aldo-keto reductase expression.
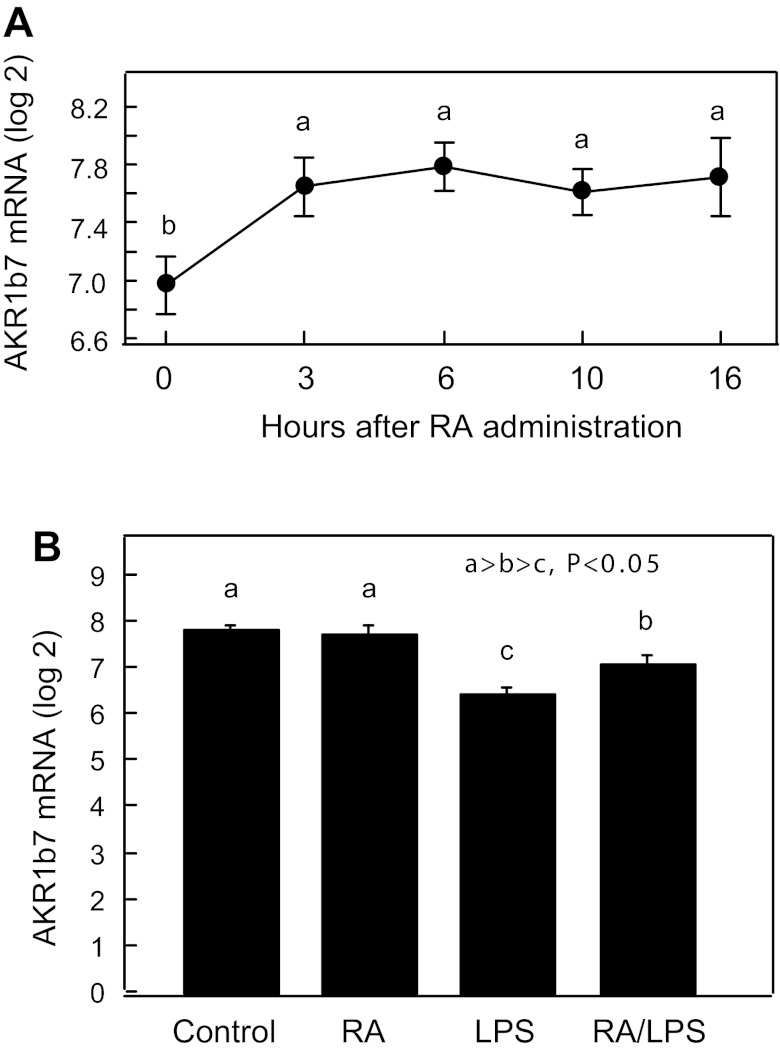
Several aldo-keto reductase (AKR) enzymes, which are cytoplasmic proteins, from subfamily 1B and 1C have been reported to have catalytic activity toward the reduction of retinal to retinol (36). Among them, human AKR1B10 and chicken AKR1B12 have been shown to have much higher catalytic activities toward reduction of all-trans-retinal, whereas human AKR1C3 have been reported to have much higher preference for the reduction of 9-cis-retinal, as the retinoid substrate. However, rat AKR1B10, whether or not it is considered the human ortholog, has been shown to have slight catalytic activity toward reduction of all-trans-retinal (11). It is not known whether AKR1B7 has catalytic activity toward reduction of retinals. In our gene expression profile assay, we found that, among the AKR genes, only the expression level of AKR1B7 mRNA is increased by >70% (P < 0.05) by RA in the liver of vitamin A-deficient rats (Fig. 6A), but not in vitamin A-sufficient animals (Fig. 6B). As a comparison to DHRS3, the AKR1B7 gene was suppressed slightly, but significantly (P < 0.05), in the liver of animals treated with LPS (Fig. 6B). We found no significant changes in the expression of AKR1B1 or AKR1B10 gene in the liver of rats treated with RA or LPS (data not shown).
Fig. 6.
Relative mRNA levels of aldo-keto reductase (AKR1B7) in liver of adult rats. A: liver samples from rats studied in Fig. 5A. B: liver samples from rats studied in Fig. 4B. Values are means ± SE for each group. Different letters indicate significant difference: P < 0.05.
