Abstract
Central to the disparate adaptive immune systems of archaea and bacteria are clustered regularly interspaced short palindromic repeats (CRISPR). The spacer regions derive from invading genetic elements and, via RNA intermediates and associated proteins, target and cleave nucleic acids of the invader. Here we demonstrate the hyperactive uptake of hundreds of unique spacers within CRISPR loci associated with type I and IIIB immune systems of a hyperthermophilic archaeon. Infection with an environmental virus mixture resulted in the exclusive uptake of protospacers from a co-infecting putative conjugative plasmid. Spacer uptake occurred by two distinct mechanisms in only one of two CRISPR loci subfamilies present. In two loci, insertions, often multiple, occurred adjacent to the leader while in a third locus single spacers were incorporated throughout the array. Protospacer DNAs were excised from the invading genetic element immediately after CCN motifs, on either strand, with the secondary cut apparently produced by a ruler mechanism. Over a 10-week period, there was a gradual decrease in the number of wild-type cells present in the culture but the virus and putative conjugative plasmid were still propagating. The results underline the complex dynamics of CRISPR-based immune systems within a population infected with genetic elements.
Introduction
Adaptive immune systems of most archaea and many bacteria primarily target invading viruses and conjugative plasmids and have recently been classified into major classes, denoted types I, II, IIIA and IIIB (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005; Makarova et al., 2011). Operationally, these CRISPR (clustered regularly interspaced palindromic repeats) systems share three main functional steps, adaptation, processing of CRISPR transcripts and interference, each associated with specific Cas proteins. Adaptation, the most conserved step mechanistically, involves the cleavage and uptake of foreign DNA as new spacers at, or near, the leader-adjacent repeat of CRISPR arrays (Barrangou et al., 2007; Deveau et al., 2008; Held and Whitaker, 2009; Lillestøl et al., 2009). Proteins Cas1, Cas2 and Cas4 have been implicated in this process, primarily on the basis of comparative genomic studies (reviewed in Garrett et al., 2011a; Makarova et al., 2011). CRISPR transcripts initiating within the leader region are cleaved specifically within repeat sequences to yield guide crRNAs either by the Cas6 protein for type I and III systems, or by a combination of a tracrRNA and RNase III in the bacteria-specific type II system (Tang et al., 2002; 2005; Brouns et al., 2008; Deltcheva et al., 2011). In the type IIIA and IIIB systems, targeting DNA and RNA respectively, guide crRNAs undergo further maturation at their 3′ ends (Hale et al., 2009; 2012; Hatoum-Aslan et al., 2011; Zhang et al., 2012). Functional interference complexes, consisting of guide crRNAs associated with a group of Cas proteins, target and cleave the nucleic acid components of invading genetic elements, and these modules generate most of the structural and functional diversity prevalent among CRISPR-based immune systems (Jore et al., 2011; Lintner et al., 2011; Zhang et al., 2012).
Although considerable progress has been made in elucidating details of the different CRISPR RNA processing and maturation mechanisms, and in determining the protein composition and structures of the different interference complexes involved in DNA and RNA targeting of type I and type IIIB systems, respectively, we still have limited insight into the nature and mechanism of the adaptation step. In early studies it was demonstrated that closely related strains of Mycobacterium tuberculosis carried different spacer sequences at one end of their CRISPR arrays, consistent with new spacers having been added (Hermans et al., 1991; Groenen et al., 1993) and this was later exploited as a typing strategy, spoligotyping (SPacer OLIGOnucleotide TYPING), where the variable region of the CRISPR array was characterized by PCR amplification and sequencing (Aranaz et al., 1996; Kamerbeek et al., 1997). Studies on archaeal CRISPR arrays, linked to the more complex CRISPR-based immune systems of different Sulfolobus species, underpinned this result by demonstrating the accumulation of multiple new spacers at the leader end of the CRISPR arrays but they also provided evidence for a complex picture of dynamic changes, including indels and rearrangements, occurring within the repeat arrays (Lillestøl et al., 2006; 2009; Held and Whitaker, 2009). Furthermore, environmental studies of CRISPR arrays of bacteria and archaea within biofilms provided evidence for a dynamic interplay between viruses and the spacer contents of CRISPR arrays. The results were consistent with mutations occurring in viral genomes to avoid targeting by guide crRNAs which resulted in the periodic uptake of new matching spacers at one end of the CRISPR arrays (Andersson and Banfield, 2008; Tyson and Banfield, 2008).
Successful uptake of spacers was first observed in the laboratory for the bacteria-specific type II CRISPR system of Streptococcus thermophilus. This process was induced by single phages and insertions occurred adjacent to leaders for two of three CRISPR loci which, in turn, led to phage resistance of the host. Single spacer inserts were detected in 39 phage-resistant mutants with a few carrying a further one to three spacer inserts (Barrangou et al., 2007; Deveau et al., 2008; Horvath et al., 2008). Very recently, unspecific uptake of chromosomal and plasmid vector protospacer DNA into CRISPR loci of Escherichia coli was shown to be induced by overexpression of two of the adaptation-associated proteins Cas1 and Cas2 (Yosef et al., 2012). Important for the adaptation mechanism are the short sequence PAM motifs (protospacer associated motif) located at the end of the protospacer that becomes leader proximal in the CRISPR array for all archaeal type I and III systems, and leader distal in the bacteria-specific type II system (Barrangou et al., 2007; Lillestøl et al., 2009; Mojica et al., 2009).
