Abstract
Most of the DNA in eukaryotes is packaged in tandemly arrayed nucleosomes that, together with numerous DNA- and nucleosome-associated enzymes and regulatory factors, make up chromatin. Chromatin modifying and remodeling agents help regulate access to selected DNA segments in chromatin, thereby facilitating transcription and DNA replication and repair. Studies of nucleotide excision repair (NER), single strand break repair (SSBR), and the homology-directed (HDR) and non-homologous end-joining (NHEJ) double strand break repair pathways have led to an ‘access-repair-restore’ paradigm, in which chromatin in the vicinity of damaged DNA is disrupted, thereby enabling efficient repair and the subsequent repackaging of DNA into nucleosomes. When damage is extensive, these repair processes are accompanied by cell cycle checkpoint activation, which provides cells with sufficient time to either complete the repair or initiate apoptosis. It is not clear, however, if base excision repair (BER) of the ~20,000 or more oxidative DNA damages that occur daily in each nucleated human cell can be viewed through this same lens. Until recently, we did not know if BER requires or is accompanied by nucleosome disruption, and it is not yet clear that anything short of overwhelming oxidative damage (resulting in the shunting of DNA substrates into other repair pathways) results in checkpoint activation. This review highlights studies of how oxidatively damaged DNA in nucleosomes is discovered and repaired, and offers a working model of events associated with BER in chromatin that we hope will have heuristic value.
The occurrence and repair of oxidative damage in DNA
Reactive oxygen species (ROS) occur naturally in the cell and play essential roles in oxidative metabolism, signal transduction, cross-linking of cell wall components in plants, and the killing of ingested microorganisms by phagocytes (e.g. (Apel and Hirt, 2004; Fang, 2004). Although most excess ROS are neutralized by superoxide dismutases, catalases, and glutathione peroxidases, residual ROS can damage both proteins and nucleic acids. In DNA, ROS can generate an array of base damages, sites of base loss (AP sites) and single strand breaks (Breen and Murphy, 1995). Exogenous radiation, which can generate ROS via hydrolysis of water, produces a similar array of DNA damages (Dizdaroglu, 2012; Ward, 1985; Ward, 1988). ROS are small and able to gain access to nucleosomal DNA, as demonstrated by free radical-mediated footprinting experiments. Specifically, in nucleosomes where the DNA adopts a preferred helical orientation relative to the underlying histone octamer, the susceptibility of DNA to free radical damages exhibits a ~10 base pair periodicity, coincident with both the helical periodicity of DNA and the periodic compression of major and minor grooves that occurs when DNA winds about the histone octamer (e.g. (Hayes et al., 1990)). Typically, however, there is only a few-fold difference between the least and most susceptible DNA segments in nucleosomes, with the most susceptible segments as vulnerable to oxidative damage as naked DNA (Enright et al., 1996).
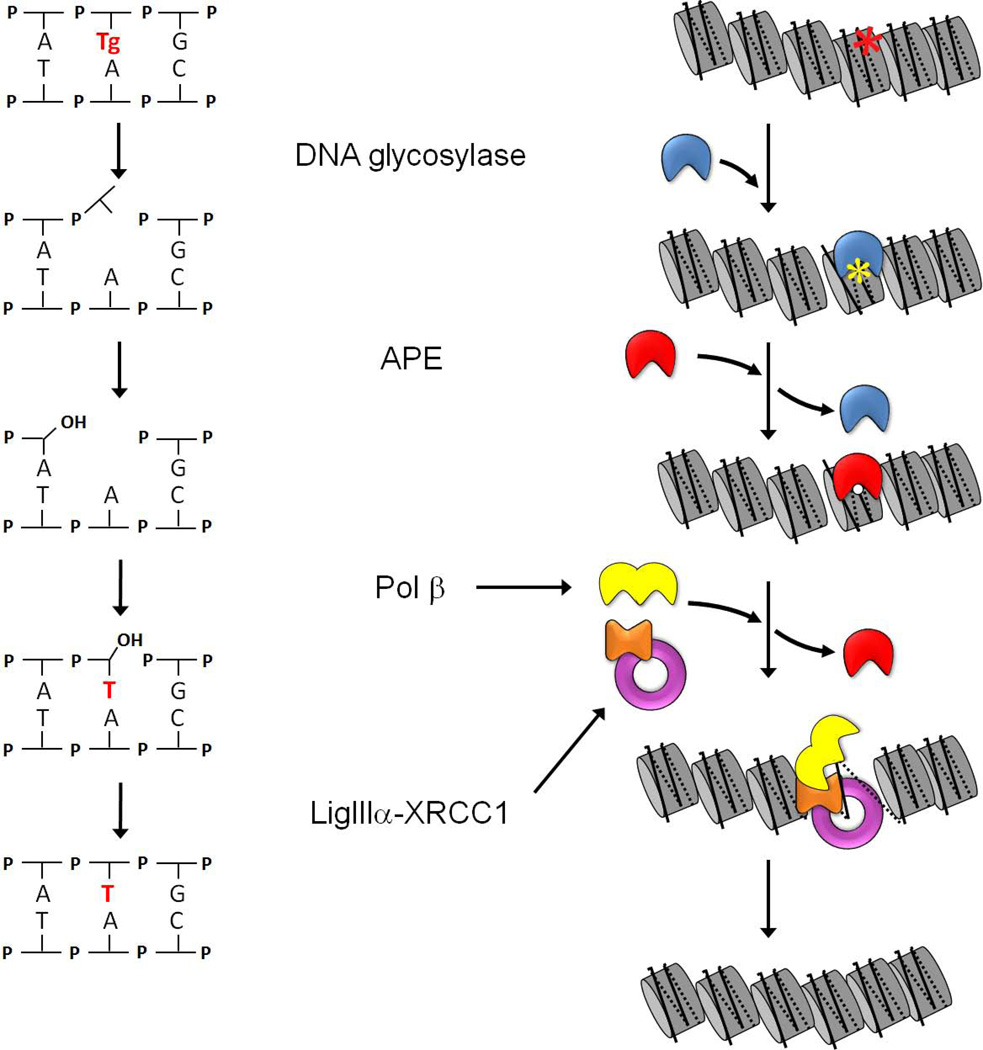
Most oxidative damages are repaired in an error-free fashion, via base excision repair (BER) (reviewed by (David et al., 2007; Duclos et al., 2012; Fromme et al., 2004; Hazra et al., 2007; Hegde et al., 2008a; Lindahl and Wood, 1999; Meira et al., 2005; Robertson et al., 2009)). Defects in BER can have lethal or mutagenic consequences, thereby increasing the risk of oncogenesis (Wallace et al., 2012). In its simplest “short-patch” form, BER entails four enzymes acting in a step-wise fashion (Figure 1). Bifunctional DNA glycosylases excise oxidatively damaged bases and then cleave the phosphodiester backbone 3’ of the damaged base. This leaves a blocking residue attached to the deoxyribose upstream of the nick, which is removed by either AP endonuclease 1 (APE) or polynucleotide kinase (PNK). The resulting single nucleotide gap is filled in by DNA polymerase β (Pol β) and the nick is sealed by DNA ligase III, bound to the presumptive scaffold protein XRCC1. In addition to repairing oxidative damages, BER glycosylases remove bases damaged by alkylation, mispaired bases generated by spontaneous deamination of 5-methylcytosine (producing thymine), and uracil residues, generated by spontaneous deamination of cytosine or misincorporated during DNA replication. Such damages are recognized and removed by monofunctional DNA glycosylases that lack the DNA lyase activity present in bifunctional DNA glycosylases (reviewed in (Friedberg, 2006)). AP sites generated in this manner, or directly by ROS-induced base loss, are incised by APE; this leaves blocking residues attached to the deoxyribose downstream of the nick that are removed by the lyase activity of Pol β. In the event that the Pol β lyase is unable to remove downstream blocking residues, DNA substrates are shunted into the long-patch BER pathway, which entails the further addition of dNTPs and displacement of a short segment of DNA attached to the blocking moiety. The displaced DNA is then removed by the structure-specific FLAP endonuclease (FEN1), and the resulting nick is sealed by DNA ligase I or III (Klungland and Lindahl, 1997).
