Abstract
The ATP-binding cassette (ABC) transporter TAP translocates peptides from the cytosol to awaiting MHC class I molecules in the endoplasmic reticulum. TAP is made up of the TAP1 and TAP2 polypeptides, which each possess a nucleotide binding domain (NBD). However, the role of ATP in peptide binding and translocation is poorly understood. We present biochemical and functional evidence that the NBDs of TAP1 and TAP2 are non-equivalent. Photolabeling experiments with 8-azido-ATP demonstrate a cooperative interaction between the two NBDs that can be stimulated by peptide. The substitution of key lysine residues in the Walker A motifs of TAP1 and TAP2 suggests that TAP1-mediated ATP hydrolysis is not essential for peptide translocation but that TAP2-mediated ATP hydrolysis is critical, not only for translocation, but for peptide binding.
Major histocompatibility complex (MHC) class I molecules bind 8- to 10-aa peptides in the endoplasmic reticulum (ER). Candidate class I binding peptides are translocated from their site of generation in the cytosol into the lumen of the ER by the TAP (transporter associated with antigen processing) transporter (reviewed in refs. 1 and 2), an integral ER membrane protein whose two subunits, TAP1 and TAP2, are encoded within the MHC. Together with tapasin and the ER resident chaperones calreticulin and ERp57, TAP forms the nucleus of the class I “loading complex,” a macromolecular structure that potentiates peptide binding to newly synthesized class I molecules (3).
TAP is a member of the ATP-binding cassette (ABC) superfamily of transporters (reviewed in refs. 4 and 5), which use ATP hydrolysis to move a variety of solutes across cellular membranes or to regulate the opening of associated ion channels. ABC transporters share a common architecture consisting of two hydrophobic, polytopic transmembrane domains and two hydrophilic nucleotide binding domains (NBDs). The hydrophobic domains are specialized to interact with the transported solute or ion channel and generally share little homology. The cytosolic NBDs act as molecular motors that hydrolyze ATP and are highly conserved (4, 5). Both TAP subunits consist of an N-terminal hydrophobic domain followed by a C-terminal NBD. The peptide-binding site is composed of residues located on the membrane-spanning domains (6) and can accommodate a wide variety of peptides, ranging in length from 8–16 residues (7).
The NBDs of both TAP1 and TAP2 contain sequence motifs that are common to all ABC transporters. These include the Walker A and B motifs, sequences involved in ATP hydrolysis, and the ABC signature motif (or C-loop; ref. 4). Whereas TAP function clearly requires ATP hydrolysis, because the non-hydrolyzable analogue ATPγS blocks peptide translocation (8), relatively little is known about the process of peptide translocation across the ER membrane. ATP may play a role in peptide binding because, although ATPγS has no effect on peptide binding, mutations in the NBDs of TAP1 and TAP2 that abolish nucleotide binding were found to prevent it (9). Here we have used two approaches to investigate the role of ATP in peptide binding and translocation. Using the ATPase inhibitor sodium orthovanadate, we find evidence of a cooperative interaction between the NBDs of TAP1 and TAP2. Furthermore, by comparing TAP function in cell lines expressing mutant TAP1 and/or TAP2 polypeptides, we present evidence that the NBDs of TAP1 and TAP2 play different roles in peptide binding and translocation.
Materials and Methods
Cells and Cell Culture.
The T2, T1, T2 transfectants and Pala cell lines were grown in RPMI 1640 medium with 10% bovine calf serum (GIBCO/BRL-Life Technologies).
Plasmids.
The TAP1-pCDM8, TAP2-pCDM8, and lacZ-pCDM8 plasmids have been described previously (10–12). The pMCFR-puro and pMCFR-neo vectors were a gift from Dr. Tom Novak (Wyeth-Ayerst, Princeton, NJ). The K>A mutants were generated by using the ExSite PCR-Based Site-Directed Mutagenesis Kit (Stratagene). Primers encoding the desired mutation (TAP1 and TAP2: GGA CCC AAT GGG TCT GGC GCC AGC ACA GTG GCT GCC) and appropriate reverse primers (TAP1: CAC CAG CGC CGT CAC CTC GCC AGG GCG TAG GGT GAA; TAP2: CAC CAG CGC CGT CAC CTC ACC AGG ACG TAG GGT AAA) were used to generate mutant plasmids. These primers were designed to change the Walker lysine to an alanine and to introduce a unique NarI restriction site. The PCR amplified material remaining after Dpn1 digestion was treated with Pfu polymerase and T4 ligase to regenerate circular plasmids and then electroporated into MC1061/P3 bacteria (InVitrogen). Plasmids were sequenced across the entire TAP cDNA. The change to the Walker A sequence is GPNGSGKS to GPNGSGAS.
Antibodies.
The following antibodies were used: 148.3, an anti-TAP1 mAb (13); R.RING4C, a rabbit anti-peptide antibody to the C terminus of TAP1 (14); 435.3, an anti-TAP2 mAb (15); w6/32, a β2-microglobulin-dependent anti-human class I mAb; HC10, a mAb recognizing free class I heavy chains (16); 4E, a mAb reacting predominantly with HLA-B class I molecules; and AF8, an anti-calnexin mAb (17).
Peptides.
The peptides RRYQNSTEL and the photopeptide analogue KB11-HSAB were described previously (18). The peptides were iodinated to specific activity of 75–90 cpm/fmol and 15–25 cpm/fmol, respectively (19). SRYWAIRTR is an HLA-B27 binding peptide. ICP47 (a.a 1–35) comprises the active domain of the HSV ICP47 protein (20).
Flow Cytometric Analysis.
Flow cytometry was performed as previously described (21).
Immunoblot Analysis and TAP Quantification.
Blots were performed as described (22), and reactive bands were detected by chemiluminescence (Pierce). TAP levels in the T2 transfectants were quantitated by fluorimagery as described (21) using calnexin as control for cell equivalents and gel loading.
Generation of T2 Transfectant Cell Lines.