Here we investigated activation of the adaptation reaction in the model crenarchaeon Sulfolobus solfataricus P2 by viral infection. The organism is an excellent host for a variety of archaeal viruses and conjugative plasmids (Zillig et al., 1994; 1998). It carries six CRISPR loci, A to F, two gene cassettes encoding the adaptation-associated proteins Cas1, Cas2 and Cas4 (Garrett et al., 2011b) and three gene cassettes associated with type I and IIIB interference modules that target DNA and RNA respectively (Gudbergsdottir et al., 2011; Manica et al., 2011; Zhang et al., 2012). The six CRISPR arrays fall into two main crenarchaeal subfamilies on the basis of the sequences of their repeats, leaders, PAM motifs and associated Cas1 proteins (Lillestøl et al., 2009; Shah et al., 2009). Loci A and B belong to subfamily II and loci C to F belong to the more common subfamily I. Loci E and F carry repeats differing at one base pair from those of loci C and D and, whereas locus E contains a different type of leader, locus F has no leader and loci E and F are not physically proximal to adaptation-associated genes (Lillestøl et al., 2009). Infecting the Sulfolobus cells with a purified environmental virus mixture produced hyperactive adaptation of subfamily I CRISPR arrays C, D and E by two different mechanisms.
Results
Activation of adaptation by viruses
Initial experiments were performed to induce new spacer uptake into the six CRISPR arrays A to F of S. solfataricus P2 (Fig. 1A) by infecting with single purified archaeal viruses, the rudivirus SIRV2, the bicaudavirus ATV and a tailed-fusiform virus STSV2. Only STSV2 (a variant of STSV1 –Xiang et al., 2005) propagated stably over longer periods but examination of the sizes of PCR products generated from the leader-proximal ends of the six CRISPR loci, tested over several weeks, failed to yield evidence for adaptation. Therefore, experiments were performed with an environmental sample containing archaeal viruses using the same PCR approach. An enrichment culture was established of a sample taken from a hot spring in Yellowstone National Park and virus-like particles were isolated from the supernatant. The main viral morphotypes present in the mixture were shown by electron microscopy to resemble closely those of crenarchaeal viruses, including single-tailed STSV1 and HAV2, two-tailed ATV and rod-shaped HAV1 (Fig. 1B) (Häring et al., 2005; Xiang et al., 2005; Garrett et al., 2010). This virus mixture was used to infect S. solfataricus P2. Examination of the viral content of the S. solfataricus P2-infected culture by electron microscopy 6 days post infection revealed that primarily the single-tailed fusiform virions were present with very few two-tailed virions and no rod-shaped virions (Fig. 1C).
Fig. 1.
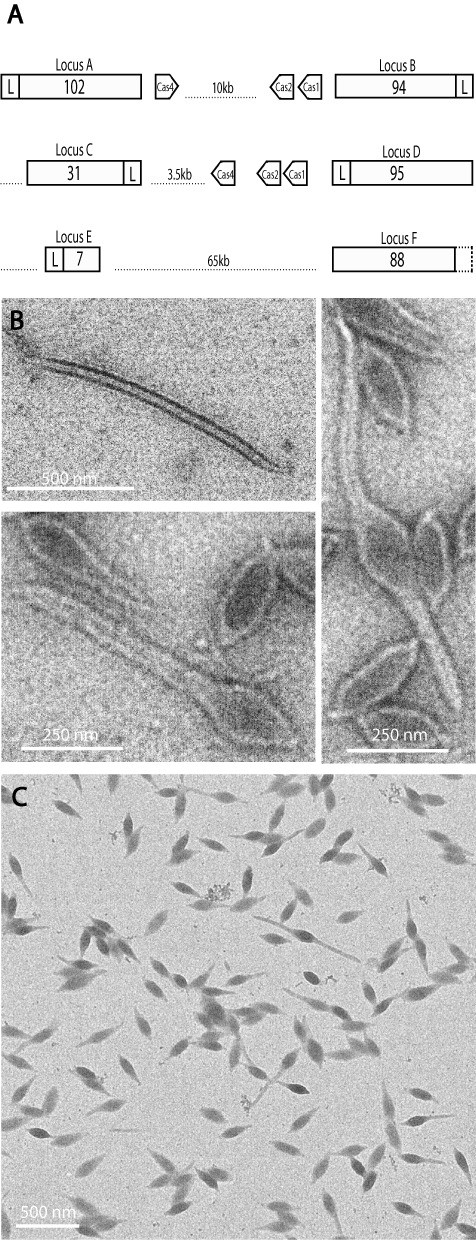
CRISPR loci and viruses infecting S. solfataricus P2.
A. Scheme of six CRISPR loci of S. solfataricus P2, their associated leader regions (L), and genes encoding adaptation-associated Cas1, Cas2 and Cas4. Numbers of repeat-spacer units within CRISPR arrays are given.
B and C. Electron micrographs of virus particles isolated from (B) supernatant of the enrichment culture, and (C) S. solfataricus P2 6 days post infection with the virus mixture in (B). Samples were negatively stained with 1% uranyl acetate and size bars are included.
Initially, cultures of uninfected and virus-infected S. solfataricus P2 produced similar growth curves (Fig. 2A). In contrast a mutant P2 strain lacking CRISPR loci A to D and the adaptation-associated cas genes (Gudbergsdottir et al., 2011) showed retarded growth immediately after infection, consistent with a CRISPR-based defence operating only in the wild-type strain (Fig. 2A). Cultures were successively diluted to an A600 of 0.05 when stationary growth was reached every 3 days, with no addition of fresh virus mixture. Growth retardation of the infected wild-type cells was first observed 10 days post infection (Fig. 2B). This change preceded formation of larger amplified products from CRISPR loci indicative of the uptake of new spacers (Fig. 2C). Single larger fragments were observed after 12 days for loci C, D and E but not for loci A, B and F. Over 12–20 days, these larger bands increased in yield and, in addition, multiple bands appeared for loci C and D but not for locus E (Fig. 2C). When these experiments were repeated the onset of growth retardation varied in the range 8–20 days with detectable spacer uptake following 2–3 days later.