DISCUSSION
In the present study, we first found that DHRS3 expression is highly regulated by RA in THP-1 mononuclear cells, a model used in numerous studies of monocytic cell differentiation (4). Among the RAR- and RXR-selective retinoids used, only Am580, a retinobenzoic acid analog of RA that selectively binds to and activates RARα (9) and also has been shown to induce cell differentiation (9, 40), was highly effective in increasing the expression of DHRS3 mRNA. Whether this occurs directly and is mediated by ligand-activated RARα or indirectly through other genes has not been established; however, the induction of DHRS3 by RA seems to occur at the transcriptional level, because 1) the induction was relatively fast, exemplified in our studies by the nearly eightfold increase in DHRS3 mRNA within 2 h after treatment of THP-1 cells with a physiological concentration of RA (26) and within 3 h after treatment of vitamin A-deficient rats with RA in vivo and 2) the induction of DHRS3 mRNA expression by RA in THP-1 cells, as well as in the liver of rats, was inhibited completely by treatment with actinomycin D, a known inhibitor of transcription. Similar results have been reported in neuroblastoma cell lines when doxorubicin, another inhibitor of the transcriptional process, was used (3). On the basis of physical analysis of the DNA sequence of DHRS3 gene, we found a direct repeat-2-like RA response element located ∼1.4 kbp upstream of the ATG codon. This direct repeat-2 element is apparently conserved in the human, mouse, and rat. Whether such an element has any responsive function to vitamin A remains to be determined.
To compare the results observed in THP-1 cells with a physiologically relevant in vivo model, we examined the expression of this gene in the rat. We first cloned the full-length cDNA from the liver and compared it with that of human DHRS3, which was previously reported (15). Although rat DHRS3 mRNA appears to be longer than human DHRS3 mRNA, both code for proteins of similar size, with highly identical (95%) amino acid residues (http://www.ncbi.nlm.nih.gov/homologene) (Fig. 3B). As shown by quantitative RT-PCR, the rat DHRS3 mRNA is highly expressed in the adrenal gland, liver, and ovary but is less abundant in our rat tissue than in the previously reported human tissue blot for a few tissues, including the pancreas, which showed stronger expression for human DHRS3 mRNA (3, 15). This enzyme is likely to play an important role in vitamin A and β-carotene metabolism, which differ somewhat between the human and rat in their digestive processing. Alternatively, retinal reductases other than DHRS3 may be present in the rat digestive system. In this regard, a substantial retinal reductase catalytic activity was reported in the homogenate of intestinal tissues of young rats (27). DHRS3 is apparently expressed in hepatocytes, although not as exclusively as RBP4 (Fig. 3D) and CYP26A1 (32), each considered hepatocyte-specific genes. As demonstrated during embryonic development (12), DHRS3 and CYP26A1 may act defensively to protect against a buildup of RA in hepatocytes, where both are expressed.
As in THP-1 cells, DHRS3 mRNA expression in the liver of RA-treated vitamin A-adequate rats was upregulated by RA. However, the induction of DHRS3 expression in this case was ∼2-fold, compared with 30- to 40-fold in THP-1 cells. Part of this difference may be due to the higher basal expression of DHRS3 mRNA in the liver. The physiological state of the animal could also be a significant factor, as rats made moderately deficient in vitamin A responded rapidly and strongly to RA administration, with a significant increase in DHRS3 mRNA within 3 h, a plateau to 6 h, and a decline to the baseline by 16 h (Fig. 5). RA, which is known to turn over rapidly in vivo, is not stored; thus it is not surprising that the induction of DHRS3 mRNA was transient. However, the data also suggest that the half-life of DHRS3 mRNA is relatively short. The more dramatic regulation of DHRS3 expression by RA in the vitamin A-deficient liver is consistent with that of other retinoid-metabolizing genes, such as cytochrome P-450 genes, especially CYP26A1 and CYP26B1, which are downregulated in the vitamin A-deficient state but expressed at relatively higher levels in the vitamin A-adequate state and respond rapidly to RA (32, 33). Thus RA may be cleared very quickly upon entry into the liver of the vitamin A-adequate host, thereby limiting the response of potentially RA-regulated genes. Indeed, metabolism of RA in the liver of rats that were treated with a bolus intravenous dose of [3H]RA in vivo was significantly slower in vitamin A-deficient than vitamin A-adequate rats (7). The mRNA for LRAT, the enzyme ultimately responsible for vitamin A storage (32), increased in a similar manner to DHRS3 mRNA in vitamin A-deficient, as well as vitamin A-sufficient, rats in this study. The correlation between DHRS3 mRNA (Fig. 5) and LRAT mRNA (32) also was very strong and significant. These data also support the possibility of a concerted regulation of BCMO1, DHRS3, and LRAT (Fig. 4) that, together, could promote retinyl ester formation and ensure the sequestration of any excess retinol produced from the cleavage of β-carotene to retinal, followed by the actions of DHRS3 on retinal to form retinol and LRAT to form retinyl esters.