Figure 1. Short-patch base excision repair (BER; left) and a working model of BER in chromatin (right).
The left-hand schematic illustrates the enzymatic steps in short-patch BER, and the right-hand schematic depicts the corresponding steps in chromatin. Repair begins with excision of an oxidized base (in this case, thymine glycol) by a bifunctional DNA glycosylase (blue), followed by cleavage of the phosphodiester bond. APE (red) displaces the product-bound DNA glycosylase and removes the 3’ blocking moiety, leaving a gap that can be filled by DNA polymerase β (yellow) and sealed by DNA ligase IIIα (purple). The scaffolding protein XRCC1 (orange) contains separate binding sites for Pol β and DNA ligase IIIα. The complex of XRCC1 with DNA ligase IIIα disrupts DNA gap- and nick-containing nucleosomes and thus, is able to facilitate both the polymerization and ligation steps. Although not yet documented, the model depicts the re-assembly of the newly repaired DNA into a nucleosome, with the aid of histone chaperones.
When BER is not enough
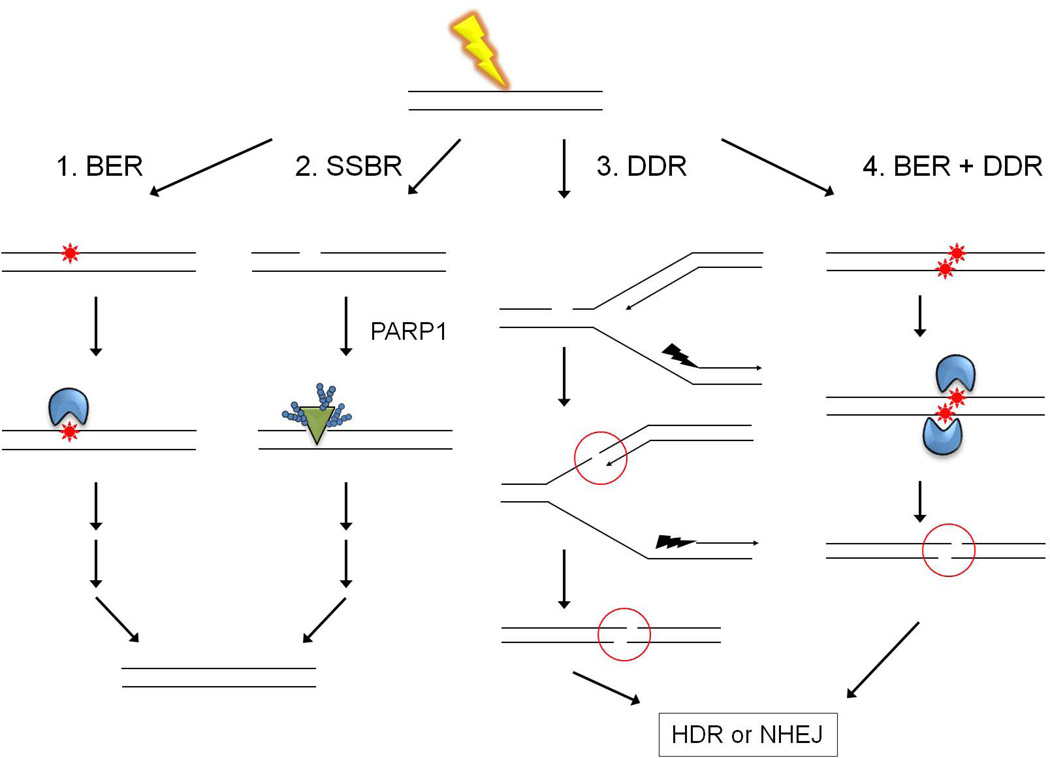
Oxidative damages include single strand DNA breaks (SSBs) and, as described below, can lead to formation of double strand DNA breaks (DSBs). These damages are channeled into single and double strand break repair pathways, as depicted in Figure 2. Molecular mechanisms employed in these pathways to clear nucleosomes from the site of damage and harness flanking nucleosomes to help recruit regulatory and repair factors are relatively well understood (reviewed in (Polo and Jackson, 2011; Sinha and Peterson, 2009; van Attikum and Gasser, 2009)). However, there is no compelling evidence that these same mechanisms act in short-patch BER, although numerous studies have been interpreted as if they might. Accordingly, before discussing the effect of chromatin on short-patch BER, it is important to discuss circumstances in which oxidative damages engage these other repair pathways. SSB repair (SSBR) (pathway #2 in Figure 2) begins with the binding of PARP1 (poly (ADP ribose) polymerase 1) to SSBs, and employs many of the same factors used in long-patch BER (reviewed by (Caldecott, 2008)). Once bound to DNA, PARP1 adds multiple ADP ribose moieties to itself and numerous other proteins, forming long, branched chains of poly (ADP-ribose) (PAR). These induce PARP1 to dissociate from DNA, at which point the PAR chains are removed by PAR glycohydrolase (PARG), freeing up PARP1 to once again bind SSBs. Depletion and inhibition studies indicate that PARP1 promotes SSBR, probably by multiple mechanisms. PARP1 can bind to, and is thought to help recruit, XRCC1 and associated repair factors. PARP1 is also able to bind to nucleosomes, and appears to block the binding of histone H1 to linker DNA between nucleosomes (reviewed by (Kraus, 2008)). This may promote or stabilize an open chromatin configuration that is more amenable to repair. Finally, PARP1 promotes the recruitment of chromatin remodeling factors, the Polycomb histone-modifying complex, and macroH2A, a histone variant associated with transcriptional repression (Chou et al., 2010; Timinszky et al., 2009).
Figure 2. Channeling of oxidatively damaged DNA into multiple DNA repair pathways.
Reactive oxygen species (ROS), in this case generated by exposure to γ-radiation (yellow lightning), can produce single and multiple clustered base damages (red stars), and single strand DNA breaks (SSBs). As depicted in the left-most diagram and in Figure 1, most oxidatively damaged bases and AP sites are repaired in an error-free manner by short-patch BER (pathway 1). SSBs generated as intermediates during BER appear to remain bound by DNA glycosylases (blue) or APE until displaced by the next enzyme in the BER pathway, and are thus protected from PARP1 (green triangle) binding. Binding of PARP1 to de novo-generated SSBs channels substrates into the single-strand break repair pathway (pathway 2). Pathway 3 depicts the generation of a double strand DNA break (DSB) via replication up to a SSB. DSBs are also produced during the attempted, near simultaneous BER of closely opposed lesions (pathway 4). In either case, DSBs trigger what is now commonly called the DNA damage response (DDR), which encompasses both homology-directed repair (HDR) and non-homologous end joining (NHEJ) pathways.