Fig. 2.
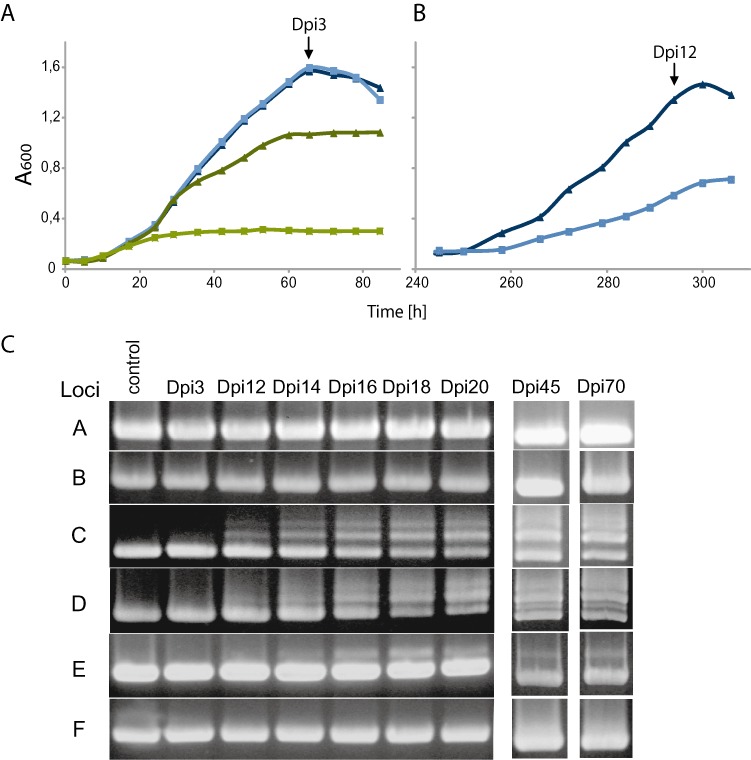
Growth curves of virus-infected S. solfataricus P2 and spacer uptake in CRISPR loci.
A. Growth curves for uninfected (dark blue) and virus-infected (light blue) wild-type cells, and for uninfected (dark green) and infected (light green) CRISPR-minus strain.
B. Growth curves for uninfected (dark blue) and virus-infected (light blue) wild-type cells at onset of spacer uptake (10–12 days). A600 measurements were made every 6 h.
C. PCR products amplified from leader proximal regions of CRISPR loci A to F of wild-type strain and matched to growth curves (Dpi3 and Dpi12). Dpi, days post infection.
DNA from the larger PCR products was cloned and transformed into E. coli, and DNA from single colonies was sequenced through the leader proximal regions of loci C and D (five spacers each) and locus E (seven spacers). Surprisingly, large numbers of newly incorporated, unique, spacer sequences were found at each locus (Table 1). For loci C and D, 263 and 200 clones yielded 160 and 164 new unique spacers, respectively, and about 25% of the clones carried two to four new spacers. For locus E, 128 clones showed 94 unique single spacer inserts (Table 2). Moreover, a few identical spacers were taken up in different loci indicating that the adaptation process was not specific for a given CRISPR locus (Table 1). To screen for insertions or deletions within the whole of locus C, PCR analyses were performed throughout the locus (2 kb) and no changes were detected after 70 days of continuous culture.
Table 1.
New spacers inserted into CRISPR loci C, D and E.
| Unique inserts of single spacers or unique combinations of spacers | |||||||
|---|---|---|---|---|---|---|---|
| Locus | Total clones (expt. 1/2) | Total spacers | Unique spacers | Single | Two | Three | Four |
| C | 263 | 333 | 160 | 124 | 39 | 5 | 0 |
| D | 200 | 260 | 164 | 126 | 39 | 3 | 1 |
| E | 128 | 128 | 94 | 94 | 0 | 0 | 0 |
Total numbers of clones sequenced for each locus are given. Some spacers unique for a given locus were shared: loci C + D (15 pairs), loci C + E (7 pairs), loci D + E (3 pairs) and loci C + D + E (1 triple) and a few spacers within multiple spacer inserts were identical to single spacer inserts.
Table 2.
Distribution of new spacer inserts in CRISPR locus E where repeat 1 is leader proximal.
| Locus E repeat number | Total clones | Unique spacers at a given repeat |
|---|---|---|
| 1 | 18 | 16 |
| 2 | 1 | 1 |
| 3 | 13 | 13 |
| 4 | 85 | 53 |
| 6 | 10 | 10 |
| 7 | 1 | 1 |
| Total | 128 | 94 |
In total 128 independent clones were sequenced carrying a total of 94 different spacer sequences. Four pairs of spacers occurred at different repeats.
Two different adaptation mechanisms
There is a major difference in the adaptation mechanisms of the different CRISPR loci. Loci C and D accrued one to four new spacers adjacent to the leader region, consistent with insertion occurring at the first repeat, although for clones exhibiting multiple new spacers the insertion order could not be ascertained. In contrast, only single spacer insertions were found for locus E and, moreover, they occurred at six of the eight repeats with a bias to repeat 4 (Table 2). These differences with respect to both the spacer insertion position and number of integrated spacers may reflect that locus E carries a different type of leader sequence from loci C and D (Gudbergsdottir et al., 2011). Moreover, the results indicate that for locus E, the repeats themselves are also important mechanistically for spacer insertion.