The finding that DHRS3 mRNA is severely downregulated in the liver of LPS-treated rats is also of physiological interest. The model of LPS-induced inflammation used in our studies is relatively mild, having been shown to result in a small increase of ∼1°C in core body temperature within 6 h, reduced food intake, and moderate shivering (31); however, rats recovered well from this treatment within 2–3 days. The dose we have used, 500 μg/kg body wt, is much lower than that used in many studies of LPS-induced liver pathology. In our in vivo studies in normal rats, treatment with LPS nearly completely extinguished DHRS3 mRNA expression in the liver, with or without cotreatment with RA. This result contrasted to the result obtained in THP-1 cells in which LPS had no effect on DHRS3, despite its well-documented effects on other processes (4).
The results with LPS are also of interest regarding β-carotene metabolism. The liver is a recognized site of significant postintestinal β-carotene cleavage (41). If BCMO1 is considered a sole provider of substrate, retinal, for the DHRS3 enzyme, then it is possible that, under conditions of LPS-induced inflammation, retinal would accumulate, and perhaps similarly in states of natural infection or inflammation. It is well known that free retinal is a very unstable compound that may result in formation of free radicals, which are capable of combining with proteins and phospholipids (38) to form compounds that are believed to be highly damaging to host tissues, unless the activity of BCMO1 is also suppressed or the activity of retinal dehydrogenase(s) is at least maintained upon LPS treatment. In the present study, the level of BCMO1 mRNA in the liver of LPS-treated rats was also reduced, although not to the extent to which DHRS3 mRNA was reduced (Fig. 4). It is possible that differences in the rate of decline of DHRS3 and BCMO1 mRNAs after LPS treatment could be due to intrinsic differences in the turnover rates of the preexisting mRNAs for each gene or to other differences in mRNA. The results of our study suggest that LPS could induce an increase in the turnover of DHRS3 and BCMO1 mRNAs in rat liver. The results also suggest that there could be some significant buildup of retinal during LPS treatment, unless retinal is further oxidized to RA through retinal dehydrogenase(s) or catabolized in some other manner.
The strong correlation between DHRS3 mRNA and LRAT mRNA we observed seems to suggest a coordinated process involved in DHRS3 and LRAT gene expression in the liver during LPS treatment. DHRS3 has been shown to be expressed in only those breast cancer cell lines that were capable of esterifying retinol, but not in other cell lines that lacked retinol esterification capacity (3). Moreover, incubation of stably DHRS3-transfected neuroblastoma cell lines or RA-treated neuroblastoma cell lines with retinal or retinol resulted in more esterification of retinol (3). Since LRAT is recognized as the major enzyme responsible for retinol esterification, this may indicate coordination of the functions of DHRS3 and LRAT. Finally, DHRS3 may not be the only enzyme in the liver to catalyze the reduction of retinal to retinol. There may be others, including AKR1B or AKR1C, as well as other retinol dehydrogenases/reductases (29, 35, 36). Among AKR1B subfamily enzymes, we found that only the AKR1B7 gene is induced by RA in the liver of vitamin A-deficient, but not vitamin A-sufficient, diet-fed rats. Compared with the DHRS3 gene, which is suppressed almost entirely in the liver of rats treated with LPS, the AKRB1B7 mRNA level is slightly but significantly reduced. The AKR1B7 enzyme is not known, however, to have catalytic activity toward reduction of retinal.
