Abstract
Misregulation of DNA repair is associated with genetic instability and tumorigenesis. To preserve the integrity of the genome, eukaryotic cells have evolved extremely intricate mechanisms for repairing DNA damage. One type of DNA lesion is a double-strand break (DSB), which is highly toxic when unrepaired. Repair of DSBs can occur through multiple mechanisms. Aside from religating the DNA ends, a homologous template can be used for repair in a process called homologous recombination (HR). One key step in committing to HR is the formation of Rad51 filaments, which perform the homology search and strand invasion steps. In S. cerevisiae, Srs2 is a key regulator of Rad51 filament formation and disassembly. In this review, we highlight potential candidates of Srs2 orthologues in human cells, and we discuss recent advances in understanding how Srs2’s so-called “anti-recombinase” activity is regulated.
Keywords: Homologous recombination, Srs2, Rad51, PARI, RTEL, Rad51 paralogues, Shu complex, Rad55-Rad57, Cancer, Genetic Instability
1. Introduction: Overview of DSB repair by HR
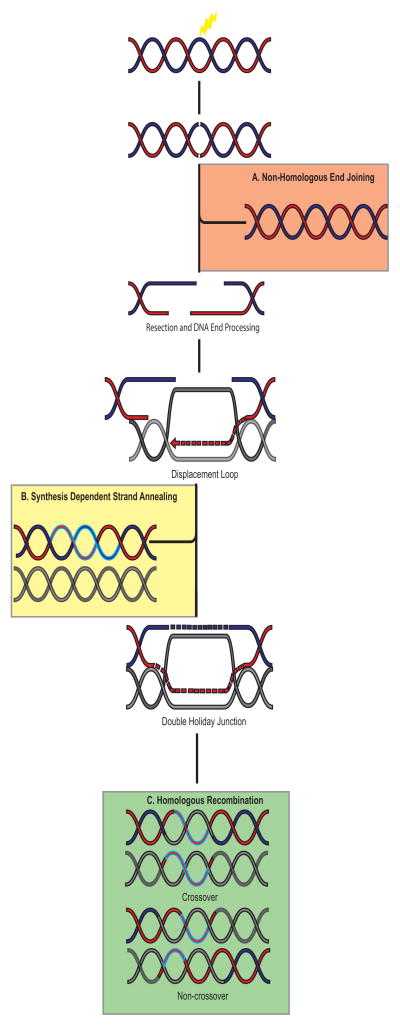
Repair of DNA damage is essential to preventing mutations and chromosomal rearrangements. The DNA repair process involves many proteins working together in a tightly regulated system to fix the damage. Double-strand breaks (DSBs) are one of the most lethal types of DNA damage, and can arise from both endogenous (i.e. replicative damage or reactive oxygen species) and exogenous sources (i.e. radon). Cells must repair approximately 50 DSBs per day, which correlates with a frequency of one DSB per 108 bp per cell cycle [1]. Mutations in genes important for DSB repair have been implicated in many cancer predisposition diseases such as ataxia telangiectasia, Nijmegen breakage syndrome, and Bloom syndrome [2]. The proteins required for DSB repair are highly conserved in eukaryotes from yeast to humans, which highlight the importance of DNA repair throughout evolution. Consequently, cells have evolved many different pathways in the repair of DSBs. For example, the DNA ends can be re-ligated together during non-homologous end joining (NHEJ; Figure 1A) and micro-homology mediated end joining or, alternatively, a homologous template could be used for repair as in homologous recombination (HR; Figure 1B and 1C)[3].
Figure 1.
Summary of the major double-strand break (DSB) repair pathways. A. Non-homologous end joining (NHEJ) is the simplest form of DNA DSB repair, because it does not require a homologous template. Broken ends are religated together. The advantages of this process are that it is quick and efficient. The disadvantage is that it is a potentially error-prone repair mechanism since no template is used for repair and genetic information can be lost at the breaksite. B. Synthesis dependent strand annealing (SDSA) is a form of non-crossover homologous recombination, in that it uses a homologous sequence as a template for DNA repair. However, this process does not involve second end capture of the DNA end. After end processing one strand is synthesized using a homologous template and then is re-ligated to the broken end. The newly synthesized strand is then used as a template and base pairs to the complimentary sequence, consequently resolving the break. C. Homologous recombination uses a homologous template for repair. After the DNA ends are resected, the ssDNA 3′ overhang invades a homologous sequence and restores any missing information at the break site. The second end of the DNA is captured resulting in a double-Holliday junction that can be resolved into a crossover or non-crossover product depending upon where the junctions are cut. Legend: Red-Blue double-helix is the broken molecule of DNA. Grey DNA is the homologous sequence. Blue highlighted segments of DNA are newly synthesized pieces. Figure is not to scale.
