Summary
Carbapenems such as meropenem are being investigated for their potential therapeutic utility against highly drug-resistant tuberculosis. These β-lactams target the transpeptidases that introduce interpeptide cross-links into bacterial peptidoglycan thereby controlling rigidity of the bacterial envelope. Treatment of M. tuberculosis (Mtb) with the β-lactamase inhibitor clavulanate together with meropenem resulted in rapid, polar, cell lysis releasing cytoplasmic contents. In Mtb it has been previously demonstrated that 3-3 cross-linkages (involving two diaminopimelate (DAP) molecules) predominate over 4-3 cross-linkages (involving one DAP and one D-alanine) in stationary-phase cells. We purified and analyzed peptidoglycan from Mtb and found that 3-3 cross-linkages predominate throughout all growth phases and the ratio of 4-3/3-3 linkages does not vary significantly under any growth condition. Meropenem treatment was accompanied by a dramatic accumulation of unlinked pentapeptide stems with no change in the tetrapeptide pools, suggesting that meropenem inhibits both a D,D-carboxypeptidase and an L,D-transpeptidase. We purified a candidate D,D-carboxypeptidase DacB2 and showed that meropenem indeed directly inhibits this enzyme by forming a stable adduct at the enzyme active site. These results suggest that the rapid lysis of meropenem-treated cells is the result of synergistically inhibiting the transpeptidases that introduce 3,3-cross-links while simultaneously limiting the pool of available substrates available for cross-linking.
Introduction
For patients infected with highly drug-resistant Mycobacterium tuberculosis (Mtb) treatment options are often very limited, toxic and of questionable efficacy (Yew & Leung, 2008). There are currently over 500,000 cases of multidrug-resistant tuberculosis (MDR-TB) and these are increasing in many areas of the world (Wright et al., 2009). Although fewer than one in ten patients with MDR-TB have access to current second-line therapies, resistance to these is rapidly developing giving rise to extensively drug-resistant TB (XDR-TB), an emerging epidemic of difficult to determine magnitude (Cohen et al., 2008). For patients with XDR-TB, treatment options are even more limited and as a result mortality is high (Johnston et al., 2009). A recent report that the β-lactam meropenem, in combination with clavulanic acid, has activity against both actively replicating and non-replicating XDR-TB strains has given hope of repurposing this class of agents for XDR-TB patients (Hugonnet et al., 2009). Meropenem has also shown activity in the murine model of Mtb infection (England et al., 2012). Meropenem, however, requires parenteral administration that greatly increases the difficulty of therapy and seriously limits utility to the most extreme cases. Expanding the therapeutic utility of the carbapenems would require development of an oral agent, a task that would be made significantly easier with identification of the molecular target of meropenem.
Peptidoglycan (PG) is an essential component of the bacterial cell wall and its biosynthesis represents the site of action of all known β-lactams. In all eubacteria its basic structure consists of linear glycan strands cross-linked to each other by short peptides that branch out from repeating glycan units. In Mtb the glycan strand is a polymer of alternating units of β-(1–4) linked N-acetylglucosamine (NAG) and N-glycolylmuramic acid (NGM) to which a short stem peptide of L-Ala-D-iGlu-meso-2,6-diaminopimelate[DAP]-D-Ala-D-Ala is attached through a lactic acid moiety. This stem, lacking one or both D-Ala residues, is cross-linked to a stem in a neighboring glycan strand, forming a mesh responsible for the strength and rigidity of the whole structure (Van Heijenoort, 1994). Cross-linking is catalyzed by D,D-transpeptidases (also referred to as penicillin-binding proteins) and by L,D-transpeptidases (Ldt) resulting in 4-3 (D-Ala-DAP) and 3-3 (DAP-DAP) cross-links, respectively (Mainardi et al., 2005). Most bacterial species contain predominantly 4-3 cross-links, however 3-3 cross-links have been reported in several bacteria including Streptomyces albus G, Clostridium perfringens (Leyh-Bouille et al., 1970), C. difficile (Peltier et al., 2011), M. smegmatis, M. bovis BCG (Wietzerbin et al., 1974), M. abscessus (Lavollay et al., 2011) and Mtb strain H37Rv (Lavollay et al., 2008).
The basic structure of PG is highly conserved in eubacteria (Goffin & Ghuysen, 2002), however, significant species-specific as well as growth-phase and condition-dependent variations in fine structure have been reported (Tuomanen & Cozens, 1987, Glauner et al., 1988, Driehuis & Wouters, 1987). Mycobacterial PG contains several unique modifications such as presence of N-glycolylmuramic acid (NGM) on the glycan strand in place of N-acetylmuramic acid (NAM) (Kotani et al., 1970, Mahapatra et al., 2005, Raymond et al., 2005), amidation of free carboxyl groups at Glu and DAP residues (Mahapatra et al., 2008, Lavollay et al., 2008), and Gly replacing the terminal D-ala of stem peptides in M. leprae (Mahapatra et al., 2008). A previous report determined that PG prepared from stationary-phase cells of Mtb contained an unusually high content (~80 %) of 3-3 type cross-links suggesting this might be a physiologic adaptation of Mtb to stationary phase. These authors also identified a penicillin-resistant but carbapenem-sensitive Ldt (Rv0116c) in Mtb (Lavollay et al., 2008). In the present study we aimed to establish the baseline cross-linking status of Mtb PG and identify any growth-phase or condition-specific alterations. This allowed us to assess the impact of challenge with β-lactams on cross-linking and to further clarify the mechanism of meropenem-induced cell lysis.
RESULTS
Meropenem induces rapid lytic killing of Mtb
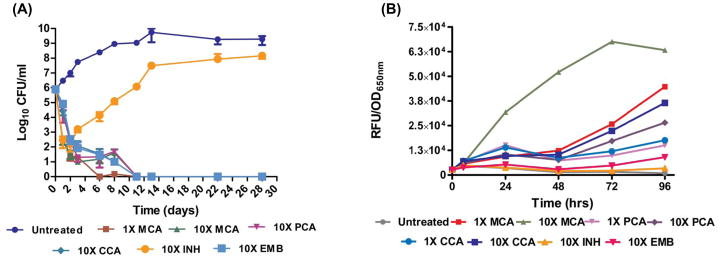
To understand better the mechanism of β-lactam-induced bactericidal activity we treated log-phase cultures of Mtb with a penicillin (penicillin G), a cephalosporin (cephaloridine) and a carbapenem (meropenem) in combination with clavulanate at ten times their MIC90 and compared the kill rates to two other cell-wall active antibiotics, isoniazid (INH) and ethambutol (EMB) (Fig. 1A). Clavulanate alone was not toxic at the concentration used (100 μM) data not shown). All of these agents induced initial cell killing with similar kinetics compared to untreated controls and sterilized the culture within two weeks. INH-treated cultures decreased in viability for the first two days but were rapidly overgrown with INH-resistant mutants as has been previously described (Bergval et al., 2009). INH treatment has been previously shown to impair the production of new cell wall but not to result in the immediate release of cytoplasmic contents, instead protoplasmic viscosity increases to a critical point gradually as other macromolecules continue to be produced (Takayama et al., 1975). To evaluate the effects of killing by β-lactams further with respect to cell wall integrity we employed an Mtb strain harboring pMSP12:GFP plasmid (Ramakrishnan et al., 2000) over-expressing cytoplasmic green-fluorescent protein (GFP). We monitored GFP in culture supernatant on exposure to β-lactams and noted that all of the β-lactams induced a rapid increase in culture supernatant associated fluorescence with meropenem inducing this cell wall lysis the most rapidly (Fig. 1B). In contrast, neither INH nor EMB induced significant cell lysis within the first two days of incubation.
Fig. 1.
Sensitivity of Mtb to various cell-wall damaging agents. (A) Kill curve of Mtb in presence of various agents. (B) Lysis of Mtb upon treatment with these agents measured by quantitating release of cytosolic GFP in cell-free culture supernatant (from a GFP-expressing Mtb/pMSP12 strain). As indicated in the graphs following agents were used either at 1-time (1X) or 10-times (10X) their corresponding MIC: Meropenem-clavulanate (MCA, MIC for meropenem 2 μM), penicillin G-clavulanate (PCA, MIC for penicillin G 2 μM), cephaloridine-clavulanate (CCA, MIC for cephaloridine 1 μM), clavulanate was used at 100 μM throughout, isoniazid (INH, 2 μM) and ethambutol (EMB, 62.5 μM).