Transfectants were generated by electroporation (Gene Pulser, Bio-Rad; Ref. 21). T2 cells (107) were transfected with 20 μg of wild-type or mutant TAP1-pCDM8 and 1 μg of pMCFR-puro and selected on irradiated HeLa cell feeders using puromycin (500 ng/ml). Transfection of the T2.TAP1 (wild-type) and T2.TAP1 (K>A) cell lines with the TAP2 cDNAs used 20 μg of either wild-type or mutant TAP2-pCDM8 and 1 μg of pMCFR-neo, selecting in 1 mg/ml G418. Transfectants were identified by immunoblotting.
Transient Expression of Individual TAP Polypeptides.
TAP cDNAs were transiently expressed in HeLa cells by infecting with a T7 polymerase encoding recombinant vaccinia virus (vTF7-3) followed by transfection with the plasmid (23). The lacZ-pCDM8 plasmid was used as a negative control and to monitor transfection efficiencies.
ATP Photolabeling of TAP.
Cells (1, 2, or 4 × 106 cells/sample) were washed in PBS, and solubilized in 30 μl of TBS (0.15 M NaCl, 0.01 M Tris, pH 7.0), containing 1% digitonin, 5 mM MgCl2, 0.5 mM PMSF, 0.1 mM N-tosyl-l-lysyl-chloromethyl ketone (TLCK), and 5 mM iodoacetamide (IAA). Aliquots from the postnuclear supernatant were placed in the wells of a prechilled 96-well plate. The [α-32P]8N3ATP or 8N3ATPγbiotin (Affinity Labeling Technologies, Lexington, KY) was lyophilized in the dark and resuspended in TBS (pH 7.0). Ten microliters of diluted 8-azido-ATP was incubated with the cell lysate for 15 min to allow ATP binding. For assays with sodium orthovanadate (Sigma), the vanadate was preincubated for 2.5 min before the addition of the 8-azido-ATP. The 96-well plate was exposed to short-wave UV (254 nm, Spectroline model XX-15F) for 2.5 min, and 2 μl of 200 mM DTT added. After 5 min, TBS containing 1% digitonin and 5 mM IAA was added. After immunoprecipitation with either the 148.3 mAb or the R.RING11 antiserum, the samples were analyzed by SDS/PAGE and either autoradiography ([α-32P]8N3ATP) or immunoblot analysis (8N3ATPγbiotin) using Extravidin-HRP (Sigma).
ATP-Agarose Binding Assay.
TAP1 and TAP2 binding to ATP-agarose was assayed by using HeLa cells infected with the vTF7-3 vaccinia virus and transfected with the indicated plasmid. Cells (106) were solubilized in 50 μl of TBS containing 1% digitonin, 200 mM l-arginine (Sigma), 5 mM MgCl2, 0.5 mM PMSF, 0.1 TLCK, and 5 mM IAA. Glycerol (50 μl) was added to the postnuclear supernatant before incubation at 4°C for 72 h. Fifty microliters of ATP-agarose (N-6 attachment; Sigma) was added, and the sample was rotated for 24 h at 4°C. After centrifugation for 30 min at 15,000 × g, the supernatant was stored at 4°C while the pellet was washed three times in TBS containing 0.1% digitonin, 200 mM l-arginine and 5 mM MgCl2. Samples were solubilized in SDS sample buffer with 10 mM EDTA and analyzed by immunoblotting with the 148.3 mAb. For the T2 transfectants, the cells were solubilized as described above, and 50 μl of ATP-agarose was incubated with the postnuclear supernatant for 1 h at 4°C. After washing, the proteins were eluted and analyzed as above.
Radiolabeling and Endo H Digestion.
Cells were labeled with l-[35S]methionine (Amersham Pharmacia; 100 μCi per 106 cells for 15 min) and extracted with TBS (pH 7.4) containing 1% digitonin, 0.5 mM PMSF, 0.1 mM TLCK, and 5.0 mM IAA. Immunoprecipitations with HLA-B-specific (4E) or TAP1-specific (148.3) mAbs and endo H (Boehringer Mannheim) treatments were performed as previously described (19) before separation by SDS/PAGE and PhosphorImager analysis (Cyclone, Packard).
Membrane Preparation and TAP Photoaffinity Labeling.
Membranes were prepared by freeze–thaw lysis of 108 cells and photoaffinity labeled as described (21).
Peptide Translocation Assay.
Peptide translocation was carried out on Streptolysin O (2 units/ml; Murex, Norcross, GA) permeabilized cells as previously described (19, 21).
Chemical Cross-Linking of TAP in Membrane.
Membranes were prepared from 107 cells by freeze-thaw lysis (21) in 150 mM NaCl, 20 mM bicine containing 10 mM IAA. The membrane pellet was resuspended in 500 μl of ICT buffer (50 mM Hepes/78 mM KCl/4 mM MgCl2/8.37 mM CaCl2/10 mM EGTA/1 mM DTT, pH 7.0). One hundred-microliter (2 × 106 cell equivalents) aliquots were incubated for 1 h on ice in the presence of 10 μM SRYWAIRTR or ICP47 peptide. The homobifunctional cross-linker ethylene glycol bis (succinimidyl succinate) (EGS; Pierce) was added to 1 mM, and the samples were rotated for 30 min at 4°C (24). The reaction was quenched by the addition of glycine (100 mM). The cross-linked membranes were washed twice at 4°C with ICT buffer (without BSA), solublized in SDS sample buffer, and analyzed by SDS/PAGE and immunoblot analysis with the 148.3 mAb.
Results
Vanadate-Enhanced Binding of ATP Reveals Cooperative Interaction Between the NBDs of TAP1 and TAP2.