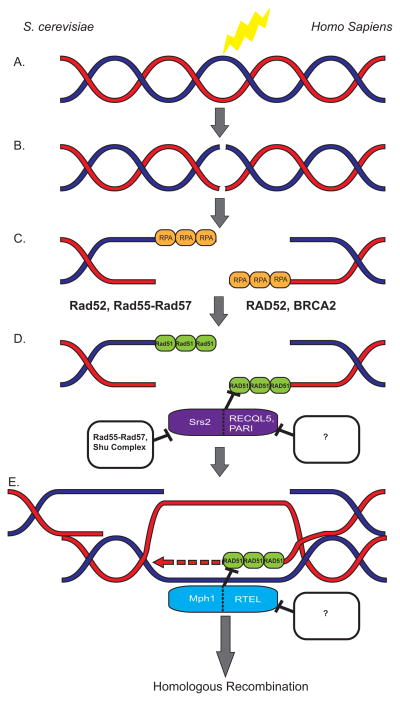
How does a cell decide which repair pathway to utilize? This has been the topic of much investigation, and recently some pieces of the puzzle have been illuminated by the discovery of novel regulators of the DNA repair pathways. During HR, when a DSB is induced in a cell, the DNA ends are resected and replication protein A (RPA) binds the single-stranded DNA (ssDNA) overhangs that are produced and serves as a general marker for ssDNA in a cell (Figure 2A–C)[4, 5]. The multimeric RPA filaments on the ssDNA also serve to protect the unstable ssDNA from further damage. In order for HR to occur, the recombinase protein Rad51 must displace RPA on the ssDNA and form its own filaments. This process is facilitated, in part, by Rad52 in yeast [6], and BRCA2 and RAD52 in humans (Figure 2C and 2D)[7–9]. In mammalian cells, the five RAD51 paralogues (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3) are required for RAD51 focus formation [10, 11]. Formation of Rad51 filaments is in essence the crux of the DSB repair pathway, because the Rad51 nucleo-protein filament is essential for all subsequent homology search and strand invasion steps of HR.
Figure 2.
Regulation of homologous recombination in yeast and humans. A,B. A double-strand break is induced. Some of the most common causes of DSBs include radon, radiation, reactive oxygen species, cisplatin, etc. C. After DNA end resection, the 3′ overhangs are coated with RPA, which forms a filament and is a general marker for ssDNA in the cell. D. Replacing RPA with RAD51 is an important step in initiating HR. In yeast Rad52, as well as Rad55-Rad57, helps load Rad51 onto ssDNA thus promoting HR. In humans, RAD52 and BRCA2 function as positive regulators of HR by facilitating the disassembly of RPA filaments and the nucleation and expansion of RAD51 filaments. Both pre-synaptic Rad51 regulation (D.) and D-loop disassembly (E.) is mediated by Srs2 in yeast. Srs2 is negatively regulated by both the Shu complex and the Rad55-Rad57 heterodimer. In humans presynaptic regulation of RAD51 is accomplished by RECQL5 and PARI. Both of these proteins interact with PCNA, but how they are regulated has yet to be fully elucidated. The next step after Rad51 filament formation is the homology search, which requires less than 16 bp for pairing and 80 bp for strand exchange [72]. E. The disassembly of the D-loop is another way to regulate homologous recombination. In yeast, D-loop disassembly is mediated by Srs2 while in humans, RTEL performs an analogous function. Figure is not to scale.
After the DNA ends are resected and Rad51 filaments are formed, a cell is committed to perform HR in order to repair the damaged DNA template (Reviewed in [4, 12]). Rad51 mediates the search for the homologous DNA sequence and, once the homologous sequence is found, Rad51 filaments facilitate the invasion of the ssDNA overhang into the homologous double-stranded DNA (dsDNA) sequence (Figure 1 and Figure 2). Thus, one strand of the duplex DNA is displaced leaving the complementary strand to serve as a template for repair. This specific recombination structure is referred to as a displacement-loop (D-loop; Figure 1). The step in the HR pathway when the D-loop is formed is referred to as a synapsis; consequently, the homologous recombination steps that occur before D-loop formation are referred to as pre-synaptic whereas the latter steps are post-synaptic. The invading end of the D-loop can be extended by the DNA polymerase, which would then copy any information that might be missing at the breaksite. Resolution of the D-loop structure can occur by two different mechanisms. The invading strand of the DNA can be displaced and reanneal to the other broken chromosome end in a process called synthesis dependent strand annealing (SDSA), which leads to only non-crossover products (Figure 1B). Alternatively, the second end of the DSB can be captured, giving rise to a structure called a double-Holliday junction (dHJ; Figure 1). Resolution of the dHJs can result in a crossover or non-crossover product (Figure 1C), with the non-crossover product being favored in mammalian somatic cells. Too much or too little HR can be toxic to a cell. For example, a cell that undergoes too much HR is defined as having a hyper-recombinant phenotype. This can result in gross chromosomal rearrangements including duplications, deletions, and translocations [13]. On the other hand, homologous recombination has the highest fidelity in the repair of dsDNA damage because it utilizes a homologous template. HR is also needed for proper chromosome segregation during meiosis. Therefore, too little HR can lead to an accumulation of mutations [3, 4, 12]. In this review, we will focus on the key regulators of Rad51 filament formation through evolution from yeast to humans.
