Abstract
Pancreatic adenocarcinoma (PAC) is among the most lethal malignancies. While research has implicated multiple genes in disease pathogenesis, identification of therapeutic leads has been difficult and the majority of currently available therapies provide only marginal benefit. To address this issue, our goal was to genomically characterize individual PAC patients to understand the range of aberrations that are occurring in each tumor. Because our understanding of PAC tumorigenesis is limited, evaluation of separate cases may reveal aberrations, that are less common but may provide relevant information on the disease, or that may represent viable therapeutic targets for the patient. We used next generation sequencing to assess global somatic events across 3 PAC patients to characterize each patient and to identify potential targets. This study is the first to report whole genome sequencing (WGS) findings in paired tumor/normal samples collected from 3 separate PAC patients. We generated on average 132 billion mappable bases across all patients using WGS, and identified 142 somatic coding events including point mutations, insertion/deletions, and chromosomal copy number variants. We did not identify any significant somatic translocation events. We also performed RNA sequencing on 2 of these patients' tumors for which tumor RNA was available to evaluate expression changes that may be associated with somatic events, and generated over 100 million mapped reads for each patient. We further performed pathway analysis of all sequencing data to identify processes that may be the most heavily impacted from somatic and expression alterations. As expected, the KRAS signaling pathway was the most heavily impacted pathway (P<0.05), along with tumor-stroma interactions and tumor suppressive pathways. While sequencing of more patients is needed, the high resolution genomic and transcriptomic information we have acquired here provides valuable information on the molecular composition of PAC and helps to establish a foundation for improved therapeutic selection.
Introduction
Pancreatic cancer is a malignant carcinoma that is currently the fourth leading cause of cancer-related deaths in the United States [1]. In 2011, an estimated 44,030 new patients were diagnosed, and the one- and five-year survival rates were approximately 26% and 6%, respectively [1]. Current standard treatment options for patients include surgical removal of the tumor, radiation therapy, chemotherapy, and targeted/biologic therapy. However, due to late diagnoses and the associated low survival rate, improved treatments are needed.
Significant effort by a number of groups has led to the identification of genomic alterations in pancreatic cancer. Heavily implicated genes include KRAS (v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog ) [2], [3], TP53 (tumor protein p53) [4], [5], SMAD4/DPC4 (SMAD family member 4/deletion target in pancreatic carcinoma 4 homolog) [6], [7], CDKN2A (cyclin-dependent kinase inhibitor 2A; p16) [8], [9], [10], and BRCA2 (breast cancer 2, early onset) [11], [12]. However, FDA approved therapies that exploit these genomic alterations in pancreatic cancer are currently not available. As a result, standard agent therapy for advanced stage and metastatic pancreatic adenocarcinoma (PAC) patients commonly target tumor DNA replication, cell division, and proliferation, or specific receptors that help to mediate signaling cascades. While PAC patients commonly have mutations in the previously mentioned genes, low survival rates for PAC patients are associated with difficulty in identifying effective treatments beyond standard therapies. Such difficulty associated with finding effective treatments demonstrates that our understanding of pancreatic cancer remains limited. In order to address these challenges, one strategy is to first individually characterize patients to fully understand the range of alterations in separate tumors. In doing so, we acquire valuable information on each patient's disease, as well as PAC as a whole, and are also able to identify druggable targets that may provide additional therapeutic options on a patient-specific basis. This approach is particularly relevant because although certain mutations are common across patients, each patient's tumor demonstrates divergent aberrations. As we acquire more tumor DNA and RNA sequence information from actual patients, we will also be able to delineate the key biological processes that are central to PAC and develop improved therapies for patients.
To carry out unbiased whole genome analyses in actual patients, we performed whole genome sequencing (WGS) of tumor biopsy DNA and matched normal DNA from blood from three separate PAC patients to identify somatic events in each patient's tumor. Our primary aim is to separately characterize each of these patients to evaluate the molecular background of each tumor. To understand the possible implications of identified genomic events and to evaluate transcriptional alterations in the tumor, we also performed RNA sequencing (RNAseq) for 2 of the patients for which RNA was available. Lastly, for patients 1 and 2, we performed comparative genomic hybridization (CGH) analyses to validate copy number changes identified through sequencing. The use of next generation sequencing (NGS) and the combined analysis of separate sets of data help to create a detailed picture of the disease in each patient and contribute to our understanding of the disease. We present here very detailed genomic characterizations of three separate PAC patients.
Materials and Methods
Detailed supplementary methods are described under Supporting Information (Methods S1). A summary of methods is presented here.
Ethics statement
All patients were treated on protocols approved by the Mayo Clinic Institutional Review Board (MCIRB) and the Western Institutional Review Board (WIRB). This study was conducted in accordance with the 1996 Declaration of Helsinki. Written informed consent was obtained from all patients.
Eligibility Criteria
For this study, patients had to be ≥18 years of age and provided signed informed consent. These patients included those with a pathologic or clinical diagnosis of a pancreatic malignant neoplasm, or who were undergoing a medically indicated procedure to obtain tissue or to resect their pancreatic tumor. Other eligibility criteria included: Karnofsky performance status (PS) ≥80%, life expectancy >3 months, baseline laboratory data indicating acceptable bone marrow reserve, liver, and renal function. Patients were allowed to participate on another clinical trial involving treatment prior to or during participation on this study. Main exclusion criteria included: symptomatic central nervous system (CNS) metastasis, untreated CNS metastases, known active infections requiring intravenous antimicrobial therapy, known HIV, HBV or HCV infection requiring antiviral therapy, pregnant or breast feeding women, or inaccessible tumor for biopsy.
Sample assessment
Tumor samples were obtained under institutional review protocols and were preserved as fresh frozen. Normal DNA was obtained from peripheral blood mononuclear cells. Percent tumor cellularity of patient 1's biopsy (tumor content) was assessed as 60% tumor, patient 2 50% tumor, and patient 3 40–50% tumor. Direct visualization of samples collected from all three patients was obtained to estimate tumor content and extent of tissue heterogeneity by a board certified pathologist (GH).
Genomic DNA isolation
Tissue was disrupted and homogenized in Buffer RLT plus (Qiagen AllPrep DNA/RNA Mini Kit), using the Bullet Blender™, Next Advance, and transferred to a microcentrifuge tube containing Buffer RLT plus and 1.6 mm stainless steel beads (patient 1), or 0.9 mm–2.0 mm RNase free stainless steel beads (patients 2 and 3). Blood leukocytes (buffy coat) were isolated from whole blood by centrifugation at room temperature and resuspended in Buffer RLT plus. All samples were homogenized, centrifuged at full speed, and lysates were transferred to the Qiagen AllPrep DNA spin column. Genomic DNA was purified following the manufacturer's protocol. DNA was quantified using the Nanodrop spectrophotometer and quality was accessed from 260/280 and 260/230 absorbance ratios.
RNA Isolation
Tissue was disrupted and homogenized in Buffer RLT plus using the Bullet Blender, and transferred to a microcentrifuge tube containing Buffer RLT plus and 0.9 mm–2.0 mm RNAse free stainless steel beads. The tissue was homogenized in the Bullet Blender, and centrifuged at full speed. The supernatant was transferred to the QiagenAllPrep DNA spin column. 70% ethanol was added to the flow-through and the mixture was applied to an RNeasy spin column. Total RNA purification was conducted as directed by the AllPrep DNA/RNA Mini Handbook. FirstChoice normal human pancreatic RNA was purchased from Ambion (catalog#AM7954) and used as the RNAseq control. RNA was quantified using the Nanodrop spectrophotometer and quality was assessed using the Agilent Bioanalyzer.
Whole genome library preparation
3 µg of genomic DNA from each sample was fragmented to a target size of 300–350 base pairs (bp). Overhangs in the fragmented samples were repaired and adenine bases were ligated on. Diluted paired end Illumina adapters were then ligated onto the A-tailed products. Following ligation, samples were run on a 3% TAE gel to separate products. Ligation products at 300 bp and 350 bp were selected for each sample, isolated from gel punches, and purified. 2× Phusion High-Fidelity PCR Master Mix (Finnzymes; catalog#F-531L) was used to perform PCR to enrich for these products. Enriched PCR products were run on a 2% TAE gel and extracted. Products were quantified using Agilent's High Sensitivity DNA chip (catalog#5067-4626) on the Agilent 2100 Bioanalyzer (catalog#G2939AA).
Whole transcriptome library preparation
All RNA samples were analyzed on the Agilent Bioanalyzer RNA 6000 Nano Chip to validate RNA integrity (RIN≥7.0). 10 ng of total RNA was used to generate whole transcriptome libraries for RNA sequencing. Using the Nugen Ovation RNA-Seq System (cat#7100-08), total RNA was used to generate double stranded cDNA, which was amplified using Nugen's SPIA linear amplification process. Amplified cDNA was input into Illumina's TruSeq DNA Sample Preparation Kit – Set A (cat#FC-121-1001) for library preparation. In summary, 1 µg of amplified cDNA was fragmented to a target insert size of 300 bp and end repaired. Samples were then adenylated and indexed paired end adapters were ligated. Ligation products were run on a 2% TAE gel and size selected at 400 bp. Ligation products were isolated from gel punches and purified. Cleaned ligation products were input into PCR to enrich for libraries. PCR products were cleaned and quantified using the Agilent Bioanalyzer.
PE next generation sequencing
Tumor and normal libraries were prepared for paired end sequencing. Clusters were generated using Illumina's cBot and HiSeq Paired End Cluster Generation Kits (catalog#PE-401-1001) and sequenced on Illumina's HiSeq 2000 using Illumina'sHiSeq Sequencing Kit (catalog#FC-401-1001).
Array CGH (aCGH) for patient 1
Samples were run with the SurePrint G3 Human aCGH Microarray 1 M (Agilent Technologies, Palo Alto, CA). The digestion, labeling, and hybridization steps were performed as previously described with minor modifications [13]. Briefly, 1.2 ug of tumor and reference DNA were independently digested with Bovine DNase I (Ambion, Austin, TX) for 12 minutes at room temperature. DNA samples from a pool of nine human, female, lymphoblastoid cell lines from the Coriell repository (NA18517, NA19240, NA18555, NA18537, NA18980, NA18972, NA12878, NA12156, and NA15510) were used as the normal reference in the hybridization experiments. Tumor samples were labeled with Cy5 dye, and the normal reference was labeled with Cy3 dye. Labeled reactions were cleaned up and hybridized at 65°C for 40 hours. Microarrays were scanned and features were extracted with Feature Extraction software (Agilent Technologies). Log2 ratio data was analyzed using Genomic Workbench software version 5.0.14 (Agilent Technologies).
Flow cytometry CGH for patient 2
DNA content based flow assays were used to identify and purify proliferating 2N (G1) populations, 4N(G2/M), and aneuploid populations from the biopsy. The biopsy was minced in the presence of NST buffer and DAPI according to published protocols [14], [15]. Nuclei were disaggregated immediately before analysis with a 25-gauge needle and then filtered through a 40-µm mesh filter and analyzed using an Influx cytometer (Becton-Dickinson Cytopeia, San Jose CA), with ultraviolet excitation and DAPI emission collected at >450 nm. DNA content and cell cycle were analyzed using the software program WinCycle (Phoenix Flow Systems, San Diego, CA). DNAs were extracted using Qiagen micro kits (Qiagen Valencia, CA). For hybridization, 100 ng of genomic DNA from each sample and of pooled commercial 46XX reference (Promega) were amplified using the GenomiPhi amplification kit (G.E. Healthcare, Piscataway, NJ). 1 ug of amplified sample and 1 ug of amplified reference template were digested with DNaseI and labeled with Cy-5 dUTP and Cy-3 dUTP respectively, using a BioPrime labeling kit (Invitrogen, Carlsbad, CA). All labeling reactions were assessed using a Nanodrop assay (Nanodrop, Wilmington, DE) prior to mixing and hybridization to a CGH array (Agilent Technologies, Santa Clara, CA).
Sequencing data analysis
Raw sequence data in the form of .bcl files were generated by the Illumina HiSeq 2000. These data were converted to .qseq files, which were used to generate .fastq files. Fastq files were validated to evaluate the distribution of quality scores and to ensure that quality scores do not drastically drop over each read. Validated fastq files were aligned to the human reference genome (build 36) using the Burrows-Wheeler Alignment (BWA) tool. Following alignment,.sai files were used to create .sam (sequence alignment map) files [16], which were input into SAMtools to create binary sequence (.bam) files. PCR duplicates were flagged for removal using Picard. Indels were realigned and base quality scores were recalibrated using GATK (Genome Analysis Toolkit) [17]. Mutation analysis was performed to identify SNPs, indels, and CNVs. Circos plots were generated for each patient to summarize results from all variant analyses (Figures 1, 2, and 3). NCBI (National Center for Biotechnology Information) SRA (Sequence Read Archive) accession numbers for each patient are as follows—patient 1 DNA: SRS334038, SRS334039; patient 2 DNA: SRS334040, SRS334041; patient 2 RNA: SRS348787; patient 3 DNA: SRS334042, SRS334045; patient 3 RNA: SRS348788; pancreas RNA control: SRS334047).
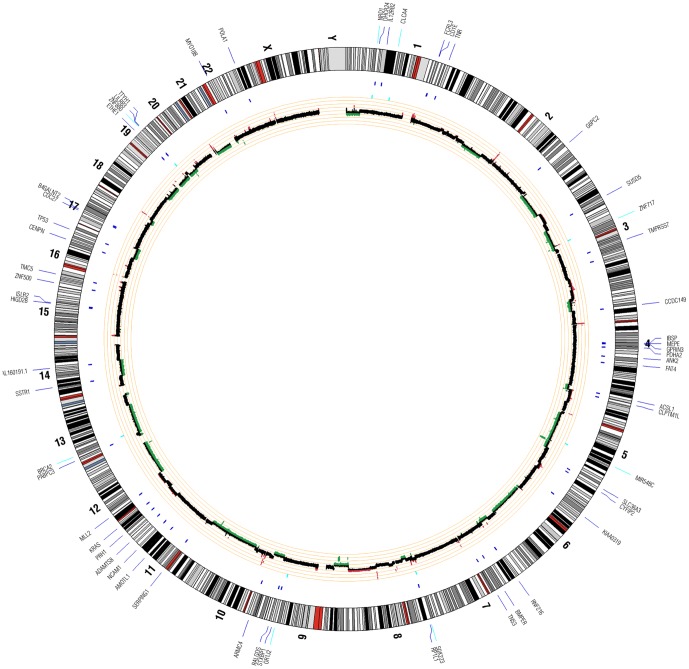
Figure 1. Patient 1 Circos Plot.
