Abstract
CYR61 is one of the six proteins of the CCN family of proteins known to play diverse roles in angiogenesis, cellular proliferation, survival, migration and wound healing. However, the specific function of CYR61 in cancer is unclear and the literature remains controversial. We used quantitative real-time PCR (qRT-PCR) to establish the expression profile of CYR61 and integrin αVβ5 in three non-small cell lung cancer (NSCLCA), five colorectal cancer (CRCA), one breast cancer (BCA) and one esophageal squamous carcinoma (ESCA) cell lines. We showed that the levels of CYR61 were significantly increased in ESCA cell line along with the enhanced levels of αVβ5 integrin. Further, we investigated whether tumor cell-secreted CYR61 can facilitate cell migration by interacting with the αVβ5 integrin. Using tumor cell lines with low, intermediate and high CYR61 expression and their isogenic variants as a cellular model, we determined that integrin αVβ5 expressed on these tumor cells is required for cell migration. Moreover, we showed that modulation of expression levels of CYR61 in these cancer cells affected their capacity for migration. These results represent an advance to the understanding of the role of CYR61 and αvβ5 integrin as proteins that cooperate to mediate cancer cell migration.
Keywords: CYR61, stable over-expression, stable knock down, integrin αvβ5, migration
Introduction
The angiogenic factor cysteine-rich angiogenic protein 61 (CYR61, CCN1) is a member of the highly conserved CCN family which includes CTGF (connective tissue growth factor, CCN2), Nov (nephroblastoma over-expressed protein, CCN3), WISP-1 (Wnt-1-induced secreted protein 1, CCN4), WISP-2 (CCN5) and WISP-3 (CCN6)1. All CCN proteins share four conserved domains with sequence homology to insulin-like growth factor binding protein, von Willebrand factor type C repeat, thrombospondin type 1 and growth factor cysteine knots2. The four modular domains of CYR61 may explain the diverse functions of the protein. Indeed, CYR61 has been reported to mediate cell adhesion, including cell migration, stimulation of chemotaxis, enhancement of growth factor-induced DNA synthesis, cell survival and angiogenesis3,4. Previous studies have suggested that CYR61 may be a marker for more aggressive phenotypes5 probably occurring through the ability of CCN proteins to bind and activate cell surface integrins1. Moreover, CYR61 has been found to be differentially expressed in many cancers and to be involved in the development and progression of various cancers6. The level of CYR61 expression has been found to be increased in rhabdomyosarcomas, malignant melanomas, colon adenocarcinomas, bladder papillomas, and pancreatic cancer7–9. Over-expressed CYR61 stimulates the progression of breast cancers10,11. High levels of CYR61 were found in malignant gliomas and CYR61 was shown to enhance the tumorigenicity through the integrin-linked kinase signaling pathway12. Moreover, up-regulation and involvement of CYR61 expression was recognized in prostate cancer cells13 and recently in esophageal squamous cell carcinoma14. Paradoxically, CYR61 is down-regulated in lung cancers and forced expression of CYR61 inhibits tumorigenicity of lung cancer cells15. Finally, it was also reported that CYR61 inhibits the growth of endometrial cancer16 and leiomyomas17.
CCN proteins, like some other extracellular matrix and matricellular proteins, regulate cell adhesion, migration, differentiation, proliferation, and survival through their interactions with cell adhesion receptors, including integrins18. Indeed, the CCN-integrin connection was first demonstrated by the direct binding of CYR61 to integrin αvβ3 to mediate endothelial cell adhesion19. CCNs utilize distinct integrins depending on the target cell types and the activities mediated. For example, in fibroblasts, CYR61 stimulates cell adhesion, migration, and DNA synthesis through α6β1, αvβ5, and αvβ3, respectively18. By contrast, CYR61 stimulates cell migration in endothelial cells and vascular smooth muscle cells through binding to αvβ3 and α6β1, respectively20. Moreover, CYR61 mutants that disrupt its α6β1 binding sites specifically abrogate α6β1-dependent CYR61 activities without affecting αvβ3-mediated angiogenic functions21. Similarly, mutation that impairs the CYR61 αvβ3 binding site abolishes αvβ3- but not α6β1-mediated functions, further establishing that these distinct integrin binding sites and their cognate signaling pathways can act independently of one another22. On the other hand, CYR61 must interact with both α6β1 and αvβ5 to mediate its synergism with TNFα, indicating that different integrin pathways can also function in concert to elicit distinct CCN activities23. The interaction of CCNs with multiple receptors may contribute to their unique activities and functions.
The specific functions of CYR61 and its interactions with integrin αvβ5 in cancer are still unclear and the literature remains controversial. In our studies we have evaluated the expression levels of CYR61 and integrin subunits αV and β5 in colorectal adenocarcinoma, lung carcinoma, breast carcinoma, esophageal squamous cell carcinoma and squamous cell adenocarcinoma cancer cell lines. Our data indicate that there are low expression levels of CYR61 in the colorectal adenocarcinoma SW620, negligible levels in the lung carcinoma H460 and high expression levels in the esophageal squamous cell carcinoma TE-7 cell line as determined using quantitative real-time PCR and Western blotting techniques. Furthermore, CYR61 as a part of the extracellular matrix has been shown to be a secreted ligand of several integrins. Thus, we examined the integrin expression in these three cancer cell lines using DNA microarray analysis24 and confirmed that these three tumor cell lines express both subunits of the CYR61 integrin receptor, αV and β5. Interestingly, the colorectal adenocarcinoma cells express the integrin αVβ5 subunits but not CYR61 and do not show invasion through matrigel24. These observations strongly suggest that CYR61 may be involved in the mechanism of tumor cell migration.
Materials and Methods
Cell Culture
Colorectal cancer cells (SW837, HT-29, HCA-7 and HCT116), lung carcinoma cells (H1155 and H2122), breast cancer cells (MCF-7) and esophageal squamous cell carcinoma TE-7 cells were all obtained from ATCC. SW620 cells were originally obtained as BIC-1 esophageal adenocarcinoma cell line and H460 as SEG-1 esophageal adenocarcinoma cell line from Dr. David Beer, University of Michigan. However, Boonstra et al., 2010 recently verified the authenticity of all available esophageal adenocarcinoma cell lines and genotyping by short tandem repeat profiling revealed that BIC-1 is not esophageal adenocarcinoma as it was considered for decades but SW620 colorectal adenocarcinoma cell line, SEG-1 is lung carcinoma cell line H46025.