Although PARP1 appears to be integral to SSBR, recent observations suggest that PARP1 binding to SSBs that form as intermediates during BER may actually hinder short-patch BER. Specifically, Helleday and colleagues found that RNAi-mediated depletion of PARP1 had no effect on BER, whereas treatment of cells with an agent that blocks the cyclical binding of PARP1 to SSBs interfered with BER (Strom et al., 2011). While this study did not exclude the possible involvement of PARP1 in long-patch BER, it is noteworthy that PARP1 failed to co-localize with DNA glycosylases during repair of damage in cells treated with low doses of oxidative damaging agents (Hanssen-Bauer et al., 2011). This observation suggests that low or constitutive levels of oxidative damage do not trigger the DNA damage responses associated with single and double strand break repair. The SSBs that form as intermediates during BER may normally be protected from PARP1 binding by the propensity of BER enzymes to remain bound to their products until displaced by the next enzyme in the repair pathway (Prasad et al., 2010; Wilson and Kunkel, 2000). This enzyme handoff phenomenon, first observed in studies with naked DNA substrates, also occurs with nucleosome substrates (Hinz et al., 2010; Odell et al., 2011).
Exposure to ionizing radiation, such as x-rays and γ-rays, can produce a cluster of oxidative lesions in DNA. When lesions within a cluster reside on opposing strands, the near simultaneous attempted BER of such lesions can produce a DSB, as depicted in pathway #4 of Figure 2 (Harrison et al., 1999). In vivo studies strongly suggest this phenomenon accounts for many of the potentially lethal double strand DNA breaks that occur each day in cells (Blaisdell et al., 2001; Yang et al., 2004). Specifically, cells depleted of DNA glycosylases that initiate BER were more tolerant of ionizing radiation and accumulated fewer DSBs than wild type cells. Conversely, overexpression of DNA glycosylases increased rates of mutagenesis and rendered cells hypersensitive to ionizing radiation. Importantly, these phenotypes were evident only when ionizing radiation was used to generate clusters of oxidative damage. In cells treated with hydrogen peroxide, which primarily induces single DNA lesions, overexpression of BER enzymes exerted a protective effect.
Whether generated as described above or by other means (e.g. pathway #3 in Figure 2), DSBs are repaired either by non-homologous DNA end-joining (NHEJ) or homology directed repair (HDR) pathways. Activation of either pathway triggers a series of events, collectively known as the DNA damage response (DDR). These include replacement of histone H2A in nucleosomes that flank DSBs with the histone variant H2AX, and concurrent or subsequent phosphorylation of a C-terminal serine in H2AX (by ATM, ATR or DNA-PK protein kinases). Phosphorylated H2AX helps recruit proteins containing phospho-specific interaction domains, such as BRCT (breast cancer C-terminal) and FHA (forkhead associated). In turn, these proteins help recruit repair factors such as TP53BP1, BRCA1 and NBS1, and the checkpoint mediator protein, MDC1. MDC1 helps recruit the ATM kinase, thereby further amplifying the DDR (reviewed by (Luijsterburg and van Attikum, 2011; Polo and Jackson, 2011)).
Is short-patch BER accompanied by activation of checkpoint or other signaling events?
Cells possess multiple mechanisms to sense their oxidative state, and employ ROS in redox-dependent signaling cascades (Finkel, 2011; Kobayashi and Suda, 2012). In response to oxidative stress, cells trigger an adaptive response that helps ameliorate the effects of the damaging agent. This response includes an increase in the nuclear concentration of APE, likely by inhibiting its export from nuclei (Mitra et al., 2007; Ramana et al., 1998), and increased expression of certain DNA repair enzymes. For example, exposure of colon cancer cells to ROS increased the transcription of the NEIL1 gene by two- to four-fold (Das et al., 2005). In both of the above examples, the increase in nuclear concentration of APE and NEIL1 lagged considerably (6–9 hours) behind the increase in intracellular ROS levels, meaning these changes probably reflect the cellular response to changes in oxidative state rather than signaling triggered by oxidative DNA damage.
High ROS levels, such as those typically generated experimentally by treatment of cells with peroxide, result in a broad array of oxidative DNA damages, some of which are channeled into the SSBR and DDR pathways described earlier. The DDR-associated activation of ATM and ATR dependent checkpoint pathways has made it difficult to determine if oxidatively damaged DNA bases, or intermediates that form during short-patch BER, also activate DNA damage checkpoints. A recent report (Boldogh et al., 2012) provides what is arguably the most compelling evidence of a link between oxidative base damage and checkpoint signaling. Specifically, a complex between the DNA glycosylase OGG1 and the oxidative lesion 8-hydroxyguanine (8oxoG) appears to act as a guanine nucleotide exchange factor that activates a Ras signaling pathway, which culminates in translocation of phosphorylated ERK1/2 into nuclei. Separate studies indicate that ERK1/2 activation, in response to UV irradiation or non-oxidative DNA damaging agents, induces cell cycle arrest (Mebratu and Tesfaigzi, 2009). However, the OGG1-8oxoG mediated signaling reported in (Boldogh et al., 2012) was induced by exogenous treatment of cells with 8oxoG. It has yet to be determined if 8oxoG containing genomic DNA can provide an equivalent trigger. In summary, considerably more study will be needed to demonstrate the existence of BER-specific triggers of regulatory networks.
Is BER of oxidative damage in nucleosomal DNA coupled to other cellular processes that destabilize or disrupt nucleosomes?
As described above, some oxidative damages are channeled into repair pathways in which nucleosomes are either harnessed to help recruit regulatory and repair factors or disrupted to permit access to damaged DNA. However, the majority of endogenously generated oxidative damages are not channeled into these repair pathways but rather into short patch BER. Even so, before we can conclude that short-patch BER enzymes must contend directly with nucleosomes in vivo, it is important to evaluate the possibility that short patch BER is obligatorily coupled to either DNA replication or transcription, as either or both processes might enable BER enzymes to circumvent the ‘nucleosome problem.’
‘Simple’ oxidative damages that result from treatment of cells with peroxide are repaired within minutes; more complex damages that require the recruitment of additional factors or involve entirely different repair pathways are repaired more slowly (e.g. (Mitra et al., 2001). This observation argues against the notion that BER in dividing cells is linked solely to DNA replication. Nevertheless, several lines of evidence suggest that at least some oxidative damages are repaired during DNA replication. The most compelling observations center on NEIL1, a DNA glycosylase that increases in abundance during S-phase (Hazra et al., 2002), co-immunoprecipitates with the DNA replication factors PCNA and FEN-1 (Dou et al., 2008; Hegde et al., 2008b), and is able to process lesions in single- as well as double-stranded DNA (Dou et al., 2003; Liu et al., 2010; Takao et al., 2009). These observations suggested that NEIL1 either travels with replication fork machinery or is recruited to forks that have stalled at sites of oxidative damage (e.g. thymine glycol, Tg). This notion is appealing, partly because NEIL1 might provide an error-free means of removing polymerase-blocking lesions, but further studies will be needed to determine if and how NEIL1 gains access to such lesions. Presumably, this requires the dissociation of replicative polymerases, as occurs during the recruitment of specialized DNA polymerases that are able to bypass blocking lesions (reviewed in (Zahn et al., 2011)). As well, the capacity of NEIL1 to process lesions in single-stranded DNA at a replication fork would seem to have a critical downside, in that its lyase activity would generate a DNA break. Such breaks might be repaired via the SSBR pathway. Alternatively, replication up to a NEIL1- or APE-generated nick would likely produce a double strand break that would be channeled into the DDR pathway (as depicted in pathway #3 in Figure 2).