Sequences of the virus mixture
DNA was extracted from the virus mixture that had been isolated from an infected culture of S. solfataricus P2 and purified on a CsCl gradient. The DNA was subjected to a round of Illumina sequencing with reads averaging about 90 bp. The sequences were assembled automatically using the CLC genomics workbench and the analysis yielded two sets of larger contigs one with a very high sequence coverage averaging 20 000-fold and another with a low coverage of five- to seven-fold. The contigs were analysed using Artemis (Rutherford et al., 2000) and several predicted ORFs for the high coverage element(s) showed best sequence matches mainly to ORFs of the bicaudavirus ATV and the tailed fusiform virus STSV1 (Häring et al., 2005; Xiang et al., 2005; Prangishvili et al., 2006b) summarized in Table 3. We inferred that these sequences derived from the dominant single-tailed virus that resembles STSV1 morphologically (Fig. 1C). Sequence alignments of these viral contigs with spacers of the six CRISPR loci of S. solfataricus P2 revealed in total eight perfect matches located in loci A (spacer 38) and D (spacers 24, 26, 34, 35, 37, 39 and 40 – all numbered from the leader) and specific TCN or CCN PAM motifs respectively. Moreover, spacers showing one to four mismatches which may have been active in interference (Gudbergsdottir et al., 2011; Manica et al., 2011) were present in loci B (spacer 34) and F (spacers 20, 21, 47, 59, 62 and 82).
Table 3.
Assembled contigs of the high sequence coverage tailed fusiform virus.
| Best match | ||||||
|---|---|---|---|---|---|---|
| Contig | Size (bp) | Orf size | Gene | Orf size | e value | Putative function |
| 1 | 19325 | 588 | ATV_66 | 618 | 1e_48 | MoxR ATPase |
| 349 | ATV_67 | 545 | 5e_05 | Membrane protein | ||
| 222 | ATV_35 | 220 | 0.006 | – | ||
| 242 | ATV_72 | 241 | 2e_128 | Integrase | ||
| 105 | ARV1_05 | 102 | 2e_13 | – | ||
| 248 | pNOB8_16 | 246 | 2e_134 | – | ||
| 137 | ATV_19 | 98 | 1e_23 | – | ||
| 307 | ATV_55 | 286 | 2e_28 | Acetyl transferase | ||
| 319 | ATV_34 | 330 | 3e_125 | |||
| 2 | 10093 | 1642 | STSV1_34 | 2308 Nter | 4e_18 | Structural |
| 126 | ATV_42 | 145 | 4e_07 | – | ||
| 279 | ATV_56 | 277 | 6e_07 | – | ||
| 3 | 6753 | 143 | Ahos_980 | 135 | 5e_53 | GtrA family |
| 757 | ATV_60 | 710 | 6e_97 | Transmembrane | ||
| 153 | ATV_62 | 131 | 3e_10 | Coat protein | ||
| 373 | ATV_71 | 1334 Cter | 6e_23 | Structural | ||
Best sequence matches to the predicted contig ORFs are given together with expectancy values.
Analysis of the contigs with low sequence coverage showed that most predicted ORFs yielded best matches to ORFs of conjugative plasmids of the Sulfolobales with the Acidianus plasmid pAH1 and the Sulfolobus plasmid pARN3 dominating (Table 4) (Greve et al., 2004; Basta et al., 2009). In addition, there were a few best matches to putative conjugative plasmid regions integrated within chromosomes of members of the Sulfolobales. In contrast to the results for the virus, no significant sequence matches were observed between the contigs of the putative conjugative plasmid and the spacers of the six CRISPR loci of S. solfataricus P2. The best match was to spacer 49 of locus A with six mismatching nucleotides.
Table 4.
Contigs assembled for the low sequence coverage putative conjugative plasmid.
| Contig | Size (bp) | Matching spacers | Unique spacers | Forward/reverse | ORFs | Orf match | e value | Putative function |
|---|---|---|---|---|---|---|---|---|
| 1 | 7277 | 151 | 125 | 60/65 | 200 | pARN3_06 | 7e_62 | Conserved plasmid |
| 472 | pARN3_05 | 0.0 | Membrane protein | |||||
| 622 | pAH1_04 | 0.0 | TrbE-like | |||||
| 223 | pARN3_02 | 2e_85 | Conserved plasmid | |||||
| 712 | pARN3_01 | 0.0 | Conserved plasmid | |||||
| 2 | 6723 | 141 | 123 | 66/57 | 192 | pAH1_09 | 1e_50 | Membrane protein |
| 182 | pAH1_p11 | 2e_93 | Membrane protein | |||||
| 132 | pARN3_10 | 2e_55 | Conserved plasmid | |||||
| 1029 | pAH1_p13 | 0.0 | TraG-like | |||||
| 3 | 2469 | 63 | 50 | 21/29 | 95 (Nter.) | pNOB8_19 | 2e_33 | Conserved plasmid |
| 93 | pNOB8_18 | 2e_09 | Conserved plasmid | |||||
| 196 | pARN3_21 | 2e_45 | Conserved plasmid | |||||
| 174 | pSOG1_16 | 1e_11 | DNA binding | |||||
| 4 | 2003 | 20 | 19 | 12/7 | 95 | pAH1_31 | 1e_54 | Conserved plasmid |
| 91 | pKEF9_29 | 2e_29 | Conserved plasmid | |||||
| 71 | pSOG1_27 | 1e_21 | Conserved plasmid | |||||
| 94 | pNOB8_01 | 0.030 | Conserved plasmid | |||||
| 158 | pAH1_23 | 1e_63 | Conserved plasmid | |||||
| 5 | 1780 | 31 | 25 | 17/8 | 435 | pHVE14_20,21 | 0.0 | OrfA/B IS200/605 |
| 6 | 1426 | 17 | 16 | 8/8 | 83 | pHVE14_41 | 8e_44 | PlrA protein |
| 228 | M. sedula Msed_2202 | 3e_161 | Unnamed protein | |||||
| 7 | 1314 | 14 | 11 | 7/4 | 361 (Cter.) | pKEF9_31 | 0.0 | Integrase |
| 8 | 412 | 10 | 7 | 4/3 | 102 | pARN3_15 | 6e_22 | Conserved plasmid |
| 9 | 185 | 9 | 8 | 5/3 | partial | A. hospitalis Ahos_0414 | 2e_84 | Transporter Orf393 |
| 10 | 152 | 23 | 21 | 12/9 | partial | Ahos_0414 | 8e_18 | Transporter Orf393 |
The sizes are given together with the number and directions of the matching spacers. The best sequence matches to the predicted contig ORFs are given together with their expectancy values.