SW620, SW480 and SW837 cell lines were maintained in a 37°C, non-CO2 equilibrated environment in Leibovitz’s L-15 medium (L-15; Mediatech, Inc., Manassas, VA) containing 10% FBS and 1% penicillin/streptomycin. HCT116 cell line was maintained at 37°C in the presence of 5% CO2 in Iwakata and Grace modified medium (McCoy’s 5A; Mediatech, Inc., Manassas, VA) containing 10% FBS and 1% penicillin/streptomycin. All non-small cell lung cancer cell lines were incubated at 37°C in 5% CO2 and cultured with RPMI-1640 medium (RPMI-1640; Mediatech, Inc., Manassas, VA) containing 10% FBS and 1% penicillin/streptomycin. Remaining cell lines were maintained in Dulbecco's modified essential medium (DMEM; Gibco-BRL, Rockville, MD) containing 10% FBS and 1% penicillin/streptomycin and cultured at 37°C in the presence of 5% CO2.
Lentiviral Particle and Stable Cell Line Generation
To generate stable CYR61 over-expressing cell lines, full length CYR61 cDNA clone along with PCR primers for amplification and modification of the resulting product for TOPO directional cloning were obtained from the ATCC and Biosynthesis. The CYR61 cDNA was stably transduced into SW620, H460 and TE-7 cancer cell lines using ViraPower Lentiviral Expression kit (Invitrogen, Carlsbad, CA). CYR61 cDNA was PCR amplified from the original ATCC vector with Pfx polymerase to generate blunt-end PCR products for directional cloning into the expression pLenti6/V5-D-TOPO vector (Invitrogen, Carlsbad, CA) which was designed to facilitate rapid, directional TOPO cloning and high level expression of PCR products in mammalian cells.
For CYR61 expression knock-down, two different shRNAs directed against the CYR61 mRNA were designed using Invitrogen’s proprietary design software. shRNAs were stably transduced by using the Block-iT Lentiviral RNAi Expression kit (Invitrogen, Carlsbad, CA). Two strands of shRNA sequences targeting CYR61 mRNA were synthesized (CYR61GS-1 targeting sequence: 5’ – GCCACACGAGTTACCAATGATT – 3’; CYR61GS-2 targeting sequence: 5’ – GCATCCTATACAACCCTTTAC – 3’), annealed and cloned into the entry pENTR/U6 vector (Invitrogen, Carlsbad, CA) which contains attL sites to facilitate transfer of the U6 RNAi cassette into the destination pLenti6/BLOCK-iT-DEST vector to generate an expression clone. To obtain pLenti6/BLOCK-iT expression clone, the LR clonase reaction between entry and destination construct was performed.
Lentiviral particles were generated by individually transfecting 293FT cells with the CYR61 expression constructs pLenti6/V5 for over-expression or pLenti6-GW/U6 for silencing and the ViraPower Packaging Mix with Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). Stable CYR61-expressing/silencing cells were generated by transduction with lentiviral particles at a 1:100 dilution in growth media with 6 µg/ml polybrene (Sigma-Aldrich, St. Louis, MO). Selection of stably expressing/silencing CYR61 cell lines was conducted with 2µg/ml blasticidin for H460 and TE-7 cell lines and 10µg/ml for SW620 cell line (Invitrogen, Carlsbad, CA). pLenti6/V5-GW/lacZ (Invitrogen, Carlsbad, CA) was used as a positive expression control vector for CYR61 over-expression and pLenti6-GW/U6-laminshRNA plasmid (Invitrogen, Carlsbad, CA) was used as a positive control for CYR61 gene silencing.
RNA Extraction and Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
RNA extraction from all isogenic variants was performed using RNAeasy kit (Qiagen, Valencia, CA). Human CYR61 (Hs00155479_A1), integrin alpha V (Hs00233808_m1), integrin beta 5 (Hs00174435_m1) and GAPDH (Hs99999905_A1) primer/probes were obtained from ABI (Applied Biosystems, Branchburg, NJ). cDNA was synthesized from 500 ng of total RNA in a 50µl reaction with master mix containing 10×RT buffer, 5.5 mM MgCl2, 2 mM dNTPs, 2.5 µM random hexamers, 2 Units of RNase Inhibitor and 62.5 Units of Multi Scribe Reverse Trascriptase. All MasterMix reagents were purchased from ABI (Applied Biosystems, Branchburg, NJ). Reactions were performed in MJ Thermocycler PTC-200 (MJ Research, Inc., Watertown, Massachusetts) followed by these conditions: 25°C for 10 minutes, 48°C for 30 minutes and 95°C for 5 minutes. 2µl of 5ng/µl cDNA was then used as a template from which to amplify the human CYR61 sequence. The conditions for PCR reactions were: 10 minutes at 95°C followed by 15 seconds at 95°C, 1 minute at 60°C for 40 cycles by using ABI7000. GAPDH amplification was included as a reference for a relative quantification of the target genes. Non-template controls were included on each PCR plate. CYR61, integrin alpha V and integrin beta 5 levels were normalized to the GAPDH control. Amplification plots were generated and the Ct value (cycle number at which fluorescence reaches threshold) recorded.
Western Blot Analysis
Cell lysates were prepared from all cell variants using the radio-immunoprecipitation assay lysis buffer (RIPA) containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton N-100, 1% sodium deoxycholate and 0.1% sodium dodecyl sulfate and protease inhibitor mixture (Leupeptin, Aprotinin, PMSF). 30µg of total protein was separated by 7.5% SDS-polyacrylamide gel electrophoresis followed by transfer to PVDF membranes (Bio-Rad, Hercules, CA). Rabbit polyclonal anti-CYR61 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology, Santa Cruz, CA) and mouse monoclonal anti-actin clone AC-40 antibody was obtained from Sigma Aldrich (St. Louis, MO). Goat anti-rabbit (for CYR61) and goat anti-mouse (for β-actin) HRP secondary antibodies (Santa Cruz Biotechnology, santa Cruz, CA) were used for detection. Blots were incubated with ECLWestern Blotting Detection Reagents (Amersham Biosciences, Piscataway, NJ) and were exposed to BioMax XAR film (Kodak, Rochester, NY). The same blot was used for both, CYR61 and β-actin detection. In details, after the CYR61 analysis was performed, the stripping buffer (2% SDS, 62.5 mM TRIS pH6.8, 100 mM β-mercaptoethanol) was used to strip the original western blot by incubating the membrane in 60°C for 30 minutes. Then the stripping buffer was poured off and the membrane was washed 2 times in PBST (3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, 0.05% Tween 20, pH 7.4) for 5 minutes and 2 times with PBST/milk for 5 minutes. Then the membrane was re-probed to detect β-actin.