In summary, the present experiments have demonstrated that the expression of DHRS3 mRNA is differentially regulated in vitro in THP-1 cells and in vivo in the liver of adult rats. Whereas RA markedly induced DHRS3 mRNA expression in THP-1 cells, the upregulation of DHRS3 in rat liver in vivo depended quantitatively on the retinoid nutritional status of the host. Inflammation, which is recognized as a central factor in many, if not all, chronic diseases, resulted in the almost complete suppression of the expression of DHRS3 mRNA in the liver of adult rats, even opposing the induction by RA. The suppression of DHRS3 mRNA in the liver in states of inflammation, such as were induced by LPS in the present study, may contribute to the perturbation of vitamin A metabolism, including a loss of stored vitamin A (22) and a reduced rate of secretion of retinol from liver into plasma (14), as has been shown to occur during infection and inflammation.
GRANTS
This work was supported in part by National Institutes of Health Grants CA-90214 and DK-41479.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
R.Z., Q.C., and A.C.R. are responsible for conception and design of the research; R.Z. and Q.C. performed the experiments; R.Z., Q.C., and A.C.R. analyzed the data; R.Z., Q.C., and A.C.R. interpreted the results of the experiments; R.Z. and A.C.R. prepared the figures; R.Z. drafted the manuscript; R.Z., Q.C., and A.C.R. edited and revised the manuscript; R.Z., Q.C., and A.C.R. approved the final version of the manuscript.
REFERENCES
- 1. Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328: 1–16, 2004 [DOI] [PubMed] [Google Scholar]
- 2. Biesalski HK, Chichili GR, Frank J, von Lintig J, Nohr D. Conversion of β-carotene to retinal pigment. Vitam Horm 75: 117–130, 2007 [DOI] [PubMed] [Google Scholar]
- 3. Cerignoli F, Guo X, Cardinali B, Rinaldi C, Casaletto J, Frati L, Screpanti I, Gudas LJ, Gulino A, Thiele CJ, Giannini G. retSDR1, a short-chain retinol dehydrogenase/reductase, is retinoic acid-inducible and frequently deleted in human neuroblastoma cell lines. Cancer Res 62: 1196–1204, 2002 [PubMed] [Google Scholar]
- 4. Chen Q, Ma Y, Ross AC. Opposing cytokine-specific effects of all trans-retinoic acid on the activation and expression of signal transducer and activator of transcription (STAT)-1 in THP-1 cells. Immunology 107: 199–208, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Q, Ross AC. Retinoic acid regulates CD1d gene expression at the transcriptional level in human and rodent monocytic cells. Exp Biol Med (Maywood) 232: 488–494, 2007 [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Q, Ross AC. Retinoic acid regulates cell cycle progression and cell differentiation in human monocytic THP-1 cells. Exp Cell Res 297: 68–81, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cifelli CJ, Ross AC. All-trans-retinoic acid distribution and metabolism in vitamin A-marginal rats. Am J Physiol Gastrointest Liver Physiol 291: G195–G202, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deisenroth C, Itahana Y, Tollini L, Jin A, Zhang Y. p53-Inducible DHRS3 is an endoplasmic reticulum protein associated with lipid droplet accumulation. J Biol Chem 286: 28343–28356, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delescluse C, Cavey MT, Martin B, Bernard BA, Reichert U, Maignan J, Darmon M, Shroot B. Selective high affinity retinoic acid receptor α or βγ ligands. Mol Pharmacol 40: 556–562, 1991 [PubMed] [Google Scholar]
- 10. Dowling JE. The retina: an approachable part of the brain. Structure 15: 905–915, 1987 [Google Scholar]
- 11. Endo S, Matsunaga T, Kuragano T, Ohno S, Kitade Y, Tajima K, El-Kabbani O, Hara A. Properties and tissue distribution of a novel aldo-keto reductase encoding in a rat gene (Akr1b10). Arch Biochem Biophys 503: 230–237, 2010 [DOI] [PubMed] [Google Scholar]