2. Srs2 and the negative regulators of Rad51
2.1 Yeast regulators of Rad51
Rad51 plays a central role in facilitating recombination by performing the homology search and strand invasion steps of HR. Therefore, regulating Rad51 filament formation is important for promoting error-free DNA repair. In yeast, Srs2 is one of the most important antagonists of Rad51, thereby helping to protect the cell from inappropriate HR. Srs2 removes Rad51 from ssDNA ends, thereby preventing the homology search of HR (Figure 2D)[14]. After strand invasion, Srs2 is unable to disassemble d-loops. The presynaptic regulator of Rad51 in yeast is Mph1, which functions by disassembling D-loops formed during strand invasion of the homologous template (Figure 2E)[15]. In higher eukaryotes, such as mammals, functional homologues of Srs2 have not been described until recently. Here, we will discuss how higher eukaryotes have evolved multiple “Srs2-like” proteins that genetically interact and perform related functions to the yeast Srs2 protein.
To appreciate how the regulation of HR evolved in higher eukaryotes, it is important to understand how Srs2’s structure and protein interactions impact its function in regulating Rad51. Srs2 is a member of the UvrD family of DNA helicases. [4, 16]. Srs2 directly interacts with Rad51 and stimulates Rad51’s intrinsic ATP-hydrolysis activity leading to the subsequent release of Rad51 monomers from the ssDNA end (Figure 3)[4]. It is proposed that Srs2’s helicase functions to move itself down to the adjacent Rad51 protein to perpetuate the Rad51 disassembly process [17–19]. It is important to note that Srs2 disrupts Rad51 filament formation using a mechanism that is independent of its helicase function [17, 20]. Consistent with a fundamental role for Srs2 in DSB repair, cells with disruption in SRS2 exhibit a hyper-recombinant phenotype leading to genomic instability [21, 22].
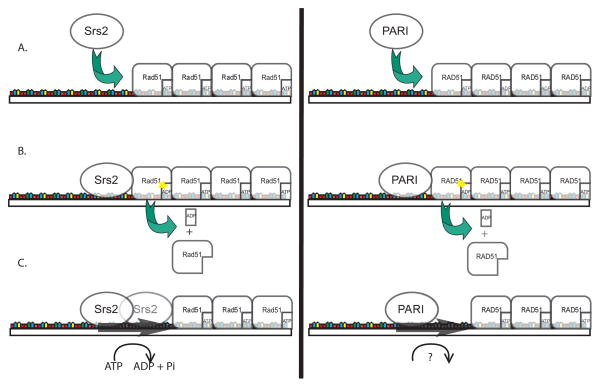
Figure 3.
Proposed mechanisms of Srs2 and PARI. PARI has many similarities to the yeast anti-recombinase Srs2, suggesting that PARI is a strong candidate for being a mammalian homologue of Srs2. A. Both Srs2 and PARI are recruited to the replication fork by PCNA where they can regulate illegitimate homologous recombination during DNA synthesis. Both proteins use their Rad51/RAD51 and DNA binding sites to physically interact with Rad51/RAD51 filaments on ssDNA. B. Rad51’s conformation when it is bound to ATP favors filament formation while its ADP bound form favors disassociation. The direct interaction with Rad51/RAD51 by Srs2 and PARI stimulates Rad51’s intrinsic ATP hydrolyzing activity and the disassociation of one monomer of Rad51 from the filament. C. Srs2 uses its helicase activity to move down ssDNA to interact with the next unit of Rad51, thus perpetuating filament disassembly. On the other hand, PARI lacks Walker A and Walker B motifs, and therefore it cannot hydrolyze ATP on its own. Without an active helicase functionality, it is unclear how PARI can move down ssDNA and facilitate the disassembly of additional RAD51 monomers from the filament. Figure not to scale.
In addition to Srs2’s function relating to the repair of DSBs, it also plays a predominant role in the repair of replicative damage through its interaction with the replication fork clamp, Proliferating Cell Nuclear Antigen complex (PCNA). PCNA is post-translationally modified at lysine 164 either by ubiquitylation or sumoylion. Srs2 is actively recruited to the replication fork by sumoylated PCNA through interactions with its PCNA interacting protein motif (PIP) and its SUMO interacting motif (SIM) [23, 24]. Because PCNA is constitutively SUMOylated during S-phase, the Srs2-PCNA interaction serves as a regulatory mechanism favoring alternative repair pathways instead of HR [25, 26].
Given the importance of Srs2 in genomic integrity and regulation of key steps of HR in yeast, it is surprising how a clear Srs2 homologue has remained elusive in other eukaryotes. Over the past ten years a number of different mammalian genes have been identified that exhibit many, but not all, of the different characteristics of this important regulator. In the following section we will provide evidence that higher eukaryotes have evolved multiple proteins to perform the important role of regulating Rad51 filament formation and strand invasion steps during HR and in response to replicative damage.
2.2 The original suspect of human regulators of RAD51: FBH1 and RECQL5
Srs2’s vital role in regulating HR in S. cerevisiae has spurred a search for its homologue in mammalian cells. A prime candidate for an Srs2 homologue in mammals is the UvrD-like helicase FBH1. FBH1 negatively regulates RAD51 focus formation, and over-expression of FBH1 leads to reduced HR [27, 28]. Importantly, yeast cells where SRS2 has been disrupted can be partially rescued by expressing FBH1 [29]. However, although depletion of FBH1 in vertebrate cells causes an increase in RAD51 foci at sites of DNA damage, these cells show no deficiency in repair of chromosomal breaks [30]. It is possible that other repair pathways can compensate for FBH1 depletion. Furthermore, FBH1 is present in other yeast species, like S. pombe, that also express Srs2, suggesting that it is not a true homologue. Taken together, FBH1 could be thought of as a paralogue of Srs2 with overlapping functionality. However, as the depletions studies suggest, there are other systems in place that share a role in regulating RAD51 in higher eukaryotes.