Conditioned Media Preparation
Untreated or CYR61 transfected cells were grown to around 80% confluence and medium was replaced with serum-free medium for a further 48-hours. 10 ml of conditioned media was centrifuged at 3,000×g for 10 min and supernatant was stored at -80°C. Conditioned media were incubated with rabbit polyclonal anti-CYR61 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C overnight. Protein A/G PLUS Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) were then added to the suspension and incubated at 4°C for 1 hour to remove CYR61 from the conditioned media. Beads with bound CYR61 were washed 3 times with cold PBS. 2× Tris-Glycine SDS Sample Loading buffer (125 mM Tris HCl, 20% Glycerol, 4% SDS, 0.001% Bromophenol Blue, 0.2% β-mercaptoethanol, pH 6.8) was added to each bead pellet. To release the CYR61 from the beads to the loading dye, samples were denaturated at 95°C for 5 minutes and then 10µl of each sample was separated by 7.5% SDS-polyacrylamide gel and subsequently the western blot analysis was performed as described above. The equal volumes were used for immunoprecipitation and western blot analysis.
Epithelial Cell Transwell Migration Assay
Migration assays were performed using isogenic variants of the H460, TE-7, and SW620 cell lines. 8.0 um pore size, 1.0×105 pore density translucent transwell migration chambers (BD Biosciences, Bedford, MA) in 24-well plate were used for migration analysis. Briefly, 600 ul of invasion buffer (DMEM media with 0.5% FBS, 0.1%BSA and without antibiotics) was added to each well. 5×104 of esophageal squamous carcinoma TE-7 cells or 1.5×105 of colorectal adenocarcinoma SW620 and lung carcinoma H460 cells re-suspended in 100 ul of invasion buffer were seeded on the top of the membrane. After overnight incubation (18 hours) at 37°C, 5% CO2, the membranes were stained with 0.5% crystal violet (Sigma Aldrich, St. Louis, MO) in 20% methanol for 1 minute. The stain was rinsed off thoroughly with water, cells remaining in the top of the migration chamber were removed by lightly swabbing, and stained cells adhering to the bottom of the chamber were counted.
Cell Migration Assay using functional blocking antibodies
All isogenic cell line variants with altered CYR61 expression were used for the migration-blocking assays. To block cell migration with antibodies, the cells were treated with 50µg/ml of specified blocking anti-αVβ5 monoclonal antibody, clone P1F6 (Millipore, Billerica, MA) for 1 hour at 4°C before the cells were seeded on the top of the membrane.
Image Analysis of transwell migrations
The 24 well BD Falcon Cell Culture Companion Plates for Inserts (BD Biosciences, San Diego, CA) were analyzed for migration of cells through the translucent transwell membrane (8.0 um pore size, 1.0×105 pore density). Images of 14 fields per transwell were captured from each well using a 10X objective in combination with a 1.5X optivar. The image capture was performed with an Olympus IMT-2 microscope (Olympus America Inc., Center Valley, PA), a Hamamatsu ORCA-100 greyscale CCD camera (Hamamatsu USA, Bridgewater, NJ), and a Ludl motorized XY stage (Ludl Electronic Products, Hawthorne, NY). The camera and stage was computer controlled by SimplePCI software (ver 6.2, Compix Inc., Sewickley, PA). After the images were captured, the SimplePCI software performed an intensity threshold, and size exclusion to avoid measuring the membrane pores, and the remaining binary image was measured for area.
Statistical analysis
Data were collected and analyzed to obtain the mean and S.E.M. for three independent experiments. Statistical significance between any two groups was determined by the two-tailed Student’s t-test using Microsoft Excel, P values less than 0.05 were considered to be significant.
Results
Expression levels of CYR61 in cancer cells
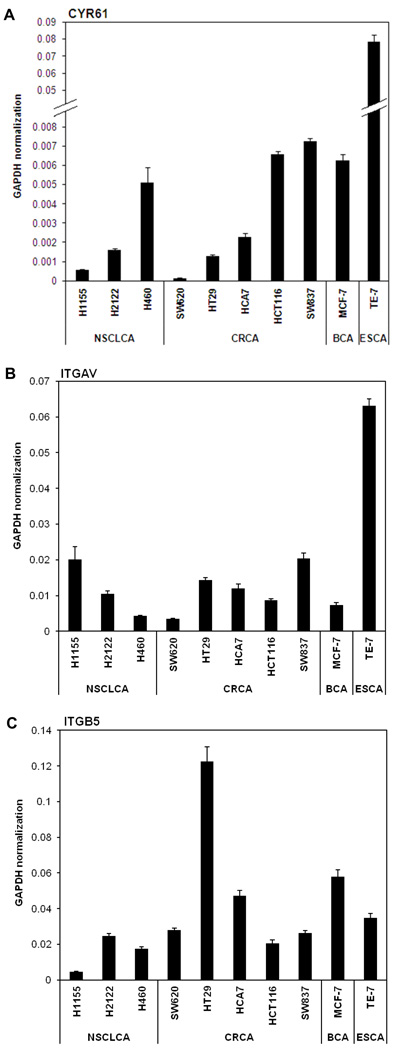
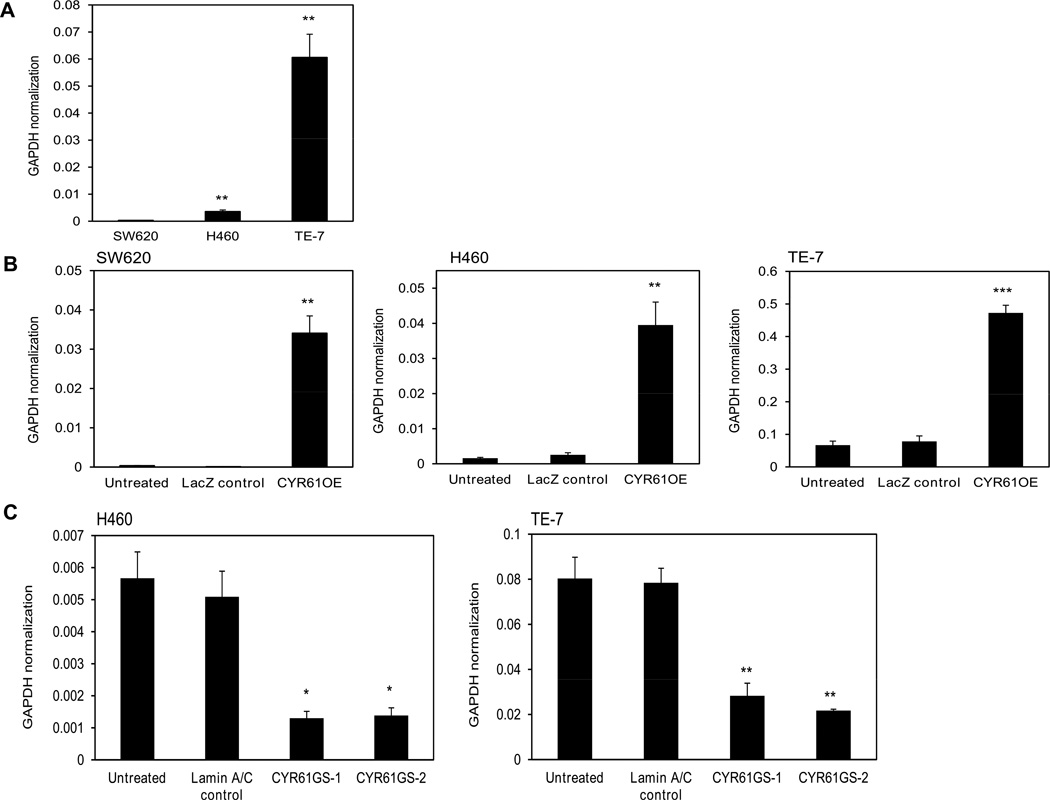
Real time PCR analysis (Fig. 1A) confirmed expression levels of CYR61 in several cancer cell lines. Overall the data indicated that CYR61 is expressed at almost undetectable levels in SW620, intermediate in H460 and high in TE-7 cell lines (Fig. 2A). The expression levels of CYR61 in H460 cells were 20 times higher and in TE-7 cells 250 times higher compared to the levels of CYR61 in SW620 cells.