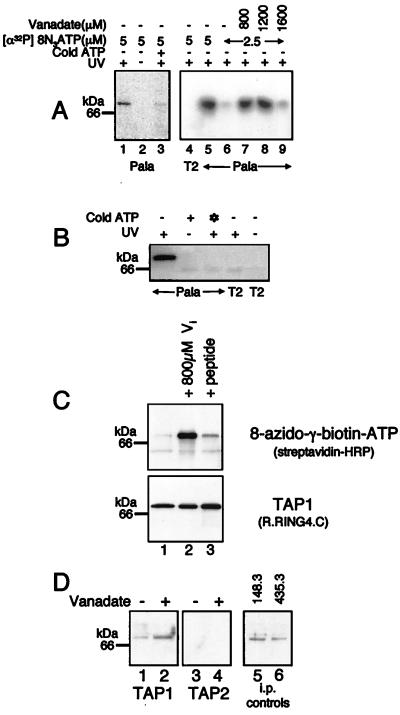
We used the ATP analog [α-32P]8N3ATP to show site-specific nucleotide photolabeling of TAP. Extracts of the Pala B cell line were incubated with [α-32P]8N3ATP. After UV irradiation, TAP proteins were immunoprecipitated, and 32P labeling was assessed by autoradiography. A single 70-kDa band consistent with TAP photolabeling was observed in Pala cells, but not in TAP-negative T2 cells (Fig. 1A). Labeling by 8-azido-[α-32P]ATP depended on the presence of Mg2+ ions (data not shown), required UV-activation, and was inhibited by 1 mM ATP (Fig. 1A).
Figure 1.
Vanadate and peptide enhance photoaffinity labeling of TAP by 8-azido ATP analogues. (A) Vanadate enhances photoaffinity labeling of TAP with [α-32P]8N3ATP. Digitonin cell lysates from T2 (1.2 × 106 cells, lane 4) and Pala cells (1.2 × 106 cells in lanes 1–5 and 4.4 × 106 cells in lanes 6–9) were labeled with [α-32P]8N3ATP at the concentrations indicated in the presence (+) or absence (−) of UV-light, cold ATP (1 mM), and vanadate. TAP was immunoprecipitated by using the TAP1 mAb and separated by 10% SDS/PAGE before PhosphorImager exposure. (B) 8N3ATPγbiotin specifically labels TAP. Digitonin lysates of T2 and Pala cells (1.0 × 106 cells/lane) were labeled with 51 μM 8N3ATPγbiotin in the presence (+) or absence (−) of UV light, cold ATP (136 μM), and cold 8N3ATPγbiotin (✻) (136 μM). Immunoprecipitated TAP was separated by SDS/PAGE and transferred to poly(vinylidene difluoride) (PVDF) membranes. Biotinylated bands were detected by chemiluminescence. (C) Vanadate and peptide enhance photoaffinity labeling of TAP with 8N3ATPγbiotin. Digitonin lysates of Pala cells (2.5 × 106 cells/lane) were preincubated with either vanadate (800 μM), or peptide (20 μM) (SRYWAIRTR) and photolabeled with 34 μM 8N3ATPγbiotin. Immunoprecipitated TAP was separated by SDS/PAGE, and biotinylated bands were detected by chemiluminescence. (C Lower) A parallel immunoprecipitation with anti TAP1 (148.3) mAb from unlabeled cell lysates was probed with TAP1 (R.RING4C) antiserum. (D) Vanadate enhances photoaffinity labeling of TAP1 with 8N3ATPγbiotin. Digitonin extracts from Pala cells (2.5 × 106 cells/lane) were preincubated with (+) or without (−) vanadate (800 μM) and photolabeled with 34 μM 8N3ATPγbiotin, and the TAP heterodimer was dissociated by incubation at 37°C for 15 min in 0.5% SDS before immunoprecipitation with either the TAP1 mAb (148.3; lanes 1 and 2) or the TAP2 mAb (435.3; lanes 3 and 4). Immunoprecipitates were separated by SDS/PAGE, and biotinylated bands were detected by chemiluminescence. Parallel immunoprecipitations with the TAP1 mAb (148.3; lane 5) or the TAP2 mAb (435.3; lane 6) were probed with TAP1 (R.RING4C) antiserum.
To assess whether the NBDs of TAP interact, we investigated the effect of vanadate on 8-azido-ATP photolabeling. Vanadate is a potent inhibitor of the ATPase activity of most ABC transporters (4). It traps ADP in the catalytic site by mimicking the pentacovalent phosphorus formed as a transition state intermediate. The resulting stable vanadate-containing complex (NBD–ADP–Vi) resembles the catalytic intermediate (NBD–ADP–Pi; refs. 25 and 26). Extracts of Pala cells were preincubated with vanadate before the addition of [α-32P]8N3ATP. Vanadate dramatically enhanced photolabeling of the TAP heterodimer by [α-32P]8N3ATP (Fig. 1A). Enhancement was maximal at 800 μM vanadate and was sensitive to the number of cell equivalents and the concentration of [α-32P]8N3ATP used. A suboptimal concentration of [α-32P]8N3ATP (2.5 μM) and a high number of cell equivalents (4.4 × 106/sample) were necessary to observe enhancement (Fig. 1A).
Vanadate-enhanced photolabeling can occur by two nonexclusive mechanisms. Vanadate may trap the hydrolysis product of the photolabel, [α-32P]8N3ADP, in the NBD, or, if a cooperative interaction exists between two NBDs, the effect may be indirect. For many ABC transporters, binding of ADP to one NBD leads to an increase in ATP binding by the second (27–29). By trapping an ADP at one NBD, vanadate can enhance photolabeling of the second by this increase in affinity. To determine the mechanisms responsible for vanadate-enhanced photolabeling of TAP, we used a non-hydrolyzable 8-azido-ATP analogue, 8N3ATPγbiotin, in which a biotin group is attached to the terminal phosphate of ATP. Specific labeling of TAP with this compound is shown in Fig. 1B. Lysates of Pala cells were prepared as for [α-32P]8N3ATP, but photolabeling of TAP by the 8N3ATPγbiotin was assayed by immunoblotting with streptavidin-horseradish peroxidase (HRP). Labeling with 8N3ATPγbiotin, although not saturable in the range tested (2.5–70 μM; data not shown), required UV-activation, was inhibited by both ATP and 8-azido-ATP, and was not seen in TAP-negative T2 cells (Fig. 1B).
Vanadate strongly enhanced TAP labeling by 8N3ATPγbiotin (Fig. 1C, lanes 1 and 2). Because ATPγ-biotin cannot be hydrolyzed, there must be at least two NBDs involved, one where vanadate has stabilized ADP (NBD–ADP–Vi) and a second where 8N3ATPγbiotin has bound (NBD–ATPγbiotin). To determine which polypeptide was binding the ATPγbiotin, we denatured TAP in 0.5% SDS before immunoprecipitating with TAP1- and TAP2-specific antibodies. ATPγbiotin exclusively labeled TAP1, and the labeling was strongly enhanced by vanadate (Fig. 1D). These results suggest that [assuming a simple 1:1 stochiometry of TAP1:TAP2 (30)] vanadate is binding predominantly to TAP2, which promotes binding of ATPγbiotin to TAP1.