One of the defining characteristics of Srs2 is that SRS2-disrupted cells exhibit a synthetic lethal phenotype when combined with mutations in the RecQ helicase gene, SGS1 [31]. The synthetic interaction observed between these two genes has led to a model where Srs2 and Sgs1 have overlapping roles in regulating HR. Interestingly, a human homologue of Sgs1, RECQL5, exhibits some similar functions to Srs2 [32]. Like Srs2, RECQL5 directly interacts with PCNA as it contains a PIP motif, suggesting a role for RECQL5 in repair of replicative damage [33]. RECQL5 physically interacts with RAD51 [32, 34] and, like Srs2, RECQL5 disrupts RAD51 filaments on ssDNA before strand invasion and D-loop formation (Figure 2D)[32]. RAD51 displacement from ssDNA requires RECQL5’s ability to hydrolyze ATP [32]. Unlike Srs2, RECQL5 has no effect on preformed D-loops [32]. Importantly, Recql5-deficient mice develop a higher prevalence of cancer than their wild-type littermates, though life spans are similar. Furthermore, Recql5-deficient mouse endothelial fibroblasts exhibit a higher incidence of sister chromatid exchanges, greater number and longer duration of RAD51 and γH2AX foci (markers of DSBs), higher incidence of gross chromosomal rearrangements, and increased susceptibility to replicative stress [32]. These results highlight how disruption of RECQL5 leads to genetic instability similar to that observed in srs2Δ yeast cells.
2.3 RTEL: A new player in meiotic, telomeric, and mitotic recombination
To identify other potential Srs2 homologues, investigators have taken advantage of the observation that srs2Δ cells exhibit a synthetic lethal phenotype with disruption of the RecQ helicase gene, SGS1. The Boulton group screened C. elegans him-6 mutants, the Sgs1 homologue in worms, for synthetic lethal interacting genes. They identified a gene, called regulator of telomere length 1 (rtel-1), as synthetic lethal with him-6 mutants [35]. In mouse studies, RTEL has been found to be an important regulator of telomere lengths [36]. Moreover, the disruption of murine Rtel leads to reduced proliferation and chromosomal abnormalities, suggesting an important function for RTEL in genomic integrity [36]. RTEL is also conserved in humans where knockdown of RTEL levels by siRNA leads to a four-fold increase in HR, hyper-recombination, and genetic instability [35]. Subsequent analysis has revealed that human RTEL serves as a functional homologue for Srs2 in its regulation of RAD51 during HR.
Like yeast Srs2, worms with mutations in rtel-1 exhibit some similar synthetic lethal interactions with certain genes. For example, an embryonic lethal phenotype is observed in rtel-1 mutants in conjunction with other genes knockouts in the DNA repair pathways such as rcq-5 (a RECQL5 homologue), dog-1 (a FANCJ homologue), and mus-81 (a MUS81 homologue). However, RTEL-1 does not exhibit synthetic lethal interactions with mutations in other DNA repair genes such as mre-11, rad-50, rad-54, rad-27 and others, as would be expected by a true Srs2 homologue [35]. These results indicate that rtel-1 likely has a similar, but more limited, function to Srs2 in its genetic interactions.
The mechanism of action for rtel-1, and its homologues in mammals, differs substantially from that of Srs2. Unlike Srs2, which can inhibit Rad51 nucleoprotein filament formation by directly interacting with Rad51 before D-loop formation [14, 16], rtel-1 only acts on preformed D-loops similar to yeast Mph1 (Figure 2E)[15, 35, 37]. Disassembly of preformed D-loops by rtel-1 is RPA dependent and specific to D-loops with a 3′ invasive end [35, 37]. Unlike Srs2 mutants, which are sensitive to a broad range of DNA damaging agents, rtel-1 mutants are specifically sensitive to lesions that affect replication fork progression, like camptothecin. These results suggest that rtel-1 likely has a specific role during replicative stress [35].
In addition to rtel’s function in DNA repair following replication damage, rtel-1 also plays a prominent role during meiosis. Using similar mechanisms to those described above, rtel-1 promotes non-crossover repair by favoring SDSA. Importantly, when rtel-1 is disrupted, all DSB events lead to crossover [37]. Unexpectedly, these cells do not exhibit a statistically significant increase in rad-51 foci [37]. One explanation for this is that rad-51 directs D-loop formation, which rtel-1 then acts on. Furthermore, it was shown that rtel-1 functions to regulate the distribution and number of meiotic crossovers in a pathway distinct from dpy28, which was previously found to have similar functionality [37]. Clearly, rtel-1 has an important role in the regulation of crossover activity during meiosis.