Figure 1. CYR61, ITGAV and ITGB5 mRNA expression levels in cancer cell lines measured by quantitative real-time PCR.
Bar graphs showing mRNA expression levels of A) CYR61 B) ITGAV and C) ITGB5 in three lung, five colorectal, one breast and one esophageal squamous carcinoma cell lines. The relative expression levels of all genes were calculated by normalization to GAPDH. Data were obtained from three independent experiments are shown as mean ± SEM (each experiment was done in triplicate parallels).
Figure 2. CYR61 mRNA expression levels in colorectal adenocarcinoma SW620, lung carcinoma H460 and esophageal squamous cell carcinoma TE-7 cells measured by quantitative real-time PCR.
A) Bar graph showing relative CYR61 mRNA expression levels for untreated cell lines. Asterisks indicate significantly higher expression of CYR61 in H460 and TE-7 cells compared to SW620 cells. B) Bar graph showing relative CYR61 mRNA expression levels for all three cell lines after stable over-expression where LacZ was used as a control. Asterisks indicate significant increase in expression of CYR61 in all three cell lines after their stable CYR61 over-expression compared to their LacZ controls. C) Bar graph showing relative CYR61 mRNA expression levels for H460 and TE-7 cell lines after CYR61 was stably knocked down using two independent shRNA sequences, Laminin A/C was used as a control. Asterisks indicate significant decrease in expression of CYR61 in both cell lines after their CYR61 knockdown compared to Lamin A/C controls. The relative CYR61 expression levels were calculated by normalization to GAPDH. Data were obtained from three independent experiments (three independent RNA samples from three individual clones of cells were used within each experiment) are shown as a mean ± SEM (*p<0.05, **p<0.0005, *** p<0.005).
Next, we modeled the isogenic variants of all three cancer cell lines with stably over-expressed and stably knocked-down CYR61 expression. We transduced the cells using the ViraPower Lentiviral Expression System, which allowed creation of a replication-incompetent, HIV-1-based lentivirus that was used to deliver and express the gene of interest in these cancer cell lines. To evaluate the efficiency of stable transfection, mRNA expression levels of CYR61 were then measured by quantitative real time PCR (Fig. 2B) Stable over-expression of CYR61 was obtained in all three cancer cell lines. The expression levels of CYR61 in SW620 (95-fold), H460 (25-fold) and TE-7 (7-fold) cell lines were increased significantly after the stable over-expression. Similarly, lentiviral particles containing the expression clone with shRNA targeted against CYR61 mRNA were used to stably knockdown Cyr61 expression in the H460 and TE-7 cancer cell lines. Quantitative real time PCR was used to measure the levels of stable CYR61 knock down in both cancer cell lines and revealed that there was more than 70% inhibition of CYR61 expression in both cell lines using two independent shRNA sequences against CYR61 (Fig. 2C). CYR61 protein expression levels in parental SW620, H460 and TE-7 cell lines were assessed also by Western blot analysis (Fig. 3A). As predicted by the DNA microarray24 (Supplemental tab. 1) and RT-PCR results, CYR61 expression was undetectable in SW620 cells, intermediate in H460 cells and high in TE-7 cells. Western blot analysis also confirmed the forced stable over-expression (OE) of CYR61 or the CYR61 gene silencing (GS) in each cancer cell line (Fig. 3B). Finally, to confirm that CYR61 is a secreted protein, the amount of CYR61 protein in conditioned media from untreated lung carcinoma H460 cells and H460 cells over-expressing CYR61 was analyzed by immuno-precipitation and Western blot analysis (Fig. 3C). The results showed there was higher level of CYR61 protein in conditioned medium collected from H460 cells over-expressing CYR61 in comparison with parental H460 cells, and confirmed that CYR61 is a secreted protein.
Figure 3. Western blot analysis showing CYR61 protein expression levels in colorectal adenocarcinoma SW620, lung carcinoma H460 and esophageal squamous cell carcinoma TE-7 cell lines.
A) CYR61 protein expression in untreated cell lines. As predicted by the microarray results, CYR61 expression was undetectable in SW620 cells, immediate in H460 cells and high in TE-7 cells. β-actin was used as a loading control. B) WB analysis as a confirmation of high/low expression levels after stably forced CYR61 over-expression/knock-down for all three cell lines. Treatment lanes: untreated cells; cells over-expressing CYR61 (OE), cells after stable transfection with two independent shRNAs directed against CYR61mRNA (GS-1, GS-2). C) Immuno-precipitation and WB analysis of CYR61 expression in conditioned medium as a confirmation that CYR61 is secreted into the media. Conditioned medium was collected from untreated H460 and H460 cells over-expressing CYR61 after they were grown in serum-free medium for 48 hours. Experiments were performed at least three times using freshly isolated protein samples from independent cell lines.