This plot summarizes all significant genomic events that were identified in patient 1 using WGS. Copy number changes are shown in the inner circle plot with red marking amplifications and green marking deletions. SNVs are indicated with dark blue tick marks and indels are indicated with light blue tick marks.
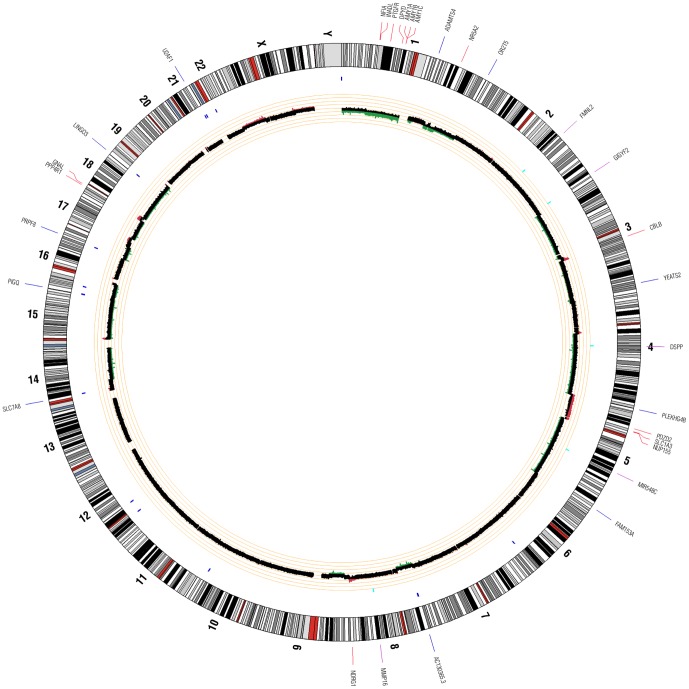
Figure 2. Patient 2 Circos Plot.
This plot summarizes all significant genomic events that were identified in patient 2 using WGS. Copy number changes are shown in the inner circle plot with red marking amplifications and green marking deletions. SNVs are indicated with dark blue tick marks and indels are indicated with light blue tick marks.
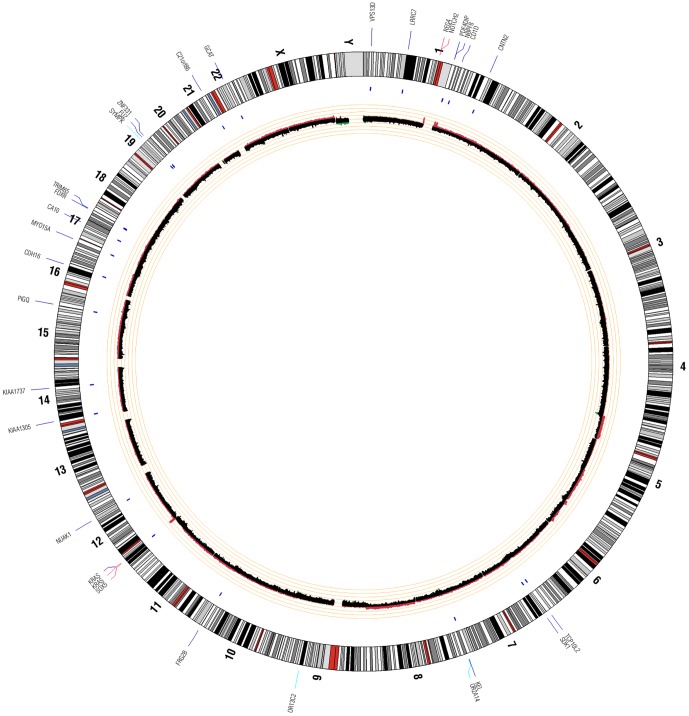
Figure 3. Patient 3 Circos Plot.
This plot summarizes all significant genomic events that were identified in patient 3 using WGS. Copy number changes are shown in the inner circle plot with red marking amplifications and green marking deletions. SNVs are indicated with dark blue tick marks and indels are indicated with light blue tick marks.
SNP (single nucleotide polymorphism) calling was performed using SolSNP (http://sourceforge.net/projects/solsnp/) and Mutation Walker, a tool developed in house and that incorporates variant discovery tools from GATK. SNPs that were called using both tools were compiled and visually examined for false positives to create a final filtered list of true SNVs (single nucleotide variants). Indel (insertion/deletion) calling was performed using GATK and a somatic indel detection tool developed in house. SIFT (Sorting Intolerant From Tolerant) or PolyPhen-2 (Polymorphism Phenotyping v2) was used to determine the effect of coding SNV's and indels on protein function. Copy number analysis was completed by determining the log2 difference of the normalized physical coverage (or clonal coverage) for both germline and tumor samples separately across a sliding 2 kb window of the mean. CREST (Clipping Reveals Structure) was used on WGS data to identify structural variations [18].
RNAseq data was aligned against human reference genome (build 36) with TopHat 1.2; RNAseq reads were only aligned against the autosomes and sex chromosomes. Mitochondrial DNA and annotations were removed from the genome and annotation references prior to alignment. Cuffdiff was used to identify differentially expressed genes and isoforms. Differential analysis was performed on FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) expression values calculated for gene and isoform. P-values were corrected for multiple testing using the Benjamini and Hochberg method. ChimeraScan [19] was used for fusion transcript detection.
Pathway analysis
Integrative analysis of whole genome and transcriptomic data was performed using the Functional Ontology Enrichment Tool in MetaCore from GeneGo, Inc. (v6.8; Thomson Reuters Business, Philadelphia, PA). Pathway analysis specific to pancreatic cancer was performed using the MetaMiner (Oncology) Pancreatic Cancer Disease Module add-on. P-values associated with each analysis are calculated in MetaCore using a hypergeometric distribution.
Results and Discussion
Whole genome sequencing
Our study was performed on a set of fresh pancreatic tumor specimens and whole blood samples from three patients diagnosed with PAC. Clinical information is listed in Table 1. For each patient, we sequenced both tumor DNA, as well as germline DNA isolated from whole blood in order to identify somatic changes in the tumors. Read alignment was performed with BWA using build 36 of the human reference genome. WGS metrics and summary statistics for each of the three patients are shown in Table 2. Using sequencing by synthesis technology and 100 bp paired end chemistry, we generated nearly 8 billion total reads from WGS for average mapped coverages ranging from 31× to 54×. SNP calling was performed using two separate callers to reduce the false negative rate. To evaluate the overall quality of variant data, germline SNPs were called and the transition to transversion and dbSNP (Single Nucleotide Polymorphism Database) [20] 129 concordance ratios were calculated. For all three patients, the transition/transversion ratios were in the range of 2.01 to 2.24, and the dbSNP 129 concordance ratios were approximately 87% (Table 2). These analyses indicate that no biases were encountered with respect to nucleotide substitutions, that SNPs identified in the data strongly correlate with common genetic variations, and that high quality variant calling was performed.
Table 1. Patient clinical information.
| Patient 1 | Patient 2 | Patient 3 | |
| Age at diagnosis (years) | 55 | 76 | 57 |
| Gender | male | female | male |
| Ethnicity | Caucasian | Caucasian | Caucasian |
| Diagnosis | adenocarcinoma w/liver metastases | adenocarcinoma w/no metastasis | adenocarcinoma w/liver metastases |
| Tumor stage | IV | IIB | IV |
| Tumor grade | poorly differentiated | moderately differentiated | poorly differentiated |
| Tumor content | 60% | 50% | 40–50% |
| Sequenced biopsy | liver metastasis | primary tumor | liver metastasis |
| Clinical status | Received treatmenta: deceased | Did not receive treatment: no recurrence after 24 months | Received treatmentb: deceased |
Clinical benefit with FOLFOX (folinic acid, fluorouracil, oxaliplatin) systemic therapy for 24 weeks with 98% maximal serum CA19-9 reduction and partial metabolic response by EORTC PET criteria.
Transient clinical benefit with FOLFOX systemic therapy for 10 weeks with maximal serum CA19-9 reduction of 36% and RECIST (Response Evaluation Criteria in Solid Tumors) reduction of 21% in sum of largest diameters.
Table 2. WGS and RNAseq metrics.
| WGS metrics | Patient 1 | Patient 2 | Patient 3 | Normal human pancreas | |
| Total amount of data generated (GB) | 271.75 | 315.80 | 420.3 | - | |
| Q30 data generated (GB) | 207.80 | 237.67 | 352.7 | - | |
| Average Total cluster densities (K/mm2) | 381.36 | 681.31 | 569.21 | - | |
| Average PF cluster densities (K/mm2) | 348.80 | 521.76 | 466.84 | - | |
| Average PF rate | 76.43 | 76.61 | 83.4 | - | |
| Total number of reads | 2256848363 | 2767484751 | 2878046795 | - | |
| Aligned Reads - Normal | 1052366015 | 1441444310 | 1271057635 | - | |
| Aligned Reads - Tumor | 1204482348 | 1326040441 | 1606989160 | - | |
| Aligned Bases - Normal | 96863052455 | 1.4991E+11 | 1.3219E+11 | - | |
| Aligned Bases - Tumor | 1.11483E+11 | 1.37908E+11 | 1.67127E+11 | - | |
| Average coverage depth - Normal | 31.31 | 48.46 | 42.73 | - | |
| Average coverage depth - Tumor | 36.04 | 44.58 | 54.03 | - | |
| Variant Analysis | BWA | BWA | BWA | - | |
| Germline SNPs called | 2013281 | 2129857 | 3610297 | - | |
| Transition/Transversion Ratio | 2.24 | 2.17 | 2.01 | - | |
| dbSNP 129 rate | 87.59 | 87.65 | 87.29 | - | |
| Non-synonymous germline variants | 504 | 10151 | 12830 | - | |
| Somatic SNVs called (strict lists) | 20323 | 714 | 25 | - | |
| False Positives (in dbSNP or 1000 Genomes) (strict lists) | 0.107 | 0.41 | 0.36 | - | |
| Somatic indels called (CODING and UTR) | 8 | 5 | 3 | - | |
| RNAseq metrics | |||||
| Total amount of data generated (GB) | - | 25.7 | 22.4 | 14.8 | |
| Q30 data generated (GB) | - | 21.1 | 18.4 | 12.2 | |
| Average Total cluster densities (K/mm2) | - | 810.0 | 694.0 | 1063.0 | |
| Average PF cluster densities (K/mm2) | - | 680.4 | 589.2 | 533.6 | |
| Average PF rate | - | 84.0 | 84.9 | 50.2 | |
| Total number of reads | - | 272694175 | 247382440 | 377376444 | |
| Total mapped reads | - | 124914613 | 104693716 | 98290756 |
Variant Analysis
Aligned reads for both tumor and normal libraries were evaluated to identify genomic events including non-synonymous SNVs (nsSNVs), indels, and copy number variants (CNVs). Summaries of identified variants in each patient are shown in Figures 1, 2, and 3. Across all 3 patients, 142 coding genomic events were identified. A total of 101 events were identified in patient 1, 17 in patient 2, and 24 in patient 3. Of these events, we identified 11 indels (Table 3), 69 nsSNVs (Table 3), and 62 focal/chromosomal CNVs (Table 4). These 62 CNVs encompass approximately 4,576 genes across all 3 patients. 119 COSMIC (Catalogue of Somatic Mutations in Cancer) genes that fall within these CNV regions are listed in Table 4. Using CREST, we did not identify any significant somatic structural variants in the 3 patients.
Table 3. Indels and SNVs identified through WGS.