Recently there have been exciting advances in understanding the mouse RTEL’s role as a DNA anti-recombinase during telomere maintenance [38, 39]. In mammalian cells, the telomere functions to preserve the end of linear chromosomes. Its genetic component is composed of TTAGGG tandem repeat sequences that end with a 3′ single stranded DNA overhang. Importantly, numerous proteins, like the shelterin complex, associate with the telomere and are essential in maintaining its structure and function [40]. Interestingly, telomeres can adopt a lasso-like structure when the 3′ overhang invades the double-stranded DNA portion [41, 42]. Importantly, these telomere loops (T-loops) resemble a D-loop structure at the site of strand invasion. Since T-loops and D-loops are structurally similar, Vannier et al. investigated whether RTEL could dissemble T-loops in a similar manner to D-loops thereby connecting RTEL1’s role in HR with telomere maintenance. In addition to RTEL’s role in displacing D-loops, it can disrupt T-loops in an ATP-dependent manner. Importantly, when RTEL1 expression is reduced, telomere length heterogeneity and telomere loss is observed, consistent with the fragile telomere phenotype previously observed [38, 43]. Unexpectedly, the fragile telomere phenotype is a result of accumulation of unresolved T-loops. The T-loops formed are subsequently cleaved by SLX4, a protein in the Fanconi Anemia pathway that resolves Holliday junctions in the latter steps of HR. Cleaved T-loops result in extra-chromosomal sequences called telomere circles, which are a consequence of telomere deletion. Importantly, a new and exciting role for RTEL has been described in maintaining telomere length by dismanteling T-loops and preventing subsequent telomere loss [38].
2.4 PARI
Recently, a new mammalian protein, PARI, has been identified as a key regulator of RAD51 filament formation [44]. PARI is related to the UvrD family of helicases and contains a RAD51 binding site as well as PIP and SIM motifs. Therefore, PARI represents a potential structural and functional mammalian homologue to Srs2. It is important to note, however, that unlike Srs2, PARI does not contain either Walker A or B motifs, which are required for ATP hydrolysis. Therefore, PARI is not an active helicase. The current understanding of Srs2’s regulation of Rad51 holds that its direct interaction with Rad51 activates Rad51’s intrinsic ATP-hydrolyzing activity, thus releasing Rad51 from its close association with ssDNA (Figure 3A and 3B). However, it is thought that Srs2 uses its helicase function to progress down the ssDNA and interact with the next Rad51-ATP molecule (Figure 3C). The lack of active helicase activity in PARI suggests that PARI needs an accessory helicase in order to function, or that its mechanism differs from that of Srs2 (Figure 3C).
In vitro experiments show PARI plays an important role in maintaining genome stability. PARI knockdown by siRNA leads to sensitivity to mitomycin C (MMC), a DNA cross-linking agent, and to chromosomal aberrations. Consistent with a role in HR, PARI-depleted cell lines have slight increase in homologous recombination observed by direct repeat recombination, which is similar to, but not as extensive as RTEL. This modest but significant increase suggests that there are redundant pathways to regulate aberrant HR in higher eukaryotes. Alternatively, PARI’s function may be limited to repair of replicative damage.
The mechanisms in place to regulate PARI specifically at replicative damage sites are tied to its interaction with PCNA through its PIP and SIM motifs. PARI, like Srs2, shows strong associations with sumoylated-PCNA. While PARI is maintained at substoichiometric concentrations in vivo, PCNA interactions increase the local concentration of PARI at the replication fork to stoichiometric levels, which pushes the cell towards other repair pathways when under replicative stress. Taken together, it is clear that this newly discovered protein, PARI, is a significant player in the overall regulation of RAD51 (Figure 2D). Combined with the other RAD51 regulators that we have previously discussed, this paints a complicated picture of negative regulation of HR. How these different regulators of RAD51 interact with each other remains an open question.
3. Regulators of Srs2
The formation of Rad51 filaments is a crucial step for the initiation of HR. There are systems in place that either facilitate this step or negatively regulate it. The balance between assembly and disassembly reactions dictate whether the conditions at the site of repair will favor HR or other repair processes. So far we have covered Srs2 and its functional homologues in humans. These proteins carry out the key functions of negatively regulating Rad51 filament formation both presynaptically and after D-loop formation. On the other hand, the positive regulators can act in two non-mutually exclusive processes: they can actively facilitate the loading and elongation of Rad51 filaments onto ssDNA, or they can negatively regulate the anti-recombinase Srs2. Quintessential proteins of the first group include Rad52 and the breast cancer associated protein, BRCA2 (Figure 2D). Here we will focus on the second group of proteins, specifically on exciting new research that has shed light on the importance of regulating Srs2 during HR.
3.1 Rad55-57, new players in regulating Srs2
Electron microscopy experiments have shown that Rad51 frequently nucleates ssDNA, but with limited extension [36, 37]. Therefore, accessory proteins are needed to stabilize the Rad51 filament to enable elongation and simultaneously prevent filament dismantling by Srs2. The Rad51 paralogues, Rad55 and Rad57, have long been thought to be positive regulators of HR. These proteins, which function as a heterodimer, act in a manner similar to Rad52 in that they promote nucleation and stabilization of the Rad51 filament (Figure 2D) [45–49]. However, recent advances have demonstrated that they also have a role in the regulation of Srs2. Surprisingly, it was found that Rad55-Rad57 can interact with Srs2 in a 1:1 ratio. Interestingly, Rad55-Rad57 can simultaneously bind both Rad51 and Srs2 [50]. These results suggest that Rad55-Rad57 blocks Srs2 translocation protecting Rad51 filaments containing Rad55-Rad57 from disruption (Figure 2D). This model is strengthened by the observation that Rad55-Rad57 heterodimers bind more strongly to Srs2 than Rad51. Furthermore, Rad55-Rad57 indirectly inhibit Srs2 by preventing Rad51’s stimulatory effect of Srs2’s helicase and translocase activity [50]. In summary, Rad55-Rad57 are incorporated onto the ssDNA to stabilize Rad51 filaments while negatively regulating Srs2.