Role of CYR61 in cancer cell migration
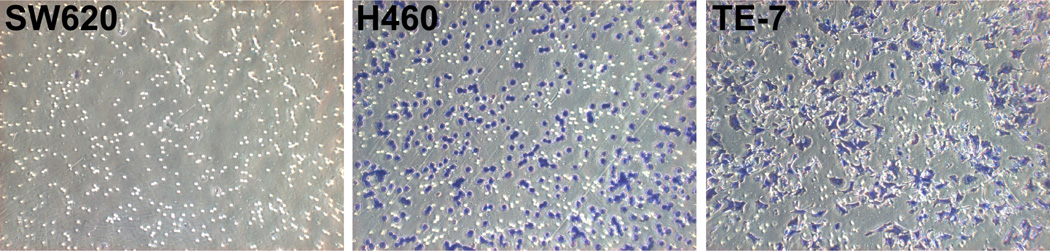
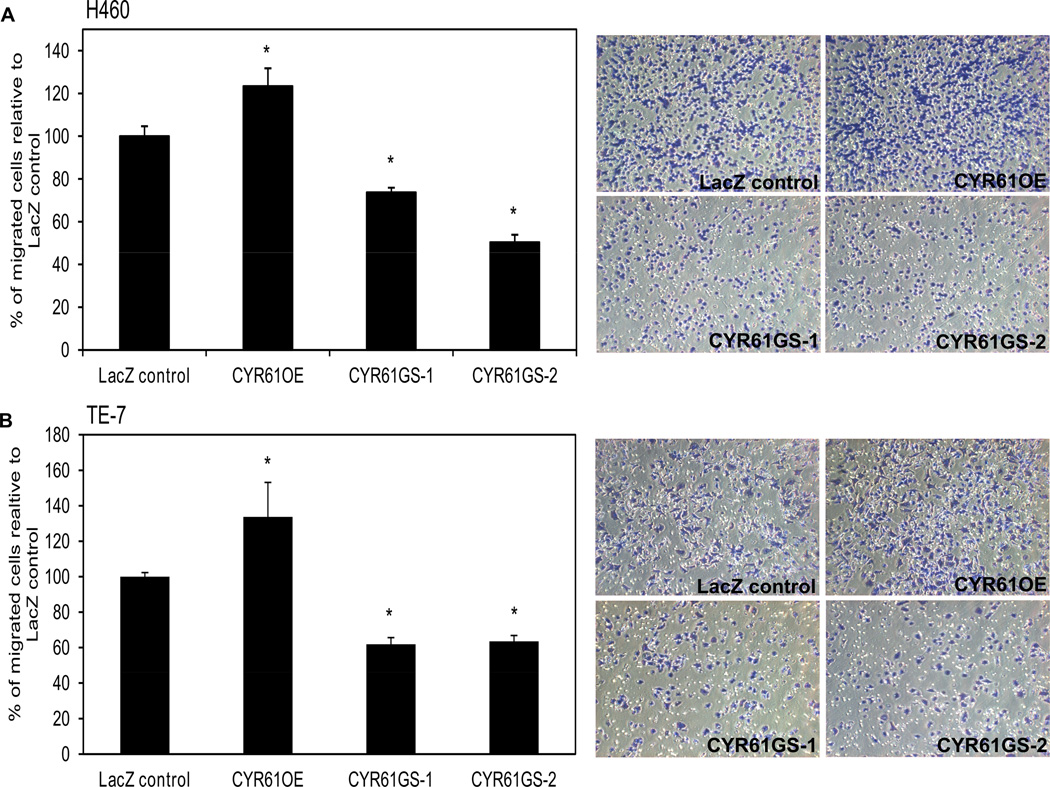
CYR61 has been shown to be a secreted protein that acts as a ligand of several integrins18. Thus, we examined the integrin expression levels in all three cancer cell lines. Data obtained from qRT-PCR (Fig.1B and C) confirmed that the three tumor cell lines expressed both subunits of the CYR61 integrin receptor, αV as well as β5. The highest expression levels of integrin αV and β5 subunits were detected in esophageal squamous cell carcinoma TE-7 cell line. These cells also showed the highest migration ability through the transwell membrane. Interestingly, SW620 cells express the integrin αVβ5 subunits but not CYR61 (Fig. 4) and do not show migration through the uncoated membrane. Fig. 4 shows the migration of all three cancer cell lines under the optimal experimental conditions, where TE-7 cells had to be seeded at much lower concentration (50,000 cells per insert) than both SW620 and H460 cells, which were seeded at higher concentration of 150,000 cells per insert. These observations suggested that CYR61 may be involved in the mechanism of cancer cell migration through the specific interaction with integrin αVβ5. To test this hypothesis, we measured the migratory capabilities of H460 and TE-7 cancer cell lines and their isogenic variants with altered CYR61 expression to determine if the expression of integrin αVβ5 was required for the cell migration on CYR61. Both H460 and TE-7 cancer cell lines exhibited significantly reduced (30–50% respectively) migration capabilities through uncoated transwell filters after stable knockdown of CYR61. H460 and TE-7 cancer cells showed increased migration (25–30% respectively) through the uncoated transwell insert after stable over-expression of CYR61 (Fig. 5).
Figure 4. CYR61 plays a role in cancer cell migration.
Migration assay through uncoated transwell inserts for parental SW620, H460 and TE-7 cancer cell lines. SW620 cells did not show any migration through the untreated transwell inserts as compared to H460 and TE-7 cell lines. Microscopic pictures of random fields of transwell insert at 20× magnification (purple - migrated cells, white - pores of the membrane).
Figure 5. Transwell migration assay for all CYR61 isogenic variants of lung carcinoma H460 and esophageal squamous cell carcinoma TE-7 cell lines.
A) Migration assay with H460 cells or B) TE-7 cells exhibiting significantly reduced migration through uncoated membrane after stable transfection with 2 independent shRNAs directed against CYR61 mRNA (GS-1 and GS-2) or a control shRNA targeting LacZ. There is also shown slightly increased migration of H460 and TE-7 cells stably over-expressing CYR61 (OE). All results are compared to LacZ control. Representative microscopic pictures are shown as random fields of transwell insert at 20× magnification. Three membrane filters were used for each condition within one experiment. Data were obtained from triplicate independent experiments and are shown as a mean ± SEM (*p<0.05). Asterisk indicates statistical difference in migration between control LacZ cells and cells with forced stable CYR61 over-expression or knock down.