Ultrastructural analysis of meropenem-treated Mtb reveals polar lysis
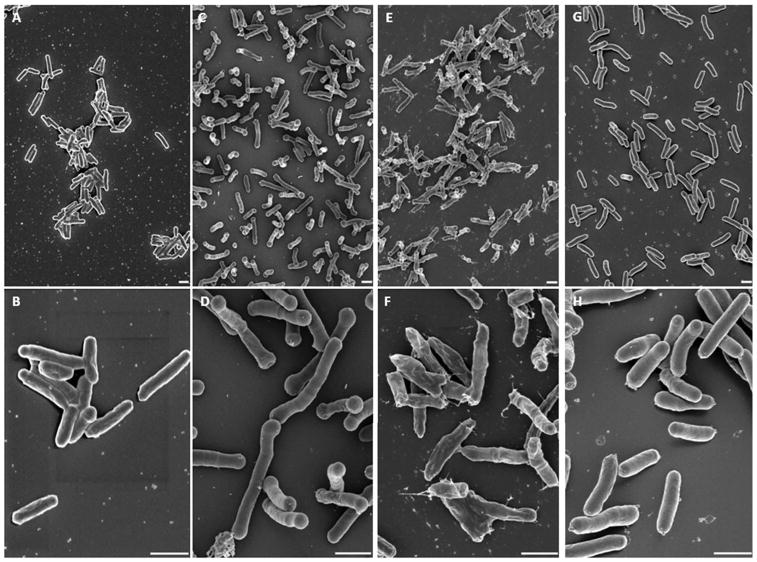
To better understand the mechanism of meropenem-induced lysis we examined Mtb treated cultures using scanning electron microscopy. Normal, logarithmically growing Mtb cells are roughly cylindrical rods about 2 μm in length (Fig. 2A and B). Meropenem treatment at the MIC90 for 24 hours induces substantial polar swelling of these rods (Fig. 2C and D) and treatment for the same time with ten times higher concentration of meropenem induces profound leakage of material from the poles of these cells leaving what appear to be empty sacculi (Fig. 2E and F). In contrast, INH treatment results in shorter, thicker cells (Fig. 2G and H). To quantify this effect we measured 200 individual cells from each of these conditions and showed that untreated cells have an average length of 1.98 ± 0.48 μm, which was only slightly changed by meropenem-treatment (2.33 ± 0.51 μm), in contrast to treatment with both INH and EMB which significantly decreased the overall length to 1.01 ± 0.38 and 1.28 ± 0.42 μm respectively (Fig. S1). These results are consistent with the rapid release of GFP from the cytoplasm of cells treated with meropenem and support that the increase in fluorescence of the supernatant from treated cultures is a direct result of a rapid lytic event induced by inhibition of peptidoglycan assembly.
Fig. 2.
SEM images showing effects of meropenem exposure on Mtb cell shape. (A & B) untreated controls, (C & D) treated with 1X MCA, (E & F) with 10X MCA or (G & H) with 10X INH (Bar=1 μm). The lower magnification pictures shown above (A, C, E, G) were captured at 8,000 X and corresponding higher magnification pictures shown below (B, D, F, I) were captured at 25, 000 X.
Preparation and characterization of mycobacterial peptidoglycan
To analyze and compare alterations in structure of mycobacterial PG that occurred with meropenem treatment, an efficient method was needed for simultaneous isolation of large amounts of PG from multiple samples. Earlier methods required use of either a French press or a bead-beater (Lavollay et al., 2008, Rezwan et al., 2007) for breaking open mycobacterial cells. Both methods were unsuitable for simultaneous processing of multiple samples in a BSL-3 environment. To circumvent this limitation we autoclaved cultures in the presence of 4% SDS (wt/vol) to allow rapid and simultaneous isolation of PG from multiple samples. PG prepared by autoclaving was compared with that prepared by bead-beating, showing that these resulted in similar muropeptide profiles (data not shown). A typical yield of PG from a 500 ml culture harvested at OD650 = 0.6 was about 10 mg.
A combination of the three muramidases (Chalaropsis muramidase, lysozyme and mutanolysin) was used to maximize the hydrolysis of PG into muropeptides for HPLC analysis. The muramidase from Chalaropsis sp. (ATCC 16003) was purified as described previously (Hash, 1963, Yagi et al., 2003, Hash & Rothlauf, 1967). To assess the efficiency of enzymatic hydrolysis of complex higher-order muropeptides from Mtb PG, we radiolabeled whole cultures of Mtb for three hours with [1-14C] D-alanine, a D-amino acid found primarily in PG. Comparison of the UV-absorbance profile with the radioprofile (Fig. S2A vs Fig. S2B) indicated that [1-14C] D-alanine was incorporated very quickly and efficiently into Mtb PG. The hydrolysis of 3 mg of labeled PG with lysozyme and mutanolysin released about 50% of the radioactivity as soluble muropeptides, addition of the Chalaropsis muramidase resulted in a further 20% release of soluble muropeptides. This result was supported by sequential digestion of unlabeled PG with individual muramidases and analysis by RP-HPLC. After complete digestion with lysozyme followed by mutanolysin (Fig. S3 A & B), the addition of Chalaropsis muramidase resulted in further release of additional soluble muropeptides (Fig. S3C). Similarly, the sequential digestion initiated with Chalaropsis muramidase alone was also incomplete (Fig. S3G) and the addition of lysozyme and mutanolysin resulted in a significant increase in soluble muropeptides (Fig. S3 H & I). However, in PG predigested with mutanolysin (Fig. S3D) and Chalaropsis muramidase (Fig. S3E), addition of lysozyme did not solublize significant amounts of additional PG. This data suggests that mutanolysin and Chalaropsis muramidase are absolutely required to maximize PG hydrolysis but lysozyme may not be essential.
To evaluate what fraction of peptides were incorporated into structures that were completely resistant to release from PG even using these enzymes we also isolated PG from bacilli that had been labeled with radioactive D-alanine following digestion with our optimal conditions. Following HPLC separation about half of the radioactivity incorporated into PG from [1-14C] D-alanine after three hours of growth was incorporated into structures that were trimeric or more complex eluting after 50 minutes, suggesting that during logarithmic growth Mtb contains a very active cross-linking ability and that there is likely to be a pool of cross-linked structures not amenable to analysis using current methodologies.
Characterization of monomeric muropeptides
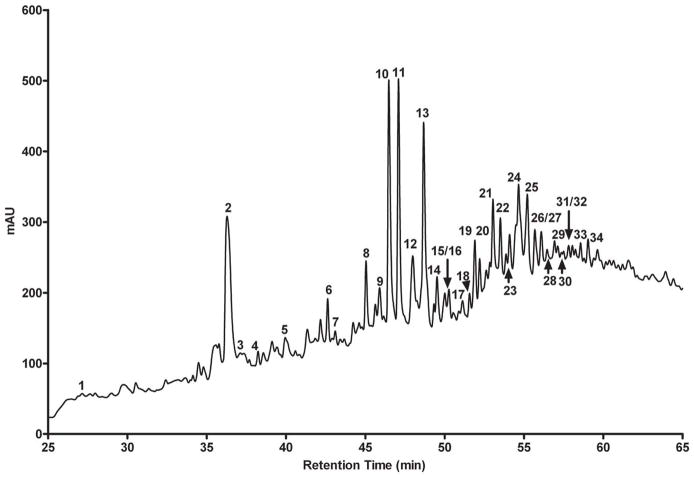
Structural analysis of PG prepared from exponentially (OD650 = 0.12) growing Mtb was performed using HPLC and mass spectrometry. The RP-HPLC trace showed 34 distinct UV active peaks (Fig. 3), and these peaks were subjected to high-resolution mass spectrometry for assignment of accurate structures. The most abundant early-eluting peak (Peak # 2, Fig. 3) had a monoisotopic mass of 532.2, which corresponded to 1 amu less than the expected mass for the canonical monomeric tetrapeptide. MS/MS data suggested that this mass defect was associated with DAP since the daughter ion corresponding to loss of DAP (m/z 272) had the calculated mass of the remainder of the expected peptide. Amidation of both the α-carboxylate of glutamate and the ε-carboxylate of DAP have been described in the stem peptides of the related organism Corynebacterium diphtheriae (Kato et al., 1968). Therefore we propose amidation at the ε-carboxylate of DAP to account for the observed ion, suggesting that the authentic tetrapeptide structure is D-Lac-L-Ala-D-iGln-mDAPNH2-D-Ala (Tetra, Table 1, Fig. 4A).
Fig. 3.
RP-HPLC profile of muropeptides prepared by muramidase digestion and ammonium hydroxide treatment from PG isolated from exponentially growing (OD650 = 0.12) Mtb. The absorbance in mAU (absorbance unit × 103) was recorded at 210 nm.