3.2 The Shu complex, containing Rad51 paralogues, negatively regulates Srs2
Similar to Rad55-Rad57, the Shu complex, consisting of Psy3, Csm2, Shu1, and Shu2, is composed of Rad51 paralogues and may function in regulating Srs2 [51–53]. Both in vivo and in vitro studies show that these proteins form a stable complex [52, 54]. This complex promotes homologous recombination while suppressing error-prone DNA repair [54–56]. Recently, the crystal structure of Psy3 and Csm2 has been solved and revealed that these proteins have a similar structure to Rad51, indicating that they are indeed Rad51 paralogues despite the lack of sequence conservation [52]. These results suggest that the structure, but the not sequence, of these proteins has been conserved.
Interestingly, this complex has also been shown to be an important regulator of Srs2 [53]. Shu2 physically interacts with Srs2 by yeast-two-hybrid, and this interaction is also conserved in the fission yeast S. pombe [51, 57]. Disruption of either shu1 or shu2 results in more spontaneous Srs2-YFP fluorescent foci which correlates with a decrease in the percentage of cells exhibiting Rad51 foci [53]. Further analysis has revealed that the Shu complex promotes Rad51 filament formation by inhibiting Srs2’s recruitment at inducible double-strand break sites at multiple genomic loci (i.e. the rDNA and at the URA3 locus) [53]. These in vivo data can be explained by two different, but not mutually exclusive, models, one where the Shu complex directly inhibits Srs2 through its interaction with Shu2 similar to the model proposed for Rad55-Rad57 or one in which the Shu complex functions as a stabilizer of Rad51 filaments. However, a predominant role in Rad51 filament nucleation is unlikely because Rad51 foci form in the absence of the Shu complex, and even more Rad51 foci are observed when srs2 is also disrupted [53].
The Shu complex is conserved in mammalian cells, although there is a discrepancy regarding which orthologues correspond to its yeast counterparts [51, 58]. For example, Martin et al. used sequence homology to show that Shu2, Shu1, and Psy3 in yeast correlates to SWS1, XRCC2, and RAD51D in humans, respectively [51]. Importantly, XRCC2 and RAD51D are both RAD51 paralogues. The identity of the Csm2 homologue remains unknown. In contrast, Liu et al. purified SWS1 and its associated proteins from 293T cells but did not identify XRCC2 or RAD51D to be stably associated [58]. However, they did identify a new interacting partner, which they named SWS-AP1, for SWS associated protein 1 [58]. Perhaps the difference between these two studies can be explained by the interaction between XRCC2 or RAD51D with SWS1 only occurring in the presence of DNA damage or on a DNA template. Further studies will elucidate the components of the mammalian Shu complex and if its interaction with the human Srs2-like proteins is conserved.
4. Conclusions and Future Directions
Regulation of recombination is critical to prevent genomic instability and chromosomal rearrangements observed in cancer. Commitment to HR, an error-free DNA repair pathway, requires formation of Rad51 filaments. In yeast, Srs2, the DNA helicase “anti-recombinase” removes Rad51 from ssDNA ends, and Mph1 disassembles D-loops. In mammalian cells, there are multiple proteins that perform similar functions. For example, both PARI and RECQL5 can remove RAD51 from ssDNA whereas RTEL can disassemble D-loops. The substrate specificity for FBH1, another potential Srs2 orthologue, is unknown. These results suggest that higher eukaryotes have evolved multiple proteins to perform the analogous functions of Srs2. Perhaps these proteins are substrate specific, for example working on dissembling RAD51 from replication intermediates in the case of PARI or from functioning to resolve meiotic D-loops and T-loop structures in the case of RTEL. It is interesting that many of the Srs2 orthologues have links to the replication fork. Perhaps so many of these proteins associate with the replication fork because of the ssDNA created during DNA replication. This is a prime target for removal by the HR machinery, which would then create illegimate Rad51 filaments. Srs2, PARI, and RECQL5 could prevent detrimental recombination at the replication fork thus enabling replication to proceed normally.
How can we manipulate regulators of recombination as cancer therapeutic targets? It is interesting that mutations in genes important in regulating DNA repair and homologous recombination are associated with cancer predisposition. For example, mutations in BRCA2 can lead to breast or ovarian cancer [2, 59, 60], RTEL in gliomas [61–65], and XRCC2 in breast cancer [66–71]. Interestingly, in RTEL and XRCC2 there are also polymorphisms that are protective against disease [63, 67]. These results highlight how regulators of recombination can be used as important therapeutic tools in treatment of cancer. Clearly, regulation of the amount of recombination is important in preventing genetic instability observed during tumorigenesis. It is therefore plausible that drugs that either increase or decrease these proteins function in regulating HR could be developed in the future and offer a personalized approach to cancer treatment.