Migration of cancer cells is CYR61 and αVβ5 dependent
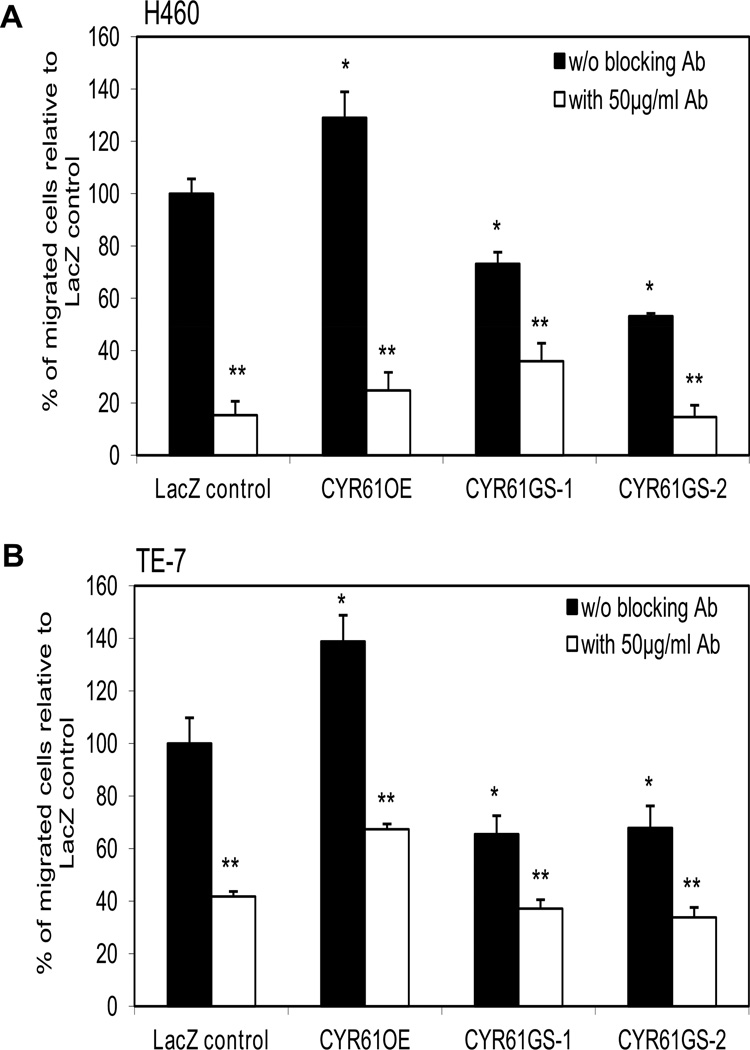
To test the functional role of integrin αVβ5 in mediating the pro-migratory effects of CYR61, we performed transwell migration assays using functional blocking antibody against the integrin αVβ5. As documented in Fig. 6, blocking antibody against integrin αVβ5 was able to diminish migration of all CYR61 isogenic variants of H460 and TE-7 cancer cell lines. The migration were reduced about 80% in all isogenic variants of H460 cells and about 60% in all TE-7 isogenic variants after 1 hour pre-treatment with integrin αVβ5 blocking antibody.
Figure 6. Blocking integrin αvβ5 antibody diminished migration capabilities of all CYR61 isogenic variants of lung carcinoma H460 and esophageal squamous cell carcinoma TE-7 cell lines.
A) H460 cells or B) TE-7 cells were treated for 1 h at 4°C with specified monoclonal (50 µg/ml) anti-αvβ5 antibody before plating on the top of the membrane. Transwell inserts were then incubated for 18 hours at 37°C, 5% CO2. Data were obtained from four independent experiments (three inserts for three individual clones of cells were used within each experiment) are shown as a mean ± SEM (p<0.05). *Asterisk indicates statistical difference in migration between untreated control LacZ cells and untreated cells with forced stable CYR61 over-expression or knockdown. **Asterisks indicate statistical differences in migration between control LacZ cells and cells with forced stable CYR61 over-expression or knock down after the cells were pre-treated with αvβ5 blocking antibody.
Discussion
In this study, we explored the functions of CYR61 in various cancer cell lines and have determined that CYR61 facilitates the migration of these cells through the integrin αVβ5. Our study confirmed that both CYR61 and integrin αVβ5 and their interactions are required for the migration of cancer cells, which has never been shown before.
CYR61 is a secreted, heparin-binding and extracellular matrix-associated angiogenic inducer. It regulates cell adhesion, migration, proliferation and is involved in tumor growth. CYR61 has been found to be differentially expressed in many cancers and is involved in the development and progression of various cancers6. Using SW620, H460 and TE-7 cancer cell lines and their isogenic variants with altered expression levels of CYR61 we determined that suppressing/increasing expression and secretion of CYR61 in various tumor cells affects their capacity for migration. We have found that the higher the CYR61 expression levels within these cancer cells, the more advanced the migratory abilities observed. We showed slightly increased migratory capabilities of H460 and TE-7 cells with CYR61 over-expression, as well as, significantly decreased migratory capabilities after stable knockdown of CYR61. Previously, it was shown that CYR61 is expressed in about 30% of invasive breast carcinomas26. Recently, Haque et al. published that CYR61 is consistently found to be over-expressed in pancreatic cancer and the expression intensifies with disease progression27. They showed that aggressive pancreatic ductal adenocarcinoma cell lines exhibited higher levels of CYR61 compared to less aggressive types and that CYR61 plays an important regulatory role in migration of these cells27. Moreover, recent findings observed high levels of CYR61 in EC109 esophageal squamous cell carcinoma cell lines and it was shown that CYR61 knockdown by RNAi leads to a significant suppression of migration, invasiveness, adhesion, formation of colonies and cell growth14. These observations confirm our findings of decreased migratory abilities of esophageal squamous cell carcinoma TE-7 cells with stably down-regulated expression of CYR61. On the other hand, lower expression levels of CYR61 were reported in non-small cell lung cancers28. Nevertheless, Tong et al. observed that the expression levels of CYR61 can alter dramatically among various lung cancer cell lines. They found high expression levels of CYR61 in three and none in five lung cancer cell lines, including H460 non-small cell lung carcinoma line29. However, we have found intermediate levels of CYR61 in H460 cells what is most likely due to using more sensitive and accurate quantitative RTPCR analysis while Tong et al. used Northern blot analysis which provides only qualitative or semi-quantitative information of mRNA levels. Moreover, they showed that H460 cells with over-expressed CYR61 formed fewer colonies and had decreased proliferation rates in comparison to control cells transfected with empty vector29. Their findings suggesting a tumor suppressor role for CYR61 in non-small cell lung cancer cells is contradictory to our results identifying CYR61 as a promoter of migration in H460 cells.
CYR61 is a well-established ligand for several integrins18,19, including αVβ5. From the diverse CYR61 functions, integrin binding has been demonstrated to affect invasion and survival4,26. Examination of integrin expression in SW620, H460 and TE-7 cancer cell lines using microarrays confirmed that these tumor cell lines express both members of the CYR61 receptor, αV and β5. Interestingly, the colorectal adenocarcinoma SW620 cells express both αV and β5 integrin subunits but do not express CYR61 and do not show migration through uncoated membranes, in compliance with findings of Watts et al. where no invasion of these cells through the matrigel was obtained24. The negative migration of SW620 cells could be explained by the presence of high levels of integrin subunits α6 and β4, the major pro-adhesion integrin combination30. TE-7 squamous cell carcinoma cell line showed the highest expression levels of CYR61 and integrin αVβ5 and demonstrated the strongest migration capabilities. In previous studies it was reported that the cell line SEG-1 (aka H460 cells25) showed strong invasiveness through matrigel, while the BIC-1 cell line (aka SW62025) did not invade at all24,31. We have reproduced these results and further demonstrate the importance of CYR61 within this process.
Integrin αVβ5 was shown to mediate CYR61-stimulated fibroblast migration32, promote resistance to apoptosis in MCF-7 cells4 and is essential for CYR61-induced metastasis of tumors growing within a pre-irradiated field33. In our experiments when a functional blocking antibody against integrin αVβ5 was included, the migration of H460 and TE-7 cells was significantly inhibited. These results confirm for the first time that tumor cell-secreted CYR61 may facilitate cancer cell migration by interacting with the αVβ5 integrin in cells. The data presented in this study suggest that suppressing/increasing expression and secretion of CYR61 in cancer cells affects their capacity for migration and that there is a requirement of CYR61 for cancer cell migration by interactions with integrin αVβ5 expressed on tumor cells.