Peptide Binding to TAP Enhances Photolabeling with ATP.
Because vanadate strongly enhanced the labeling of TAP by 8N3ATPγbiotin, it was important to determine whether peptide had a similar effect. Although less dramatic than vanadate, the TAP-binding peptide (20 μM; SRYWAIRTR) also enhanced 8-azido-γ-biotin-ATP photolabeling of TAP (Fig. 1C, lanes 1 and 3).
TAP1 and TAP2 Polypeptides Lacking the Walker A Lysine Retain the Ability to Bind ATP.
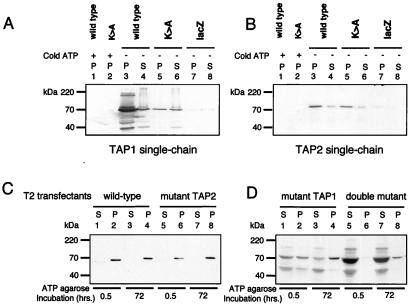
The NBDs of ABC transporters contain a conserved sequence motif (GxxGxGKT/S), known as the Walker A motif or phosphate-binding loop (P-loop) (31, 32). The Walker A motif is involved in holding the γ- and β-phosphoryl groups of nucleotides in the optimal position for hydrolysis (32, 33). The conserved lysine residue participates in the positioning of the terminal phosphate of ATP and is thought to stabilize the reaction transition state (34). Mutating the lysine inhibits hydrolytic activity without, in most cases, affecting ATP binding (32, 35). To explore the role of ATP hydrolysis in peptide translocation, we generated mutant polypeptides in which the “Walker lysine” was mutated to an alanine (K544A TAP1 and K509A TAP2). To confirm that ATP binding had been retained, wild-type and mutant TAP1 and TAP2 polypeptides were transiently expressed in HeLa cells by using a vaccinia virus-based system (23). The faint band seen in the lacZ-transfected control lanes (Fig. 2 A and B) represents the low level expression of endogenous TAP in HeLa cells. We failed to demonstrate ATP binding to the mutated TAP1 or either of the TAP2 polypeptides (data not shown). We suspected this problem might be due to slow dissociation of prebound nucleotides and/or poor solvent accessibility of the TAP2 NBD. Accordingly, we assayed binding by using a glycerol-based procedure developed to displace tightly bound and solvent-inaccessible nucleotides from the mitochondrial F1-ATPase (36). We found that both the wild-type and mutant TAP1 and TAP2 bound to ATP-agarose (Fig. 2 A and B). Binding was inhibited by 5 mM ATP (Fig. 2 A and B) and did not occur on unmodified or Protein-A-conjugated agarose (data not shown). The difficulty showing ATP binding by mutant TAP1 is probably because it is less stable than the wild type (data not shown). We consistently recovered less mutant TAP1 in our HeLa cell lysates (Fig. 2A, lanes 3 and 5). We cannot, however, exclude the possibility that the K>A mutation, against our expectations, has significantly reduced ATP binding affinity.
Figure 2.
Mutant TAP1 and TAP2 polypeptides retain the ability to bind ATP. (A and B) Single-chain TAP polypeptides can bind to ATP-agarose. Subconfluent HeLa cells were infected with vTF7-3 and then transfected with vectors encoding wild-type and mutant TAP polypeptides and a control lacZ (as indicated). After overnight incubation, the cells were extracted in 1% Triton X-100 and incubated with 50 μl of glycerol for 4 days and ATP-agarose for a further 24 h. Equal cell equivalents of the ATP-agarose bound pellet (P) and unbound supernatant (S) fractions were separated on a 10% SDS/PAGE gel, transferred onto poly(vinylidene difluoride) (PVDF) membranes, and probed with (A) TAP1 (148.3) mAb and (B) TAP2 (435.3) mAb. (C and D) Wild-type and mutant TAP-transfected T2 bind ATP-agarose. T2 transfectants were extracted in 1% Triton X-100, and, after incubation with ATP-agarose for 0.5 and 72 h, the bound (P) and unbound (S) proteins were separated by 10% SDS/PAGE and probed with the TAP1 (148.3) mAb. Reactive bands were detected by chemiluminescence; the exposure in D was 10 times longer than in C.
The majority of TAP2 bound to the ATP-agarose, whereas significant amounts of TAP1 remained in the supernatant (Fig. 2A). TAP1 may have a lower affinity for ATP than TAP2, but this possibility seems at odds with our own and published photolabeling results (37, 38). It seems more likely that TAP1 is in rapid and reversible equilibrium with the ATP-agarose, whereas TAP2 is not. This result is reminiscent of the F1-ATPase, in which one NBD can engage in rapid and reversible binding of nucleotides whereas the other exchanges bound nucleotides slowly and only in the presence of glycerol (36).
Reconstitution of T2 with Wild-Type and Mutant TAP Polypeptides.
Four T2 transfectants were generated, expressing all possible combinations of mutant and wild-type TAP1 and TAP2 genes (Table 1). The reconstituted cell lines do not express equivalent amounts of TAP. Analysis of more than 100 transfectants expressing mutant TAPs showed that these were consistently expressed at lower levels than their wild-type counterpart. The relative levels of TAP were quantitated by fluorimagery (Table 1). Equivalent numbers of each cell type were used in all subsequent experiments.
Table 1.