Acknowledgments
We thank Wolf Heyer, Dominique Bratton-Palmer, Cheryl Clauson, Emma Satlof-Bedrick, Stephen Godin, and Faiz Kabbinavar for providing helpful comments and edits to the manuscript. This work is supported by the National Institutes of Health grant R00GM088413 to KAB, the PNC, UPCI Director Distinguished Scholar Award to KAB and the Dean’s Summer Research Program at the University of Pittsburgh School of Medicine to YK.
Footnotes
Conflict of interest
The authors confirm there is no conflict of interest, financial or otherwise, in this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci U S A. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duker NJ. Chromosome breakage syndromes and cancer. Am J Med Genet. 2002;115:125–129. doi: 10.1002/ajmg.10688. [DOI] [PubMed] [Google Scholar]
- 3.Hiom K. Coping with DNA double strand breaks. DNA Repair (Amst) 2010;9:1256–1263. doi: 10.1016/j.dnarep.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair. 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 6.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 7.Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol. 2011;18:748–754. doi: 10.1038/nsmb.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 9.Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U S A. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyer WD. Biochemistry of eukaryotic homologous recombination. Top Curr Genet. 2007;17:95–133. doi: 10.1007/978-3-540-71021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome Instability and Defective Recombinational Repair in Knockout Mutants of the Five Rad51 Paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasparek TR, Humphrey TC. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol. 22:886–897. doi: 10.1016/j.semcdb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 15.Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, Ira G. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 17.Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seong C, Colavito S, Kwon Y, Sung P, Krejci L. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. J Biol Chem. 2009;284:24363–24371. doi: 10.1074/jbc.M109.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colavito S, Macris-Kiss M, Seong C, Gleeson O, Greene EC, Klein HL, Krejci L, Sung P. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 2009;37:6754–6764. doi: 10.1093/nar/gkp748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mankouri HW, Chu WK, Hickson ID. A novel antirecombinase gains PARIty. Mol Cell. 2012;45:6–7. doi: 10.1016/j.molcel.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Hegde V, Klein H. Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 2000;28:2779–2783. doi: 10.1093/nar/28.14.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rong L, Palladino F, Aguilera A, Klein HL. The hyper-gene conversion hpr5–1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics. 1991;127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moldovan G-L, Pfander B, Jentsch S. PCNA, the Maestro of the Replication Fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D’Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 26.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, Bartek J, Lukas J, Mailand N. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. The Journal of Cell Biology. 2009;186:655–663. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA Helicase Fbh1 Prevents Rhp51-Dependent Recombination without Mediator Proteins. Mol Cell Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiolo I, Saponaro M, Baryshnikova A, Kim J-H, Seo Y-S, Liberi G. The Human F-Box DNA Helicase FBH1 Faces Saccharomyces cerevisiae Srs2 and Postreplication Repair Pathway Roles. Mol Cell Biol. 2007;27:7439–7450. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laulier C, Cheng A, Huang N, Stark JM. Mammalian Fbh1 is important to restore normal mitotic progression following decatenation stress. DNA Repair. 2010;9:708–717. doi: 10.1016/j.dnarep.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 32.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5Œ≤ helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P. Physical Interaction of RECQ5 Helicase with RAD51 Facilitates Its Anti-recombinase Activity. J Biol Chem. 2010;285:15739–15745. doi: 10.1074/jbc.M110.110478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MIR, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ. RTEL1 Maintains Genomic Stability by Suppressing Homologous Recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, Tam PPL, Nagy A, Lansdorp PM. Regulation of Murine Telomere Length by Rtel: An Essential Gene Encoding a Helicase-like Protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 37.Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, ONeil NJ, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 Enforces Meiotic Crossover Interference and Homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 Dismantles T Loops and Counteracts Telomeric G4-DNA to Maintain Telomere Integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 39.Uringa EJ, Lisaingo K, Pickett HA, Brind’amour J, Rohde JH, Zelensky A, Essers J, Lansdorp PM. RTEL1 contributes to DNA replication, repair and telomere maintenance. Mol Biol Cell. 2012 doi: 10.1091/mbc.E12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 41.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian Telomeres End in a Large Duplex Loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 42.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, Paoletti J, Gilson E, Giraud-Panis M-J. A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 43.