Table 1.
Structure and relative abundance of muropeptides in logarithmically growing Mtb (OD650=0.12)
| Peak | Muropeptide | Mass (Da) | Cross-link | (%) |
|---|---|---|---|---|
| 1 | Tri | 461.5 | NA | 1.1 |
| 2 | Tetra | 532.2 | NA | 15.3 |
| 3 | TetraOH | 533.2 | NA | 1.5 |
| 4 | Tetra(E) | 533.2 | NA | 0.5 |
| 5 | Penta | 603.2 | NA | 1.6 |
| 6 | Tri-Tri | 903.4 | 3-3 | 1.6 |
| 7 | Tri-TriOH | 904.4 | 3-3 | 0.8 |
| 8 | Tetra-Tri | 974.4 | 4-3 | 2.4 |
| 9 | Tri(Anh) | 865.5 | NA | 2.2 |
| 10 | Tetra-Tri | 974.4 | 3-3 | 11.6 |
| 11 | Tetra-TriOH | 975.4 | 3-3 | 10.2 |
| 12 | TetraOH-TriOH | 976.4 | 3-3 | 2.7 |
| 13 | Tetra-Tetra | 1045.6 | 4-3 | 10.2 |
| 14 | Tetra-TetraOH | 1046.6 | 4-3 | 3.1 |
| 15 | Tetra-TetraOH | 1046.6 | 4-3 | 1.5 |
| 16 | Tetra-Penta | 1116.6 | 4-3 | 1.5 |
| 17 | Tri-Tetra(Anh) | 1377.7 | ND | 1.2 |
| 18 | Tri-Tetra-Tetra | 1487.6 | ND | 0.8 |
| 19 | Tri-Tri-Tetra | 1416.7 | ND | 2.7 |
| 20 | TriOH-Tri-Tetra | 1417.7 | ND | 1.4 |
| 21 | Tri-Tetra-Tetra | 1487.6 | ND | 3.1 |
| 22 | Tri-Tetra-Tetra | 1488.6 | ND | 2.2 |
| 23 | Tri-Tri-Tetra | 1418.7 | ND | 2.0 |
| 24 | Tetra-Tetra-Tetra | 1558.5 | ND | 7.0 |
| 25 | Tri-Tetra-Tetra | 1489.3 | ND | 4.3 |
| 26 | Tri-Tetra(Anh) | 1378.7 | 3-3 | 1.9 |
| 27 | Tri-Tetra(Anh) | 1379.7 | 3-3 | 1.3 |
| 28 | TriOH-Tri-Tetra-Tetra | 1931.9 | ND | 0.4 |
| 29 | TriOH-Tetra-Tetra-Tetra | 1001.6 (2x) | ND | 1.0 |
| 30 | Tetra-Tetra-Tetra | 1561.3 | ND | 0.4 |
| 31 | Tetra-Penta(Anh) | 1449.6 | ND | 0.5 |
| 32 | ? | 1369.8 | ND | 0.6 |
| 33 | Tri-Tri-Tetra(Anh)-Tetra | 1179.2(2x) | ND | 0.5 |
| 34 | Tri-Tetra(Anh)-Tetra | 1892.7 | ND | 1.0 |
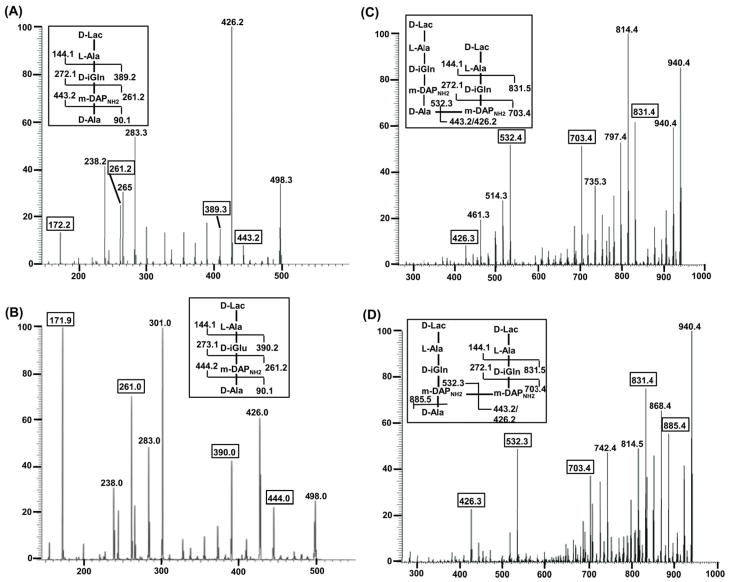
Fig. 4.
Characterization of amidation sites in monomeric and dimeric muropeptides by MS/MS analysis. The expected MS/MS fragmentation pattern is shown in the boxes above each m/z chromatogram. (A) MS/MS profile of [M+H]+ ion at m/z 532.2, the observed ions (443.2, 389.2, 272.1, MS/MS/MS of 172.2 showed it corresponded to DAPNH2: data not shown) matched with the expected ions for Tetra, D-Lac-L-Ala-D-iGln-mDAPNH2-D-Ala. (B) MS/MS profile of ion m/z 533.2 corresponding to Tetra(E), D-Lac-L-Ala-D-iGlu-mDAPNH2-D-Ala, MS/MS/MS of 171.9 showed it corresponded to DAPNH2: data not shown. (C) MS/MS profile ion at m/z 974.4 in peak 8 (Table 1) showing 4-3 cross-linked Tetra-Tri. (D) MS/MS profile ion at m/z 974.4 in peak 10 (Table 1) showing 3-3 cross-linking Tetra-Tri.
Two other smaller apparent tetrapeptide peaks were also observed (# 3 and # 4, Fig. 3) each having a mass of 533.2 Da suggesting one of the two amide groups was absent on each. Analysis of the MS/MS fragmentation of these two LC peaks (Fig. S4 & Fig. 4B) showed that peak 3 lacked amidation at the ε-carboxylate of DAP (TetraOH, Fig. S4) while peak 4 lacked amidation at the α-carboxylate of D-iso-Gln (Tetra(E), Fig. 4B). A total of 6 peaks appeared to contain monomeric structures with Tetra being by far the predominant moiety. The analysis also detected a small quantity of pentapeptide (D-Lac-L-Ala-D-iGln-mDAPNH2-D-Ala-D-Ala) that was also amidated at both carboxylates (# 5, Fig. 3, Table 1). Overall these data show that the vast majority of monomeric peptides available as substrates for transpeptidases are amidated at both the α-carboxyl group of iso-Glu and the ε-carboxyl group of DAP.
Characterization of cross-linked peptides
Twelve peaks were assigned dimeric structures based on MS/MS data (Table 1), and individual peaks included either 3-3 (DAP to DAP) or 4-3 (D-Ala to DAP) cross-linked dimers. Two of the three most prominent peaks among the dimeric moieties (# 10 and # 11, Fig. 3) were found to be cross-linked by 3-3 linkage, but differed from each other by amidation at ε-carboxyl of DAP (Table 1). Peak # 10 showed full amidation of both stem peptides (Tetra-Tri, mass 974.5 Da, Fig. 4D), whereas peak # 11 lacked amidation at the ε-carboxyl of DAP from the donor stem peptide (Tetra-TriOH, mass 975.5 Da, Fig. S5). Given the strong bias in amidated monomers available, these data strongly suggest that one or more of the L,D-transpeptidases shows a substrate preference for an unamidated ε-carboxyl donor stem in forming a 3-3 cross-link.
Five peaks with 4-3 cross-linkages were observed (# 8 and # 13 to # 16, Fig. 3). Peak # 8 was determined to be the diamidated Tetra-Tri 4-3 cross-linked dimer (mass 974.5 Da, Fig. 4C), whereas # 13 to # 15 were Tetra-Tetra dimers with differential amidation similar to that observed in 3-3 cross-linked dimers (Table 1). Overall among dimeric peptides about 65% were fully amidated on both stem peptides. Approximately, 50% of 3-3 cross-linked peptides lacked amidation on at least one of the stem peptides; whereas over 75% of 4-3 cross-linked dimers were fully amidated on both stem peptides. These results revealed that overall 60% of the dimeric muropeptides in logarithmically growing Mtb were cross-linked by 3-3 linkages. We also detected peaks corresponding to more complex higher order cross-links, which were assigned structures but the nature of cross-links and amidations of these species were not fully determined.