It had been demonstrated that downstream effectors of αVβ5 integrin as integrin-linked kinase (ILK) and small GTPase (RhoB) might be promising candidate targets for improving the efficiency of radiotherapy34. Recently, it was reported that CCN3-stimulated migration of chondrosarcoma cells is mediated through the αvβ3/αvβ5 integrin receptor, FAK, PI3K, Akt, p65, and NF-κB signal transduction pathway35. Also, it was shown that integrin αvβ5 directly interacts with PAK4 (serine/threonine p-21 activating kinase) and regulates carcinoma cell motility in an integrin-specific manner36. In breast cancer cells, integrin αvβ5 mediates activation of PAK4 after the cells are attached to vitronectin37. Thus, besides of CYR61 as a possible target for cancer therapy, further investigation of signaling pathways that are activated downstream of integrin αvβ5 might also bring us new possible targets for therapeutic interventions and improving the efficiency of treatment of various cancers.
Supplementary Material
Raw data for three individual cell lines are indicated. Affymetrix chips were executed for each cell lines in triplicates as described previously24.
Acknowledgment
This work was supported in part by the NCI Cancer Center Support Grant P30 CA023074 (CCSG), The GI SPORE grant CA95060, and a pilot grant from the American Cancer Society.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Brigstock DR. The CCN family: a new stimulus package. J Endocrinol. 2003;178:169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- 2.Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- 3.Grzeszkiewicz TM, Lindner V, Chen N, Lam SC, Lau LF. The angiogenic factor cysteine-rich 61 (CYR61, CCN1) supports vascular smooth muscle cell adhesion and stimulates chemotaxis through integrin alpha(6)beta(1) and cell surface heparan sulfate proteoglycans. Endocrinology. 2002;143:1441–1450. doi: 10.1210/endo.143.4.8731. [DOI] [PubMed] [Google Scholar]
- 4.Lin MT, Chang CC, Chen ST, Chang HL, Su JL, Chau YP, Kuo ML. Cyr61 expression confers resistance to apoptosis in breast cancer MCF-7 cells by a mechanism of NF-kappaB-dependent XIAP up-regulation. J Biol Chem. 2004;279:24015–24023. doi: 10.1074/jbc.M402305200. [DOI] [PubMed] [Google Scholar]
- 5.D'Antonio KB, Schultz L, Albadine R, Mondul AM, Platz EA, Netto GJ, Getzenberg RH. Decreased expression of Cyr61 is associated with prostate cancer recurrence after surgical treatment. Clin Cancer Res. 2010;16:5908–5913. doi: 10.1158/1078-0432.CCR-10-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleau AM, Planque N, Perbal B. CCN proteins and cancer: two to tango. Front Biosci. 2005;10:998–1009. doi: 10.2741/1594. [DOI] [PubMed] [Google Scholar]
- 7.Babic AM, Kireeva ML, Kolesnikova TV, Lau LF. CYR61, a product of a growth factor-inducible immediate early gene, promotes angiogenesis and tumor growth. Proc Natl Acad Sci U S A. 1998;95:6355–6360. doi: 10.1073/pnas.95.11.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genini M, Schwalbe P, Scholl FA, Schafer BW. Isolation of genes differentially expressed in human primary myoblasts and embryonal rhabdomyosarcoma. Int J Cancer. 1996;66:571–577. doi: 10.1002/(SICI)1097-0215(19960516)66:4<571::AID-IJC24>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 9.Holloway SE, Beck AW, Girard L, Jaber MR, Barnett CC, Jr, Brekken RA, Fleming JB. Increased expression of Cyr61 (CCN1) identified in peritoneal metastases from human pancreatic cancer. J Am Coll Surg. 2005;200:371–377. doi: 10.1016/j.jamcollsurg.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Xie D, Miller CW, O'Kelly J, Nakachi K, Sakashita A, Said JW, Gornbein J, Koeffler HP. Breast cancer. Cyr61 is overexpressed, estrogen-inducible, and associated with more advanced disease. J Biol Chem. 2001;276:14187–14194. doi: 10.1074/jbc.M009755200. [DOI] [PubMed] [Google Scholar]
- 11.Xie D, Nakachi K, Wang H, Elashoff R, Koeffler HP. Elevated levels of connective tissue growth factor, WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res. 2001;61:8917–8923. [PubMed] [Google Scholar]
- 12.Xie D, Yin D, Tong X, O'Kelly J, Mori A, Miller C, Black K, Gui D, Said JW, Koeffler HP. Cyr61 is overexpressed in gliomas and involved in integrin-linked kinase-mediated Akt and beta-catenin-TCF/Lef signaling pathways. Cancer Res. 2004;64:1987–1996. doi: 10.1158/0008-5472.can-03-0666. [DOI] [PubMed] [Google Scholar]
- 13.Sun ZJ, Wang Y, Cai Z, Chen PP, Tong XJ, Xie D. Involvement of Cyr61 in growth, migration, and metastasis of prostate cancer cells. Br J Cancer. 2008;99:1656–1667. doi: 10.1038/sj.bjc.6604712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie JJ, Xu LY, Xie YM, Du ZP, Feng CH, Dong H, Li EM. Involvement of Cyr61 in the growth, invasiveness and adhesion of esophageal squamous cell carcinoma cells. Int J Mol Med. 2011;27:429–434. doi: 10.3892/ijmm.2011.603. [DOI] [PubMed] [Google Scholar]
- 15.Tong X, O'Kelly J, Xie D, Mori A, Lemp N, McKenna R, Miller CW, Koeffler HP. Cyr61 suppresses the growth of non-small-cell lung cancer cells via the beta-catenin-c-myc-p53 pathway. Oncogene. 2004;23:4847–4855. doi: 10.1038/sj.onc.1207628. [DOI] [PubMed] [Google Scholar]
- 16.Chien W, Kumagai T, Miller CW, Desmond JC, Frank JM, Said JW, Koeffler HP. Cyr61 suppresses growth of human endometrial cancer cells. J Biol Chem. 2004;279:53087–53096. doi: 10.1074/jbc.M410254200. [DOI] [PubMed] [Google Scholar]
- 17.Sampath D, Zhu Y, Winneker RC, Zhang Z. Aberrant expression of Cyr61, a member of the CCN (CTGF/Cyr61/Cef10/NOVH) family, and dysregulation by 17 beta-estradiol and basic fibroblast growth factor in human uterine leiomyomas. J Clin Endocrinol Metab. 2001;86:1707–1715. doi: 10.1210/jcem.86.4.7423. [DOI] [PubMed] [Google Scholar]