- 12. Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol 338: 1–14, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fields AL, Soprano DR, Soprano K., Jr Retinoids in biological control and cancer. J Cell Biochem. [DOI] [PubMed] [Google Scholar]
- 14. Gieng SH, Green MH, Green JB, Rosales FJ. Model-based compartmental analysis indicates a reduced mobilization of hepatic vitamin A during inflammation in rats. J Lipid Res 48: 904–913, 2007 [DOI] [PubMed] [Google Scholar]
- 15. Haeseleer F, Huang J, Lebioda L, Saari JC, Palczewski K. Molecular characterization of a novel short-chain dehydrogenase/reductase that reduces all-trans-retinal. J Biol Chem 273: 21790–21799, 1998 [DOI] [PubMed] [Google Scholar]
- 16. Harty MW, Papa EF, Huddleston HM, Young E, Nazareth S, Riley CA, Ramm GA, Gregory SH, Tracy TFJ. Hepatic macrophages promote the neutrophil-dependent resolution of fibrosis in repairing cholestatic rat livers. Surgery 143: 667–678, 2008 [DOI] [PubMed] [Google Scholar]
- 17. Kam RK, Chen Y, Chan SO, Chan WY, Dawid IB, Zhao H. Developmental expression of Xenopus short-chain dehydrogenase/reductase 3. Int J Dev Biol 54: 1355–1360, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kane MA. Analysis, occurrence, and function of 9-cis-retinoic acid. Biochim Biophys Acta 1821: 10–20, 2012 [DOI] [PubMed] [Google Scholar]
- 19. Kang SG, Park J, Cho JY, Ulrich B, Kim CH. Complementary roles of retinoic acid and TGF-β1 in coordinated expression of mucosal integrins by T cells. Mucosal Immunol 4: 66–82, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirschner RD, Rother K, Müller GA, Engeland K. The retinal dehydrogenase/reductase retSDR1/DHRS3 gene is activated by p53 and p63 but not by mutants derived from tumors or EEC/ADULT malformation syndromes. Cell Cycle 9: 2177–2188, 2010 [DOI] [PubMed] [Google Scholar]
- 21. Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 209: 497–499, 1980 [DOI] [PubMed] [Google Scholar]
- 22. Li D, Friedman SL. Liver fibrogenesis and the role of hepatic stellate cells: new insights and prospects for therapy. J Gastroenterol Hepatol 14: 618–633, 1999 [DOI] [PubMed] [Google Scholar]
- 23. Li Y, Zhang Y, Hill J, Kim HT, Shen Q, Bissonnette RP, Lamph WW, Brown PH. The rexinoid, bexarotene, prevents the development of premalignant lesions in MMTV-erbB2 mice. Br J Cancer 98: 1380–1388, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lietz G, Lange J, Rimbach G. Molecular and dietary regulation of β,β-carotene 15,15′-monooxygenase 1 (BCMO1). Arch Biochem Biophys 502: 8–16, 2010 [DOI] [PubMed] [Google Scholar]
- 25. Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta 1821: 152–167, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Napoli JL, Pramanik BC, Williams JB, Dawson MI, Hobbs PD. Quantification of retinoic acid by gas-liquid chromatography-mass spectrometry: total versus all-trans-retinoic acid in human plasma. J Lipid Res 26: 387–392, 1985 [PubMed] [Google Scholar]
- 27. Ong DE, Lucas PC, Kakkad B, Quick TC. Ontogeny of two vitamin A-metabolizing enzymes and two retinol-binding proteins present in the small intestine of the rat. J Lipid Res 32: 1521–1527, 1991 [PubMed] [Google Scholar]
- 28. Pai T, Chen Q, Zhang Y, Zolfaghari R, Ross AC. Galactomutarotase and other galactose-related genes are rapidly induced by retinoic acid in human myeloid cells. Biochemistry 46: 15198–15207, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parés X, Farrés J, Kedishvili N, Duester G. Medium- and short-chain dehydrogenase/reductase gene and protein families: medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism. Cell Mol Life Sci 65: 3936–3949, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, Petkovich M. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res A Clin Mol Teratol 88: 883–894, 2010 [DOI] [PubMed] [Google Scholar]