If BER occurred only in the context of DNA replication, base damages would accumulate in non-dividing, terminally differentiated somatic cells. In human adults, non-epithelial cells in the intestine and intercostal skeletal muscle cells are estimated to be 15–16 years old, on average, and most occipital neurons are nearly as old as their hosts (Spalding et al., 2005). Hence, the 20,000 oxidative damages that accumulate each day in human cells, left unrepaired, could grow to involve ~1% of a cell’s total DNA in a decade, and up to 10% of DNA during a lifetime. Although this calculation does not constitute proof of ongoing BER in terminally differentiated cells, it seems inconceivable that such cells could remain viable with a damage burden of this magnitude. An untested but more plausible notion is that BER in non-dividing cells occurs only in transcribed regions, as appears to be the case for nucleotide excision repair (NER) of bulky lesions. Specifically, bulky lesions accumulate over time in non-transcribed regions of non-dividing differentiated cells, but transcription-coupled NER continues to occur (along with repair of bulky lesions in the opposing, non-transcribed DNA strand; (Nouspikel, 2007; Nouspikel and Hanawalt, 2002)). Thus, global NER appears to be attenuated in such cells, possibly providing them with a metabolic advantage. While the extent and efficiency of BER in terminally differentiated cells compared to that in dividing cells has yet to be determined, several workers have asked if BER is coupled to transcription in a manner similar to that of transcription-coupled NER. Most bulky DNA adducts subject to NER stall RNA polymerases, triggering the recruitment of NER factors, whereas most oxidative lesions do not (Hanawalt and Spivak, 2008). Recently, 8-oxoG was reported to inhibit transcription in mouse embryonic fibroblasts, but only when cells contained a functional copy of the DNA glycosylase OGG1 (Kitsera et al., 2011). This result suggests that oxidative lesions do not generally block transcription, but at least some BER processing intermediates do, which could account for several reports of preferential BER of transcribed DNA (e.g. (Reis et al., 2012); reviewed in (Izumi et al., 2003)). However, these observations do not necessarily mean that cells possess a transcription-coupled BER pathway, as it is possible that BER processing intermediates in the path of RNA polymerases are shunted into the transcription-coupled NER pathway. In summary, while the DNA replication-, transcription-, SSBR and DDR-associated disruption of nucleosomes may enhance BER of some oxidative damages, most short-patch BER events occur independently of these processes. The remainder of this review will focus on how these ‘independent’ BER events occur in chromatin.
The discovery of oxidative damages in nucleosomes
The discovery of target sites or oxidative lesions in a vast sea of undamaged DNA must occur at physiologically meaningful rates. Seminal studies by Riggs and Von Hippel and their coworkers indicated that most DNA sequence or structure-specific binding proteins first bind DNA non-specifically (after a diffusion-driven, three-dimensional search), and then locate their specific targets using restricted ‘one-dimensional’ search mechanisms (Riggs et al., 1970; von Hippel et al., 1974). Single molecule studies of DNA sequence-specific binding proteins are consistent with this two-step search mechanism, and suggest that DNA polymerases and repair factors search for their targets in the same fashion (Blainey et al., 2009). The one-dimensional search rates derived from single molecule studies also suggest that such proteins, once non-specifically bound to DNA, move in a helical fashion, tracking along either the sugar-phosphate backbone or one of the DNA grooves (Blainey et al., 2009). Importantly, the one-dimensional search process by DNA glycosylases entails both fast and slow modes (Dunn et al., 2011). The fast mode corresponds to helical tracking along DNA, as just described, whereas the slow mode likely reflects base interrogation, in which the glycosylase inserts an aromatic side chain probe into the double helix (Banerjee et al., 2006). This inference is based on finding that mutation of residues critical for interrogation eliminates the slow search behavior (Dunn et al., 2011). To our knowledge there are no published single molecule studies of target discovery in nucleosomes, although model nucleosome studies described below indicate that BER factors are able to sample bases in nucleosomes. Tracking along either the sugar-phosphate backbone or one of the grooves in nucleosomal DNA would necessarily require at least the transient disruption of histone-DNA contacts. DNA glycosylases and APE also appear able to interrogate a subset of DNA bases in intact nucleosomes; this may entail an enzyme tracking for only one or a few base pairs before steric impediments force it to dissociate and rebind at a nearby site (Berg and von Hippel, 1985; Gowers and Halford, 2003; Qi et al., 2012).
The efficiency of the above-described search mechanisms is governed largely by the concentration and specific- and non-specific DNA binding constants of the protein of interest. Measurements of these parameters for two DNA glycosylases, hNTH1 and NEIL1, both of which can excise the oxidative lesion thymine glycol (Tg), suggested that hNTH1 is much better suited for the global discovery and repair of oxidative damage in chromatin (Odell et al., 2010). These findings are consistent with the notion that NEIL1 is associated with DNA replication forks. Interestingly, NEIL1 but not hNTH1 is found in mitochondria as well as in the nucleus. Presumably, the reduced efficiency with which NEIL1 finds targets in DNA is less of a problem in mitochondria, owing to its far smaller genome size. Although it is outside the scope of this review, mitochondrial DNA is compacted into nucleoid structures (Chen and Butow, 2005; Hu et al., 2005; Ikeda et al., 2002; Takao et al., 2002) that may present accessibility problems analogous to those associated with nucleosomes.