| Patient | Chr. | Location | Gene Name | Coding event | Alteration | Sequence Change | Effecta |
| 1 | 13 | 31805365 | BRCA2 | Indel | deletion | AAAAG | NMDb; frameshift |
| 1 | 1 | 86818484 | CLCA4 | Indel | deletion | CCTACA | no NMD |
| 1 | 1 | 52078651 | NRD1 | Indel | deletion | TCT | no NMD |
| 1 | 9 | 124313206 | OR1J2 | Indel | insertion | T | NMD unknown; frameshift |
| 1 | 8 | 8272117 | SGK223 | Indel | insertion | G | NMD; frameshift |
| 1 | 19 | 57578957 | ZNF880 | Indel | deletion | A | NMD; frameshift |
| 1 | 17 | 7518264 | TP53 | SNV | R248W | G/A | damaging |
| 1 | 12 | 25289551 | KRAS | SNV | G12V | C/A | damaging |
| 1 | 4 | 55642955 | KDR | SNV | T1258M | G/A | damaging |
| 1 | 3 | 131766828 | COL6A6 | SNV | S321N | G/A | tolerated |
| 1 | 4 | 185938530 | ACSL1 | SNV | K143X | T/A | termination |
| 1 | 11 | 129794256 | ADAMTS8 | SNV | L288F | G/A | damaging |
| 1 | 11 | 94172689 | AMOTL1 | SNV | R229X | C/T | termination |
| 1 | 4 | 114415228 | ANK2 | SNV | G553R | G/A | damaging |
| 1 | 10 | 28312788 | ARMC4 | SNV | F270Y | A/T | tolerated |
| 1 | 17 | 44591549 | B4GALNT2 | SNV | T217M | C/T | damaging |
| 1 | 7 | 33976531 | BMPER | SNV | W123X | G/A | termination |
| 1 | 4 | 24419470 | CCDC149 | SNV | A336G | G/C | damaging |
| 1 | 17 | 42569603 | CDC27 | SNV | H615Q | A/T | damaging |
| 1 | 16 | 79619379 | CENPN | SNV | A305P | G/C | damaging |
| 1 | 5 | 1387414 | CLPTM1L | SNV | A294V | G/A | damaging |
| 1 | 5 | 156718707 | CYFIP2 | SNV | R232M | G/T | tolerated |
| 1 | 1 | 55090513 | DHCR24 | SNV | C511F | C/A | damaging |
| 1 | 11 | 117156429 | DSCAML1 | SNV | R118H | C/T | tolerated |
| 1 | 19 | 48702891 | ETHE1 | SNV | F239S | A/G | tolerated |
| 1 | 4 | 126592198 | FAT4 | SNV | L1824S | T/C | tolerated |
| 1 | 2 | 169467273 | G6PC2 | SNV | C97X | C/A | termination |
| 1 | 4 | 144580767 | GAB1 | SNV | P456Q | C/A | damaging |
| 1 | 4 | 90388090 | GPRIN3 | SNV | R732L | C/A | damaging |
| 1 | 1 | 67628381 | IL12RB2 | SNV | K676N | G/T | damaging |
| 1 | 15 | 72213782 | ISLR2 | SNV | R545H | G/A | damaging |
| 1 | X | 48707520 | KCND1 | SNV | R158H | C/T | damaging |
| 1 | 6 | 24664901 | KIAA0319 | SNV | D924N | C/T | tolerated |
| 1 | 19 | 59437727 | LILRA6 | SNV | E114D | C/A | tolerated |
| 1 | 4 | 88985483 | MEPE | SNV | I147F | A/T | damaging |
| 1 | 7 | 141354973 | MGAM | SNV | K109M | A/T | damaging |
| 1 | 12 | 47726693 | MLL2 | SNV | L1462F | G/A | no prediction |
| 1 | 22 | 24494160 | MYO18B | SNV | D95H | G/C | tolerated |
| 1 | 11 | 112610991 | NCAM1 | SNV | V8M | G/A | tolerated |
| 1 | 4 | 96980836 | PDHA2 | SNV | L171P | T/C | damaging |
| 1 | X | 24816075 | POLA1 | SNV | R1360H | G/A | tolerated |
| 1 | 12 | 10926455 | PRH1 | SNV | Q70H | C/A | damaging |
| 1 | 9 | 134965563 | RALGDS | SNV | V773I | C/T | damaging |
| 1 | 7 | 5658630 | RNF216 | SNV | H643P | T/G | tolerated |
| 1 | 8 | 10505716 | RP1L1 | SNV | P1101H | G/T | damaging |
| 1 | 11 | 57138427 | SERPING1 | SNV | P477T | C/A | tolerated |
| 1 | 4 | 975236 | SLC26A1 | SNV | Q86K | G/T | damaging |
| 1 | 5 | 150648782 | SLC36A3 | SNV | E89X | C/A | termination |
| 1 | 14 | 37748706 | SSTR1 | SNV | R121C | C/T | tolerated |
| 1 | 16 | 1068870 | SSTR5 | SNV | M1V | A/G | damaging |
| 1 | 9 | 129482338 | STXBP1 | SNV | V515I | G/A | tolerated |
| 1 | 3 | 33170461 | SUSD5 | SNV | L223V | G/C | tolerated |
| 1 | 16 | 19359304 | TMC5 | SNV | G148V | G/T | damaging |
| 1 | 3 | 113263420 | TMPRSS7 | SNV | K343N | G/T | tolerated |
| 1 | 1 | 173638901 | TNR | SNV | A325E | G/T | tolerated |
| 1 | 19 | 59634093 | TTYH1 | SNV | P346L | C/T | tolerated |
| 1 | 18 | 72721138 | ZNF236 | SNV | V354L | G/T | tolerated |
| 1 | 16 | 4755940 | ZNF500 | SNV | E14G | T/C | tolerated |
| 2 | 4 | 88756318 | DSPP | Indel | deletion | GACAGCAGC | no NMD; frameshift |
| 2 | 2 | 153184312 | FMNL2 | Indel | insertion | CCA | no NMD |
| 2 | 2 | 233420470 | GIGYF2 | Indel | deletion | ACA | NMD; frameshift |
| 2 | 8 | 89150850 | MMP16 | Indel | insertion | A | NMD unknown; frameshift |
| 2 | 12 | 25289551 | KRAS | SNV | G12V | C/A | damaging |
| 2 | 19 | 2242562 | LINGO3 | SNV | G72S | C/T | tolerated |
| 2 | 17 | 1508349 | PRPF8 | SNV | F1818C | A/C | damaging |
| 2 | 21 | 43397525 | U2AF1 | SNV | S34F | G/A | damaging |
| 3 | 19 | 51042923 | SYMPK | Indel | deletion | GA | no NMD |
| 3 | 12 | 25289552 | KRAS | SNV | G12R | C/G | damaging |
| 3 | 17 | 47065916 | CA10 | SNV | R295H | C/T | damaging |
| 3 | 1 | 156418469 | CD1D | SNV | A118T | G/A | tolerated |
| 3 | 16 | 65505678 | CDH16 | SNV | L241P | A/G | tolerated |
| 3 | 10 | 135290149 | FRG2B | SNV | T30S | T/A | tolerated |
| 3 | 19 | 55006332 | FUZ | SNV | L198F | G/A | possibly damaging |
| 3 | 7 | 142361153 | KEL | SNV | A313T | C/T | tolerated |
| 3 | 14 | 23954072 | KIAA1305 | SNV | A1093T | G/A | damaging |
| 3 | 14 | 76650235 | KIAA1737 | SNV | R341C | C/T | damaging |
| 3 | 1 | 70277766 | LRRC7 | SNV | A1191V | C/T | damaging |
| 3 | 7 | 4251904 | SDK1 | SNV | A2108T | G/A | tolerated |
| 3 | 1 | 12300948 | VPS13D | SNV | E2461K | G/A | tolerated |
| 3 | 19 | 58772525 | ZNF331 | SNV | E300A | A/C | damaging |
Effects were determined using SIFT/Polyphen-2.
NMD = nonsense mediated decay.
Table 4. Copy number changes identified through WGS.
| Patient | Chromosome | CNVa | Physical Position (Mb) | Patient | Chromosome | CNV1 | Physical Position (Mb) |
| 1 | 1p | Loss | 0.8–29.0 | 1 | 13q | Focal Loss | 18.6–20.7 |
| 1 | 1q | Focal Gain | 143.7–144.0 | 1 | 13q | Loss | 25.2–87.2 |
| 1 | 20p | Loss | 0.2–18.8 | 1 | 13q | Focal Loss | 111.7–114.2 |
| 1 | 21p | Focal Gain | 9.9 | 1 | 14q | Loss | 41.4–73.3 |
| 1 | 21q | Loss | 13.9–46.9 | 1 | 15q | Focal Gain | 19.3 |
| 1 | 22q | Focal Loss | 15.4–16.7 | 1 | 16p | Focal Loss | 0.5–1.3 |
| 1 | 2p | Loss | 17.6–63.3 | 1 | 16q | Focal Gain | 69.7 |
| 1 | 2q | Loss | 189.0–242.5 | 1 | 17p | Loss | 0.06–21.2 |
| 1 | 3p | Loss | 38.5–77.2 | 1 | 18p | Loss | 3.2–10.7 |
| 1 | 3q | Gain | 162.1–175.5 | 1 | 18q | Focal Loss | 71.0–76.0 |
| 1 | 4p | Loss | 0.3–20.7 | 1 | 19p | Loss | 0.2–24.1 |
| 1 | 4q | Loss | 184.0–189.4 | 1 | 19q | Loss | 34.3–59.4 |
| 1 | 5q | Loss | 52.9–133.8 | 2 | 1p | Loss | 53.3–115.0 |
| 1 | 5q | Focal Loss | 69.3–70.4 | 2 | 1q | Loss | 177.8–198.4 |
| 1 | 5q | Focal Loss | 118.3–119.0 | 2 | 3q | Focal Gain | 106.7–107.0 |
| 1 | 6p | Focal Loss | 32.1–32.1 | 2 | 5p | Focal Gain | 1.3 |
| 1 | 6q | Loss | 57.1–134.6 | 2 | 5p | Gain | 31.5–50.8 |
| 1 | 6q | Loss | 154.4–170.8 | 2 | 8q | Focal Gain | 131.2–135.7 |
| 1 | 6q | Focal Loss | 157.6–158.0 | 2 | 15q | Focal Gain | 19.8–19.9 |
| 1 | 6q | Focal Loss | 167.9–168.0 | 2 | 17p | Focal Loss | 0.09 |
| 1 | 7p | Loss | 0.5–6.0 | 2 | 18p | Gain | 9.1–14.2 |
| 1 | 7q | Focal Loss | 74.1 | 3 | 1p | Focal Loss | 1.1–3.6 |
| 1 | 8p | Focal Loss | 21.9–30.1 | 3 | 1p/q | Gain | 120.0–143.7 |
| 1 | 8q | Gain | 100.8–146.3 | 3 | 3q | Focal Loss | 121.8–121.9 |
| 1 | 9p | Loss | 0.3–27.5 | 3 | 4p | Focal Loss | 1.7–3.4 |
| 1 | 9p | Focal Loss | 19.7–22.0 | 3 | 4q | Focal Loss | 69.1 |
| 1 | 10p | Loss | 0.2–22.4 | 3 | 5p | Focal Gain | 32.4 |
| 1 | 10q | Loss | 67.6–135.3 | 3 | 9q | Focal Loss | 136.3–138.4 |
| 1 | 11p | Loss | 0.2–36.3 | 3 | 12p | Focal Gain | 23.9–26.4 |
| 1 | 12q | Loss | 60.5–132.3 | 3 | 18q | Focal Loss | 74.8–75.3 |
Focal gains/losses are defined as CNVs occurring across regions that are < = 5 Mb.
Whole transcriptome sequencing
Whole transcriptome sequencing was performed for patients 2 and 3 and normal human pancreatic RNA (Materials and Methods). RNAseq was not performed for patient 1 because tumor RNA was not available. An average of 109 million mapped reads was generated across the 3 analyzed samples. Tumor RNAseq data was compared to normal human pancreatic RNAseq data to identify expression changes in the tumor biopsies. Whole transcriptome sequencing metrics are listed in Table 2.
RNA-seq Analysis
Overall, in patient 2, 1,841 genes showed significant expression changes (q<0.05, corrected for multiple testing), whereas in patient 3, 1,939 genes showed significant changes. From these two analyses, 877 common genes/isoforms were identified as showing significant expression changes. Genes demonstrating both CNVs and significant expression changes (in patients 2 and 3) are listed in Table S1. Putative fusion transcripts identified in patients 2 and 3 are listed in Table S2.
Patient 1 analysis
Whole genome analysis
Well-established genes implicated in PAC include BRCA2, TP53, CDKN2A (p16), MYC (v-myc myelocytomatosis viral oncogene homolog), SMAD4, and KRAS. Compared to the other sequenced patients, patient 1 harbored the majority of genomic events in these genes including a deletion within and CNV loss encompassing BRCA2, an SNV in TP53 (a nonsynonymous mutation along with 17p hemizygous loss of the wildtype allele), a homozygous deletion of the CDKN2A locus, and an interstitial 8q CNV gain encompassing MYC. The deletion identified in BRCA2 in patient 1 causes a frameshift and nonsense-mediated decay of the transcript, whereas SNVs identified in TP53, KRAS, and KDR are all associated with damaging effects on the coding product. The alterations that affect BRCA2 suggest that DNA repair mechanisms may be affected, thereby, providing an explanation for the high number of somatic aberrations identified in patient 1 compared to the other sequenced patient tumors. BRCA2 germline mutations, in addition to being associated with increased risk of breast and ovarian cancers [21], [22], [23] also occurs in a small subset of both familial pancreatic cancer cases [24], [25]. Although BRCA2 mutations have been identified in PAC, the deletion we identify here in exon 10 of BRCA2 has not been previously reported.
The R248 SNV identified in TP53 has been previously reported in multiple cancers [26], [27]. TP53 also fell within a region of CNV loss in patient 1. The missense mutation is predicted to be damaging and the SNV and CNV loss suggest that tumor suppressor activity of TP53 may be compromised. Furthermore, MDM2 (Mdm p53 binding protein homolog) demonstrated a CNV loss. MDM2 is involved in regulation of TP53 activity such that the cumulative effect of its CNV loss, along with the alterations identified in TP53, suggest that regulation of TP53 and TP53's normal functions are impacted. A homozygous deletion of CDKN2A was also identified to indicate that p16 tumor suppressor functions are likely compromised. CNV loss of CDKN2A has been previously reported in PAC [28]. Patient 1 also demonstrated a previously reported mutation in KRAS for which glycine (G) is converted to valine (V) at amino acid position 12 [29], [30], [31]. The SNV in KDR, which codes for a tyrosine kinase VEGF (vascular endothelial growth factor) receptor, has not been previously reported in PAC. These genomic events identified in KDR and KRAS may lead to dysregulation of signaling cascades upstream of tumor cell proliferation to help promote tumor growth.
Copy number gains encompassing MYC indicate that this gene is likely oncogenic in patient 1. Amplification of MYC has been reported in PAC [32], [33], and one study identified a positive correlation between MYC amplification and tumor grade but not survival [34]. Aside from commonly reported genes in PAC, APC (adenomatous polyposis coli), MAP2K4 (mitogen-activated protein kinase kinase 4), FHIT (fragile histidine triad), and AKT2 (v-akt murine thymoma viral oncogene homolog 2) also fell in regions of CNV loss. Mutations in APC have been reported in PAC [35], [36], [37], and APC copy number loss has been reported in colorectal cancer [38], [39] and gastric cancer [40]. Due to APC's function as a tumor suppressor, decreased copy number of this gene in patient 1 likely represents a key inactivating event in patient 1's cancer. Similar to APC, FHIT and MAP2K4, which both may act as tumor suppressors [41], , demonstrated copy number losses and may also represent inactivating aberrations. A copy number loss in FHIT has also been previously reported in PAC [28], and mutations in MAP2K4 have been identified in pancreatic and other cancers [41], [42]. Lastly, AKT2, a putative oncogene, has been reported to be amplified in pancreatic cancer [44].
Somatic CNV losses identified using WGS also encompassed RB1 (retinoblastoma 1), another tumor suppressor. Copy number losses in TP53, AKT2, APC, MAP2K4, and RB1 represent key events likely associated with tumor progression and growth in patient 1. Additional relevant genes that fell in CNV regions identified using WGS are listed in Table 4 and include PIK3R1 (phosphoinositide-3-kinase, regulatory subunit 1 (alpha)), MLLT3 (myeloid/lymphoid or mixed-lineage leukemia), FGFR2 (fibroblast growth factor receptor 2), ALK (anaplastic lymphoma receptor tyrosine kinase), EML4 (echinoderm microtubule associated protein like 4), and HRAS (v-Ha-ras Harvey rat sarcoma viral oncogene homolog), all of which demonstrated copy number losses and all of which have not been reported in PAC. A total of 43 regions demonstrating CNV alterations, and which encompassed 4,426 genes, were identified in patient 1. Structural variant analysis did not identify any aberrations in the tumor genome of patient 1.
We further performed aCGH analysis on patient 1's tumor and validated all CNVs described here (Table S3). aCGH analysis also identified biallelic deletion of NF2 (neurofibromin 2), a tumor suppressor gene, which was initially not reported due to CNV threshold cutoffs in the WGS analysis but which was subsequently confirmed in the whole genome sequence data. This gene has not been implicated in PAC but one study on pancreatic endocrine tumors localized tumor suppressor loci to regions that include NF2 [45].