The presence of a putative conjugative plasmid in the virus preparation was unexpected given that the virus mixture was purified by buoyant density gradient centrifugation and that the plasmid was not detectable by PCR in the wild-type S. solfataricus P2 strain prior to infection with the virus mixture. To test for its presence in the purified virus mixture, the latter was treated with DNase I before extracting the DNA from the virus particles. A decrease in DNA concentration was detected, probably due to virus particles having been disrupted during the purification process, but PCR amplification of the traG-like gene after DNA digestion confirmed the presence of the putative conjugative plasmid in the virus mixture (Fig. S1).
Location and distribution of protospacer sites
Sequences of new spacer inserts were assembled automatically with all the larger contigs deriving from the virus mixture (Tables 3 and 4) and, to our surprise, they assembled exclusively on the low coverage contigs of the putative conjugative plasmid. The number of spacers associated with each contig are listed in Table 4. The spacers were distributed fairly evenly along each of the DNA contigs and they were arranged almost equally on the two strands (Table 4), consistent with earlier bioinformatic analyses (Shah et al., 2009; 2011). Only 32 of the 418 sequenced spacers could not be directly matched to the contigs and we assume they lie within gaps in the conjugative plasmid sequence.
CCN PAM motif was invariant for CRISPR loci C, D and E
Although the protospacers are distributed fairly evenly along the contigs there were a few local clusters of spacer matches. These were detected initially when we attempted to assemble the newly incorporated spacer sequences alone. They yielded a few small contigs of 80–150 bp each carrying multiple unique spacer sequences, exemplified by the 152 bp contig in Fig. 3. These provided insights into both the specificity and the variety of protospacer excision sites. The leader proximal end of the spacer sequence was adjacent to an invariant CCN protospacer adjacent motif (PAM), located on either DNA strand as predicted earlier for crenarchaeal subfamily I CRISPR arrays (Lillestøl et al., 2009; Mojica et al., 2009; Shah et al., 2009). For overlapping, near identical spacers, the PAM-associated ends were homogeneous with heterogeneities occurring only when preceded by overlapping PAM motifs including -CCC- or -CCCC-. In contrast, leader distal ends were heterogeneous with no evidence of any base or sequence specificity at the cleavage site (Fig. 3). Given that spacer lengths varied from 36 to 43 bp (Fig. 3), it is inferred that the cleavage occurs by an imprecise ruler mechanism measured from the PAM motif site. Rare examples of apparent inverted spacer insertion were detected, relative to the PAM motif site, and two are colour-coded in Fig. 3. This general pattern of protospacer cleavage was conserved throughout the contigs of the putative conjugative plasmid (Table 4).
Fig. 3.
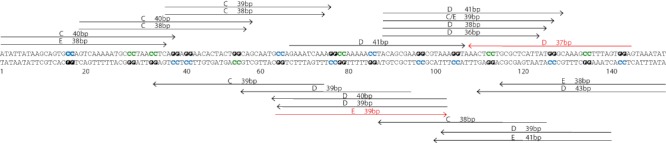
Assembly of newly inserted spacer sequences. Composition of one contig generated from overlapping spacer sequences on both DNA strands. Arrowheads indicate the direction away from the leader (corresponding to spacer crRNA sequence). Two spacers (in red) have opposite orientations. Spacers are preceded by a CCN protospacer-associated motif (PAM) (Lillestøl et al., 2009) (in blue and bold type). CCN motifs with no detected spacers (green). Loci origins and spacer lengths are indicated.
Simultaneous intracellular adaptation of three CRISPR loci and their effect on genetic elements
Single clones were generated from the culture approximately 15 days after the onset of adaptation (day 27). Fourteen clones were selected that showed PCR evidence for new spacer uptake in the CRISPR loci C, D and E. Sequencing revealed that each of the clones carried new spacer inserts. Six clones contained new spacers in either locus C or D, seven clones had acquired spacers in both loci C and D, and one clone exhibited new spacers in all three loci (Table 5). One-third of the new inserts in loci C and D were of two or three spacers. On balance locus D appeared more active than locus C carrying 21 versus 13 new spacers and this may correlate with locus D being much larger than locus C in the wild-type strain (Fig. 1A). Locus E exhibited a lower activity level.
Table 5.