Variation in PG structure during different growth stages
To understand changes in the structure of Mtb PG at different stages of growth, PG was prepared from Mtb grown to different cell densities by harvesting cells at different time points as indicated in the growth curve (Fig. S6A). Similar total numbers of cells were used to prepare each sample. Very similar profiles of lactoylpeptides derived from these cultures were seen on a RP-HPLC column (Fig. S6) suggesting an overall conservation of Mtb PG architecture irrespective of stage of growth. Virtually the same 34 peaks of muropeptides seen in early log phage (Fig. 3) were observed in PG prepared from different stages (Fig. S6 B-I). Quantification of peak areas showed a small relative decrease in monomeric peptides with a corresponding increase in dimeric and higher order structures with the increase in culture density (Table 2). The data also showed 80% cross-linking of muramyl peptides in Mtb PG in the form of dimeric or higher cross-linked species. The ratio of 3-3 and 4-3 cross-linked muramyl dipeptides was observed to be about 3:2 with no significant growth-stage specific variation. The relative abundance of all peaks is presented in Table S1.
Table 2.
Abundance of different types of muramyl peptides and cross-links in Mtb PG prepared from different stages of growth.
| Muropeptide | T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 |
|---|---|---|---|---|---|---|---|---|---|
| Monomeric (%) | 22.2 | 22.4 | 21.8 | 22.0 | 22.1 | 19.2 | 22.3 | 21.7 | 20.8 |
| Dimeric (%) | 50.4 | 50.4 | 49.4 | 49.2 | 48.6 | 50.3 | 47.6 | 48.3 | 49.8 |
| Higher (%) | 27.4 | 27.2 | 28.9 | 28.8 | 29.3 | 30.4 | 30.2 | 29.9 | 29.3 |
| Dimeric 3-3 (%) | 59.7 | 60.7 | 61.3 | 63.7 | 64.2 | 63.5 | 64.6 | 63.2 | 57.8 |
| Dimeric 4-3 (%) | 40.3 | 39.3 | 38.7 | 36.3 | 35.8 | 36.5 | 35.4 | 36.8 | 42.2 |
Variation in PG structure under non-replicating persistent conditions
In view of this surprising result, we also studied the structure of PG from Mtb under conditions of non-replicating persistence (Wayne & Sohaskey, 2001). The PG profile (Fig. S7) revealed that the overall PG structure remained constant even under these conditions of growth. As evident from the RP-HPLC profile, all major peaks were present in comparable ratio; however three additional peaks were observed indicated as peaks 6*, 7* and 8* (Fig. S7). The structure of these peaks were determined by MS-MS to consist of donor-stem peptides differing only by the presence of an asparagine residue from peaks # 6, # 7 and # 8. Incorporation of aspargine into PG may have been the result of growth on Dubos medium, which contains 2 g l−1 asparagine. Similar to what was observed under normal aerobic conditions, the integration of peak areas showed that 6% of dimeric peptides were cross-linked by L,D-transpeptidation (3-3) whereas 32% were cross-linked by D,D-transpeptidation (4-3). Overall 20% of muropeptides were monomeric and 80% were cross-linked.
Effect of meropenem/clavulanate on PG structure
We next investigated the mechanism of the cidal activity of the meropenem/clavulanate combination by studying the effect of these drugs on PG structure, the known primary target of the β-lactams (Ghuysen, 1977). The PG structure was analyzed from mid-log Mtb (OD650=0.6) exposed to a range of meropenem concentrations (0.2, 0.5, 2, 5 and 50 μM) with or without clavulanate. As expected, treatment with clavulanate (100 μM) alone did not have any effect on the structure of mycobacterial peptidoglycan (Fig. 5A), however, when used in combination with meropenem even at a sub-inhibitory concentration of 0.2 μM, one monomeric peak (# 5) increased in intensity in the chromatogram (Fig. 5B). This peak corresponded exactly to the pentapeptide peak seen in normally growing cells in low abundance (Fig. 3, Table 1). In addition, there was also a substantial increase in a Tetra-Penta (4-3 cross-linked) moiety (peak # 16, mass 1116.6, Fig. S10), which would have resulted from cross-linking of two Penta stem peptides presumably through the action of a meropenem insensitive D,D-transpeptidase (Peak # 16 in Fig. 5B, Fig. S8 and S9).
Fig. 5.
Effect of meropenem/clavulanic acid on PG structure. RP-HPLC profile of muropeptides from PG isolated from Mtb exposed to the following concentrations of meropenem and clavulanic acid: (A) 100 μM clavulanic acid only (B) 0.2 μM meropenem and 100 μM clavulanic acid (C) 0.5 μM meropenem and 100 μM clavulanic acid (D) 2.0 μM meropenem and 100 μM clavulanic acid (E) 5.0 μM meropenem and 100 μM clavulanic acid (F) 50 μM meropenem and 100 μM clavulanic acid.
A further increase in these peaks was observed at higher concentrations of meropenem in the presence of clavulanate (Fig. 5C–E) while at the highest concentration tested (50 μM, Fig. 5F) a decrease was observed probably due to rapid cell lysis. Treatment with meropenem alone was sufficient to induce accumulation of the pentapeptide (#5, Fig. S8 A–D), although relatively higher concentrations were required to effect comparable accumulation of pentapeptide to treatment with a combination of the carbapenem and β-lactamase inhibitor. Surprisingly, there was no significant impact on the abundance of tetrapeptide or cross-linked dimeric species. The increase in pentapeptide at 0.5, and 2.0 μM meropenem was relatively small in comparison to that observed with the combination of meropenem and clavulanate and a striking increase was seen only at 5 and 50 μM meropenem.
The exposure of Mtb at an early log-phase of growth (OD650 = 0.2) to meropenem and clavulanic acid for longer period of time (48 hr, Fig. S9D) showed a remarkably high accumulation of Penta (# 5) and 4-3 cross-linked Penta-Tetra stem peptides resulting in an almost equivalent abundance of 3-3 and 4-3 cross-links (52 and 48% respectively. The treatment of an early stationary phase culture (OD650 = 3.2) for 72 hr with meropenem and clavulanic acid also resulted in accumulation of pentapeptide (# 5, Fig. S9F), indicating an active synthesis of PG in the early stationary phase of growth.
Analysis of D,D-Carboxypeptidases from Mtb
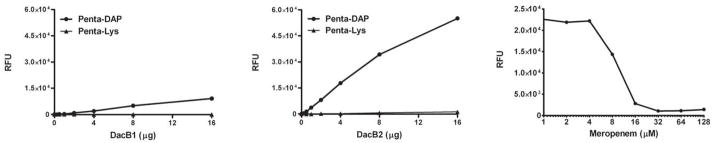
The accumulation of pentapeptide suggested that meropenem was inactivating a D,D-carboxypeptidase whose activity was required to produce substrate for the L,D-transpeptidase responsible for formation of 3-3 cross linkages. The Mtb genome has two annotated homologs of D,D-carboxypeptidases, DacB1 (Rv3330) and DacB2 (Rv2911) belonging to class C, type-5 and type-7 PBPs, respectively, based on predicted structural domains (Sauvage et al., 2008). DacB1 is 405 amino acids in length and is characterized by C-terminal transmembrane domain in addition to an N-terminal signal peptide and a peptidase S11 domain (PfamA). DacB2 is 291 amino acids in length and possesses an N-terminal signal peptide in addition to a Peptidase S11 domain (PfamA). DacB2 was genetically characterized recently and its deletion resulted in altered colony morphology and a conditional growth defect (Bourai et al., 2012). We cloned, expressed and purified the catalytic domains of DacB1 (aa 36–374) and DacB2 (aa 26–291) with his6 tags at the C-terminus. D,D-carboxypeptidase activity in these proteins was determined using a coupled fluorescent assay with a synthetic pentapeptide substrate as shown in Fig. 6. DacB1 showed very poor activity (Fig. 6A); possibly due to deletion of the C-terminal transmembrane domain. Efforts to purify the protein with the C-terminal domain proved futile due to formation of inclusion bodies; hence we cannot rule out that DacB1 may still have carboxypeptidase activity. In contrast, DacB2 showed significant D,D-carboxypeptidase activity as evident from D-Ala release from the synthetic Penta-DAP substrate (Fig. 6B). Mass-spectrometric analysis of meropenem-treated DacB2 showed that this protein formed a covalent adduct with meropenem as indicated by the formation of a peak with a mass corresponding to the expected mass of the DacB2+meropenem adduct (+383, Table 3). We also observed that meropenem alone was also a potent inhibitor of the carboxypeptidase activity of DacB2 in vitro (Fig. 6C).
Fig. 6.