Summary
Many patients who are treated with gemcitabine and 5-FU based treatments often fail and are thus interested in and positioned to try additional agents that might offer benefit. Knowledge of the specific mutations in a patient's cancer may indicate targetable drivers and an oncologist and physician may decide to empirically treat the tumor based off the hypothesis that targeting the mutant may offer benefit. Our WGS findings thus provide insight into potential therapeutic options as well as patients' responses to treatments. For patient 1, based off the deletion and copy number loss identified for BRCA2, potential therapies include platinum compounds (cisplatin/carboplatin), mitomycin C, or alkylators. Following the collection of the tumor biopsy for sequencing, patient 1 was treated with a platinum compound (oxaliplatin) as a part of FOLFOX (folinic acid, fluorouracil, oxaliplatin) treatment. Patient 1 showed a complete response, but subsequently developed resistance 6 months later. Furthermore, the copy number loss identified for AKT2 may be associated with patient 1's initial response to gemcitabine prior to biopsy as a recent study showed that AKT2 inhibition is associated with increased gemcitabine sensitivity [46]. Other studies also show that inhibition or silencing of AKT2 may block the growth of tumor cells and tumor formation [47], [48]. Patient 1's partial response was measured by EORTC PET (European Organisation for Research and Treatment of Cancer positron emission tomography) criteria along with normalization of CA19-9 (after six months, the cancer progressed and developed elevation in CA19-9). Overall, the BRCA2 deletion is likely the driving mutation in this patient as the loss of DNA repair functions permits the occurrence of mutations that, in this patient, affected numerous genes including tumor suppressors. Given this finding, the use of PARP (poly ADP ribose polymerase) inhibitors may have represented a viable therapeutic option. Lowery et al. reported treatments and responses of pancreatic cancer patients with BRCA mutations and demonstrated the utility of using PARP (poly (ADP-ribose) polymerase) inhibitors for these patients. This finding and association provides evidence of the utility of performing whole genome analyses of patients in order to identify less common mutations that may be relevant for therapeutic selection. Our identification of copy number losses in EML4 and ALK, as well as the absence of an EML4-ALK fusion, also provides evidence that crizotinib, an ALK inhibitor typically used to treat non-small cell lung cancer, would not be an option for this patient. Lastly, potential therapies that may be considered based on the KDR mutation include sunitinib, a tyrosine kinase inhibitor, and bevacizumab, which blocks the action of VEGFA (vascular endothelial growth factor A).
Patient 2 analysis
Whole genome analysis
Patient 2 did not harbor any events in BRCA2, TP53, CDKN2A, SMAD4, or MYC. Like patient 1, patient 2 also demonstrated a mutation in KRAS at the same position (G12V). Overall, patient 2 demonstrated much fewer genomic aberrations compared to patient 1 and did not demonstrate aberrations affecting DNA repair genes. Structural variant analysis using CREST did not identify any significant somatic events in patient 2.
Aside from U2AF1 (U2 small nuclear RNA auxiliary factor 1), SNVs and indels identified in patient 2 affect genes that have not been previously reported in PAC or COSMIC. U2AF1 functions as a part of the spliceosome and mutations in this gene have been identified in myeloid hematopoietic cancers including chronic myelomonocytic leukemia [49], [50]. The SNV in U2AF1 identified in patient 2 is predicted to be damaging such that proper splicing of transcripts may be affected. A nine base pair deletion, causing a frameshift, was identified in DSPP (dentin sialophosphoprotein), which has been reported in oral squamous cell carcinoma [51].This gene codes for tooth extracellular matrix proteins so its potential role in PAC is unclear. We identified a frameshift insertion in FMNL2 (formin-like 2), which normally functions to regulate processes requiring actin, including cytokinesis, invasion, and cell motility. Although FMNL2 mutations have not been reported in PAC, it may have roles in colorectal carcinoma [52], [53], [54] and hepatocellular carcinoma [55]. For GIGYF2 (GRB10 interacting GYF protein 2), we identified a frameshift deletion. GIGYF2 was shown to interact with RQCD1 (RCD1 required for cell differentiation1 homolog) and may be involved in regulating the activity of AKT in the EGFR pathway in breast cancer [56],[57]. Although GIGYF2 has not been described in PAC, the deletion and resulting frameshift in patient 2 may affect normal functions associated with AKT regulation. Interestingly, MMP16 (matrix metallopeptidase 16 (membrane-inserted)), which shows a single base insertion in patient 2, was previously found to be the target of a micro-RNA whose over-expression inhibited migration and invasion of the MIA PaCa-2 pancreatic cancer cell line [58]. This finding suggests that MMP16 may be involved with migration and invasion of pancreatic cancer cells. In a recent exome sequencing study of intraductal papillary mucinous neoplasms of the pancreas, PRPF8 (PRP8 pre-mRNA processing factor 8 homolog) was recently found to garner a mutation (A1842V) resulting from a SNV (C>T) [59]. This mutation differs from the SNV we identified in patient 2, and has not been reported in PAC or other cancers, but provides evidence of a potential role of this gene, which functions in pre-mRNA splicing, in PAC.
In patient 2, we identified 9 regions, covering 114 genes that demonstrate copy number alterations (Table 4). These regions encompass CBLB (Cbl proto-oncogene, E3 ubiquitin protein ligase B), IL7R (interleukin 7 receptor), LIFR (leukemia inhibitory factor receptor alpha), and NDRG1 (N-myc downstream regulated 1), all of which showed copy number gains. CBLB has not been implicated in PAC, but mutations in this gene have been identified in leukemias [60], [61]. IL7R also has not been previously reported in PAC, but has been found to demonstrate activating mutations in lymphoblastic leukemias [62], [63], [64]. LIFR has been reported in other malignancies including colorectal and hepatocellular carcinomas [65], [66], and is also suggested to have a role in tumor growth in pancreatic cancer [67]. Lastly, NDRG1 has not been reported in pancreatic cancer but is suggested to arrest metastasis in prostate and colon cancers [68], [69]and it was also shown that NDRG1 expression suppresses tumor cell growth [70].
Copy number validation was performed using flow sorted aCGH which involves flow sorting nuclei from the tumor biopsy to identify aneuploid populations. The sorted aneuploid population is then separately analyzed using aCGH. Using this analysis, we validated CNV gains identified using WGS in CBLB, IL7R, LIFR, and NDRG1 (Table S3).
Whole transcriptome analysis
1,841 genes demonstrating significant expression changes (q<0.05, corrected for multiple testing) in the tumor were identified. COSMIC genes demonstrating significant expression changes are listed in Table 5. Genes showing significantly altered expression in the tumor and that also fall in regions of copy number change are listed in Table S1. Putative fusion transcripts identified in patient 2, of which 2 contributing genes showed significantly altered expression, are listed in Table S2. As structural variant analysis did not detect significant somatic aberrations, the fusion transcripts detected in patient 2 are not correlated with genomic data.
Table 5. Selecteda differentially expressed genes identified using RNAseqb.
| Patient | Gene | ln (fold change) | q-value (corrected) | Patient | Gene | ln (fold change) | q-value (corrected) |
| 2 | AKT3 | −4.37 | 2.92E-03 | 3 | ABL1 | 5.51 | 5.50E-03 |
| 2 | ATM | −3.84 | 2.25E-02 | 3 | AKT2 | 4.42 | 4.52E-02 |
| 2 | ATRX | −3.19 | 4.89E-02 | 3 | ATRX | −5.32 | 3.12E-05 |
| 2 | ATRX | −5.68 | 5.53E-06 | 3 | BCL3 | 4.43 | 4.40E-03 |
| 2 | BCL3 | 4.42 | 4.77E-03 | 3 | BCL3 | 3.93 | 1.40E-02 |
| 2 | BCL3 | 4.25 | 1.05E-02 | 3 | BIRC3 | 6.64 | 2.06E-07 |
| 2 | BIRC3 | 5.86 | 1.67E-05 | 3 | BRCA1 | 5.22 | 2.58E-04 |
| 2 | BRCA2 | 3.48 | 2.31E-02 | 3 | CDH1 | −3.38 | 1.93E-02 |
| 2 | CBLB | 4.54 | 1.74E-03 | 3 | CDH11 | 5.94 | 1.10E-03 |
| 2 | COL1A1 | 3.42 | 3.88E-02 | 3 | CREBBP | 3.94 | 4.65E-03 |
| 2 | CREB1 | 3.75 | 1.79E-02 | 3 | DNM2 | −3.27 | 4.73E-02 |
| 2 | ERBB2 | 3.96 | 1.39E-02 | 3 | EML4 | −3.26 | 1.87E-02 |
| 2 | ERBB4 | −5.98 | 4.53E-08 | 3 | ERCC4 | −4.05 | 2.27E-03 |
| 2 | ERCC4 | −5.50 | 1.37E-05 | 3 | FGFR1 | −2.98 | 4.97E-02 |
| 2 | FGFR1 | −4.56 | 1.17E-02 | 3 | FSTL3 | 4.23 | 5.35E-03 |
| 2 | FGFR1 | −6.44 | 4.06E-02 | 3 | GOLGA5 | −2.82 | 4.28E-02 |
| 2 | FLT3 | −4.82 | 3.02E-03 | 3 | HERPUD1 | −3.21 | 1.67E-02 |
| 2 | FLT3 | −4.94 | 2.17E-03 | 3 | IL7R | −4.38 | 2.09E-03 |
| 2 | FOXP1 | −3.23 | 3.85E-02 | 3 | KRAS | 4.35 | 1.61E-03 |
| 2 | FUS | 3.89 | 1.16E-02 | 3 | MAML2 | 3.47 | 3.65E-02 |
| 2 | GNAS | −3.81 | 5.77E-03 | 3 | MDM4 | −3.22 | 2.03E-02 |
| 2 | GSK3B | 3.33 | 1.60E-03 | 3 | MLH1 | 4.12 | 6.01E-03 |
| 2 | HERPUD1 | −4.86 | 1.40E-04 | 3 | MLL3 | 2.99 | 4.69E-02 |
| 2 | KTN1 | −3.84 | 5.91E-03 | 3 | MLL3 | −3.13 | 1.86E-02 |
| 2 | MAML2 | 3.45 | 4.62E-02 | 3 | MLLT6 | 3.08 | 3.75E-02 |
| 2 | MLL3 | 3.46 | 1.77E-02 | 3 | NDRG1 | 4.05 | 3.35E-03 |
| 2 | MLL3 | −3.47 | 6.01E-03 | 3 | NFIIB | −3.10 | 3.29E-02 |
| 2 | NDRG1 | 5.71 | 2.73E-05 | 3 | NOTCH2 | 4.12 | 7.57E-03 |
| 2 | NFIIB | −3.23 | 2.54E-02 | 3 | PALB2 | 4.41 | 5.20E-03 |
| 2 | NFIIB | −3.43 | 1.17E-02 | 3 | PICALM | 3.28 | 3.40E-02 |
| 2 | PIM1 | 4.90 | 1.42E-03 | 3 | PIM1 | 3.84 | 2.57E-02 |
| 2 | PPARG | 3.82 | 3.95E-02 | 3 | PPARG | 4.44 | 2.54E-02 |
| 2 | PRDM1 | 4.36 | 3.80E-03 | 3 | REG4 | 6.92 | 2.04E-07 |
| 2 | PRDM1 | 3.66 | 2.19E-02 | 3 | REG4 | 6.49 | 5.05E-07 |
| 2 | REG4 | 4.57 | 1.85E-03 | 3 | REG4 | 4.77 | 3.84E-04 |
| 2 | REG4 | 3.43 | 1.76E-02 | 3 | RUNX1 | 3.61 | 8.25E-03 |
| 2 | RUNX1 | 3.92 | 6.18E-03 | 3 | TOP2A | 8.99 | 2.97E-04 |
| 2 | SET | 4.04 | 6.85E-03 | 3 | TOP2A | 8.30 | 1.24E-03 |
| 2 | SOX2 | 3.57 | 2.75E-02 | 3 | TPM4 | 3.99 | 8.54E-03 |
| 2 | TFPT | 3.87 | 1.21E-02 | ||||
| 2 | TFRC | 4.71 | 6.57E-04 | ||||
| 2 | TOP2A | 9.15 | 2.05E-04 | ||||
| 2 | TP53 | 4.24 | 5.10E-03 | ||||
| 2 | TPM4 | 4.24 | 3.63E-03 | ||||
| 2 | ZNF384 | −5.56 | 1.57E-05 |
Selected genes are genes that are reported in COSMIC.
RNAseq was performed on patients 2 and 3.
Transcriptomic analysis led to the identification of significantly altered expression of genes that have been previously implicated in cancer. Significantly up-regulated genes in the tumor include GSK3β (glycogen synthase kinase 3 beta), BRCA2, TP53, TOP2A (topoisomerase II alpha 170 kDa), BCL3 (B-cell CLL/lymphoma 3), and REG4 (regenerating islet-derived family, member 4). Mutations in GSK3β have not been reported in PAC, but its increased expression in patient 2 may have a role in contributing to tumor malignancy and proliferation through SEMA3A [71]. TOP2A and REG4 have been previously found to be associated with pancreatic cancer [70], [71], whereas BCL3 has been found to be associated with other cancers [72], [73]. Additional up-regulated genes that fall in the COSMIC database include MLL3 (myeloid/lymphoid or mixed-lineage leukemia 3), BIRC3 (baculoviral IAP repeat containing 3), ERBB2/HER2 (v-erb-b2 erythroblastic leukemia viral oncogene homolog 2), PPARG (peroxisome proliferator-activated receptor gamma), and CBLB, for which we also identified a copy number change. Mutations in MLL3 have been previously identified in pancreatic cancer [72], [73], and MLL3 was also identified as a candidate pancreatic cancer gene using a mutagenic screen in mice [74]. BIRC3, which acts to block apoptosis, was also previously reported to show increased expression in pancreatic cancer [75] and was found to be amplified in 22 pancreatic cancer cell lines [76]. ERBB2/HER2, an EGFR family tyrosine kinase that is involved in cell proliferation, has been frequently reported as demonstrating increased expression in pancreatic cancer [77], [78]. PPARG over-expression has also been previously identified in PAC and its over-expression was also found to be correlated with shorter survival [79]. Interestingly, inhibition of PPARG has been shown to block liver metastasis in a xenograft mouse model and motility of pancreatic cancer cells in vitro [80], and may thus represent a therapeutic target.