Dynamic properties of nucleosomes facilitate detection and excision of oxidized bases from nucleosomal DNA
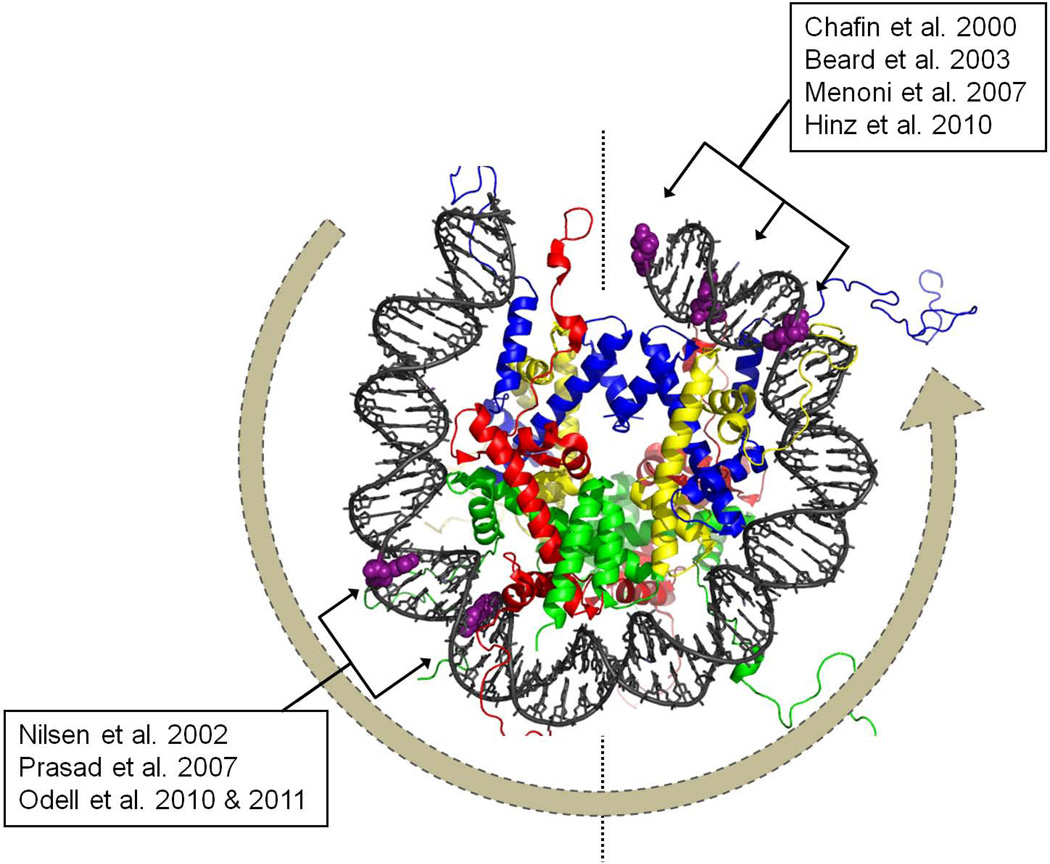
Canonical nucleosomes contain 147 bp of DNA, wrapped ~1.65 times in a left-handed toroidal helix around a histone core (Figure 3; (Luger et al., 1997)). The histone core contains two copies each of histones H2A, H2B, H3 and H4, and interacts closely with the minor groove at regular ~10 base pair intervals on each side of a pseudo-2-fold axis of symmetry called the dyad. Nucleosomes assemble with newly replicated or repaired DNA in a step-wise fashion, beginning with the wrapping of DNA about a histone H3-H4 tetramer, followed by the addition of two histone H2A-H2B dimers that help organize the outermost wraps of nucleosomal DNA. Thermal denaturation studies during the late 1970’s, in conjunction with nuclease accessibility and electron microscopy studies, suggested that DNA near the edges of the nucleosome (associated mainly with H2A-H2B dimers) more readily dissociates from the underlying histone octamer than does DNA in the central wrap (reviewed in (van Holde, 1988)). Fifteen years later, Widom and colleagues reported that restriction enzymes could partially cleave target sites in nucleosomal DNA, with an efficiency proportional to the distance between the target site and the dyad axis, and noted that partial and transient DNA unwrapping from the nucleosome edges could account for this phenomenon (Anderson and Widom, 2000; Polach and Widom, 1995). Concurrent high resolution DNA footprinting and transcription studies suggested that partial unwrapping of nucleosomal DNA occurs in vivo as well, and may in some cases facilitate the binding of transcription factors (Geraghty et al., 1998). Widom and colleagues also conducted FRET studies which indicated that, on average, DNA unwrapping events occur several times each second at physiological temperatures, with unwrapped configurations persisting for ~25 ms (Li and Widom, 2004). Later studies, suggesting that unwrapping events are far more frequent, were eventually discovered to have been influenced by photoblinking, a phenomenon in which the acceptor dye transiently shifts to a non-emitting state (Tomschik et al., 2009). A recent evaluation of published FRET-based studies of nucleosome dynamics in (Buning and van Noort, 2010) lent further support to the original kinetic estimates of Widom and colleagues.
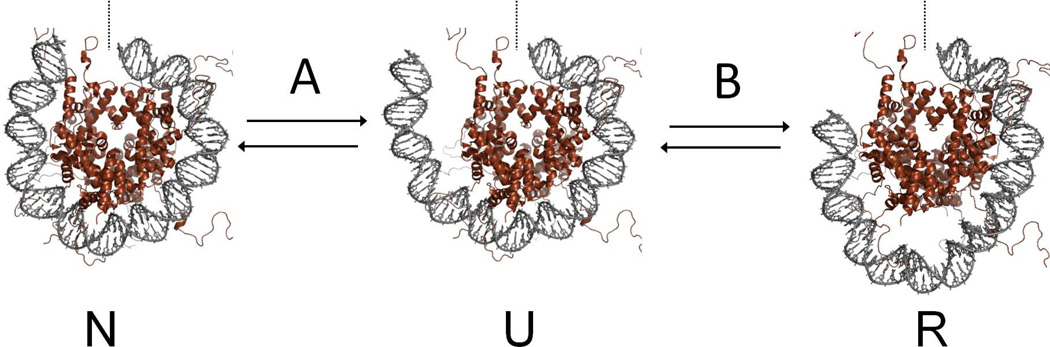
Figure 3. Dynamic properties of nucleosomes that permit BER.
(A) Location of DNA base damages in model nucleosomes used to study BER. The nucleosome structure is adapted from (Davey et al., 2002), pdb 1KX5. Histones H2A, H2B, H3, and H4 are colored red, green, blue, and yellow, respectively. For clarity, only one wrap of the DNA is shown, with outermost DNA on the left. Symbols: dotted line, dyad axis; purple spheres, DNA base lesions; tan arrow, direction of spontaneous, transient partial DNA unwrapping that facilitates processing of sterically-occluded lesions. (B) Transitions between nucleosome states. Spontaneous, partial unwrapping of DNA from the histone octamer transforms the canonical nucleosome (N) to an unwrapped configuration (U) that enables the BER of sterically occluded lesions. Out of register re-wrapping (possibly facilitated by a chromatin remodeling agent) may generate configuration R, containing a DNA loop that can lead to a shift in nucleosome position. Although such shifts have not been observed in in vitro studies of BER on model nucleosomes, they may contribute to lesion exposure and BER in vivo.
Most nucleosome-sized or longer DNA segments exhibit a preferred helical orientation relative to the histone octamer. This has made it possible to assemble well-defined model nucleosomes to investigate their impact on a number of processes, including BER. Four laboratories (including ours) have assembled nucleosomes that contain defined oxidative lesions at discrete sites, to investigate how BER occurs in nucleosomes (Figure 3). Results from these studies are largely consistent with a model in which the accessibility of substrates in nucleosomes is governed both by enzyme structure and three rules that reflect the structure and dynamic properties of nucleosomes. The first rule is is that the efficiency with which oxidatively damaged bases and AP sites in nucleosomes are processed depends on their helical orientation relative to the histone octamer. Both DNA glycosylases and APE bind the lesion-containing strand of DNA, and extrude the damaged base or AP-site from the duplex into an extra-helical recognition pocket (Fromme and Verdine, 2003; Gorman et al., 1997; Mol et al., 2000; Parikh et al., 1998; Slupphaug et al., 1996). Both DNA glycosylases and APE can readily process substrates in nucleosomes, provided those substrates are located such that they can flip into an exta-helical configuration without clashing with the histone octamer (Beard et al., 2003; Hinz et al., 2010; Odell et al., 2011; Odell et al., 2010; Prasad et al., 2007).
The second rule is that spontaneous, reversible partial unwrapping of nucleosomal DNA facilitates BER of oxidative lesions in nucleosomes, particularly those that are, nominally, sterically occluded. (Prasad et al., 2007) reported that, at an enzyme concentration sufficient for a maximal rate of excision of optimally oriented (“outward facing”) lesions from nucleosomes by the DNA glycosylase hNTH1, the efficiency of excision of occluded (“inward facing”) lesions was poor. However, further increases in the concentration of hNTH1 substantially increased the excision of inward-facing lesions. This result was reminiscent of observations by Widom and colleagues on the accessibility of restriction sites in nucleosomes. (Prasad et al., 2007) reported as well that, regardless of enzyme concentration, lesion excision occurred without irreversibly altering the lesion-containing nucleosome or its translational position. Importantly, the concentration of hNTH1 required for the efficient excision of inward-facing lesions was no higher than its estimated in vivo concentration (Liu et al., 2003; Odell et al., 2010). The high hNTH1 concentrations needed to process lesions at positions that are nominally inaccessible can be related to the frequency and duration of DNA unwrapping episodes that expose these lesions to solvent (Prasad, Rizvanova, Maher, Wallace and Pederson, manuscript in revision). Later studies indicated that enzyme concentrations have similarly dramatic effects on the efficiency with which other DNA glycosylases, APE, and Pol β process occluded substrates in nucleosomes (Hinz et al., 2010; Odell et al., 2011; Odell et al., 2010).