Sequences of newly integrated spacers identified in 14 single clones (S1 to S14) generated 15 days after the onset of adaptation (day 27)
 |
Three of the single clones, S11 with an insertion in locus C, S3 with an insertion in loci C and D and S10 with insertion in loci C, D and E (Table 5), and an additional purified clone R1 devoid of additional spacers were cultured, with the wild-type strain as control, and reinfected with the purified virus mixture. Growth rates of each of the four single clones did not differ significantly from those of the wild-type control and no new spacer uptake was detected 25 days post infection.
PCR amplification of orf174 within an operon in viral contig 1 (Table 3) showed that each of the three clones S3, S10 and S11, as well as R1, could be reinfected with the tailed fusiform virus (Fig. 4). Amplification of the traG-like gene located in contig 2 of the putative conjugative plasmid by PCR (Table 4), showed that the plasmid was not detectable after about 20 days post infection in cultures of the single clones S3, S10 and S11 as a result of being out-diluted by successive dilutions every 3 days. However it was propagating in the wild-type culture (Fig. 4). In contrast, the single clone R1 was still sensitive to the putative conjugative plasmid but yields were constantly lower than for the wild-type strain over a 30-day period (data not shown). Thus, only the single clones with newly acquired spacers were resistant to the putative conjugative plasmid and we infer, therefore, that the latter was confined to cells for which adaptation had not been activated in the infected wild-type culture.
Fig. 4.
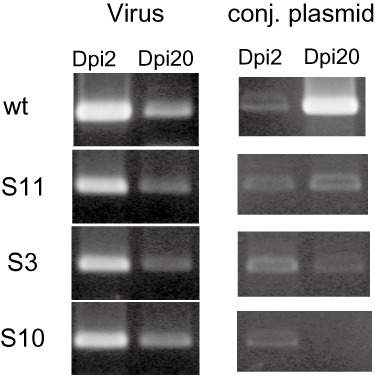
Yields of virus and putative conjugative plasmid in single clones carrying new spacers. Reinfection with the purified virus mixture of isolated single clones containing new spacers in locus C (S11), loci C and D (S3) and in loci C, D and E (S10). PCR products of orf174 within an operon in viral contig 1 (Table 3) and of the traG-like gene in contig 2 of the putative conjugative plasmid (Table 4) were PCR-amplified from DNA isolated from cultures 2 and 20 days post infection (Dpi).
Challenging a CRISPR-minus mutant with the virus mixture
Growth curves demonstrated that the mutant which lacked CRISPR loci A to D and the adaptation-associated genes showed retarded growth immediately after infection with the purified virus mixture (Fig. 2A). However, in two out of three cultures the growth recovered and, after several days of culturing, CRISPR loci A to D were detected in the culture (Fig. S2), implying that a small residual population of CRISPR loci-containing cells had survived in the original mutant (Gudbergsdottir et al., 2011). After 12 days multiple PCR bands were generated from the cells. The bands were cloned and sequenced and revealed the presence of new spacers in each of loci C, D and E, many of which were identical in sequence and all matches were to the putative conjugative plasmid. Of 55 clones, 29 yielded one unique spacer sequence, 10 produced another, and in total there were only 17 unique sequences. This contrasts with the high percentage of unique spacers observed in the infected wild-type culture (Tables 1 and 2). We infer that adaptation occurred in the small percentage of cells carrying CRISPR loci A to D and the adaptation-associated proteins (Fig. 1A), and that these then rapidly outgrew the CRISPR-minus cells and dominated the culture.
Longer-term viability of the genetic elements
The infected wild-type culture was continued beyond 74 days, with successive dilutions every 3 days at stationary phase. The growth rate recovered after about 35 days and PCR amplification of CRISPR loci C to E, after 45 days and 70 days, showed no substantial changes in the intensity ratio between the wild-type band and the larger PCR products carrying new spacer inserts (Fig. 2C). Single clones examined after culturing for 30 days (14 with insertions and 24 without), after 37 days (13 with insertions and 6 without) and after 70 days (23 with insertions and 7 without) revealed a progressive increase in the percentage of clones carrying new spacer inserts in any CRISPR locus. Moreover, PCR products obtained under the same conditions from culture samples taken at regular intervals throughout the 74-day period showed that the level of viral and plasmid DNA oscillated, although the level of plasmid DNA was generally very low (Fig. 5). The presence of the virus after 70 days was also confirmed by electron microscopy (data not shown).
Fig. 5.
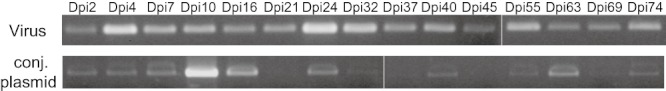
Yields of virus and putative conjugative plasmid in the wild-type culture over a 74-day period. PCR products are shown for orf174 of the virus and the traG-like gene of the putative conjugative plasmid isolated from cultures 2–74 days post infection (Dpi). Template DNA was isolated from 2 ml of infected cells and concentrations were adjusted to be the same for each sample prior to PCR analysis.
Discussion
Archaeal viruses are extremely diverse both morphologically and in their genomic properties and they have been classified into several new viral families, distinct from those of bacteria and eukarya (Prangishvili et al., 2006a; Porter et al., 2007; Pina et al., 2011). A distinguishing property of many of the viruses, and especially those found in high-temperature environments is that they propagate in carrier-like states with their hosts, often together with other viruses, and very few have been shown to induce cell lysis (Prangishvili et al., 2006a; Pina et al., 2011). This may reflect a tendency to minimize contact with harsh extracellular conditions. Moreover, it has been suggested that CRISPR-based immune systems, which are often complex and disparate in hyperthermophilic archaea, may play a regulatory role in maintaining viruses in low copy numbers (Lillestøl et al., 2009; Garrett et al., 2011a, b).