Down-regulated genes include ERBB4 (v-erb-a erythroblastic leukemia viral oncogene homolog 4), ERCC4 (excision repair cross-complementing rodent repair deficiency, complementation group 4), and FGFR1 (fibroblast growth factor receptor 1). ERCC4 has been reported to possibly be associated with risk of developing PAC [74], whereas FGFR1 has been implicated in lung cancer [75], [76] and bladder carcinoma [77]. Decreased expression of ERBB4 has been found in non-metastatic pancreatic cancer [81] and was reported to potentially influence metastasis of pancreatic cancer cells [82].
Of the fusion transcripts identified in patient 2, 2 genes that were identified as part of fusions also demonstrated statistically significant expression changes (q-value<0.05, corrected; Table S2). These genes include LMO2 (LIM domain only 2 (rhombotin-like 1)) and BACH1 (BTB and CNC homology 1, basic leucine zipper transcription factor 1), which were both identified in 1 putative fusion each. LMO2 was identified as the 5′ gene in an interchromosomal fusion with ACVR2A (activin A receptor, type IIA). Interestingly, LMO2 has been implicated in B-cell lymphoma [83] and prostate cancer [84] and is proposed to be a prognostic marker of longer survival in pancreatic cancer based on expression and immunohistochemical analyses [85]. Furthermore, a mutagenic screen aimed at identifying candidate pancreatic cancer genes led to the identification of point mutations in ACVR2A, in addition to other genes [74]. While only 2 non-junction-spanning reads support this chimera, this transcript may have relevant implications in patient 2's disease. BACH1 was identified as the 3′ gene in an intrachromosomal fusion with C21Orf109 (LINC00189; long intergenic non-protein coding RNA 189). 18 reads spanned the fusion junction to demonstrate increased confidence in this fusion. BACH1 has been found to bind and inhibit TP53 such that its increased expression [86] and potential transcript fusion in patient 2 may influence tumor suppressor functions of TP53. While the exact function of C21orf109 is unknown, long non-coding RNAs are known for their roles in transcriptional regulation. The putative chimera reported here may thus affect normal functions of this transcript and of BACH1. 2 additional predicted fusions were identified (FAM18B2-CDRT4 and SLC35A3-HIAT1) with 17 and 15 reads spanning the junctions, respectively, but none of these genes have been reported in PAC or other cancers.
Summary
Following resection of the tumor, patient 2 was treated with chemoradiation followed by gemcitabine and erlotinib, and at 16 months post-resection, has not experienced a recurrence. The absence of somatic events affecting DNA repair genes and genes including BRCA2, TP53, CDKN2A, SMAD4, and MYC, as well as increased expression of BRCA2 and TP53 in the tumor, may all contribute to the status of this patient. If a recurrence were to occur, the increased expression of TOP2A indicates that topoisomerase inhibitors may be a possible treatment option. While additional studies are needed, up-regulated expression of BIRC3 may provide evidence that sorafenib, a small molecule inhibitor of tyrosine and RAF kinases, and TRAIL (tumor necrosis factor-related apoptosis inducing ligand) may represent possible therapeutic options. Ricci et al. found that sorafenib down-regulates BIRC3 and MCL1 (myeloid cell leukemia sequence 1) expression and in doing so, causes TRAIL-resistant colon cancer cells to become sensitive to TRAIL, which promotes apoptosis [87] (we did not however identify statistically significant expression changes for MCL1 and TRAIL). Lastly, the identification of over-expression of ERBB2/HER2 provides evidence that trastuzumab, a monoclonal antibody that interferes with signaling through ERBB2/HER2, and/or lapatinib, which blocks ERBB1/EGFR and ERBB2 to obstruct cell growth and division, may be possible treatment options. The combined use of cetuximab and trastuzumab was found to be more beneficial than gemcitabine with regards to regression and survival when treating human pancreatic cancer xenografts [88]. Another study showed that a combined treatment of trastuzumab and matuzumab (anti-EGFR monoclonal antibody) on human pancreatic cancer xenografts demonstrated therapeutic benefit [89], whereas the use of multiple anti-ERBB2 antibodies targeting different ERBB2 epitopes also showed therapeutic benefit in mice [90].
Patient 3 analysis
Whole genome analysis
Patient 3 did not harbor any events in BRCA2, TP53, CDKN2A, SMAD4, or MYC. However, patient 3 demonstrated a KRAS mutation for which glycine (G) is converted to arginine (R) at amino acid position 12 and also showed a somatic CNV gain of 1.38 (log2 scale) in KRAS. The missense G12R mutation has been reported in pancreatic cancer [91], [92] and other cancers [30], [93]. Outside of KRAS, we identified 13 additional SNVs and indels, of which 7 are predicted to be damaging or potentially damaging (Table 3).
FUZ (fuzzy homolog (Drosophila)), KIAA1305 (NYNRIN; NYN domain and retroviral integrase containing), and KIAA1737 (uncharacterized) have not been implicated in any cancers. CA10 has been reported in chondroblastoma [94] and was identified as a putative methylation marker in bladder cancer [95], but has not been reported in PAC. ZNF331 (zinc finger protein 331) may have a role in follicular thyroid adenomas [96], and has also been implicated as a potential tumor suppressor in gastric cancer [97]. Because the SNV identified in ZNF331 is predicted to be damaging, its putative role as a tumor suppressor may represent a key event in this patient. Lastly, mutations in LRRC7 (leucine rich repeat containing 7) have been identified in multiple cancers, including skin, ovarian, and breast cancer, but not in PAC.
Overall, CNV analysis of patient 3 led to the identification of 10 regions, covering 34 genes that demonstrated CNV alterations (COSMIC genes falling within these regions are listed in Table 4). Aside from a gain in KRAS, other key affected genes include NOTCH2 (notch homolog 2), which also showed CNV gains, PDE4DIP (phosphodiesterase 4D interacting protein; myomegalin), and FGFR3 (fibroblast growth factor receptor 3) and MLLT4/AF6 (myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4), which both showed CNV losses. Interestingly, one animal study showed that KRAS(G12D)/NOTCH2 knockout mice survived longer and demonstrated no progression of pancreatic intraepithelial neoplasms (PanINs) compared to KRAS(G12D) and KRAS (G12D)/NOTCH1 knockout mice, thereby, showing that NOTCH2 may have a significant role in tumor malignancy and development [98]. PDE4DIP and FGFR3 have not been reported in PAC but PDE4DIP was identified as a tumor marker for esophageal squamous cell carcinoma [99], and mutations in FGFR3 have been found in pancreatic endocrine tumors [100] as well as bladder cancer [101], [102]. Lastly, MLLT4 has also not been reported in PAC but down-regulated expression of this gene is reported to be associated with increased likelihood of relapse in 14.5 to 15% of breast carcinoma cases as well as unfavorable prognosis [103], [104]. As previously mentioned, no significant somatic structural variants were identified for patient 3's tumor.
Whole transcriptome analysis
In patient 3, 1,939 genes were found to demonstrate significant expression changes (q<0.05, corrected for multiple testing) in the tumor. Selected genes are listed in Table 5 and genes that demonstrated both copy number changes and significant expression changes are listed in Table S1. Fusion transcripts identified in patient 3 are listed in Table S2. Like patient 2, somatic translocations were not identified so the fusion transcripts detected in patient 3 do not directly correlate with the tumor genome sequence.
Similar to patient 2, significant up-regulated expression was identified for TOP2A, BCL3, BIRC3, MLL3, PPARG, and REG4, and down-regulated expression was identified for ERBB4, ERCC4, and FGFR1. Patient 3's biopsy also demonstrated increased expression of PTCH1 (patched 1), BRCA1 (breast cancer 1, early onset), DNM2 (dynamin 2), MDM4 (p53 binding protein homolog (mouse)), NOTCH2, and KRAS. Although mutations in PTCH1, a tumor suppressor, have not been reported in PAC, its increased expression may influence tumor proliferation through the Sonic hedgehog pathway [105]. Up-regulated BRCA1 expression suggests that patient 3's tumor may boast increased genomic stability; such an increase in expression has also been identified in putative tumor-initiating cells isolated from multiple pancreatic cancer cell lines compared to bulk cells [106]. Interestingly, up-regulated expression of DNM2 has been reported in pancreatic cancer and was shown to be associated with increased tumor cell migration and invasion in human pancreatic cancer cells in vitro [107], and may thus represent a new therapeutic target for PAC. MDM4 normally acts to block TP53's tumor suppressor functions such that its increased expression in patient 3's tumor may be a key malignant event in this patient. Increased expression of MDM4 has been identified in a number of other cancers that have wild type p53, including head and neck squamous carcinoma [108], breast cancer, and lymphoblastic leukemia [109]. One study described MDM4 as an oncogene upon identifying the development of spontaneous tumors in conditional transgenic mice overexpressing MDM4, along with an increase in tumorigenesis in offspring when these mice were crossed with TP53+/− mice [110]. While no mutations were identified in TP53 for this patient, up-regulated expression of MDM4 and the other genes described here provide valuable information for the identification of new therapeutic targets and also provide insight on the biological processes that are occurring within the tumor.
In patient 3, we detected putative fusion transcripts supported by the identification of reads spanning the transcript breakpoint (Table S2). With the exception of CAV1 (caveolin-1), the genes identified in these fusions have not been reported in PAC. Over expression of CAV1, which also demonstrated increased expression (q<0.05, corrected) in our study, has been found to be associated with disease recurrence in pancreatic cancer patients [111]. Its increased expression and potential role in an interchromosomal fusion transcript in our results indicates that these events may influence tumor progression in this patient. Additional fusions, that did not harbor junction-spanning reads, but have been previously reported in PAC, were also identified. BCL3, which we've previously described in this patient, was detected as the 3′ gene in a fusion with PHLPPL (PH domain and leucine rich repeat protein phosphatase 2). Another gene that was identified in our transcriptomic analyses in this patient and that was found to be a fusion gene is REG4, for which multiple fusions were predicted. These fusions include REG4-SLC23A2 (solute carrier family 23 (nucleobase transporters), member 2) and REG4-LARP1 (La ribonucleoprotein domain family, member 1). Although the 3′ genes in these fusions have not been reported in PAC, an in vitro study showed that LARP1 may have a key role in cell migration [112]. Other genes of interest that were found in separate putative fusions include MAP4K4 (mitogen-activated protein kinase kinase kinase kinase 4), S100A4 (S100 calcium binding protein A4), MMP7 (matrix metallopeptidase 7), and IER3/IEX1 (immediate early response 3). MAP4K4 was detected as the 3′ gene in a fusion with APLP2 (amyloid beta (A4) precursor-like protein 2), which codes for a protein that was found in pancreatic cancer cell line supernatant [113]. While the effect of this putative fusion is unclear, MAP4K4 over expression, which was also identified here, was reported in stage II PAC patients and was found to be correlated with negative prognosis in these patients [114]. S100A4 was found to be in predicted intrachromosomal fusion with LZIC (leucine zipper and CTNNBIP1 domain containing). One study showed a relationship between S100A4 inhibition and increased gemcitabine sensitivity in PAC cell lines [115]. While implications for the predicted S100A4-LZIC fusion are not known, the increased expression of S100A4 that we identified in our RNAseq analyses indicates that this gene may be a relevant therapeutic target.
We also identified MMP7, which demonstrated significant increased expression in patient 3's tumor, as the 3′ gene in a putative fusion with EPHX1 (epoxide hydrolase 1, microsomal), which was shown to not play a role in pancreatic cancer [116]. Over expression of MMP7, which has roles in cell proliferation and differentiation, has been reported to be correlated with poor prognosis in PAC and tumor stage [117], [118], [119] and has also been reported specifically in liver metastases of pancreatic cancer [117]. Given these findings, the possible presence of the MMP7 fusion transcript may not significantly affect MMP7's normal functions given the diagnosis and outcome of patient 3. Another predicted chimera was an IER3-SERPINA6 (serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6) fusion—both genes in this fusion also showed statistically significant over expression in the tumor. While SERPINA6 has not been reported as having a role in PAC, studies have shown that IER3 expression is linked to both poor prognosis [120] and improved prognosis [121] in PAC patients. While additional experiments are necessary for clarifying the discrepancy across these findings, the presence of an IER3 fusion may have had implications on this patient's prognosis. Because the effect of the fusions detected here are unclear, additional sequencing and compilation of chimeric transcripts are needed so that we can begin to unveil the role of these species on pancreatic tumorigenesis.
Summary
Prior to biopsy, patient 3 was first treated with TH-302, an investigational drug that activates nitroazole under hypoxic conditions plus gemcitabine as part of a phase I clinical trial. He had transient clinical benefit at first but progressed and was then treated with gemcitabine and nab-paclitaxel, but the disease continued to progress. Our identification of a copy number gain in and increased expression of NOTCH2 may provide some explanation for the patient's responses to his first two treatments. Up-regulated expression of NOTCH2 was identified in gemcitabine-resistant pancreatic cancer cells to suggest its possible involvement in chemotherapy resistance [122]. Increased expression of NOTCH2 in patient 3 may be associated with disease progression following gemcitabine treatments. NOTCH2 inhibitors are thus a possible therapeutic option for this patient. A trial is currently recruiting stage IV pancreatic cancer patients, for whom tumor resection is not an option, to evaluate the efficacy of a combined therapy of MK0752, a NOTCH inhibitor, and gemcitabine hydrochloride. While additional analyses are needed, increased expression of S100A4 in patient 3 also suggests that this may be key target as S100A4 inhibition may be associated with increased sensitivity to gemcitabine. Another potential treatment is topoisomerase inhibitors given up-regulated TOP2A expression in the tumor. Similar to patient 2 and although additional studies are required, sorafenib and TRAIL may represent future options for patients whose tumors over express BIRC3. Lastly, increased expression of MDM4 in patient 3's tumor indicates that MDM4 inhibitors may also be a possible future option for patients. This option is preceded by Wang et al., who identified a benzofuroxan derivative that acts as an MDM4 inhibitor and showed that this small molecule inhibitor acts to promote apoptosis in a breast cancer cell line [123].
Pathway Analysis
While the goal of this study is to perform patient-specific analyses, we also performed pathway analysis across all patients to evaluate affected biological processes. This type of analysis is preceded by Jones et al. who performed whole genome and expression analyses on 24 pancreatic ductal adenocarcinoma cell lines and xenografts [72]. In this study, mutations, copy number changes, deletions, and expression changes were identified using targeted sequencing of exons, microarrays, and mRNA sequencing using SAGE (Serial Analysis of Gene Expression) tags. Using this approach, the authors identified 12 core signaling pathways for pancreatic cancer. For our analyses, results from WGS were integrated with RNAseq data to identify pathways that may be affected across all 3 patients. The 142 identified genomic events, including all genes falling in regions demonstrating CNVs, were evaluated alongside significant expression changes (q<0.05, corrected) in patients 2 and 3.