Expression of TAPs in T2 cells
| Name of cell line | TAP1 | TAP2 | Relative TAP levels |
|---|---|---|---|
| Wild-type TAP | Wild type | Wild type | 1.0 |
| Mutant TAP2 | Wild type | K>A | 0.6 |
| Mutant TAP1 | K>A | Wild type | 0.2 |
| Double mutant | K>A | K>A | 0.7 |
We examined the ATP-binding capacity of TAP in the reconstituted cell lines. Although all bound to ATP-agarose, TAP heterodimers containing a wild-type TAP1 polypeptide reached steady-state levels of binding within 30 min, whereas TAP heterodimers containing a mutant TAP1 did not reach steady state within 72 h (Fig. 2 C and D). The rate of ATP-agarose binding was slowest for the TAP heterodimer composed of both a mutant TAP1 and TAP2, indicating that hydrolysis at TAP2 does have some influence on ATP binding by TAP. This effect is minor, however, in the presence of a wild-type TAP1 polypeptide (compare Fig. 2C, lanes 2 and 6). Consistent with our photolabeling experiments, these results indicate that the TAP heterodimer binds to ATP primarily via the TAP1 polypeptide. This process is affected by the Walker lysine substitution, suggesting that rapid interaction with ATP involves ATP hydrolysis, perhaps of prebound ATP molecules. Differential reactivity with both 8N3ATP and ATP-agarose shows that the NBDs of TAP1 and TAP2 are biochemically distinct. TAP1 appears to be engaged in rapid interaction with ATP in the solvent, whereas TAP2 has a low affinity for ATP, a much slower rate of hydrolysis, or is relatively solvent inaccessible and therefore has a slow rate of nucleotide binding and exchange.
Rescue of HLA-B51 Expression in T2 Cells Requires a Wild-Type TAP2, but Not TAP1 Polypeptide.
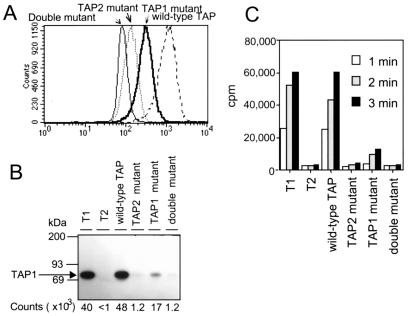
To assess the functional consequences of the TAP (K>A) mutation, we analyzed cell surface HLA-B51 expression in the reconstituted cell lines. As expected, T2 expressing a fully wild-type transporter expressed HLA-B51 at high levels (Fig. 3A). The cell line expressing a wild-type TAP1 and mutant TAP2 had very low levels of surface HLA-B51 and the double mutant cell line showed no rescue of class I expression. Surprisingly, the mutant TAP1 cell line expressed significant levels of surface HLA-B51 (Fig. 3A). Pulse-chase analysis confirmed that the TAP1 mutant cell line assembles HLA-B51 molecules at a decreased, but measurable rate (data not shown). This unexpected result suggests that nucleotide hydrolysis by TAP1 is less critical for TAP function than hydrolysis by TAP2.
Figure 3.
Reconstitution of T2 cells with wild-type and mutant TAP polypeptides. (A) MHC class I expression in wild-type and mutant TAP-transfected T2 cells. Wild-type and mutant TAP-transfected T2 cells were analyzed for MHC class I expression (HLA-B alleles) by flow cytometry, using the monoclonal antibody 4E. (B and C) Peptide binding and translocation require a catalytically active NBD on TAP2. (B) Photolabeling of TAP from wild-type and mutant TAP-transfected T2 cells with the [125I]KB11-HSAB peptide was performed as described in Materials and Methods. After solubilization in 1% digitonin, labeled TAP molecules were immunoprecipitated with the anti-TAP1 mAb. 148.3 and analyzed by SDS/PAGE and autoradiography, and bands corresponding to 125I labeled TAP1 were quantitated by PhosphorImager analysis. (C) SLO permeabilized cells from wild-type and mutant T2 TAP transfectants were incubated with the iodinated reporter peptide RRYQNSTEL at 37°C for the indicated time periods, and the reaction stopped by lysis with cold 3% Triton X-100, as described in Materials and Methods. Translocation into the ER was assessed by binding of the glycosylated reporter peptide to Con A-Sepharose beads and counting on a γ-counter.
Peptide Binding Requires a Catalytically Active TAP2.
Peptide binding to TAP expressed in the reconstituted cell lines was quantitated by using an iodinated, photoactivatable reporter peptide, [125I]KB11-HSAB (19). Cell membranes were incubated with the reporter peptide for 15 min at 25°C, placed on ice, and illuminated with UV light. The photolabeled membranes were solubilized in detergent, and TAP-associated counts were quantitated after immunoprecipitation with anti-TAP1 antibodies. TAP-specific labeling was seen in the T1 parent cell line as well as the fully wild-type TAP-reconstituted T2, but was absent from the TAP-negative T2 cell line. TAP complexes containing a mutant TAP2 did not detectably bind peptide (Fig. 3B), whereas those containing wild-type TAP2 and mutant TAP1 did. Interestingly, once differences in the level of TAP expression are taken into account (Table 1), TAP isolated from the TAP1 mutant cell line appears to bind peptide better than fully wild-type TAP.
Low Levels of Peptide Translocation Can Be Detected in Cells Expressing Catalytically Inactive TAP1.
TAP-mediated translocation of peptides into the ER was assessed by using an iodinated reporter peptide containing an N-linked glycosylation motif (8). The iodinated reporter was introduced into the cytosol of Streptolysin O (SLO)-permeabilized cells, translocation allowed to proceed at 37°C for 1 to 3 min, and glycosylated reporter peptide recovered from cell extracts by using Con A-Sepharose. TAP containing a mutant TAP2 does not translocate peptide, which is not surprising because this TAP cannot bind peptide (Fig. 3C). Translocation was detectable but low in the TAP1 mutant cell line, consistent with the class I cell surface expression data (Fig. 3A). This level of translocation is particularly significant in that the TAP1 mutant cell line expresses 80% less TAP than the wild-type cell line (Table 1).
Peptide Dependent Cross-Linking of TAP1 and TAP2 Occurs Only in the Presence of a Wild-Type TAP2 Polypeptide.