Our initial attempts to induce CRISPR adaptation by infecting S. solfataricus P2 singly with the rudivirus SIRV2, the bicaudavirus ATV and the tailed-fusiform virus STSV2 were unsuccessful which suggested that activation of the adaptation mechanism might be more complex in Sulfolobus than in the bacteria-specific system of S. thermophilus where single phage infections induced adaptation (Barrangou et al., 2007; Deveau et al., 2008). Successful uptake of spacers could only be induced when employing an environmental sample containing viruses and a putative conjugative plasmid. Selective targeting of the low level putative conjugative plasmid but not of the viral DNA was unexpected given the 1:3300 DNA weight ratio estimated from sequence coverage. Nevertheless, previous analyses of Sulfolobus CRISPR loci identified many spacers predicted to arise from conjugative plasmids (Lillestøl et al., 2006; Shah et al., 2009) and this may reflect that these plasmids can propagate and spread rapidly and efficiently in Sulfolobus cultures if unchallenged (Prangishvili et al., 1998; Zillig et al., 1998). Co-migration of the putative plasmid with virions in bouyant density gradients, and its resistance to DNase I treatment, suggested that it may have been encapsulated by viral coat proteins and transferred as a virus satellite, as has been observed for cryptic plasmids of Sulfolobus (Arnold et al., 1999; Wang et al., 2007). Another possibility is that it was extruded from S. solfataricus cells within vesicle particles (Soler et al., 2008).
Non-targeting of the virus by the activated adaptation mechanism of the host was not anticipated although it is consistent with many crenarchaeal viruses coexisting in stable carrier relationships with their hosts (Prangishvili et al., 2006a). However, this observation became more intriguing when we discovered that the viral contigs carried eight perfect sequence matches to spacers within loci A (one) and D (seven) as well as specific PAM motifs. This was surprising first because the host was isolated from Pisciarelli near Naples whereas the viruses were sampled in Yellowstone National Park, USA, and it suggested that the virus and or host are more mobile geographically than previously imagined. Second, of the three single viruses employed unsuccessfully in preliminary adaptation studies, SIRV2 and ATV infected the CRISPR-minus mutant but did not propagate in wild-type S. solfataricus P2 which carries perfect spacer matches against each virus (Shah et al., 2009; Gudbergsdottir et al., 2011). In contrast STSV2, carrying no significant spacer matches, propagated in wild-type S. solfataricus P2 but did not induce adaptation (S.E. and R.A.G., unpublished). However, exceptionally for crenarchaeal viruses, SIRV2 and ATV, in contrast to STSV2 and the Yellowstone virus, have been shown to induce cell lysis (Prangishvili et al., 2006b; Bize et al., 2009). Possibly, they elicit a strong response from the CRISPR immune system at the onset of lysis. Nevertheless, it is evident that the tailed-fusiform virus described here is not extinguished by the wild-type S. solfataricus P2 CRISPR interference response (Gudbergsdottir et al., 2011; Zhang et al., 2012), nor is it susceptible to the host adaptation apparatus despite the latter having been activated. At present we do not understand the mechanism underlying the resistance but it is unlikely that the virus interferes with expression of the adaptation gene cassette by integration in a csa3 gene encoding a putative transcriptional regulator, as has been inferred for another Sulfolobus virus (Shah et al., 2011), because this would have blocked all uptake of protospacers, although it could have played a role in specifically blocking adaptation of CRISPR loci A and B.
Induction of spacer uptake in Sulfolobus required infection by a virus mixture and was highly specific for (i) the Sulfolobus subfamily I CRISPR arrays and (ii) a single genetic element in the mixture; moreover, no spacers derived from the S. solfataricus P2 chromosome (She et al., 2001). This contrasts with the recent demonstration that CRISPR adaptation in E. coli, induced by overexpression of adaptation-associated Cas1 and Cas2, was relatively unspecific in acquiring many spacers from the host chromosome although the targeting of chromosomal DNA could reflect the absence of an active interference system in these strains (Yosef et al., 2012). The regulation of adaptation in Sulfolobus is likely to be complex but possibly the third adaptation-related protein Cas4, which is generally encoded in gene cassettes together with Cas1 and Cas2 in archaea (Shah and Garrett, 2011), facilitates specific recognition and fragmentation of invading genetic elements.
Wild-type S. solfataricus appeared resistant to the virus mixture with no reduction in growth over the first 10 days post infection, in contrast to the CRISPR-minus mutant. Subsequently growth was retarded and this coincided, 12 days post infection, with CRISPR loci C, D and E rapidly acquiring hundreds, and probably thousands, of new spacers, often in multiple unique copies per CRISPR locus, and in multiple CRISPR loci for a given cell. In total, we sequenced about 420 unique spacers deriving from the putative conjugative plasmid. Given that the CCN PAM motifs occur on average every 10 bp in the contigs (see Fig. 3), this probably constitutes a small proportion of those newly acquired spacers present in the population. Diversity of the protospacer sequences is also enhanced by the variable cleavage distance, 39 (± 4) nucleotides, from the PAM motif although this sequence heterogeneity probably produces minimal benefits for the effectivity of the CRISPR system, a supposition that is strengthened by the observation that overlapping spacers rarely occur within CRISPR loci found in natural Sulfolobus strains (Lillestøl et al., 2006; 2009; Shah et al., 2009).