D, D-Carboxypeptidase activity assay. The D-ala release assay performed using penta-peptide substrates (Penta-DAP: L-Ala-γ-D-Gln-DAP-D-Ala-D-Ala; Penta-Lys: L-Ala-γ-D-Glu-L-Lys-D-Ala-D-Ala). (A) D-Ala release by DacB1 (B) D-Ala release by DacB2. (C) Inhibition of enzymatic activity of DacB2 by meropenem.
Table 3.
Detection of DacB2-meropenem adduct by mass-spectrometry.
| Protein | Expected | Observed | Remarks |
|---|---|---|---|
| DacB2 | 28946 | 28944 | |
| + Mero | + (383.5) | 29282/29329 | + 336/+ 383 |
DISCUSSION
The structure of the pentapeptide precursor of PG in Mtb has been determined to be L-Ala-γ-D-Glu-meso-DAP-D-Ala-D-Ala by direct isolation of the nucleotide-linked precursor from the cytosol (Mahapatra et al., 2005). Analysis of monomeric stem peptides isolated from mature PG in Mtb showed widespread amidation of the free carboxyl groups of Glu and DAP residues resulting in L-Ala-D-Glu(NH2)-meso-DAP(NH2)-D-Ala as the most abundant monomeric peptide in Mtb PG (Mahapatra et al., 2008). Similarly, this tetrapeptide was shown to be the most abundant monomeric peptide in stationary phase cultures of Mtb grown under non-shaking conditions (Lavollay et al., 2008). Earlier reports have shown existence of DAP-DAP (3-3) cross-links in mycobacterial PG (Wietzerbin et al., 1974), and a recent study revealed that almost 80% of dimeric peptides from stationary-phase cells were cross-linked by a 3-3 mechanism (Lavollay et al., 2008). In this study, we also report that the majority of the monomeric stem peptides are tetrapeptides (Fig. 3, Table 1). Pentapeptide (Fig. 3, Table 1) was also detected albeit in very low abundance, hinting at a highly efficient carboxypeptidase activity resulting in effective processing.
Among dimeric peptides we determined the ratio of 3-3 to 4-3 cross-linked peptides to be 3:2; a ratio that did not vary significantly with growth phase or condition. It can be inferred therefore that 3-3 cross-links predominate in mycobacterial PG under all phases of growth. It was interesting to note that the α-carboxylate of Glu and ε-carboxylate of DAP were fully amidated in majority of monomeric peptides and also among majority of 4-3 cross-linked dimeric peptides, however, in 3-3 cross-linked dimeric peptides about half of donor stem peptides lacked amidation at the ε-carboxylate of DAP. The differential amidation of stem peptides suggests the intriguing possibility that the transpeptidases may selectively recognize potential substrates based on differential amidation and offers a potential regulatory mechanism for cross-linking in PG. Amidation of both glutamine and DAP in PG has also been shown to influence recognition by specific mammalian proteins such as NOD1 and NOD2 and may modulate inflammation induced during infection (Chamaillard et al., 2003, Inohara et al., 2005). Little attention has focused on variation in PG structure in circulating clinical isolates of Mtb or in understanding the potential contribution of PG modification to virulence.
In many other bacteria like Bacillus subtilis (Atrih & Foster, 1999) and E. coli (Driehuis & Wouters, 1987), the PG has been shown to alter with stage of life cycle and under different growth conditions. In slowly growing E. coli changes in PG composition have been reported showing a decrease in the major monomeric species, decreases in the average chain length of stem peptides, and a significant increase in DAP-DAP containing cross-links (Tuomanen & Cozens, 1987). It is therefore somewhat surprising that Mtb PG does not show any significant growth phase-dependent modulation under similar in vitro conditions of growth. 3-3 cross-linkages in the peptidoglycan layer of Mtb have been shown to be essential for maintenance of normal colony morphology and impacts growth of the organism in mice (Gupta et al., 2010). The peptidoglycan layer in mycobacteria however, serves as an anchor for arabinogalactan, which is, in turn, linked to the mycolic acids of the outer lipid layer (Barry et al., 2007). This basement role as the innermost polymer of a macromolecular complex may not lend itself to dramatic structural reorganization during growth or stress. The mycolic acids bound to arabinogalactan that form the primary permeability barrier of the mycobacterial cell do show variation in fine structure during growth in vivo and in response to oxygen deprivation (Yuan et al., 1998). Likewise, many other peripheral lipids and glycolipids are environmentally responsive such as phthiocerol dimycocerosates (PDIMs) and triacylglycerides (TAGs) (Flores-Valdez et al., 2009, Barry et al., 1998). It is worth emphasizing, however, that our sampling of the architecture of PG in Mtb may be limited to only a subset of cross-links (approximately 70% of the total) that are accessible to muramidase digestion and analysis using HPLC as even in a relatively short time frame (several hours) nearly one-third of newly incorporated radioactive D-Ala is converted into structures that are not released by current analytical techniques (Fig. S2 and data not shown).
The essentiality of PG for cellular integrity has proven a valuable target for the development of a number of antibacterials such as β-lactams, cycloserine and vancomycin. β-lactams primarily target penicillin binding proteins (PBPs) that include D,D-transpeptidases and D,D-carboxypeptidases. It has been reported that 3-3 cross-links are installed by an ampicillin insensitive L,D-transpeptidase in Enterococcus faecium (Mainardi et al., 2000, Sacco et al., 2010). The acquisition of β-lactam resistance in this organism was not associated with changes in the expression level of the corresponding transpeptidase but rather was associated with production of a D,D-carboxypeptidase leading to an increase in the amount of tetrapeptide carrying substrate for formation of 3-3 cross-links (Mainardi et al., 2002). In Mtb a penicillin-insensitive L,D-transpeptidase Rv0116c has also been described (Lavollay et al., 2008), which in contrast to the E. faecium enzyme, proved sensitive to meropenem and other carbapenems.
Our results show that meropenem treatment resulted in the accumulation of pentapeptide stems in mature PG (Fig. 5) suggesting that meropenem may be directly inhibiting a D,D-carboxypeptidase as well as other transpeptidase targets in vivo. Characterized L,D-transpeptidases show a preference for tetrameric substrates whose production requires D,D-carboxypeptidase activity. Inhibition of such a D,D-carboxypeptidase has recently been shown to affect L,D-transpeptidases preferentially over D,D-transpeptidase in Corynebacterium jeikeium and these authors report a similar accumulation of pentapeptide precursors in cells treated with ampicillin (Lavollay et al., 2009). However, unlike what these authors report in C. jeikeium, we do not observe a significant decrease in the quantity of tetrapeptide precursors available for cross-linking in Mtb (Lavollay et al., 2009). This supports that both the L,D-transpeptidase which cross-links tetrapeptide stems and the D,D-carboxypeptidase that processes pentapeptide stems to substrates are both exquisitely sensitive to meropenem in Mtb.
We biochemically characterized the D,D-carboxypeptidase DacB2 for its D,D-carboxypeptidase activity (Fig. 6B) and showed that it forms a covalent adduct with meropenem (Table 3) in addition to inhibiting enzymatic activity (Fig. 6C). Our results therefore suggest that while meropenem likely has an L,D-transpeptidase target, the D,D-carboxypeptidase DacB2 is also highly sensitive. Preventing processing of pentapeptide precursors to tetrapeptides while simultaneously inhibiting the transpeptidases capable of utilizing such precursors to form 3-3 or 4-3 cross-linkages would be expected to be synergistic and may contribute to the uniquely rapid lytic activity of the carbapenems. Of note, one of the L,D-transpeptidases encoded in the Mtb genome (Rv0116c) has been shown to form an adduct with meropenem in vitro and to be broadly susceptible to other carbapenems (Lavollay et al., 2008). Also consistent with the idea that meropenem simultaneously inhibits multiple targets is that we have been unable to generate spontaneously resistant mutants regardless of the number of bacteria plated, despite replenishing drug concentration daily (data not shown).
The surprising lack of structural variation of mycobacterial PG may provide a unique point of vulnerability for targeting using existing β-lactams such as meropenem. The ability of these agents to kill even non-replicating cells (Hugonnet et al., 2009) is also surprising in this context but presumably hints that peptidoglycan synthesis, recycling and remodeling may be important processes at all stages of growth of the organism. The difficulty of parenteral administration of existing carbapenems may provide an impetus to revive attempts to make an orally bioavailable agent in this class. Because of the apparent redundancy in L,D-transpeptidase enzymes, the apparent vulnerability of the D,D-carboxypeptidase may also offer an attractive alternative target for the development of alternative chemotherapeutic approaches.