Using GeneGo's Metaminer Pancreatic Cancer Disease module, we evaluated the extent to which 21 annotated pancreatic cancer pathways are affected in the three patients (Table S4). The top pathway maps (minimum mapping p-value<0.05) that demonstrated the lowest probability of genes mapping to the specified map by chance are summarized in Figure 4. Genes demonstrating both mutations and expression changes in the top maps are listed in Table S5. As expected, integrated analysis of WGS and RNAseq data indicated that the most highly affected pathway is KRAS signaling in pancreatic cancer. Affected genes include those that solely demonstrate mutations or expression changes, as well as those that demonstrate both mutations and expression changes. Known cancer genes that fall in the KRAS signaling pathway and that demonstrated alterations include KRAS, TP53, MYC, PTEN (phosphatase and tensin homolog), and AKT2.
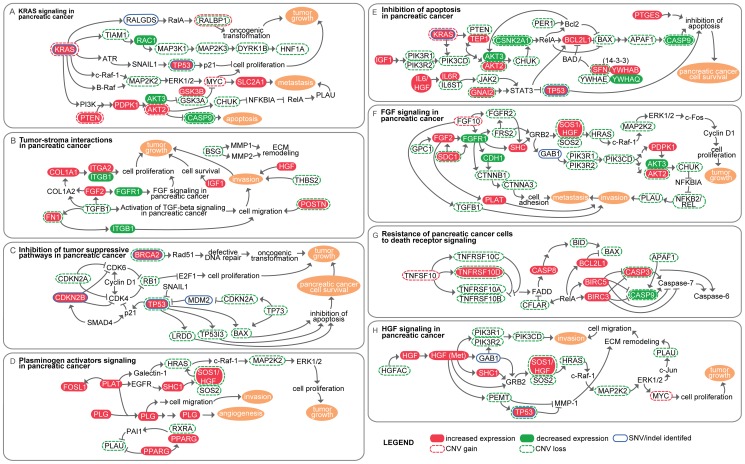
Figure 4. Pathway analysis of WGS and RNAseq results.
Whole genome and RNAseq data were integrated and analyzed using GeneGo's Metaminer Pancreatic Cancer Disease module to identify pathways that may be affected by mutations and/or significant expression changes (q-value<0.05, corrected). The top pathways (minimum mapping p-value across all WGS and RNAseq datasets <0.05) are summarized based off of GeneGo maps. Breakdown of affected pathways in each patient are shown in Table S4.
Genomic events and expression changes were also analyzed across the entire GeneGo pathway database in order to perform an unbiased global analysis and to identify processes that may not be captured in the Pancreatic Cancer Disease module. The top map categories that were identified as demonstrating the largest number of alterations include prostatic neoplasms, hepatocellular carcinoma, and pancreatic neoplasms (Table S4 and Figure 4). Additional map categories outline hallmark processes of cancer and tumorigenic pathways. Identification of pathways that are implicated in other cancers (prostate and liver) provides insight as to possibly novel interactions that are not reported or less common in pancreatic cancer. Consistent with pathway analysis against MetaMiner's Pancreatic Cancer Disease Module, the most highly affected pathway was KRAS signaling in pancreatic cancer (Figure 4), followed by ligand-independent activation of androgen receptor. The latter map is annotated with cell progression, cell proliferation, and survival pathways in prostate cancer, and may provide clues into processes that may drive tumorigenesis in PAC.
As expected, processes in the top pathways (minimum mapping p-value<0.05) identified through NGS analyses overlap with the 12 core signaling pathways previously reported by Jones et al., who also used GeneGo for pathway analysis. Overlapping processes and pathways include KRAS signaling, apoptosis, cell adhesion, and invasion. While apoptosis, cell adhesion, and invasion processes represent hallmark features of pancreatic cancer and other human cancers, the identification of KRAS signaling across multiple PAC samples, as well as the identification of previously reported KRAS mutations in the 3 patients analyzed here, emphasizes the tumorigenic role of KRAS signaling in pancreatic cancer. The high incidence of KRAS mutations in PAC [31], [124], along with the finding that patients who have a KRAS mutation have negative clinical outcomes when treated with a commonly prescribed combination of erlotinib, an EGFR inhibitor, and gemcitabine [125], further indicates that processes surrounding KRAS represent relevant therapeutic targets.
Although patient 1's tumor demonstrated the highest number of mutations, RNAseq data from patients 2 and 3 showed widespread pathway overlap with these genomic events. Patient 1 also uniquely harbors mutations that affect DNA repair pathways with respect to mismatch and nucleotide excision repair, DNA damage-induced responses, and BRCA1 as a transcription regulator. The larger number of identified genomic events in patient 1 may be associated with alterations in genes involved in DNA repair pathways and likely represents passenger mutations. Although tumors from patients 2 and 3 demonstrated fewer mutations, RNAseq data from these two patients suggest that common pathways are affected across all 3 patients.
Pathway analysis of WGS and RNAseq data allows us to understand which tumorigenic processes are present across the three patients. However, it is also important to recognize several caveats including: (1) mutations that are detected in larger genes (such as MAP2K4, NCAM1, LAMA1, and LAMC1, which are all over 100 kb) have a greater probability of representing a random mutation; the presence of such random events may bias pathway analysis; (2) alterations may influence additional key processes that are not annotated in the map database; (3) the tumor contents are 50% and 40 to 50% for patients 2 and 3, respectively, so that smaller, but potentially important, expression changes in tumor cells may not be readily identifiable; and (4) patient 2 was chemotherapeutically naïve and had her primary tumor sequenced whereas patients 1 and 3 were treated prior to biopsy collection and had their metastases sequenced. Despite these differences, pathway analysis allows us to evaluate commonly affected pathways across all 3 patients. KRAS is the only gene that harbored mutations (SNVs and a CNV) across all three patients and that also demonstrated a focal CNV gain and significant increased expression in patient 3.
Conclusion
Due to the lack of effectiveness of current treatments for PAC patients, we are tasked with improving our understanding of genomic aberrations and processes that drive PAC tumorigenesis, tumor progression, and malignancy in order to identify and develop efficacious treatments. Our approach involves individually characterizing patients to fully understand the range of molecular events associated with this disease. In this study, we report our findings of 3 individual genomic characterizations of tumors collected from 3 separate patients. In 2 of the 3 patients, we additionally performed RNAseq on the same whole genome sequenced biopsies to identify significant expression changes and fusion transcripts that may be associated with tumorigenesis and that may be linked to the genomic events identified from WGS. With this patient-specific characterization, we identified potentially actionable therapeutic targets and contribute our findings to the research and clinical communities. Using this approach, we also detected aberrations that have not been previously reported in PAC, but may represent viable targets in other patients who also carry the same alteration. While further studies are needed to determine which aberrations are passenger and driver mutations, these results contribute valuable information to our understanding of the disease.
The utility of RNAseq data is clear when considering our analyses of patients 2 and 3, compared to patient 1. While WGS allowed us to identify non-synonymous mutations and copy number changes in patient 1, expression data provides more information on likely affected biological processes. As needle biopsies are most commonly performed, analyses are typically limited by the availability of tumor biopsy tissue. This limitation thereby obstructs proteomic analyses. However, by layering in RNAseq data, we acquire a more detailed picture of potentially tumorigenic events in individual patients. By evaluating these changes, our aim is to demonstrate the utility of using NGS to understand what molecular events are occurring in the tumors of separate patients and to move towards a more detailed understanding of the spectrum of aberrations that occur in this disease. In doing so, this information may point to additional therapeutic options that clinicians may consider during therapeutic selection. Furthermore, identification of targets that fall outside of FDA-approved pharmaceuticals or clinical trials serves to provide novel and relevant areas of research for drug development. One caveat here is that such analyses are dependent on the quality and tumor content of the biopsies that are collected. The percentage of tumor cells in the 3 analyzed patients' biopsies ranged from 40% to 60%, average mapped coverages ranged from 31× to 54× using WGS, and using RNAseq, over 100 million mapped reads were achieved in each of patients 2 and 3. We show that an average tumor content of approximately 50% is sufficient for NGS analysis of tumor biopsies. Under circumstances whereby only biopsies with lower tumor contents are available, NGS analyses may prove to be difficult, particularly for the identification of heterozygous mutations, and otherwise will require an increase in coverage and an increase in the number of reads needed to identify pertinent genomic events and changes in gene expression. A second caveat in this study is that mutations that were not present in the original tumor may arise while patients are undergoing therapy and potentially hinder the efficacy of the treatment. While our understanding of the details surrounding such events is limited, additional sequencing of patients at different time points before, during, and after treatments, will allow us to begin to understand the contribution of these aberrations to the disease.
Given our findings, the advantages of whole genome and transcriptome NGS in cancer patients are threefold—(1) foremost is our ability to survey the entire genome and transcriptome in order to detect abnormalities that may be missed using currently available cancer testing panels, (2) the identification of expression changes that may be associated with genomic events or that point to putative drug targets, and (3) the annotation of PAC genomes that provide insight into the molecular and cellular events involved in tumorigenesis. The utility of NGS has also been demonstrated in other sequencing studies that have used this technology to evaluate genomic rearrangements in pancreatic cancer [126] as well as differences in clonal populations between primary and metastatic pancreatic tumors [127]. Such advantages and applications are intertwined with rapid improvements in NGS technologies. The throughput for sequencing has nearly doubled within one year and is forecasted to continue to grow over the next few years. While turnaround time and the pipeline from sample collection to sequencing results are still being optimized, we demonstrate that NGS represents a compelling solution to obtaining detailed molecular information on tumor biopsies in order to provide guidance for therapeutic selection. Such an approach is applicable to all cancers for which tumor biopsy material can be acquired and is an obvious and powerful method for advancing our understanding of pancreatic cancer. Because we are still early in this process, the diversity of the findings in each of the 3 patients in this study does not come as a surprise. As we continue to sequence patients, we will acquire a better understanding of the compendium of events that have a role in the disease, determine what aberrations represent driver or passenger mutations, and strengthen our knowledge base for identifying and developing improved therapeutics.
Supporting Information
Detailed methods are listed here.
(DOCX)
Selected genes demonstrating both CNVs and expression changes in patient tumors. Selected genes that demonstrate a CNV gain or loss along with a significant (q-value<0.05) expression change are listed. aRNAseq was performed for patients 2 and 3. bCorrelation between genomic event and expression change, e.g. + indicates a positive correlation between copy number change and expression change.
(DOCX)
ChimeraScan results for patients 2 and 3. ChimeraScan was used to identify putative fusion transcripts based on RNAseq data collected from patients 2 and 3. Selected predicted fusions are listed along with significant expression changes where relevant.
(DOCX)
aCGH validation of CNVs identified using WGS. To validate CNV alterations identified using whole genome sequencing (WGS), aCGH was performed on patient 1 and flow sorted aCGH was performed on patient 2. aaCGH was performed on patient 1, flow sorted aCGH was performed on patient 2. bP-values are calculated using the ADM2 algorithm [2] which generates ADM2 scores; p-values with a 0 value results when an interval has either a large copy number change, covers a large number of probes on the array, or both. The ADM2 score represents the deviation of the average of the normalized log ratios from its expected value of zero and is proportional to the height h (absolute average log ratio) of the genomic interval, and to the square root of the number of probes in the interval.
(DOCX)
Pathway analysis: Affected genes identified within each patient. The total number of genes that fall in the specified pathway across WG and RNAseq datasets across all patients are shown along with the genes themselves and p-values associated with each patient for the specific pathway. aTotal number of objects/genes in pathway map. bNumber of genes demonstrating significant changes (q-value<0.05, corrected).
(DOCX)
Genes demonstrating mutations and expression changes in the top 10 pathways identified using GeneGo's Pancreatic Cancer Disease module. Genes listed fall within the top ten pathways of GeneGo's Pancreatic Cancer Disease module. Genes show either a somatic alteration, significant expression change, or both.
(DOCX)
Acknowledgments
We would like to thank the patients and their families for contributing to this study.
Funding Statement
The authors thank the National Foundation of Cancer Research, the Randy Pausch Scholar Fund, and the Seena Magowitz Foundation for supporting this study. This study was also supported in part by P01 CA109552 (from the United States Department of Health and Human Services), and the Mayo Clinic Comprehensive Cancer Center. The authors thank the network and computing systems division of TGen for making available the supercomputing resources funded by the United States National Institutes of Health grant 1S10RR25056-01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.(2011) Cancer Facts and Figures 2011. Atlanta: American Cancer Society.