- 31. Rosales FJ, Ritter SJ, Zolfaghari R, Smith JE, Ross AC. Effects of acute inflammation on plasma retinol, retinol-binding protein, and its mRNA in the liver and kidneys of vitamin A-sufficient rats. J Lipid Res 37: 962–971, 1996 [PubMed] [Google Scholar]
- 32. Ross AC, Cifelli CJ, Zolfaghari R, Li NQ. Multiple cytochrome P-450 genes are concomitantly regulated by vitamin A under steady-state conditions and by retinoic acid during hepatic first-pass metabolism. Physiol Genomics 43: 57–67, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ross AC, Zolfaghari R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu Rev Nutr 31: 65–87, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ross AC, Zolfaghari R. Regulation of hepatic retinol metabolism: perspectives from studies on vitamin A status. J Nutr 134: 269S–275S, 2004 [DOI] [PubMed] [Google Scholar]
- 35. Ruiz FX, Porté S, Gallego O, Moro A, Ardèvol A, Del Río-Espínola A, Rovira C, Farrés J, Parés X. Retinaldehyde is a substrate for human aldo-keto reductases of the 1C subfamily. Biochem J 440: 335–344, 2011 [DOI] [PubMed] [Google Scholar]
- 36. Ruiz FX, Porté S, Parés X, Farrés J. Biological role of aldo-keto reductases in retinoic acid biosynthesis and signaling. Front Pharmacol 3: 2017, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soprano DR, Blaner WS. Plasma retinol-binding protein. In: The Retinoids: Biology, Chemistry and Medicine, edited by Sporn MB, Roberts AB, Goodman DS. New York: Raven, 1994, p. 257–281 [Google Scholar]
- 38. Sparrow JR, Wu Y, Kim CY, Zhou J. Phospholipid meets all-trans-retinal: the making of RPE bisretinoids. J Lipid Res 51: 247–261, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Staab CA, Ceder R, Roberg K, Grafström RC, Höög JO. Serum-responsive expression of carbonyl-metabolizing enzymes in normal and transformed human buccal keratinocytes. Cell Mol Life Sci 65: 3653–3663, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tamura K, Kagechika H, Hashimoto Y, Shudo K, Ohsugi K, Ide H. Synthetic retinoids, retinobenzoic acids, Am80, Am580 and Ch55 regulate morphogenesis in chick limb bud. Cell Differ Dev 32: 17–26, 1990 [DOI] [PubMed] [Google Scholar]
- 41. Tang G, Qin J, Dolnikowski GG, Russell RM. Short-term (intestinal) and long-term (postintestinal) conversion of β-carotene to retinol in adults as assessed by a stable-isotope reference method. Am J Clin Nutr 78: 259–266, 2003 [DOI] [PubMed] [Google Scholar]
- 42. von Lintig J. Colors with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annu Rev Nutr 30: 35–56, 2010 [DOI] [PubMed] [Google Scholar]
- 43. von Lintig J, Kiser PD, Golczak M, Palczewski K. The biochemical and structural basis for trans-to-cis isomerization of retinoids in the chemistry of vision. Trends Biochem Sci 35: 400–410, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei LN. Retinoid receptors and their coregulators. Annu Rev Pharmacol Toxicol 43: 47–72, 2003 [DOI] [PubMed] [Google Scholar]
- 45. Zolfaghari R, Cifelli CJ, Lieu SO, Chen Q, Li NQ, Ross AC. Lipopolysaccharide opposes the induction of CYP26A1 and CYP26B1 gene expression by retinoic acid in the rat liver in vivo. Am J Physiol Gastrointest Liver Physiol 292: G1029–G1036, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zolfaghari R, Ross AC. An essential set of basic DNA response elements is required for receptor-dependent transcription of the lecithin:retinol acyltransferase (Lrat) gene. Arch Biochem Biophys 489: 1–9, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zolfaghari R, Ross AC. Lecithin:retinol acyltransferase from mouse and rat liver. cDNA cloning and liver-specific regulation by dietary vitamin A and retinoic acid. J Lipid Res 41: 2024–2034, 2000 [PubMed] [Google Scholar]