Experimental Procedures
Growth conditions
Mtb (strain H37Rv) was grown at 37 °C in Middlebrook 7H9 broth (Becton Dickinson) supplemented with ADC (Albumin (50 g l−1)/dextrose (20 g l−1)/NaCl (8.1 g l−1)), 0.2% glycerol and 0.05% Tween 80 in rolling bottles. A preculture was maintained below an OD650 of 0.1 for two weeks before final subculturing (diluted to OD650 0.005) to obtain an early log phase (OD650 = 0.12) culture. A similar culture was allowed to grow further to obtain higher density cultures for PG analysis. Non-replicating persistent cultures were prepared as described by Wayne (Wayne & Hayes, 1996) in Dubos medium, which consisted of Dubos broth base supplemented with 10X Dubos ADC enrichment (Albumin (50 g l−1)/dextrose (75 g l−1)/NaCl (8.5 g l−1).
Time kill kinetics
For kill curves, duplicate mid-log cultures of Mtb at an optical density (OD650) of 0.2 were diluted 100-fold. The parent cultures were then split into 15 ml aliquots and treated with the test compounds at 1-time (1X) or 10-times (10X) their corresponding MIC (penicillin G, 2 μM; cephaloridine, 1 μM; meropenem 2μM) in presence of 100 μM clavulanic acid. For β-lactam-clavulanic acid combinations clavulanate was used at 100 μM and cultures were dosed daily with both agents for 13 days. For treatment with isoniazid (10X INH, 2 μM) and ethambutol (10X EMB, 62.5 μM) cultures were dosed once at day 0. At desired time points 100 μl aliquots were removed from replicate cultures, serially diluted and plated in duplicates on Middlebrook’s 7H11 medium (Becton Dickinson) supplemented with OADC (Oleic acid (1.2 ml l−1)/Albumin (50 g l−1)/Dextrose (20 g l−1)/NaCl (8.1 g l−1)). Viable numbers were counted after 4 weeks of incubation at 37 °C.
GFP release assays
A GFP-expressing Mtb strain (Mtb harboring pMSP12-GFP plasmid) was grown to an OD650 of 0.2. The parent culture was then split into 50 ml aliquots and treated with the test compounds at 0, 1 and 10-times MIC. At each time point OD650 of the cultures was recorded. Also at each time point 1 ml cultures were centrifuged (15,700 × g, 5 min) and the supernatant was dispensed in 100 μl aliquots (in triplicates) in consecutive wells of a half area 96-well black plate (Costar item # 3694). GFP fluorescence was measured at excitation (480 nm) and emission (520 nm) using a 96-well plate reader (FluoStar Optima, BMG LabTech).
Scanning electron microscopy
Mtb was grown to an OD650 of 0.2. The cultures were then split into 100 ml aliquots and treated with MCA, EMB or INH at 1- or 10 -times MIC or left untreated for 24 hr. After 24 hr of treatment, cells were harvested by centrifugation at 2,500 × g for 10 min. The cell pellets were then washed with 1 ml 1X-phosphate buffered saline (PBS) and cells were recovered by centrifugation (11,000 × g, 5 min). The cell pellets were resuspended in 0.1 ml of 1X PBS and 50 μl cell suspension was transferred on to a silicon chip (TedPella Inc., Redding, CA) placed in a 24-well plate. The suspension was allowed to adhere to the chip for 20 min. The unadhered material was removed using a pipette and the chip was washed twice with 0.5 ml of 1X PBS by gently pipetting it into the well, incubating for 2 min and removing it. The chips were then fixed with 2.5% glutaraldehyde prepared in 0.1 M sodium cacodylate buffer (pH 6.0). The cells were post fixed with 1% osmium tetroxide/0.8% potassium ferrocyanide in 0.1 M sodium cacodylate using a Pelco Biowave laboratory microwave system at 250 W using a 2 min on, 2 min off, 2 min on cycle under 20 inches Hg vacuum. The chips were rinsed with water and dehydrated in graded ethanol series for 45 sec each. The specimen was critical point dried in a Bal-Tec cpd 030 drier (Bal-Tec AG, Balzers, Liechtenstein) and coated with a 80 °A of iridium using a IBS ion beam sputter (South Bay Technology Inc, San Clemente, CA). Images were collected on a Hitachi SU 8000 Scanning Electron Microscope (Hitachi, High-technologies, Tokyo, Japan).
Purification of Chalaropsis muramidase
The muramidase from Chalaropsis sp. (ATCC 16003) was purified as described previously (Hash, 1963, Yagi et al., 2003, Hash & Rothlauf, 1967). Briefly, Chalaropsis sp. was grown with shaking in baffled flasks at 28 °C in a growth medium containing glucose (40 g l−1) and mycological peptone (10 g l−1) for 96–120 hr until the cultures turned jet black. The muramidase from culture supernatant was purified by binding to cation exchange resin (Amberlite CG-50-H+, Sigma), followed by ammonium sulfate precipitation (0.7 saturation) and gel filteration (Superdex 75, GE Healthcare) chromatography as previously described (Yagi et al., 2003). The enzymatic activity was monitored by measuring reduction in turbidity (OD650) of a Staphylococcus aureus H (ATCC 13801) whole cell suspension, and purity was checked by SDS-PAGE gels stained with Coomassie Brilliant Blue R250.
Preparation of mycobacterial peptidoglycan
Mtb cultures were inactivated by autoclaving in presence of 4% SDS (wt/vol). Inactivated cell mass was recovered by centrifugation (28, 000 × g for 20 min at room temperature), resuspended in PBS containing 2% Triton X-100 (vol/vol) and lysed by ultrasonication (Misonix sonicator S2020, 3 cycles of 5 min, amplitude 80, 5 s on, 10 s off) at 4 °C. The cell wall fraction containing peptidoglycan was recovered by centrifugation (28, 000 × g, 20 min, 4 °C) and extracted twice with 5% SDS in PBS (30 min, 100 °C), followed by incubation with Proteinase K (30 min, 60 °C) in PBS containing 2% SDS. The pellet fraction was further extracted twice with 5% SDS in PBS (30 min, 95 °C). The purified peptidoglycan fraction was recovered by centrifugation (28, 000 × g, 20 min, RT), washed thrice with PBS, once each with water, 80% acetone and acetone, followed by drying and lyophilization for further analysis.
Structural analysis of PG
PG (3 mg) was digested with lysozyme (50 μg, Sigma), mutanolysin (50 μg, Sigma) and Chalaropsis muramidase (20 μg) in a 200 ml reaction mixture containing sodium phosphate (20 mM, pH 4.8) and sodium azide (0.02%) for 16 hr, followed by heat inactivation at 100 °C for 5 min (Glauner, 1988). The soluble muropeptides were recovered by centrifugation at 18, 000 × g for 10 min, after which they were resuspended in a total volume of 200 μl of HPLC grade water and incubated with 64 μl ammonium hydroxide (32%) at 37 °C for 5 hr to generate lactoyl peptides. The reaction mixture was neutralized with 61 μl of glacial acetic acid, lyophilized and dissolved in 100 μl of 0.05% TFA in water (Arbeloa et al., 2004). The lactoyl peptides were injected on LC/MS system (Agilent 1100, Wilmington, DE, USA) equipped with a RP-HPLC column (Strategy 3μ, C18–2, 4.6 mm × 250 mm, Interchim, France), and a 0–20% gradient was applied between 10–90 min (Solvent A: 0.05% TFA in water, Solvent B: 0.035% TFA in acetonitrile) at a flow rate of 0.5 ml min−1, UV traces were recorded at 210 nm and peak areas were used to estimate the relative abundances of muropeptides (Arbeloa et al., 2004). The MS scan range was 199–2500 in positive mode with cone voltage set to 100 V, a nitrogen flow rate of 12 L min−1 at 350 °C and a capillary voltage of 2500 V. For MS/MS analysis 0.5 ml fractions (1 min) were collected, lyophilized, resuspended in water and infused into the mass spectrometer using a Nanomate chip- based ESI system (Advion Biosciences, Ithaca, NY). The ESI voltage was 1.4 kV and all spectra were recorded in positive ion mode. The cone voltage was 30 eV. The mass spectrometer used was an LTQ linear ion trap (Thermo, Waltham, MA). In MS mode the ion trap time was 600 ms and in MS/MS mode it was 500 ms. Ions for MS/MS analysis were selected manually. The collision energy was set to 35 eV. For high resolution accurate mass analysis an LTQ-FT mass spectrometer (Thermo, Waltham, MA) was used. The resolution was 100K and both MS and MS/MS analyses were recorded using the positive ion FT mode.