- 18.Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41:771–783. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kireeva ML, Lam SC, Lau LF. Adhesion of human umbilical vein endothelial cells to the immediate-early gene product Cyr61 is mediated through integrin alphavbeta3. J Biol Chem. 1998;273:3090–3096. doi: 10.1074/jbc.273.5.3090. [DOI] [PubMed] [Google Scholar]
- 20.Lau LF. Integrin-mediated CCN functions. London: Imperial College Press; 2005. [Google Scholar]
- 21.Leu SJ, Chen N, Chen CC, Todorovic V, Bai T, Juric V, Liu Y, Yan G, Lam SC, Lau LF. Targeted mutagenesis of the angiogenic protein CCN1 (CYR61). Selective inactivation of integrin alpha6beta1-heparan sulfate proteoglycan coreceptor-mediated cellular functions. J Biol Chem. 2004;279:44177–44187. doi: 10.1074/jbc.M407850200. [DOI] [PubMed] [Google Scholar]
- 22.Chen N, Leu SJ, Todorovic V, Lam SC, Lau LF. Identification of a novel integrin alphavbeta3 binding site in CCN1 (CYR61) critical for pro-angiogenic activities in vascular endothelial cells. J Biol Chem. 2004;279:44166–44176. doi: 10.1074/jbc.M406813200. [DOI] [PubMed] [Google Scholar]
- 23.Chen CC, Young JL, Monzon RI, Chen N, Todorovic V, Lau LF. Cytotoxicity of TNFalpha is regulated by integrin-mediated matrix signaling. EMBO J. 2007;26:1257–1267. doi: 10.1038/sj.emboj.7601596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watts GS, Tran NL, Berens ME, Bhattacharyya AK, Nelson MA, Montgomery EA, Sampliner RE. Identification of Fn14/TWEAK receptor as a potential therapeutic target in esophageal adenocarcinoma. Int J Cancer. 2007;121:2132–2139. doi: 10.1002/ijc.22898. [DOI] [PubMed] [Google Scholar]
- 25.Boonstra JJ, van Marion R, Beer DG, Lin L, Chaves P, Ribeiro C, Pereira AD, Roque L, Darnton SJ, Altorki NK, Schrump DS, Klimstra DS, Tang LH, Eshleman JR, Alvarez H, Shimada Y, van Dekken H, Tilanus HW, Dinjens WN. Verification and unmasking of widely used human esophageal adenocarcinoma cell lines. J Natl Cancer Inst. 2010;102:271–274. doi: 10.1093/jnci/djp499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai MS, Hornby AE, Lakins J, Lupu R. Expression and function of CYR61, an angiogenic factor, in breast cancer cell lines and tumor biopsies. Cancer Res. 2000;60:5603–5607. [PubMed] [Google Scholar]
- 27.Haque I, Mehta S, Majumder M, Dhar K, De A, McGregor D, Van Veldhuizen PJ, Banerjee SK, Banerjee S. Cyr61/CCN1 signaling is critical for epithelial-mesenchymal transition and stemness and promotes pancreatic carcinogenesis. Mol Cancer. 2011;10:8. doi: 10.1186/1476-4598-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen PP, Li WJ, Wang Y, Zhao S, Li DY, Feng LY, Shi XL, Koeffler HP, Tong XJ, Xie D. Expression of Cyr61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PLoS One. 2007;2:e534. doi: 10.1371/journal.pone.0000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong X, Xie D, O'Kelly J, Miller CW, Muller-Tidow C, Koeffler HP. Cyr61, a member of CCN family, is a tumor suppressor in non-small cell lung cancer. J Biol Chem. 2001;276:47709–47714. doi: 10.1074/jbc.M107878200. [DOI] [PubMed] [Google Scholar]
- 30.Borradori L, Sonnenberg A. Structure and function of hemidesmosomes: more than simple adhesion complexes. J Invest Dermatol. 1999;112:411–418. doi: 10.1046/j.1523-1747.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Kuwabara Y, Iwata H, Mitani M, Shinoda N, Sato A, Mitsui A, Sugiura M, Kato J, Fujii Y. Role of matrix metalloproteinase-9 in in vitro invasion of esophageal carcinoma cells. J Surg Oncol. 2002;81:80–86. doi: 10.1002/jso.10134. [DOI] [PubMed] [Google Scholar]
- 32.Grzeszkiewicz TM, Kirschling DJ, Chen N, Lau LF. CYR61 stimulates human skin fibroblast migration through Integrin alpha vbeta 5 and enhances mitogenesis through integrin alpha vbeta 3, independent of its carboxyl-terminal domain. J Biol Chem. 2001;276:21943–21950. doi: 10.1074/jbc.M100978200. [DOI] [PubMed] [Google Scholar]
- 33.Monnier Y, Farmer P, Bieler G, Imaizumi N, Sengstag T, Alghisi GC, Stehle JC, Ciarloni L, Andrejevic-Blant S, Moeckli R, Mirimanoff RO, Goodman SL, Delorenzi M, Ruegg C. CYR61 and alphaVbeta5 integrin cooperate to promote invasion and metastasis of tumors growing in preirradiated stroma. Cancer Res. 2008;68:7323–7331. doi: 10.1158/0008-5472.CAN-08-0841. [DOI] [PubMed] [Google Scholar]
- 34.Monferran S, Skuli N, Delmas C, Favre G, Bonnet J, Cohen-Jonathan-Moyal E, Toulas C. avb3 and avb5 integrins control glioma cell response to ionizing radiation through ILK and RhoB. Int J Cancer. 2008;123:357–364. doi: 10.1002/ijc.23498. [DOI] [PubMed] [Google Scholar]
- 35.Tzeng HE, Chen JC, Tsai CH, Kuo CC, Hsu HC, Hwang WL, Fong YC, Tang CH. CCN3 Increases Cell Motility and MMP-13 Expression in Human Chondrosarcoma Through Integrin-Dependent Pathway. J Cell Physiol. 2011;12:3181–3189. doi: 10.1002/jcp.22672. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Li Z, Viklund EK, Strömblad S. P21-activated kinase 4 interacts with integrin alpha v beta 5 and regulates alpha v beta 5-mediated cell migration. J Cell Biol. 2002;7:1287–1297. doi: 10.1083/jcb.200207008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Lock JG, Olofsson H, Kowalewski JM, Teller S, Liu Y, Zhang H, Strömblad S. Integrin-mediated cell attachment induces a PAK4-dependent feedback loop regulating cell adhesion through modified integrin alpha v beta 5 clustering and turnover. Mol Biol Cell. 2010;21:3317–3329. doi: 10.1091/mbc.E10-03-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data for three individual cell lines are indicated. Affymetrix chips were executed for each cell lines in triplicates as described previously24.