Nucleosome stretching experiments, using optical tweezers, indicate that half of the DNA in a nucleosome can be unwrapped without irreversibly disrupting the nucleosome (reviewed by (Bednar and Dimitrov, 2011)). Given that both the DNA and protein components contribute to nucleosome solubility and stability at physiological ionic strengths, it is surprising that nucleosomes could survive such extensive unwrapping. Possibly, elimination of the repulsive forces between adjacent turns of the negatively charged DNA backbone, as a result of unwrapping, strengthens the histone-DNA contacts in the half of the nucleosome that remains intact (Schiessel, 2006). In any event, spontaneous DNA unwrapping appears to be sufficient, in principle, for BER enzymes to sample the entire DNA in a nucleosome. However, the exceedingly low frequency of exposure of DNA close to the nucleosome dyad (cf. below) makes it unlikely that cells rely exclusively on intrinsically-driven DNA unwrapping as a means of lesion discovery.
The third rule is that BER substrates close to the dyad axis are processed less efficiently than more distant substrates that share the same helical orientation. Much of the variability in the literature on the impact of nucleosomes on BER can be reconciled by relating enzyme accessibility to the distance between lesions and the nucleosome dyad (Figure 3A). In two studies, lesions in the outer wrap of the nucleosome (> 40 nt from the dyad) were almost fully repaired in reactions containing all the enzymes needed for short-patch BER (Nilsen et al., 2002; Odell et al., 2011). By contrast, DNA lesions near the dyad axis of the nucleosome could be excised by a DNA glycosylase, but subsequent steps were completely inhibited (Beard et al., 2003; Menoni et al., 2007). The distance-dependent excision of oxidative lesions is most easily explained by assuming that the length of DNA exposed during a transient unwrapping event is stochastic, such that the frequency with which lesions near the dyad axis become available to BER enzymes is exceedingly low.
Unlike the DNA glycosylases and APE, Pol β could use a bit of help
The above-listed rules can accommodate observations from several groups, and make sense from the standpoint of nucleosome structure, but do not entirely account for significant differences in the efficiency with which successive steps in BER are carried out in nucleosomes. The capacity of a given enzyme to bind and act on DNA in intact (fully wrapped) nucleosomes necessarily reflects the degree to which the enzyme encircles DNA upon binding and the extent to which it bends or otherwise distorts DNA in its active configuration. Both DNA glycosylases and APE bind, roughly, one-half (~180 degrees) of the DNA helix. This may account for their capacity to process nucleosomal substrates that are ‘appropriately’ oriented relative to the underlying histone octamer. At 39 kDa, Pol β is the smallest eukaryotic DNA polymerase, and similar in size to the bifunctional DNA glycosylases and APE, which range from 35–44 kDa (Demple et al., 1991; Hazra et al., 2002; Ikeda et al., 1998; Nishioka et al., 1999). Pol β also binds DNA asymmetrically, primarily making contacts with the DNA backbone near the nucleotide gap (Beard and Wilson, 2000). This is in contrast to the larger, replicative DNA polymerases, which contact a larger number of DNA base pairs and thus encircle more of the DNA helix. Consistent with its asymmetric binding to the gap-containing strand of DNA, Pol β is able to fill a single-nucleotide gap, provided the gap-containing strand of DNA faces away from the histone octamer and is located >40 nt from the dyad axis (Nilsen et al., 2002; Odell et al., 2011). Also consistent with its one sided DNA binding, the activity of Pol β was substantially reduced when the gap faced inward towards the histone octamer (Odell et al., 2011).
Whereas the DNA glycosylases induce their DNA substrates to bend anywhere from 45° to 70° (Bruner et al., 2000; Fromme and Verdine, 2003; Zharkov et al., 2002), Pol β bends DNA opposite a single base gap by 90° (Pelletier et al., 1994; Pelletier et al., 1996). This more severe bending may limit the capacity of Pol β to function without disrupting histone-DNA contacts. Conceivably, Pol β can disrupt enough histone-DNA contacts to take on an active configuration when binding to the outer wrap of nucleosomal DNA, but not when presented with a DNA gap near the nucleosome dyad, where DNA is constrained by multiple contacts on both sides. This could explain why Pol β had virtually no activity on a DNA gap near the dyad axis, irrespective of helical orientation (Beard et al., 2003; Menoni et al., 2007). Thus, at least for centrally located substrates, Pol β likely requires nucleosome remodeling or disruption to act. As described below, help for Pol β may arrive in the form of the next enzyme complex in BER, namely LigIIIα-XRCC1.
DNA ligation occurs in nucleosomes, but how?
Mammalian cells contain three DNA ligases, LigI, LigIII and LigIV (reviewed in (Ellenberger and Tomkinson, 2008)). LigI joins Okazaki fragments during DNA replication (Levin et al., 2000), whereas LigIV functions in both NHEJ and V(D)J recombination. LigIII is differentially spliced, giving rise to α and β variants (Mackey et al., 1997; Perez-Jannotti et al., 2001). LigIIIα is expressed in somatic cells and is stable in nuclei only when complexed with the scaffold protein XRCC1 (LigIIIα-XRCC1) (Caldecott et al., 1994); in mitochondria, LigIIIα interacts instead with Pol γ (De and Campbell, 2007). LigIIIβ is expressed in germ cells and does not require XRCC1. The LigIIIα-XRCC1 complex catalyzes the final step of short-patch BER, whereas LigI, by virtue of its interaction with PCNA, catalyzes the final step of long-patch BER; both LigI and LigIII function in other repair pathways as well.
LigI and LigIII are each just over 100 kDa in size, substantially larger than the enzymes that catalyze the first three steps in short-patch BER, and differ dramatically in structure as well. Each ligase contains an N-terminal DNA binding domain (DBD), followed by a catalytic core, which can be further divided into a nucleotidyltransferase domain (NTD) and an OB-fold domain (OBD) (Ellenberger and Tomkinson, 2008). In contrast to the DNA glycosylases, APE, and Pol β, DNA ligases fully encircle their DNA substrates. The DBD binds the minor groove of both strands of DNA, up and downstream of the nick, while the OBD interacts with the minor groove downstream of the nick. The three protein segments are flexible and allow the DNA ligases to open and close around their substrates. These observations led to a “jackknife” model for the binding of LigIIIα, which begins with detection of a single strand break by an N-terminal zinc finger (ZnF), followed by progressive DNA binding by the DBD, NTD, and OBD (Cotner-Gohara et al., 2010).
Because DNA ligases completely encircle their DNA substrates, the capacity of LigIIIα-XRCC1 to function during BER in nucleosomes would, at minimum, require breaking of local histone-DNA contacts. This prediction proved true, in that the activity of LigIIIα-XRCC1 on nucleosome substrates in vitro depended critically on enzyme concentration, regardless of substrate orientation (Odell et al., 2011). As this result suggested that DNA unwrapping enables LigIIIα-XRCC1 to bind substrates in nucleosomes, it was somewhat surprising to find that binding of LigIIIα-XRCC1 to either gap or nick-containing nucleosomes also led to nucleosome disruption (Odell et al., 2011). Importantly, it is an N-terminal ZnF that enables LigIIIα to bind to gapped DNA. This ZnF, which LigI lacks, may be critical to the capacity of LigIIIα-XRCC1 to bind and disrupt gap-containing nucleosomes, thereby enhancing the activity of Pol β. These findings provided the first evidence that nucleosome disruption accompanies both the polymerization and ligation steps in BER, as depicted in Figure 1.