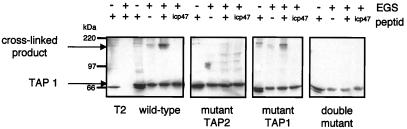
Lacaille and Androlewicz demonstrated conformational changes in TAP using the cross-linking agent, EGS (24), which cross-links TAP1 to TAP2. Formation of the cross-linked product is enhanced by transportable peptides and inhibited by the minimal ICP47 peptide. It was proposed that peptide binding leads to conformational changes that bring the solvent-exposed portions of TAP1 and TAP2 together, facilitating intramolecular cross-linking (24). To determine whether the mutant TAPs could undergo peptide-induced conformational changes, cell membranes from the reconstituted cell lines were incubated with EGS. Cross-linking of TAP was observed in both the wild-type and the mutant TAP1 cell lines (Fig. 4), and formation of the cross-linked species was strongly enhanced by the addition of peptide. This result suggests that the conformational change detected by EGS does not require ATP hydrolysis by TAP1. No cross-linked product was detected in membranes prepared from either the TAP2 mutant or the double-mutant cell line, even after addition of peptide (Fig. 4), consistent with the observed lack of peptide binding to the TAP expressed in these cell lines.
Figure 4.
Peptide-stimulated chemical cross-linking requires a catalytically active NBD on TAP2. Membranes from wild-type and mutant TAP-transfected T2 cells were incubated in the presence (+) or absence (−) of 10 μM of the HLA-B27 peptide (SRYWAIRTR) or ICP47 peptide for 1 h at 4°C before chemical cross-linking with EGS. The cross-linked membranes were resolved on a 6% SDS/PAGE gel and immunoblotted with the TAP1 (148.3) antibody.
Discussion
TAP is the conduit providing cytosolic peptides with access to the ER. Our results suggest that ATP is required for not only powering peptide transport but also for regulating transitions between different conformational states of TAP. Using the ATPase inhibitor sodium orthovanadate, we find evidence of cooperative interaction between the ATP hydrolyzing domains of the TAP1 and TAP2 subunits. The photoactivatable ATP analogue, 8-azido-γ-biotin-ATP, preferentially labeled TAP1, an effect strongly enhanced by vanadate, which acts by trapping ADP in a nucleotide binding site. Because the biotinylated ATP is non-hydrolyzable, vanadate-enhanced labeling must involve at least two nucleotide binding sites. The simplest interpretation of our data is that, after ATP hydrolysis on TAP2, vanadate trapping of the residual ADP enhances ATP binding by TAP1 via a cooperative interaction between the two NBDs. Peptide also enhanced photolabeling of TAP by ATP. Although the strength of the signal was too weak to allow TAP subunit analysis, it seems likely that peptide binding to TAP also enhances ATP binding on TAP1.
Similar cooperativity has been observed in other ABC transporters, the best characterized example being Pgp. Both NBDs of Pgp must be catalytically active to confer multidrug resistance (35), indicating cooperative function and interdependence. Inactivation of just one NBD of Pgp leads to complete loss of ATP hydrolysis. Despite this requirement for two NBDs, the two ATP-hydrolyzing domains of Pgp appear to be functionally equivalent; hydrolysis by either NBD can transport a drug molecule out of the cell (26). This functional equivalence is reflected at the biochemical level—in the presence of vanadate, photolabeling occurs with equal probability at either of the NBDs (39).
Vanadate-trapping experiments with CFTR (27), SUR1 (28), and MRP1 (29) indicate clear biochemical and functional differences between their two NBDs, in that photolabeling occurs preferentially on the N-terminal NBD (NBD1). This result is analogous to our findings, because TAP1 is homologous to the N-terminal half of these transporters. Furthermore, vanadate trapping of ADP on SUR1 and MRP1 occurs almost exclusively at NBD2 and is associated with an increased affinity for ATP at NBD1. This biochemical asymmetry suggests a functional dichotomy between the two NBDs of CFTR, SUR1, and MRP1. Functional studies have confirmed that the NBDs of these proteins are non-equivalent, each involved in a different stage of the transport or conductance cycle (29, 40–42).
Our photolabeling data suggested similar functional differences between TAP1 and TAP2. To explore this possibility, we mutated the Walker A lysine of TAP1 and TAP2 to an alanine. This substitution should eliminate ATP hydrolysis, based on results obtained with Pgp and many other ATPases (32, 35). Although critical for hydrolysis, the Walker A lysine is generally not required for ATP binding, and, as expected, both mutant TAP polypeptides retained the ability to bind ATP (Fig. 2). We assessed the ability of TAP dimers containing the mutant TAPs to bind ATP-agarose. As the photolabeling data suggested, only TAP dimers containing wild-type TAP1 bind ATP efficiently. TAP dimers containing a mutant TAP1 bound to ATP-agarose very slowly. The fact that TAP1 reacts more readily with ATP in solution than TAP2 has been observed in previous studies (37, 38, 43). C-terminal fragments of TAP, encoding just the NBD, have been expressed in Escherichia coli and insect cells. Whereas the NBD of TAP1 binds efficiently to ATP-agarose and is readily labeled by 8-azido-ATP, the TAP2 NBD shows comparatively little nucleotide binding activity. This may be because the NBD of TAP2 has a low affinity for ATP. Alternatively, TAP2 may contain a tightly bound, slowly exchanging nucleotide, or an ATP-binding site that is not fully solvent accessible, despite reacting with vanadate. Because incubation in glycerol promoted binding of TAP2 to ATP-agarose, we favor the latter explanation.
Previous experiments suggested that peptide binding to TAP did not require nucleotide (15, 18). This conclusion has recently been questioned by studies of the effect of mutations within the Walker A site that eliminate ATP binding (9). Mutant TAP heterodimers unable to bind ATP on either TAP1 or TAP2 were also unable to bind peptide (9), a result difficult to reconcile with previous findings that peptide binding was unaffected by ATP depletion. Our data suggest a possible explanation for these apparent contradictions. Although we find that ATP hydrolysis by the NBD of TAP2 is critical for peptide binding, the nucleotide binding site does not appear to be in ready equilibrium with the solvent phase. Hence, depletion of ATP is unlikely to have an effect on the TAP2 NBD. Only by mutating the ATP binding site itself was it possible to see the dependence of peptide binding on nucleotides. Because the mutants derived by Knittler et al. (9) had lost the ability to bind ATP, they could not determine whether ATP hydrolysis was involved in peptide binding. Nor could they ascertain whether the TAP1 or TAP2 mutation eliminated peptide binding.