- 2. Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, et al. (1988) Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53: 549–554. [DOI] [PubMed] [Google Scholar]
- 3. Grunewald K, Lyons J, Frohlich A, Feichtinger H, Weger RA, et al. (1989) High frequency of Ki-ras codon 12 mutations in pancreatic adenocarcinomas. Int J Cancer 43: 1037–1041. [DOI] [PubMed] [Google Scholar]
- 4. Barton CM, Staddon SL, Hughes CM, Hall PA, O'Sullivan C, et al. (1991) Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br J Cancer 64: 1076–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scarpa A, Capelli P, Mukai K, Zamboni G, Oda T, et al. (1993) Pancreatic adenocarcinomas frequently show p53 gene mutations. Am J Pathol 142: 1534–1543. [PMC free article] [PubMed] [Google Scholar]
- 6. O'Brien C (1996) New tumor suppressor found in pancreatic cancer. Science 271: 294. [DOI] [PubMed] [Google Scholar]
- 7. Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, et al. (1996) DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271: 350–353. [DOI] [PubMed] [Google Scholar]
- 8. Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, et al. (1994) Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet 8: 27–32. [DOI] [PubMed] [Google Scholar]
- 9. Naumann M, Savitskaia N, Eilert C, Schramm A, Kalthoff H, et al. (1996) Frequent codeletion of p16/MTS1 and p15/MTS2 and genetic alterations in p16/MTS1 in pancreatic tumors. Gastroenterology 110: 1215–1224. [DOI] [PubMed] [Google Scholar]
- 10. Bartsch D, Shevlin DW, Tung WS, Kisker O, Wells SA Jr, et al. (1995) Frequent mutations of CDKN2 in primary pancreatic adenocarcinomas. Genes Chromosomes Cancer 14: 189–195. [DOI] [PubMed] [Google Scholar]
- 11. Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, et al. (2003) BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst 95: 214–221. [DOI] [PubMed] [Google Scholar]
- 12. Naderi A, Couch FJ (2002) BRCA2 and pancreatic cancer. Int J Gastrointest Cancer 31: 99–106. [DOI] [PubMed] [Google Scholar]
- 13. Braggio E, Keats JJ, Leleu X, Van Wier S, Jimenez-Zepeda VH, et al. (2009) Identification of copy number abnormalities and inactivating mutations in two negative regulators of nuclear factor-kappaB signaling pathways in Waldenstrom's macroglobulinemia. Cancer Res 69: 3579–3588 Epub 2009 Apr 3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, et al. (2006) Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 38: 468–473 Epub 2006 Mar 2026. [DOI] [PubMed] [Google Scholar]
- 15. Rabinovitch PS, Longton G, Blount PL, Levine DS, Reid BJ (2001) Predictors of progression in Barrett's esophagus III: baseline flow cytometric variables. Am J Gastroenterol 96: 3071–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (1297) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J, Mullighan CG, Easton J, Roberts S, Heatley SL, et al. (2011) CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods 8: 652–654 doi: 610.1038/nmeth.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iyer MK, Chinnaiyan AM, Maher CA (2011) ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 27: 2903–2904 Epub 2011 Aug 2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wheeler DL, Chappey C, Lash AE, Leipe DD, Madden TL, et al. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 28: 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liede A, Malik IA, Aziz Z, Rios Pd Pde L, Kwan E, et al. (2002) Contribution of BRCA1 and BRCA2 mutations to breast and ovarian cancer in Pakistan. Am J Hum Genet 71: 595–606 Epub 2002 Aug 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verhoog LC, van den Ouweland AM, Berns E, van Veghel-Plandsoen MM, van Staveren IL, et al. (2001) Large regional differences in the frequency of distinct BRCA1/BRCA2 mutations in 517 Dutch breast and/or ovarian cancer families. Eur J Cancer 37: 2082–2090. [DOI] [PubMed] [Google Scholar]
- 23. Wang F, Fang Q, Ge Z, Yu N, Xu S, et al. (2011) Common BRCA1 and BRCA2 mutations in breast cancer families: a meta-analysis from systematic review. Mol Biol Rep 4: 4. [DOI] [PubMed] [Google Scholar]
- 24. Lowery MA, Kelsen DP, Stadler ZK, Yu KH, Janjigian YY, et al. (2011) An Emerging Entity: Pancreatic Adenocarcinoma Associated with a Known BRCA Mutation: Clinical Descriptors, Treatment Implications, and Future Directions. Oncologist 20: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, et al. (2003) BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst 95: 214–221. [DOI] [PubMed] [Google Scholar]
- 26. Crook T, Brooks LA, Crossland S, Osin P, Barker KT, et al. (1998) p53 mutation with frequent novel condons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene 17: 1681–1689. [DOI] [PubMed] [Google Scholar]
- 27. Pacifico A, Goldberg LH, Peris K, Chimenti S, Leone G, et al. (2008) Loss of CDKN2A and p14ARF expression occurs frequently in human nonmelanoma skin cancers. Br J Dermatol 158: 291–297 Epub 2007 Dec 2006. [DOI] [PubMed] [Google Scholar]
- 28. Birnbaum DJ, Adelaide J, Mamessier E, Finetti P, Lagarde A, et al. (2011) Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer 50: 456–465 doi: 410.1002/gcc.20870. Epub 22011 Mar 20815. [DOI] [PubMed] [Google Scholar]
- 29. Fryzek JP, Garabrant DH, Schenk M, Kinnard M, Greenson JK, et al. (2006) The association between selected risk factors for pancreatic cancer and the expression of p53 and K-ras codon 12 mutations. Int J Gastrointest Cancer 37: 139–145. [DOI] [PubMed] [Google Scholar]
- 30. Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, et al. (2010) Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466: 869–873 Epub 2010 Jul 2028. [DOI] [PubMed] [Google Scholar]
- 31. Lee J, Jang KT, Ki CS, Lim T, Park YS, et al. (2007) Impact of epidermal growth factor receptor (EGFR) kinase mutations, EGFR gene amplifications, and KRAS mutations on survival of pancreatic adenocarcinoma. Cancer 109: 1561–1569. [DOI] [PubMed] [Google Scholar]
- 32. Armengol G, Knuutila S, Lluis F, Capella G, Miro R, et al. (2000) DNA copy number changes and evaluation of MYC, IGF1R, and FES amplification in xenografts of pancreatic adenocarcinoma. Cancer Genet Cytogenet 116: 133–141. [DOI] [PubMed] [Google Scholar]
- 33. Mahlamaki EH, Barlund M, Tanner M, Gorunova L, Hoglund M, et al. (2002) Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer 35: 353–358. [DOI] [PubMed] [Google Scholar]
- 34. Nagy A, Kozma L, Kiss I, Ember I, Takacs I, et al. (2001) Copy number of cancer genes predict tumor grade and survival of pancreatic cancer patients. Anticancer Res 21: 1321–1325. [PubMed] [Google Scholar]
- 35. Abraham SC, Wu TT, Hruban RH, Lee JH, Yeo CJ, et al. (2002) Genetic and immunohistochemical analysis of pancreatic acinar cell carcinoma: frequent allelic loss on chromosome 11p and alterations in the APC/beta-catenin pathway. Am J Pathol 160: 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horii A, Nakatsuru S, Miyoshi Y, Ichii S, Nagase H, et al. (1992) Frequent somatic mutations of the APC gene in human pancreatic cancer. Cancer Res 52: 6696–6698. [PubMed] [Google Scholar]
- 37. Yashima K, Nakamori S, Murakami Y, Yamaguchi A, Hayashi K, et al. (1994) Mutations of the adenomatous polyposis coli gene in the mutation cluster region: comparison of human pancreatic and colorectal cancers. Int J Cancer 59: 43–47. [DOI] [PubMed] [Google Scholar]
- 38. Camps J, Nguyen QT, Padilla-Nash HM, Knutsen T, McNeil NE, et al. (2009) Integrative genomics reveals mechanisms of copy number alterations responsible for transcriptional deregulation in colorectal cancer. Genes Chromosomes Cancer 48: 1002–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tyson J, Majerus TM, Walker S, Armour JA (2010) Screening for common copy-number variants in cancer genes. Cancer Genet Cytogenet 203: 316–323. [DOI] [PubMed] [Google Scholar]
- 40. Fang Z, Xiong Y, Li J, Liu L, Zhang W, et al. (2012) APC gene deletions in gastric adenocarcinomas in a Chinese population: a correlation with tumour progression. Clin Transl Oncol 14: 60–65. [DOI] [PubMed] [Google Scholar]
- 41. Su GH, Hilgers W, Shekher MC, Tang DJ, Yeo CJ, et al. (1998) Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res 58: 2339–2342. [PubMed] [Google Scholar]
- 42. Teng DH, Perry WL 3rd, Hogan JK, Baumgard M, Bell R, et al. (1997) Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res 57: 4177–4182. [PubMed] [Google Scholar]
- 43. Siprashvili Z, Sozzi G, Barnes LD, McCue P, Robinson AK, et al. (1997) Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc Natl Acad Sci U S A 94: 13771–13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, et al. (1996) Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci U S A 93: 3636–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chung DC, Brown SB, Graeme-Cook F, Tillotson LG, Warshaw AL, et al. (1998) Localization of putative tumor suppressor loci by genome-wide allelotyping in human pancreatic endocrine tumors. Cancer Res 58: 3706–3711. [PubMed] [Google Scholar]
- 46. Chen D, Niu M, Jiao X, Zhang K, Liang J, et al. (2012) Inhibition of AKT2 Enhances Sensitivity to Gemcitabine via Regulating PUMA and NF-kappaB Signaling Pathway in Human Pancreatic Ductal Adenocarcinoma. Int J Mol Sci 13: 1186–1208 Epub 2012 Jan 1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nitsche C, Edderkaoui M, Moore RM, Eibl G, Kasahara N, et al. (2012) The phosphatase PHLPP1 regulates Akt2, promotes pancreatic cancer cell death, and inhibits tumor formation. Gastroenterology 142: 377–387.e371–375 Epub 2011 Oct 2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi XH, Liang ZY, Ren XY, Liu TH (2009) Combined silencing of K-ras and Akt2 oncogenes achieves synergistic effects in inhibiting pancreatic cancer cell growth in vitro and in vivo. Cancer Gene Ther 16: 227–236 Epub 2008 Oct 2024. [DOI] [PubMed] [Google Scholar]
- 49. Hahn CN, Scott HS (2011) Spliceosome mutations in hematopoietic malignancies. Nat Genet 44: 9–10 doi: 10.1038/ng.1045. [DOI] [PubMed] [Google Scholar]
- 50. Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, et al. (2012) Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ogbureke KU, Abdelsayed RA, Kushner H, Li L, Fisher LW (2010) Two members of the SIBLING family of proteins, DSPP and BSP, may predict the transition of oral epithelial dysplasia to oral squamous cell carcinoma. Cancer 116: 1709–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y, Zhu X, Zeng Y, Wang J, Zhang X, et al. (2010) FMNL2 enhances invasion of colorectal carcinoma by inducing epithelial-mesenchymal transition. Mol Cancer Res 8: 1579–1590 Epub 2010 Nov 1511. [DOI] [PubMed] [Google Scholar]
- 53. Zhu XL, Liang L, Ding YQ (2008) Overexpression of FMNL2 is closely related to metastasis of colorectal cancer. Int J Colorectal Dis 23: 1041–1047 Epub 2008 Jul 1030. [DOI] [PubMed] [Google Scholar]
- 54. Zhu XL, Zeng YF, Guan J, Li YF, Deng YJ, et al. (2011) FMNL2 is a positive regulator of cell motility and metastasis in colorectal carcinoma. J Pathol 224: 377–388 doi: 310.1002/path.2871. Epub 2011 Apr 1019. [DOI] [PubMed] [Google Scholar]
- 55. Liang L, Guan J, Zeng Y, Wang J, Li X, et al. (2011) Down-regulation of formin-like 2 predicts poor prognosis in hepatocellular carcinoma. Hum Pathol 42: 1603–1612 Epub 2011 Apr 1614. [DOI] [PubMed] [Google Scholar]
- 56. Ajiro M, Katagiri T, Ueda K, Nakagawa H, Fukukawa C, et al. (2009) Involvement of RQCD1 overexpression, a novel cancer-testis antigen, in the Akt pathway in breast cancer cells. Int J Oncol 35: 673–681. [PubMed] [Google Scholar]
- 57. Ajiro M, Nishidate T, Katagiri T, Nakamura Y (2010) Critical involvement of RQCD1 in the EGFR-Akt pathway in mammary carcinogenesis. Int J Oncol 37: 1085–1093. [DOI] [PubMed] [Google Scholar]
- 58. Lin F, Wang X, Jie Z, Hong X, Li X, et al. (2011) Inhibitory effects of miR-146b-5p on cell migration and invasion of pancreatic cancer by targeting MMP16. J Huazhong Univ Sci Technolog Med Sci 31: 509–514 Epub 2011 Aug 2017. [DOI] [PubMed] [Google Scholar]
- 59. Furukawa T, Kuboki Y, Tanji E, Yoshida S, Hatori T, et al. (2011) Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep 1: 161 Epub 2011 Nov 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Caligiuri MA, Briesewitz R, Yu J, Wang L, Wei M, et al. (2007) Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood 110: 1022–1024 Epub 2007 May 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Makishima H, Jankowska AM, McDevitt MA, O'Keefe C, Dujardin S, et al. (2011) CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood 117: e198–206 Epub 2011 Feb 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, et al. (2011) Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med 208: 901–908 Epub 2011 May 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, et al. (2011) Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet 43: 932–939 doi: 910.1038/ng.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, et al. (2012) The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481: 157–163 doi: 110.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cho YG, Chang X, Park IS, Yamashita K, Shao C, et al. (2011) Promoter methylation of leukemia inhibitory factor receptor gene in colorectal carcinoma. Int J Oncol 39: 337–344 doi: 310.3892/ijo.2011.1050. Epub 2011 May 3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Okamura Y, Nomoto S, Kanda M, Li Q, Nishikawa Y, et al. (2010) Leukemia inhibitory factor receptor (LIFR) is detected as a novel suppressor gene of hepatocellular carcinoma using double-combination array. Cancer Lett 289: 170–177 Epub 2009 Sep 2003. [DOI] [PubMed] [Google Scholar]
- 67. Kamohara H, Ogawa M, Ishiko T, Sakamoto K, Baba H (2007) Leukemia inhibitory factor functions as a growth factor in pancreas carcinoma cells: Involvement of regulation of LIF and its receptor expression. Int J Oncol 30: 977–983. [PubMed] [Google Scholar]
- 68. Bandyopadhyay S, Pai SK, Gross SC, Hirota S, Hosobe S, et al. (2003) The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer Res 63: 1731–1736. [PubMed] [Google Scholar]
- 69. Guan RJ, Ford HL, Fu Y, Li Y, Shaw LM, et al. (2000) Drg-1 as a differentiation-related, putative metastatic suppressor gene in human colon cancer. Cancer Res 60: 749–755. [PubMed] [Google Scholar]
- 70. Kurdistani SK, Arizti P, Reimer CL, Sugrue MM, Aaronson SA, et al. (1998) Inhibition of tumor cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res 58: 4439–4444. [PubMed] [Google Scholar]
- 71. Muller MW, Giese NA, Swiercz JM, Ceyhan GO, Esposito I, et al. (2007) Association of axon guidance factor semaphorin 3A with poor outcome in pancreatic cancer. Int J Cancer 121: 2421–2433. [DOI] [PubMed] [Google Scholar]
- 72. Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, et al. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 Epub 2008 Sep 1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, et al. (2007) Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res 67: 3545–3550. [DOI] [PubMed] [Google Scholar]