It is not known if ligases other than LigIIIα-XRCC1 induce nucleosome disruption, nor if the nucleosome disruption induced by LigIIIα depends on its association with XRCC1 (by itself, XRCC1 does not disrupt nucleosomes (Odell et al., 2011)). Full-length LigI was reported to function at a ~10-fold reduced rate on nucleosome substrates (Chafin et al., 2000). Interestingly, the catalytic cores of LigI and LigIII (the NTD and OBD) bind one side of the DNA duplex (Cotner-Gohara et al., 2010; Pascal et al., 2004) and are capable of ligating DNA substrates in the absence of the DBD, again at a ~10-fold reduced rate (Cotner-Gohara et al., 2008; Pascal et al., 2004). Taken together, these observations suggest DNA ligases might not invariably need to fully encircle DNA when acting on nucleosomal substrates.
Future Prospects
Virtually nothing is known about the possible involvement of histone chaperones or ATP-dependent remodeling agents in BER. The prototypical chromatin remodeling complex SWI/SNF was reported to increase the efficiency of the first three steps of BER in a model nucleosome (Menoni et al., 2007), and ISW1 and ISW2 were reported to enhance excision of uracil residues from oligo-nucleosome arrays in vitro (Nakanishi et al., 2007). However, it remains to be determined whether any of these agents act during BER in vivo and, if so, what is responsible for their recruitment to sites of oxidative damage in chromatin. Clearly, these areas represent an important avenue for future studies.
The possible impact of histone secondary modifications on the efficiency of BER in nucleosomes has not been studied in a systematic fashion. Given that acetylation of K56 in histone H3 increases DNA unwrapping events, we predict that such modifications will prove important in the regulation of BER in chromatin. So far, however, we can note only that model nucleosomes assembled with unmodified recombinant histones have yielded results very similar to those based on nucleosomes assembled by octamer transfer, using donor chromatin (containing a diverse array of histone modifications) purified from chicken erythrocytes.
Acknowledgements
We thank Drs. Jeff Bond, Sylvie Doublié, Robyn Maher, Scott Morrical, Joann Sweasy, and past and current members of the Pederson lab for helpful conversations, and Anahí Odell and current members of the Pederson lab for critical comments on the manuscript. This research was supported by grants from the NSF (MCB-0821941) and the NCI (P01-CA098993).
References
- Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. Journal of molecular biology. 2000;296(4):979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual review of plant biology. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science (New York, NY. 2006;311(5764):1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(13):7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutation research. 2000;460(3–4):231–244. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- Bednar J, Dimitrov S. Chromatin under mechanical stress: from single 30 nm fibers to single nucleosomes. The FEBS journal. 2011;278(13):2231–2243. doi: 10.1111/j.1742-4658.2011.08153.x. [DOI] [PubMed] [Google Scholar]
- Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annual review of biophysics and biophysical chemistry. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nature structural & molecular biology. 2009;16(12):1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell JO, Harrison L, Wallace SS. Base excision repair processing of radiation-induced clustered DNA lesions. Radiation protection dosimetry. 2001;97(1):25–31. doi: 10.1093/oxfordjournals.rpd.a006634. [DOI] [PubMed] [Google Scholar]
- Boldogh I, Hajas G, Aguilera-Aguirre L, Hegde ML, Radak Z, Bacsi A, Sur S, Hazra TK, Mitra S. Activation of Ras Signaling by 8-oxoguanine DNA glycosylase Bound to Its Excision Product 8-oxoguanine. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free radical biology & medicine. 1995;18(6):1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403(6772):859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- Buning R, van Noort J. Single-pair FRET experiments on nucleosome conformational dynamics. Biochimie. 2010;92(12):1729–1740. doi: 10.1016/j.biochi.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nature reviews Genetics. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Molecular and cellular biology. 1994;14(1):68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafin DR, Vitolo JM, Henricksen LA, Bambara RA, Hayes JJ. Human DNA ligase I efficiently seals nicks in nucleosomes. The EMBO journal. 2000;19(20):5492–5501. doi: 10.1093/emboj/19.20.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nature reviews Genetics. 2005;6(11):815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(43):18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotner-Gohara E, Kim IK, Hammel M, Tainer JA, Tomkinson AE, Ellenberger T. Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States. Biochemistry. 2010 doi: 10.1021/bi100503w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotner-Gohara E, Kim IK, Tomkinson AE, Ellenberger T. Two DNA-binding and nick recognition modules in human DNA ligase III. The Journal of biological chemistry. 2008;283(16):10764–10772. doi: 10.1074/jbc.M708175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Hazra TK, Boldogh I, Mitra S, Bhakat KK. Induction of the human oxidized base-specific DNA glycosylase NEIL1 by reactive oxygen species. The Journal of biological chemistry. 2005;280(42):35272–35280. doi: 10.1074/jbc.M505526200. [DOI] [PubMed] [Google Scholar]
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. Journal of molecular biology. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A, Campbell C. A novel interaction between DNA ligase III and DNA polymerase gamma plays an essential role in mitochondrial DNA stability. The Biochemical journal. 2007;402(1):175–186. doi: 10.1042/BJ20061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B, Herman T, Chen DS. Cloning and expression of APE the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(24):11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M. Oxidatively induced DNA damage: Mechanisms, repair and disease. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. The Journal of biological chemistry. 2003;278(50):49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. The Journal of biological chemistry. 2008;283(6):3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- Duclos S, Doublie S, Wallace SS. Consequences and Repair of Oxidative DNA Damage. The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carcinogens: The Royal Society of Chemistry. 2012:115–159. [Google Scholar]
- Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic acids research. 2011;39(17):7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annual review of biochemistry. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright H, Miller WJ, Hays R, Floyd RA, Hebbel RP. Preferential targeting of oxidative base damage to internucleosomal DNA. Carcinogenesis. 1996;17(5):1175–1177. doi: 10.1093/carcin/17.5.1175. [DOI] [PubMed] [Google Scholar]
- Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nature reviews Microbiology. 2004;2(10):820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species. The Journal of cell biology. 2011;194(1):7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 2006. xxix, 1118 p. 1111p. of plates p. [Google Scholar]
- Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Current opinion in structural biology. 2004;14(1):43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. The EMBO journal. 2003;22(13):3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty DS, Sucic HB, Chen J, Pederson DS. Evidence that partial unwrapping of DNA from nucleosomes facilitates the binding of heat shock factor following DNA replication in yeast. The Journal of biological chemistry. 1998;273(32):20463–20472. doi: 10.1074/jbc.273.32.20463. [DOI] [PubMed] [Google Scholar]
- Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, Hickson ID, Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. The EMBO journal. 1997;16(21):6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers DM, Halford SE. Protein motion from non-specific to specific DNA by three-dimensional routes aided by supercoiling. The EMBO journal. 2003;22(6):1410–1418. doi: 10.1093/emboj/cdg125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nature reviews. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Hanssen-Bauer A, Solvang-Garten K, Sundheim O, Pena-Diaz J, Andersen S, Slupphaug G, Krokan HE, Wilson DM, 3rd, Akbari M, Otterlei M. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environmental and molecular mutagenesis. 2011;52(8):623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. Journal of molecular biology. 1999;290(3):667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- Hayes JJ, Tullius TD, Wolffe AP. The structure of DNA in a nucleosome. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(19):7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA repair. 2007;6(4):470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(6):3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell research. 2008a;18(1):27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. The Journal of biological chemistry. 2008b;283(40):27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz JM, Rodriguez Y, Smerdon MJ. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(10):4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. The Journal of biological chemistry. 2005;280(49):40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. The Journal of biological chemistry. 1998;273(34):21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kohmoto T, Tabata R, Seki Y. Differential intracellular localization of the human and mouse endonuclease III homologs and analysis of the sorting signals. DNA repair. 2002;1(10):847–854. doi: 10.1016/s1568-7864(02)00145-3. [DOI] [PubMed] [Google Scholar]
- Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193(1–2):43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- Kitsera N, Stathis D, Luhnsdorf B, Muller H, Carell T, Epe B, Khobta A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic acids research. 2011;39(14):5926–5934. doi: 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) The EMBO journal. 1997;16(11):3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi CI, Suda T. Regulation of reactive oxygen species in stem cells and cancer stem cells. J Cell Physiol. 2012;227(2):421–430. doi: 10.1002/jcp.22764. [DOI] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Current opinion in cell biology. 2008;20(3):294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol. 2000;10(15):919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- Li G, Widom J. Nucleosomes facilitate their own invasion. Nature structural & molecular biology. 2004;11(8):763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. Quality control by DNA repair. Science (New York, NY. 1999;286(5446):1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(11):4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Choudhury S, Roy R. In vitro and in vivo dimerization of human endonuclease III stimulates its activity. The Journal of biological chemistry. 2003;278(50):50061–50069. doi: 10.1074/jbc.M309997200. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, van Attikum H. Chromatin and the DNA damage response: the cancer connection. Molecular oncology. 2011;5(4):349–367. doi: 10.1016/j.molonc.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey ZB, Ramos W, Levin DS, Walter CA, McCarrey JR, Tomkinson AE. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Molecular and cellular biology. 1997;17(2):989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: Is subcellular localization the answer? Cell Cycle. 2009;8(8):1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meira LB, Burgis NE, Samson LD. Base excision repair. Adv Exp Med Biol. 2005;570:125–173. doi: 10.1007/1-4020-3764-3_5. [DOI] [PubMed] [Google Scholar]
- Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Molecular and cellular biology. 2007;27(17):5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Boldogh I, Izumi T, Hazra TK. Complexities of the DNA base excision repair pathway for repair of oxidative DNA damage. Environmental and molecular mutagenesis. 2001;38(2–3):180–190. doi: 10.1002/em.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Izumi T, Boldogh I, Bhakat KK, Chattopadhyay R, Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA repair. 2007;6(4):461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403(6768):451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Prasad R, Wilson SH, Smerdon M. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic acids research. 2007;35(13):4313–4321. doi: 10.1093/nar/gkm436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. The EMBO journal. 2002;21(21):5943–5952. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Molecular biology of the cell. 1999;10(5):1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouspikel T. DNA repair in differentiated cells: some new answers to old questions. Neuroscience. 2007;145(4):1213–1221. doi: 10.1016/j.neuroscience.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA repair. 2002;1(1):59–75. doi: 10.1016/s1568-7864(01)00005-2. [DOI] [PubMed] [Google Scholar]
- Odell ID, Barbour JE, Murphy DL, Della-Maria JA, Sweasy JB, Tomkinson AE, Wallace SS, Pederson DS. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Molecular and cellular biology. 2011;31(22):4623–4632. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell ID, Newick K, Heintz NH, Wallace SS, Pederson DS. Non-specific DNA binding interferes with the efficient excision of oxidative lesions from chromatin by the human DNA glycosylase, NEIL1. DNA repair. 2010;9(2):134–143. doi: 10.1016/j.dnarep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. The EMBO journal. 1998;17(17):5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432(7016):473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science (New York, NY. 1994;264(5167):1891–1903. [PubMed] [Google Scholar]
- Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35(39):12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- Perez-Jannotti RM, Klein SM, Bogenhagen DF. Two forms of mitochondrial DNA ligase III are produced in Xenopus laevis oocytes. The Journal of biological chemistry. 2001;276(52):48978–48987. doi: 10.1074/jbc.M107177200. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. Journal of molecular biology. 1995;254(2):130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes & development. 2011;25(5):409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Molecular and cellular biology. 2007;27(24):8442–8453. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. The Journal of biological chemistry. 2010;285(52):40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Nam K, Spong MC, Banerjee A, Sung RJ, Zhang M, Karplus M, Verdine GL. Strandwise translocation of a DNA glycosylase on undamaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(4):1086–1091. doi: 10.1073/pnas.1111237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana CV, Boldogh I, Izumi T, Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(9):5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis AM, Mills WK, Ramachandran I, Friedberg EC, Thompson D, Queimado L. Targeted detection of in vivo endogenous DNA base damage reveals preferential base excision repair in the transcribed strand. Nucleic acids research. 2012;40(1):206–219. doi: 10.1093/nar/gkr704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. Journal of molecular biology. 1970;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66(6):981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessel H. The nucleosome: a transparent, slippery, sticky and yet stable DNA-protein complex. The European physical journal E, Soft matter. 2006;19(3):251–262. doi: 10.1140/epje/i2005-10049-y. [DOI] [PubMed] [Google Scholar]
- Sinha M, Peterson CL. Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics. 2009;1(2):371–385. doi: 10.2217/epi.09.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384(6604):87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122(1):133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic acids research. 2011;39(8):3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, Weinfeld M, Kobayashi K, Miyazaki J, Muijtjens M, Hoeijmakers JH, van der Horst G, Yasui A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. The EMBO journal. 2002;21(13):3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Oohata Y, Kitadokoro K, Kobayashi K, Iwai S, Yasui A, Yonei S, Zhang QM. Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells. 2009;14(2):261–270. doi: 10.1111/j.1365-2443.2008.01271.x. [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nature structural & molecular biology. 2009;16(9):923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Tomschik M, van Holde K, Zlatanova J. Nucleosome dynamics as studied by single-pair fluorescence resonance energy transfer: a reevaluation. Journal of fluorescence. 2009;19(1):53–62. doi: 10.1007/s10895-008-0379-1. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends in cell biology. 2009;19(5):207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- van Holde KE. In: Chromatin. Rich A, editor. Springer-Verlag; New York: 1988. [Google Scholar]
- von Hippel PH, Revzin A, Gross CA, Wang AC. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: I. The lac operon: equilibrium aspects. Proceedings of the National Academy of Sciences of the United States of America. 1974;71(12):4808–4812. doi: 10.1073/pnas.71.12.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012 doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JF. Biochemistry of DNA lesions. Radiation research. 1985;(Supplement 8):S103–S111. [PubMed] [Google Scholar]
- Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Progress in nucleic acid research and molecular biology. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nature structural biology. 2000;7(3):176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- Yang N, Galick H, Wallace SS. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA repair. 2004;3(10):1323–1334. doi: 10.1016/j.dnarep.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Zahn KE, Wallace SS, Doublie S. DNA polymerases provide a canon of strategies for translesion synthesis past oxidatively generated lesions. Current opinion in structural biology. 2011;21(3):358–369. doi: 10.1016/j.sbi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. The EMBO journal. 2002;21(4):789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]