The mutations we introduced into the NBDs were designed to allow analysis of the role of ATP hydrolysis in transporter function. By generating cell lines expressing different combinations of mutant and wild-type TAP1 and TAP2, we were able to analyze their individual contributions to peptide translocation. The studies revealed that ATP hydrolysis by TAP2, but not TAP1, is critical for transporter function. Several independent lines of evidence indicate that the ATPase activity of TAP2 is required for TAP to bind cytosolic peptides. First, peptide binding to wild-type TAP enhances photolabeling by 8-azido ATP compounds in a manner similar to vanadate (Fig. 1C). This result suggests that peptide binding is linked to the acquisition of an ADP by TAP2, i.e., peptide binding promotes ATP hydrolysis by TAP2. This type of solute-induced ATPase activity has been reported for several ABC transporters, the best characterized being Pgp, where the basal rate of ATP hydrolysis is enhanced by substrate binding. Second, TAP heterodimers containing the mutant TAP2 did not bind peptide. The mutation on TAP1 did not affect the efficiency of peptide binding, indicating that only the ATPase activity of TAP2 is required for this stage of the translocation process. Finally, peptide binding has been shown to produce conformational changes in TAP (24). In our mutant cells, loss of hydrolysis by TAP2, but not TAP1, prevents TAP from assuming the conformation detected by cross-linking. Peptide-induced conformational changes of TAP have also been demonstrated by measuring the fluorescence quenching of fluorophore-labeled peptides (44). The capture of peptide was shown to be a two-stage process—a fast bimolecular association step, followed by a slower intramolecular isomerization. We suggest that the first step is the hydrolysis-driven capture of peptide, which produces local conformational changes that are propagated to more distal portions of the transporter. The proposed early events in peptide transport by TAP are depicted in Fig. 5.
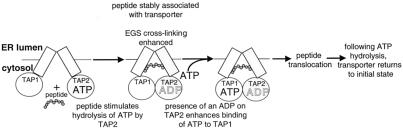
Figure 5.
Model of early events in peptide transport by TAP.
ATP hydrolysis by TAP1 is not essential for peptide translocation (Fig. 3). The observed reduction in peptide transport in the TAP1 mutant may largely be explained by reduced TAP levels (Table 1). Whereas ATP hydrolysis on TAP2 must occur early in the transport cycle, the results of the vanadate experiments indicate that ATP binding by TAP1 occurs later, secondary to hydrolysis at TAP2. Similarly, the EGS cross-linking results place ATP hydrolysis by TAP1 downstream of hydrolysis by TAP2, but as yet we are unable to further clarify the role of TAP1. ATP hydrolysis on TAP1 may be involved in later stages of the translocation cycle, such as returning the transporter to its initial state, or in regulating TAP activity in response to other signals, such as the availability of class I molecules or the intracellular nucleotide concentration.
There are several interesting structural features that may provide clues to functional differences between the TAP subunits. A recent structure of the RAD50 ATPase DNA repair enzyme, which shares sequence similarity with the NBDs of ABC transporters, established a critical role for the conserved “LSGGQ” ABC signature motif (45). In the RAD50 homodimer, the serines of each signature motif form hydrogen bonds with the γ-phosphate of the ATP bound by the opposing RAD50 subunit. Modeling of the TAP NBDs using the RAD50 structure suggests that the canonical signature motif of TAP1 (LSGGQ) forms hydrogen bonds with the γ-phosphate of the ATP bound to TAP2. TAP2, however, does not have reciprocal interactions with the ATP bound to TAP1, because the TAP2 signature motif (LAAGQ) is missing the necessary serine residue, and ATP hydrolysis at this site is predicted to be impaired (K. P. Hopfner, personal communication). Not only is the absence of this serine unique among ABC transporters, but a mutation at this site in CFTR (S549R) is responsible for some cases of cystic fibrosis (46). The lack of a serine in the TAP2 signature motif would be predicted to decrease the rate of ATP hydrolysis by TAP1, suggesting a possible mechanism underlying the functional asymmetry observed in this study.
Acknowledgments
We thank Karl-Peter Hopfner for modeling of the TAP2 signature motif, Matt Androlewicz for the photolabeled peptide, and Uyen Phan and Nancy Dometios for valuable help with manuscript preparation. This work was supported by the Howard Hughes Medical Institute (J.T.K. and P.C.) and The Wellcome Trust (J.T.K., P.J.L., S.S.G., and E.W.H.).
Abbreviations
- ER
endoplasmic reticulum
- ABC
ATP-binding cassette
- NBD
nucleotide binding domain
- IAA
iodoacetamide
- TLCK
N-tosyl-l-lysyl-chloromethyl ketone
- EGS
ethylene glycol bis (succinimidyl succinate)
- TAP
transporter associated with antigen processing
References
- 1.Abele R, Tampe R. Biochim Biophys Acta. 1999;1461:405–419. doi: 10.1016/s0005-2736(99)00171-6. [DOI] [PubMed] [Google Scholar]
- 2.Karttunen J T, Trowsdale J, Lehner P J. Curr Biol. 1999;9:R820–R824. doi: 10.1016/s0960-9822(99)80499-0. [DOI] [PubMed] [Google Scholar]
- 3.Cresswell P, Bangia N, Dick T, Diedrich G. Immunol Rev. 1999;172:21–28. doi: 10.1111/j.1600-065x.1999.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 4.Schneider E, Hunke S. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 5.Holland I B, Blight M A. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- 6.Nijenhuis M, Hammerling G J. J Immunol. 1996;157:5467–5477. [PubMed] [Google Scholar]