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian Telomeres Resemble Fragile Sites and Require TRF1 for Efficient Replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moldovan G-L, Dejsuphong D, Petalcorin MIR, Hofmann K, Takeda S, Boulton SJ, D’Andrea AD. Inhibition of Homologous Recombination by the PCNA-Interacting Protein PARI. Mol Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 46.Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 48.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 50.Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, Bowden G, Kalmyrzaev B, Warren-Perry M, Snape K, Adlard JW, Barwell J, Berg J, Brady AF, Brewer C, Brice G, Chapman C, Cook J, Davidson R, Donaldson A, Douglas F, Greenhalgh L, Henderson A, Izatt L, Kumar A, Lalloo F, Miedzybrodzka Z, Morrison PJ, Paterson J, Porteous M, Rogers MT, Shanley S, Walker L, Eccles D, Evans DG, Renwick A, Seal S, Lord CJ, Ashworth A, Reis-Filho JS, Antoniou AC, Rahman N. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernstein KA, Reid RJ, Sunjevaric I, Demuth K, Burgess RC, Rothstein R. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol Biol Cell. 2011;22:1599–1607. doi: 10.1091/mbc.E10-08-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shor E, Weinstein J, Rothstein R. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics. 2005;169:1275–1289. doi: 10.1534/genetics.104.036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mankouri HW, Ngo HP, Hickson ID. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol Biol Cell. 2007;18:4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol Microbiol. 2009;73:89–102. doi: 10.1111/j.1365-2958.2009.06748.x. [DOI] [PubMed] [Google Scholar]
- 57.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, Welch R, Lissowska J, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Bardin-Mikolajczak A, Struewing JP. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet. 2006;119:376–388. doi: 10.1007/s00439-006-0135-z. [DOI] [PubMed] [Google Scholar]
- 60.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 61.Chen H, Chen Y, Zhao Y, Fan W, Zhou K, Liu Y, Zhou L, Mao Y, Wei Q, Xu J, Lu D. Association of sequence variants on chromosomes 20, 11, and 5 (20q13.33, 11q23.3, and 5p15.33) with glioma susceptibility in a Chinese population. Am J Epidemiol. 2011;173:915–922. doi: 10.1093/aje/kwq457. [DOI] [PubMed] [Google Scholar]
- 62.Wang SS, Hartge P, Yeager M, Carreon T, Ruder AM, Linet M, Inskip PD, Black A, Hsing AW, Alavanja M, Beane-Freeman L, Safaiean M, Chanock SJ, Rajaraman P. Joint associations between genetic variants and reproductive factors in glioma risk among women. Am J Epidemiol. 2011;174:901–908. doi: 10.1093/aje/kwr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Shete S, Etzel CJ, Scheurer M, Alexiou G, Armstrong G, Tsavachidis S, Liang FW, Gilbert M, Aldape K, Armstrong T, Houlston R, Hosking F, Robertson L, Xiao Y, Wiencke J, Wrensch M, Andersson U, Melin BS, Bondy M. Polymorphisms of LIG4, BTBD2, HMGA2, and RTEL1 genes involved in the double-strand break repair pathway predict glioblastoma survival. J Clin Oncol. 2010;28:2467–2474. doi: 10.1200/JCO.2009.26.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lonn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, LaChance DH, McCoy L, O’Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park DJ, Lesueur F, Nguyen-Dumont T, Pertesi M, Odefrey F, Hammet F, Neuhausen SL, John EM, Andrulis IL, Terry MB, Daly M, Buys S, Le Calvez-Kelm F, Lonie A, Pope BJ, Tsimiklis H, Voegele C, Hilbers FM, Hoogerbrugge N, Barroso A, Osorio A, Giles GG, Devilee P, Benitez J, Hopper JL, Tavtigian SV, Goldgar DE, Southey MC. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90:734–739. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin WY, Camp NJ, Cannon-Albright LA, Allen-Brady K, Balasubramanian S, Reed MW, Hopper JL, Apicella C, Giles GG, Southey MC, Milne RL, Arias-Perez JI, Menendez-Rodriguez P, Benitez J, Grundmann M, Dubrowinskaja N, Park-Simon TW, Dork T, Garcia-Closas M, Figueroa J, Sherman M, Lissowska J, Easton DF, Dunning AM, Rajaraman P, Sigurdson AJ, Doody MM, Linet MS, Pharoah PD, Schmidt MK, Cox A. A role for XRCC2 gene polymorphisms in breast cancer risk and survival. J Med Genet. 2011;48:477–484. doi: 10.1136/jmedgenet-2011-100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krupa R, Sliwinski T, Wisniewska-Jarosinska M, Chojnacki J, Wasylecka M, Dziki L, Morawiec J, Blasiak J. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer--a case control study. Mol Biol Rep. 2011;38:2849–2854. doi: 10.1007/s11033-010-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu KD, Chen AX, Qiu LX, Fan L, Yang C, Shao ZM. XRCC2 Arg188His polymorphism is not directly associated with breast cancer risk: evidence from 37,369 subjects. Breast Cancer Res Treat. 2010;123:219–225. doi: 10.1007/s10549-010-0753-y. [DOI] [PubMed] [Google Scholar]
- 70.Silva SN, Tomar M, Paulo C, Gomes BC, Azevedo AP, Teixeira V, Pina JE, Rueff J, Gaspar JF. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol. 2010;34:85–92. doi: 10.1016/j.canep.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 71.Curtin K, Lin WY, George R, Katory M, Shorto J, Cannon-Albright LA, Smith G, Bishop DT, Cox A, Camp NJ. Genetic variants in XRCC2: new insights into colorectal cancer tumorigenesis. Cancer Epidemiol Biomarkers Prev. 2009;18:2476–2484. doi: 10.1158/1055-9965.EPI-09-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van der Heijden T, Modesti M, Hage S, Kanaar R, Wyman C, Dekker C. Homologous recombination in real time: DNA strand exchange by RecA. Mol Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]