Incorporation of [1-14C] D-alanine in mycobacterial PG
400 ml Mtb culture grown to mid log (OD650 0.6) was supplemented with 100 μCi [1-14C] D-alanine (55 mCi ml−1, American Radiolabeled Chemicals, Inc. St. Louis, USA). After 3 hr incubation at 37 °C, 16 g SDS powder was added and the culture was inactivated by autoclaving. PG was purified as described above except for using a cup horn probe (Misonix) for sonication at optimum cavitation intensity for 30 min. Purified PG (3 mg) was hydrolyzed with muramidases, heat inactivated and centrifuged at 13, 000 × g for 10 min. Radioactivity in both pellet and supernatant was quantified with a liquid scintillation counter. The muramyl peptides in the supernatant were converted into lactoyl peptides and analyzed on RP-HPLC as described above, radioactivity in the eluent (1 ml fractions) was measured to determine the distribution of radiolabel in the lactoyl peptides.
Sequential hydrolysis of PG with muramidases
PG (3 mg) was hydrolyzed independently and sequentially with each muramidase i. e. lysozyme (50 μg), mutanolysin (50 μg) or Chalaropsis muramidase (20 μg) for 16 hr each. In one such sequence PG (3 mg) was digested first with lysozyme for 16 hr. The reaction mixture was centrifuged (18, 000 × g for 10 min), the pellet was washed and resuspended in water, and further hydrolyzed with mutanolysin for 16 hr, followed by digestion with Chalaropsis muramidase. The sequence of addition of muramidases was changed in two other sets of reaction (Fig. S3). The soluble fractions after each step were heat inactivated (100 °C, 5 min) and centrifuged (13,000 × g, 10 min). The supernatants were collected and analyzed by LC/MS as described above.
Meropenem treatment
Mtb cultures (400 ml for each treatment) were grown to an OD650 of 0.6 in 2 litre roller bottles and treated with meropenem (USP, Rockville, MD, USA) at the desired concentration (0.2 μM to 50 μM) with or without 100 μM clavulanic acid (Sigma) for 18 hr. After meropenem treatment PG was prepared as described above.
Cloning, Expression and Purification of DacB1 and DacB2 from Mtb
The dacB1 gene (Rv3330) fragment (amino acid 36–374) lacking (aa 2–35) N-terminus amino acids corresponding to a signal peptide and C-terminus residues (aa 375–405) corresponding to a transmembrane domain was amplified using (Rv3330-dN35-FP-NdeI 5′ GCCGCACATATGGCGTGCCCGTACAAGGTGTCC 3 ′ and Rv3330-dC374-RP-HindIII 5′ CGGAAGCTTGGCGTCGGCGGCCGATATCCG 3′) Pfx DNA Polymerase (Invitrogen), and cloned in pET28b vector as a C-terminus his6 fusion between NdeI and HindIII sites. Similarly, dacB2 gene (Rv2911) fragment (aa 26–291) lacking N-terminus amino acids (aa 2–25) corresponding to a signal peptide was also amplified using (Rv2911-dN25-Fp-NdeI 5′ GCGCATATGGATGCCGACGTGCAGCCGGCC 3 ′ and Rv2911-RP-HindIII 5′ GCGAAGCTTGAGCGAGCCGACGCTGGCCTG 3’) Pfx polymerase (Invitrogen) and cloned in pET28b vector for over-expression. The accuracy of constructs was verified by DNA sequencing. The constructs were transformed into E. coli BL21 (DE3) strain harboring pGRO7 (Takara Mirus) plasmid. The recombinant proteins were expressed by inducing the cultures, grown to OD595 0.6 at 37 °C with IPTG (0.5 mM) at 16 °C for 16 hr. The clarified lysate was prepared and purified using Ni2+-NTA (1 ml His-Trap column, GE Healthcare) followed by size exclusion chromatography (Superdex 75 column, GE Healthcare). The proteins were pure to near homogeneity as assessed by SDS-PAGE analysis and the proteins were quantified using Coomassie Protein Assay Reagent (Thermo Scientific) as per manufacturer’s instructions.
D, D-Carboxypeptidase Assay
D, D-carboxypeptidase activity was measured using a coupled assay based on fluorescence read out, in the first part the D-Ala is released as a result of D,D-carboxypeptidase activity of the proteins and in the second part the released D-Ala is oxidized resulting in a conversion of Amplex Red reagent into a fluorescent end product (Baum et al., 2005). In an assay reaction, the penta-peptide substrate (Penta-DAP: L-Ala-γ-D-Gln-DAP-D-Ala-D-Ala or Penta-Lys: L-Ala-γ-Glu-L-Lys-D-Ala-D-Ala (Invivogen); 0.5 mM) was incubated with different amounts of DacB1 or DacB2 at 37 °C in 1X PBS for 30 min in a 40 μl reaction to allow cleavage of terminal D-Ala in a 96 well half area black plate (Costar item # 3694). The released D-Ala was quantified using 40 μl detection reagent containing (100 mM Tris-HCl pH 8.5, 0.38 units ml−1 D-amino acid oxidase, 12.5 units ml−1 Horseradish peroxidase, 6.25 μg ml−1 flavin adenine dinucleotide and 50 mM Amplex Red) at 37 °C for 30 min and the fluorescence was read using a flourometer (FlouStar Optima, BMG LabTech; excitation 544 nm and emission 590 nm). The hydrolysis rate of the pentapeptide under this condition was ~0.11 nmoles/ml/min/μg without drug, which was reduced to about half (0.06 nmoles/ml/min/μg) at 8 μM meropenem and a log reduction (0.01 nmoles/ml/min/μg) was seen at 16 μM of meropenem. Inhibition of the D,D-carboxypeptidase activity of DacB2 by meropenem was determined by preincubating 2 μg of purified DacB2 with increasing amounts of meropenem for 30 min at room temperature (Final concentration 0–128 μM), followed by the D,D-carboxypeptidase assay as described above to quantify inhibition of enzymatic activity.
Synthesis of Penta-DAP
The synthesis of L-Ala-D-isoGln-DAP-D-Ala-D-Ala was accomplished by the coupling of protected L-Ala-D-isoGln-DAP and D-Ala-D-Ala as depicted in Fig. S11. The structure of Penta-DAP was confirmed with LCMS, HRMS, 1H-NMR and 13C NMR. The detailed synthesis of Penta-DAP will be reported in the near future (Lee IY and Barry CE, unpublished). HRMS: C21H38N7O9 (532.2731)=532.2731, 13C-NMR (D2O, 75MHz): 17.8, 18.4, 22.1, 28,0, 31.1, 31.8, 32.2, 50.2, 50.6, 52.3, 53.6, 55.0, 55.6, 173.6, 174.1, 174.9, 175.7, 176.1, 176.8, 181.1, 1H-NMR (D2O, 300MHz): 1.35–1.52(m, 8H), 1.59(J=7.14, 3H), 1.74–2.23(m, 6H), 2.43–2.49(m. 2H), 3.76(t, J=6.18, 1H), 4.01–4.20(m,2H), 4.29–4.43(m, 3H).
Formation of Enzyme-Meropenem adduct
DacB1 and DacB2 (17.5 μM) were incubated with meropenem (87.5 μM) in 1X PBS at 37 °C for 1 hr. Nominal molecular weight analysis of intact proteins and adducts was performed on an Agilent (Santa Clara, CA USA) Model 1100 LC/MSD quadrupole mass spectrometer equipped with an electrospray ion source. The proteins were HPLC separated using a Michrom Bioresources (Auburn, CA USA) 4000 Å PLRP-S 2.0 × 150 mm column. Solvent A was 5% acetic acid in water and solvent B was 100% acetonitrile. The solvent gradient was 0 to 90% B in 50 minutes and the column temperature was 75 °C. Nominal molecular weights were determined with the Agilent deconvolution software. The electrospray ionization conditions were: ESI capillary 4000V, desolvation gas temperature was 300 °C at a nitrogen gas flow rate of 10 L min−1 and the fragmentor was 50V.
Supplementary Material
Acknowledgments
We would like to thank Michael Goodwin for technical assistance. We would like to thank Chandan Gope for writing the software for cell length measurements. This work was funded by the Intramural Research Program of the NIAID, NIH.