- 7.Uebel S, Tampe R. Curr Opin Immunol. 1999;11:203–208. doi: 10.1016/s0952-7915(99)80034-x. [DOI] [PubMed] [Google Scholar]
- 8.Neefjes J J, Momburg F, Hammerling G J. Science. 1993;261:769–771. doi: 10.1126/science.8342042. [DOI] [PubMed] [Google Scholar]
- 9.Knittler M R, Alberts P, Deverson E V, Howard J C. Curr Biol. 1999;9:999–1008. doi: 10.1016/s0960-9822(99)80448-5. [DOI] [PubMed] [Google Scholar]
- 10.Trowsdale J, Hanson I, Mockridge I, Beck S, Townsend A, Kelly A. Nature (London) 1990;348:741–744. doi: 10.1038/348741a0. [DOI] [PubMed] [Google Scholar]
- 11.Powis S H, Mockridge I, Kelly A, Kerr L A, Glynne R, Gileadi U, Beck S, Trowsdale J. Proc Natl Acad Sci USA. 1992;89:1463–1467. doi: 10.1073/pnas.89.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shastri N, Gonzalez F. J Immunol. 1993;150:2724–2736. [PubMed] [Google Scholar]
- 13.Meyer T H, van Endert P M, Uebel S, Ehring B, Tampe R. FEBS Lett. 1994;351:443–447. doi: 10.1016/0014-5793(94)00908-2. [DOI] [PubMed] [Google Scholar]
- 14.Ortmann B, Androlewicz M, Cresswell P. Nature (London) 1994;368:864–867. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 15.van Endert P M, Tampe R, Meyer T H, Tisch R, Bach J F, McDevitt H O. Immunity. 1994;1:491–500. doi: 10.1016/1074-7613(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 16.Stam N J, Spits H, Ploegh H L. J Immunol. 1986;137:2299–2306. [PubMed] [Google Scholar]
- 17.Hochstenbach F, David V, Watkins S, Brenner M B. Proc Natl Acad Sci USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Androlewicz M J, Cresswell P. Immunity. 1994;1:7–14. doi: 10.1016/1074-7613(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 19.Androlewicz M J, Anderson K S, Cresswell P. Proc Natl Acad Sci USA. 1993;90:9130–9134. doi: 10.1073/pnas.90.19.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galocha B, Hill A, Barnett B C, Dolan A, Raimondi A, Cook R F, Brunner J, McGeoch D J, Ploegh H L. J Exp Med. 1997;185:1565–1572. doi: 10.1084/jem.185.9.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehner P J, Surman M J, Cresswell P. Immunity. 1998;8:221–231. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
- 22.Sadasivan B, Lehner P J, Ortmann B, Spies T, Cresswell P. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 23.Arunachalam B, Lamb C A, Cresswell P. Int Immunol. 1994;6:439–451. doi: 10.1093/intimm/6.3.439. [DOI] [PubMed] [Google Scholar]
- 24.Lacaille V G, Androlewicz M J. J Biol Chem. 1998;273:17386–17390. doi: 10.1074/jbc.273.28.17386. [DOI] [PubMed] [Google Scholar]
- 25.Urbatsch I L, Sankaran B, Weber J, Senior A E. J Biol Chem. 1995;270:19383–19390. doi: 10.1074/jbc.270.33.19383. [DOI] [PubMed] [Google Scholar]
- 26.Senior A E, al-Shawi M K, Urbatsch I L. FEBBS Lett. 1995;377:285–289. doi: 10.1016/0014-5793(95)01345-8. [DOI] [PubMed] [Google Scholar]
- 27.Szabo K, Szakacs G, Hegeds T, Sarkadi B. J Biol Chem. 1999;274:12209–12212. doi: 10.1074/jbc.274.18.12209. [DOI] [PubMed] [Google Scholar]
- 28.Ueda K, Inagaki N, Seino S. J Biol Chem. 1997;272:22983–22986. doi: 10.1074/jbc.272.37.22983. [DOI] [PubMed] [Google Scholar]
- 29.Hou Y, Cui L, Riordan J R, Chang X. J Biol Chem. 2000;275:20280–20287. doi: 10.1074/jbc.M001109200. [DOI] [PubMed] [Google Scholar]
- 30.Ortmann B, Copeman J, Lehner P J, Sadasivan B, Herbert J A, Grandea A G, Riddell S R, Tampe R, Spies T, Trowsdale J, Cresswell P. Science. 1997;277:1306–1309. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 31.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saraste M, Sibbald P R, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 33.Hung L W, Wang I X, Nikaido K, Liu P Q, Ames G F, Kim S H. Nature (London) 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 34.Reinstein J, Schlichting I, Wittinghofer A. Biochemistry. 1990;29:7451–7459. doi: 10.1021/bi00484a014. [DOI] [PubMed] [Google Scholar]
- 35.Azzaria M, Schurr E, Gros P. Mol Cell Biol. 1989;9:5289–5297. doi: 10.1128/mcb.9.12.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrett N E, Penefsky H S. J Biol Chem. 1975;250:6640–6647. [PubMed] [Google Scholar]
- 37.Muller K M, Ebensperger C, Tampe R. J Biol Chem. 1994;269:14032–14037. [PubMed] [Google Scholar]
- 38.Lapinski P E, Miller G G, Tampe R, Raghavan M. J Biol Chem. 2000;275:6831–6840. doi: 10.1074/jbc.275.10.6831. [DOI] [PubMed] [Google Scholar]
- 39.al-Shawi M K, Urbatsch I L, Senior A E. J Biol Chem. 1994;269:8986–8992. [PubMed] [Google Scholar]
- 40.Carson M R, Travis S M, Welsh M J. J Biol Chem. 1995;270:1711–1717. doi: 10.1074/jbc.270.4.1711. [DOI] [PubMed] [Google Scholar]
- 41.Gunderson K L, Kopito R R. Cell. 1995;82:231–239. doi: 10.1016/0092-8674(95)90310-0. [DOI] [PubMed] [Google Scholar]
- 42.Ueda K, Matsuo M, Tanabe K, Morita K, Kioka N, Amachi T. Biochim Biophys Acta. 1999;1461:305–313. doi: 10.1016/s0005-2736(99)00157-1. [DOI] [PubMed] [Google Scholar]
- 43.van Endert P M. J Biol Chem. 1999;274:14632–14638. doi: 10.1074/jbc.274.21.14632. [DOI] [PubMed] [Google Scholar]
- 44.Neumann L, Tampe R. J Mol Biol. 1999;294:1203–1213. doi: 10.1006/jmbi.1999.3329. [DOI] [PubMed] [Google Scholar]
- 45.Hopfner K P, Karcher A, Shin D S, Craig L, Arthur L M, Carney J P, Tainer J A. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 46.Gadsby D C, Nairn A C. Physiol Rev. 1999;79:S77–S107. doi: 10.1152/physrev.1999.79.1.S77. [DOI] [PubMed] [Google Scholar]