- 74. Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, et al. (2012) Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A 109: 5934–5941 Epub 2012 Mar 5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Feldmann G, Habbe N, Dhara S, Bisht S, Alvarez H, et al. (2008) Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut 57: 1420–1430 Epub 2008 May 1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, et al. (2005) Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia 7: 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hall PA, Hughes CM, Staddon SL, Richman PI, Gullick WJ, et al. (1990) The c-erb B-2 proto-oncogene in human pancreatic cancer. J Pathol 161: 195–200. [DOI] [PubMed] [Google Scholar]
- 78. Safran H, Steinhoff M, Mangray S, Rathore R, King TC, et al. (2001) Overexpression of the HER-2/neu oncogene in pancreatic adenocarcinoma. Am J Clin Oncol 24: 496–499. [DOI] [PubMed] [Google Scholar]
- 79. Giaginis C, Katsamangou E, Tsourouflis G, Zizi-Serbetzoglou D, Kouraklis G, et al. (2009) Peroxisome proliferator-activated receptor-gamma and retinoid X receptor-alpha expression in pancreatic ductal adenocarcinoma: association with clinicopathological parameters, tumor proliferative capacity, and patients' survival. Med Sci Monit 15: BR148–156. [PubMed] [Google Scholar]
- 80. Nakajima A, Tomimoto A, Fujita K, Sugiyama M, Takahashi H, et al. (2008) Inhibition of peroxisome proliferator-activated receptor gamma activity suppresses pancreatic cancer cell motility. Cancer Sci 99: 1892–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Graber HU, Friess H, Kaufmann B, Willi D, Zimmermann A, et al. (1999) ErbB-4 mRNA expression is decreased in non-metastatic pancreatic cancer. Int J Cancer 84: 24–27. [DOI] [PubMed] [Google Scholar]
- 82. Thybusch-Bernhardt A, Beckmann S, Juhl H (2001) Comparative analysis of the EGF-receptor family in pancreatic cancer: expression of HER-4 correlates with a favourable tumor stage. Int J Surg Investig 2: 393–400. [PubMed] [Google Scholar]
- 83. Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, et al. (2004) Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med 350: 1828–1837. [DOI] [PubMed] [Google Scholar]
- 84. Ma S, Guan XY, Beh PS, Wong KY, Chan YP, et al. (2007) The significance of LMO2 expression in the progression of prostate cancer. J Pathol 211: 278–285. [DOI] [PubMed] [Google Scholar]
- 85. Nakata K, Ohuchida K, Nagai E, Hayashi A, Miyasaka Y, et al. (2009) LMO2 is a novel predictive marker for a better prognosis in pancreatic cancer. Neoplasia 11: 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nishizawa H, Ota K, Dohi Y, Ikura T, Igarashi K (2012) Bach1-mediated suppression of p53 is inhibited by p19(ARF) independently of MDM2. Cancer Sci 103: 897–903 doi: 810.1111/j.1349-7006.2012.02244.x. Epub 02012 Apr 02211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ricci MS, Kim SH, Ogi K, Plastaras JP, Ling J, et al. (2007) Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell 12: 66–80. [DOI] [PubMed] [Google Scholar]
- 88. Larbouret C, Robert B, Bascoul-Mollevi C, Penault-Llorca F, Ho-Pun-Cheung A, et al. (2010) Combined cetuximab and trastuzumab are superior to gemcitabine in the treatment of human pancreatic carcinoma xenografts. Ann Oncol 21: 98–103 Epub 2009 Nov 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Larbouret C, Robert B, Navarro-Teulon I, Thezenas S, Ladjemi MZ, et al. (2007) In vivo therapeutic synergism of anti-epidermal growth factor receptor and anti-HER2 monoclonal antibodies against pancreatic carcinomas. Clin Cancer Res 13: 3356–3362. [DOI] [PubMed] [Google Scholar]
- 90. Ben-Kasus T, Schechter B, Lavi S, Yarden Y, Sela M (2009) Persistent elimination of ErbB-2/HER2-overexpressing tumors using combinations of monoclonal antibodies: relevance of receptor endocytosis. Proc Natl Acad Sci U S A 106: 3294–3299 Epub 2009 Feb 3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. da Cunha Santos G, Dhani N, Tu D, Chin K, Ludkovski O, et al. (2010) Molecular predictors of outcome in a phase 3 study of gemcitabine and erlotinib therapy in patients with advanced pancreatic cancer: National Cancer Institute of Canada Clinical Trials Group Study PA.3. Cancer 116: 5599–5607 doi: 5510.1002/cncr.25393. Epub 22010 Sep 25397. [DOI] [PubMed] [Google Scholar]
- 92. Moore PS, Orlandini S, Zamboni G, Capelli P, Rigaud G, et al. (2001) Pancreatic tumours: molecular pathways implicated in ductal cancer are involved in ampullary but not in exocrine nonductal or endocrine tumorigenesis. Br J Cancer 84: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Winder T, Mundlein A, Rhomberg S, Dirschmid K, Hartmann BL, et al. (2009) Different types of K-Ras mutations are conversely associated with overall survival in patients with colorectal cancer. Oncol Rep 21: 1283–1287. [DOI] [PubMed] [Google Scholar]
- 94. Romeo S, Szuhai K, Nishimori I, Ijszenga M, Wijers-Koster P, et al. (2009) A balanced t(5;17) (p15;q22–23) in chondroblastoma: frequency of the re-arrangement and analysis of the candidate genes. BMC Cancer 9: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chung W, Bondaruk J, Jelinek J, Lotan Y, Liang S, et al. (2011) Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol Biomarkers Prev 20: 1483–1491 Epub 2011 May 1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Meiboom M, Murua Escobar H, Pentimalli F, Fusco A, Belge G, et al. (2003) A 3.4-kbp transcript of ZNF331 is solely expressed in follicular thyroid adenomas. Cytogenet Genome Res 101: 113–117. [DOI] [PubMed] [Google Scholar]
- 97. Yu J, Liang QY, Wang J, Cheng Y, Wang S, et al. (2012) Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene 27: 54. [DOI] [PubMed] [Google Scholar]
- 98. Mazur PK, Einwachter H, Lee M, Sipos B, Nakhai H, et al. (2010) Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A 107: 13438–13443 Epub 12010 Jul 13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Shimada H, Kuboshima M, Shiratori T, Nabeya Y, Takeuchi A, et al. (2007) Serum anti-myomegalin antibodies in patients with esophageal squamous cell carcinoma. Int J Oncol 30: 97–103. [PubMed] [Google Scholar]
- 100. Corbo V, Beghelli S, Bersani S, Antonello D, Talamini G, et al. (2012) Pancreatic endocrine tumours: mutational and immunohistochemical survey of protein kinases reveals alterations in targetable kinases in cancer cell lines and rare primaries. Ann Oncol 23: 127–134 Epub 2011 Mar 2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ahmad I, Sansom OJ, Leung HY (2012) Exploring molecular genetics of bladder cancer: lessons learned from mouse models. Dis Model Mech 5: 323–332 Epub 2012 Mar 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Juanpere N, Agell L, Lorenzo M, de Muga S, Lopez-Vilaro L, et al. (2012) Mutations in FGFR3 and PIK3CA, singly or combined with RAS and AKT1, are associated with AKT but not with MAPK pathway activation in urothelial bladder cancer. Hum Pathol 12: 12. [DOI] [PubMed] [Google Scholar]
- 103. Fournier G, Cabaud O, Josselin E, Chaix A, Adelaide J, et al. (2011) Loss of AF6/afadin, a marker of poor outcome in breast cancer, induces cell migration, invasiveness and tumor growth. Oncogene 30: 3862–3874 doi: 3810.1038/onc.2011.3106. Epub 2011 Apr 3811. [DOI] [PubMed] [Google Scholar]
- 104. Letessier A, Garrido-Urbani S, Ginestier C, Fournier G, Esterni B, et al. (2007) Correlated break at PARK2/FRA6E and loss of AF-6/Afadin protein expression are associated with poor outcome in breast cancer. Oncogene 26: 298–307 Epub 2006 Jul 2003. [DOI] [PubMed] [Google Scholar]
- 105. Yamazaki M, Nakamura K, Mizukami Y, Ii M, Sasajima J, et al. (2008) Sonic hedgehog derived from human pancreatic cancer cells augments angiogenic function of endothelial progenitor cells. Cancer Sci 99: 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Mathews LA, Cabarcas SM, Hurt EM, Zhang X, Jaffee EM, et al. (2011) Increased expression of DNA repair genes in invasive human pancreatic cancer cells. Pancreas 40: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Eppinga RD, Krueger EW, Weller SG, Zhang L, Cao H, et al. (2012) Increased expression of the large GTPase dynamin 2 potentiates metastatic migration and invasion of pancreatic ductal carcinoma. Oncogene 31: 1228–1241 doi: 1210.1038/onc.2011.1329. Epub 2011 Aug 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Valentin-Vega YA, Barboza JA, Chau GP, El-Naggar AK, Lozano G (2007) High levels of the p53 inhibitor MDM4 in head and neck squamous carcinomas. Hum Pathol 38: 1553–1562 Epub 2007 Jul 1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Han X, Garcia-Manero G, McDonnell TJ, Lozano G, Medeiros LJ, et al. (2007) HDM4 (HDMX) is widely expressed in adult pre-B acute lymphoblastic leukemia and is a potential therapeutic target. Mod Pathol 20: 54–62 Epub 2006 Nov 2024. [DOI] [PubMed] [Google Scholar]
- 110. Xiong S, Pant V, Suh YA, Van Pelt CS, Wang Y, et al. (2010) Spontaneous tumorigenesis in mice overexpressing the p53-negative regulator Mdm4. Cancer Res 70: 7148–7154 Epub 2010 Aug 7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Suzuoki M, Miyamoto M, Kato K, Hiraoka K, Oshikiri T, et al. (2002) Impact of caveolin-1 expression on prognosis of pancreatic ductal adenocarcinoma. Br J Cancer 87: 1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Burrows C, Abd Latip N, Lam SJ, Carpenter L, Sawicka K, et al. (2010) The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res 38: 5542–5553 Epub 2010 Apr 5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mauri P, Scarpa A, Nascimbeni AC, Benazzi L, Parmagnani E, et al. (2005) Identification of proteins released by pancreatic cancer cells by multidimensional protein identification technology: a strategy for identification of novel cancer markers. Faseb J 19: 1125–1127 Epub 2005 May 1124. [DOI] [PubMed] [Google Scholar]
- 114. Liang JJ, Wang H, Rashid A, Tan TH, Hwang RF, et al. (2008) Expression of MAP4K4 is associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res 14: 7043–7049. [DOI] [PubMed] [Google Scholar]
- 115. Mahon PC, Baril P, Bhakta V, Chelala C, Caulee K, et al. (2007) S100A4 contributes to the suppression of BNIP3 expression, chemoresistance, and inhibition of apoptosis in pancreatic cancer. Cancer Res 67: 6786–6795. [DOI] [PubMed] [Google Scholar]
- 116. Ockenga J, Strunck S, Post C, Schulz HU, Halangk J, et al. (2009) The role of epoxide hydrolase Y113H gene variant in pancreatic diseases. Pancreas 38: e97–e101. [DOI] [PubMed] [Google Scholar]
- 117. Fukushima H, Yamamoto H, Itoh F, Nakamura H, Min Y, et al. (2001) Association of matrilysin mRNA expression with K-ras mutations and progression in pancreatic ductal adenocarcinomas. Carcinogenesis 22: 1049–1052. [DOI] [PubMed] [Google Scholar]
- 118. Jones LE, Humphreys MJ, Campbell F, Neoptolemos JP, Boyd MT (2004) Comprehensive analysis of matrix metalloproteinase and tissue inhibitor expression in pancreatic cancer: increased expression of matrix metalloproteinase-7 predicts poor survival. Clin Cancer Res 10: 2832–2845. [DOI] [PubMed] [Google Scholar]
- 119. Yamamoto H, Itoh F, Iku S, Adachi Y, Fukushima H, et al. (2001) Expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in human pancreatic adenocarcinomas: clinicopathologic and prognostic significance of matrilysin expression. J Clin Oncol 19: 1118–1127. [DOI] [PubMed] [Google Scholar]
- 120. Hamidi T, Algul H, Cano CE, Sandi MJ, Molejon MI, et al. (2012) Nuclear protein 1 promotes pancreatic cancer development and protects cells from stress by inhibiting apoptosis. J Clin Invest 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sasada T, Azuma K, Hirai T, Hashida H, Kanai M, et al. (2008) Prognostic significance of the immediate early response gene X-1 (IEX-1) expression in pancreatic cancer. Ann Surg Oncol 15: 609–617 Epub 2007 Nov 2017. [DOI] [PubMed] [Google Scholar]
- 122. Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, et al. (2009) Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res 69: 2400–2407 Epub 2009 Mar 2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang H, Ma X, Ren S, Buolamwini JK, Yan C (2011) A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol Cancer Ther 10: 69–79 Epub 2010 Nov 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, et al. (1988) Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53: 549–554. [DOI] [PubMed] [Google Scholar]
- 125. Kim ST, Lim DH, Jang KT, Lim T, Lee J, et al. (2011) Impact of KRAS mutations on Clinical Outcomes in Pancreatic Cancer Patients Treated with First-line Gemcitabine-based Chemotherapy. Mol Cancer Ther 23: 23. [DOI] [PubMed] [Google Scholar]
- 126. Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, et al. (2010) The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467: 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yachida S, Jones S, Bozic I, Antal T, Leary R, et al. (2010) Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467: 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed methods are listed here.
(DOCX)
Selected genes demonstrating both CNVs and expression changes in patient tumors. Selected genes that demonstrate a CNV gain or loss along with a significant (q-value<0.05) expression change are listed. aRNAseq was performed for patients 2 and 3. bCorrelation between genomic event and expression change, e.g. + indicates a positive correlation between copy number change and expression change.
(DOCX)
ChimeraScan results for patients 2 and 3. ChimeraScan was used to identify putative fusion transcripts based on RNAseq data collected from patients 2 and 3. Selected predicted fusions are listed along with significant expression changes where relevant.
(DOCX)
aCGH validation of CNVs identified using WGS. To validate CNV alterations identified using whole genome sequencing (WGS), aCGH was performed on patient 1 and flow sorted aCGH was performed on patient 2. aaCGH was performed on patient 1, flow sorted aCGH was performed on patient 2. bP-values are calculated using the ADM2 algorithm [2] which generates ADM2 scores; p-values with a 0 value results when an interval has either a large copy number change, covers a large number of probes on the array, or both. The ADM2 score represents the deviation of the average of the normalized log ratios from its expected value of zero and is proportional to the height h (absolute average log ratio) of the genomic interval, and to the square root of the number of probes in the interval.
(DOCX)
Pathway analysis: Affected genes identified within each patient. The total number of genes that fall in the specified pathway across WG and RNAseq datasets across all patients are shown along with the genes themselves and p-values associated with each patient for the specific pathway. aTotal number of objects/genes in pathway map. bNumber of genes demonstrating significant changes (q-value<0.05, corrected).
(DOCX)
Genes demonstrating mutations and expression changes in the top 10 pathways identified using GeneGo's Pancreatic Cancer Disease module. Genes listed fall within the top ten pathways of GeneGo's Pancreatic Cancer Disease module. Genes show either a somatic alteration, significant expression change, or both.
(DOCX)