References
- Arbeloa A, Hugonnet JE, Sentilhes AC, Josseaume N, Dubost L, Monsempes C, Blanot D, Brouard JP, Arthur M. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J Biol Chem. 2004;279:41546–41556. doi: 10.1074/jbc.M407149200. [DOI] [PubMed] [Google Scholar]
- Atrih A, Foster SJ. The role of peptidoglycan structure and structural dynamics during endospore dormancy and germination. Antonie Van Leeuwenhoek. 1999;75:299–307. doi: 10.1023/a:1001800507443. [DOI] [PubMed] [Google Scholar]
- Barry CE, 3rd, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- Barry CE, Crick DC, McNeil MR. Targeting the formation of the cell wall core of M. tuberculosis. Infect Disord Drug Targets. 2007;7:182–202. doi: 10.2174/187152607781001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum EZ, Crespo-Carbone SM, Foleno B, Peng S, Hilliard JJ, Abbanat D, Goldschmidt R, Bush K. Identification of a dithiazoline inhibitor of Escherichia coli L,D-carboxypeptidase A. Antimicrob Agents Chemother. 2005;49:4500–4507. doi: 10.1128/AAC.49.11.4500-4507.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergval IL, Schuitema AR, Klatser PR, Anthony RM. Resistant mutants of Mycobacterium tuberculosis selected in vitro do not reflect the in vivo mechanism of isoniazid resistance. The Journal of antimicrobl chemother. 2009;64:515–523. doi: 10.1093/jac/dkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourai N, Jacobs WR, Jr, Narayanan S. Deletion and overexpression studies on DacB2, a putative low molecular mass penicillin binding protein from Mycobacterium tuberculosis H(37)Rv. Microb pathog. 2012;52:109–116. doi: 10.1016/j.micpath.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- Cohen T, Colijn C, Wright A, Zignol M, Pym A, Murray M. Challenges in estimating the total burden of drug-resistant tuberculosis. Am J Respir Crit Care Med. 2008;177:1302–1306. doi: 10.1164/rccm.200801-175PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driehuis F, Wouters JT. Effect of growth rate and cell shape on the peptidoglycan composition in Escherichia coli. J Bacteriol. 1987;169:97–101. doi: 10.1128/jb.169.1.97-101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England K, Boshoff HI, Arora K, Weiner D, Dayao E, Schimel D, Via LE, Barry CE., 3rd Meropenem-Clavulanic Acid Shows Activity against Mycobacterium tuberculosis In Vivo. Antimicrob Agents Chemother. 2012;56:3384–3387. doi: 10.1128/AAC.05690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Valdez MA, Morris RP, Laval F, Daffe M, Schoolnik GK. Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J. 2009;23:4091–4104. doi: 10.1096/fj.09-132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen JM. The concept of the penicillin target from 1965 until today. The thirteenth marjory stephenson memorial lecture. J Gen Microbiol. 1977;101:13–33. doi: 10.1099/00221287-101-1-13. [DOI] [PubMed] [Google Scholar]
- Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- Glauner B, Holtje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- Goffin C, Ghuysen JM. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol Mol Biol Rev. 2002;66:702–738. doi: 10.1128/MMBR.66.4.702-738.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat med. 2010;16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hash JH. Purification and Properties of Staphylolytic Enzymes from Chalaropsis Sp. Arch Biochem Biophys. 1963;102:379–388. doi: 10.1016/0003-9861(63)90245-5. [DOI] [PubMed] [Google Scholar]
- Hash JH, Rothlauf MV. The N,O-diacetylmuramidase of Chalaropsis species. I. Purification and crystallization. J Biol Chem. 1967;242:5586–5590. [PubMed] [Google Scholar]
- Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Johnston JC, Shahidi NC, Sadatsafavi M, Fitzgerald JM. Treatment outcomes of multidrug-resistant tuberculosis: a systematic review and meta-analysis. PLoS One. 2009;4:e6914. doi: 10.1371/journal.pone.0006914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Strominger JL, Kotani S. Structure of the cell wall of Corynebacterium diphtheriae. I. Mechanism of hydrolysis by the L-3 enzyme and the structure of the peptide. Biochemistry. 1968;7:2762–2773. doi: 10.1021/bi00848a010. [DOI] [PubMed] [Google Scholar]
- Kotani S, Yanagida I, Kato K, Matsuda T. Studies on peptides, glycopeptides and antigenic polysaccharide-glycopeptide complexes isolated from an L-11 enzyme lysate of the cell walls of Mycobacterium tuberculosis strain H37Rv. Biken J. 1970;13:249–275. [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Riegel P, Gutmann L, Mainardi JL. The beta-lactam-sensitive D,D-carboxypeptidase activity of Pbp4 controls the L,D and D,D transpeptidation pathways in Corynebacterium jeikeium. Mol Microbiol. 2009;74:650–661. doi: 10.1111/j.1365-2958.2009.06887.x. [DOI] [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, Arthur M, Mainardi JL. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by L,D-transpeptidases. J Bacteriol. 2011;193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyh-Bouille M, Bonaly R, Ghuysen JM, Tinelli R, Tipper D. LL-diaminopimelic acid containing peptidoglycans in walls of Streptomyces sp. and of Clostridium perfringens (type A) Biochemistry. 1970;9:2944–2952. doi: 10.1021/bi00817a002. [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Crick DC, McNeil MR, Brennan PJ. Unique structural features of the peptidoglycan of Mycobacterium leprae. J Bacteriol. 2008;190:655–661. doi: 10.1128/JB.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Scherman H, Brennan PJ, Crick DC. N Glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. J Bacteriol. 2005;187:2341–2347. doi: 10.1128/JB.187.7.2341-2347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem. 2005;280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. Novel mechanism of beta-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J Biol Chem. 2000;275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Morel V, Fourgeaud M, Cremniter J, Blanot D, Legrand R, Frehel C, Arthur M, Van Heijenoort J, Gutmann L. Balance between two transpeptidation mechanisms determines the expression of beta-lactam resistance in Enterococcus faecium. J Biol Chem. 2002;277:35801–35807. doi: 10.1074/jbc.M204319200. [DOI] [PubMed] [Google Scholar]
- Peltier J, Courtin P, El Meouche I, Lemee L, Chapot-Chartier MP, Pons JL. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J Biol Chem. 2011;286:29053–29062. doi: 10.1074/jbc.M111.259150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan L, Federspiel NA, Falkow S. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- Raymond JB, Mahapatra S, Crick DC, Pavelka MS., Jr Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J Biol Chem. 2005;280:326–333. doi: 10.1074/jbc.M411006200. [DOI] [PubMed] [Google Scholar]
- Rezwan M, Laneelle MA, Sander P, Daffe M. Breaking down the wall: fractionation of mycobacteria. J Microbiol Methods. 2007;68:32–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Sacco E, Hugonnet JE, Josseaume N, Cremniter J, Dubost L, Marie A, Patin D, Blanot D, Rice LB, Mainardi JL, Arthur M. Activation of the L,D-transpeptidation peptidoglycan cross-linking pathway by a metallo-D,D-carboxypeptidase in Enterococcus faecium. Mol Microbiol. 2010;75:874–885. doi: 10.1111/j.1365-2958.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- Takayama K, Keith AD, Snipes W. Effect of isoniazid on the protoplasmic viscosity in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1975;7:22–24. doi: 10.1128/aac.7.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomanen E, Cozens R. Changes in peptidoglycan composition and penicillin-binding proteins in slowly growing Escherichia coli. J Bacteriol. 1987;169:5308–5310. doi: 10.1128/jb.169.11.5308-5310.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heijenoort J. Biosynthesis of bacterial peptidoglycan unit. In: Ghuysen JM, Hakenbeck R, editors. The Bacterial CellWall. Amsterdam, The Netherlands: 1994. pp. 39–54. [Google Scholar]
- Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- Wietzerbin J, Das BC, Petit JF, Lederer E, Leyh-Bouille M, Ghuysen JM. Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry. 1974;13:3471–3476. doi: 10.1021/bi00714a008. [DOI] [PubMed] [Google Scholar]
- Wright A, Zignol M, Van Deun A, Falzon D, Gerdes SR, Feldman K, Hoffner S, Drobniewski F, Barrera L, van Soolingen D, Boulabhal F, Paramasivan CN, Kam KM, Mitarai S, Nunn P, Raviglione M. Epidemiology of antituberculosis drug resistance 2002–07: an updated analysis of the Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Lancet. 2009;373:1861–1873. doi: 10.1016/S0140-6736(09)60331-7. [DOI] [PubMed] [Google Scholar]
- Yagi T, Mahapatra S, Mikusova K, Crick DC, Brennan PJ. Polymerization of mycobacterial arabinogalactan and ligation to peptidoglycan. J Biol Chem. 2003;278:26497–26504. doi: 10.1074/jbc.M302216200. [DOI] [PubMed] [Google Scholar]
- Yew WW, Leung CC. Management of multidrug-resistant tuberculosis: Update 2007. Respirology. 2008;13:21–46. doi: 10.1111/j.1440-1843.2007.01180.x. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhu Y, Crane DD, Barry CE., 3rd The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol Microbiol. 1998;29:1449–1458. doi: 10.1046/j.1365-2958.1998.01026.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.