Loci A and B were both inert to adaptation despite their actively taking up new spacers in closely related S. solfataricus strains (Lillestøl et al., 2006; 2009) and the observation that their transcripts are constitutively expressed and processed in vivo (Lillestøl et al., 2009; Wurtzel et al., 2010; Deng et al., 2012). However, their assignment to the less common subfamily II crenarchaeal CRISPR loci, on the basis of the sequences of their leaders, repeats, adaptation-linked Cas1 proteins and differing TCN PAM motif (Lillestøl et al., 2009; Gudbergsdottir et al., 2011), suggests that their adaptation mechanism may be activated differently. We infer that the inactivity of locus F in new spacer uptake is due to its lacking a leader region (Lillestøl et al., 2006).
The adaptation mechanism for loci C and D follows the pattern seen for the bacteria-specific type II CRISPR system of S. thermophilus and very recently in the type IE system of E. coli where new spacers were added adjacent to the leader on phage infection (Barrangou et al., 2007; Deveau et al., 2008; Horvath et al., 2008; Yosef et al., 2012). Clearly, the spacer insertion mechanism operating on locus E of Sulfolobus differs both with respect to position, since few insertions occur at repeat 1 (Table 2), and in being limited to single spacer insertions. The result also undermines the consensus hypothesis that spacer order in CRISPR arrays invariably provides a chronological record of genetic element invasion. The altered mechanism may reflect that locus E is associated with a different type of leader sequence common to Sulfolobus islandicus species, from which it may derive (Garrett et al., 2011b), and it requires complementation by Cas proteins associated with other CRISPR loci, probably loci C and D carrying similar repeats (Lillestøl et al., 2009). These properties may alter the compatibility between the Cas proteins and the leader of locus E leading to a less stringent insertion mechanism. Although there is support for the leader playing an important functional role (Marraffini and Sontheimer, 2008; Lillestøl et al., 2009; Yosef et al., 2012), possibly as an assembly site for the adaptation-associated Cas proteins, for locus E the repeats themselves appear to be important mechanistic determinants of adaptation.
After this article was submitted, a second report of spacer uptake in type IE CRISPR systems of E. coli strains appeared, in which it was claimed that the 3′-nucleotide of the AAG PAM motif became part of the first repeat during adaptation (Swarts et al., 2012). For the Sulfolobus systems described here, the third nucleotide of the CCN PAM motif is not conserved and therefore the same mechanism cannot apply.
In the long-term infection experiments, continued beyond 74 days, we observed an increase in the proportion of cells carrying spacer insertions, but neither the wild-type host nor the invading genetic elements were extinguished from the culture. Thus, virus–plasmid–host conflicts are ongoing and long term. Presumably when cells are no longer disadvantaged by mutated elements there must be a very strong selection for a limited number of spacers that are especially effective, possibly corresponding to functionally critical, and relatively conserved, genomic sites such as origins of replication.
In conclusion, we have developed a natural system to induce new spacer uptake in a subfamily of CRISPR loci of the model crenarchaeon Sulfolobus which allows us to study, in detail, the molecular mechanisms involved in the adaptation process, the molecular basis for viral resistance to the CRISPR-based immune systems, and the longer-term selection processes occurring within the CRISPR-based immune systems of infected Sulfolobus populations.
Experimental procedures
Isolation of virus particles and infection of S. solfataricus P2
An aqueous mud sample was taken from an acidic hot spring (pH 2, 85°C) in Monument Geysir Basin, Yellowstone National Park. One millilitre of this sample was added to 50 ml of Sulfolobus medium supplemented with 0.2% trypton, 0.1% yeast extract and 0.2% sucrose (TYS medium) (Zillig et al., 1994) and incubated aerobically for 5 days at 78°C. Two litres of enrichment culture was then established in TYS medium at 78°C. Cells were pelleted (6000 g, 10 min) and virus particles were isolated by filtration of the supernatant through 0.2 µm pore filters (Nalgene®). This virus mixture was then used to infect S. solfataricus P2 (DSM 1617) cultured in the Sulfolobus medium (Zillig et al., 1994).
Cells of S. solfataricus P2 were harvested from fresh culture by centrifuging (6000 g, 10 min) and resuspending in 1 ml of TYS medium. Twenty microlitres of virus mixture was added at 3 PFU µl−1 and after incubating for 2 h at 80°C, infected cells were transferred to 50 ml of pre-heated (78°C) TYS medium. Infected S. solfataricus P2 was then incubated at 78°C for 3–6 days before isolating infectious particles as described above for the enrichment culture. Infections with the single viruses SIRV2, ATV and STSV2 were performed using the same procedure.
Sequencing of the virus mixture
Sulfolobus solfataricus cells were infected and grown for 3 days in 2 l of TYS medium. Cells were separated by centrifugation (6000 g, 10 min) and virus particles were isolated from the supernatant by filtration using Vivaspin 20 columns (1 000 000 Mwco, Sartorius Stedim, Aubagne, France) at 1500 g and dissolved in 10 mM Tris-HCl, pH 8.0. The virus mixture was then loaded onto 0.45 g ml−1 CsCl and centrifuged for 48 h at 38 000 r.p.m. in a SW41 rotor (Beckman, Fullerton, USA). The virus band was extracted from gradients and CsCl was removed by dialysis against 10 mM Tris-HCl, pH 8. DNA was isolated using DNeasy® Blood&Tissue Kit (Qiagen, Hilden, Germany). Sequencing by Illumina sequencing and raw data treatment was performed by Beijing Genomics Institute (Shenzhen, China). Clean data consisting of approximately 90 bp DNA fragments were assembled using the CLC genomics workbench (CLC Bio, Aarhus, Denmark). orf174 within an operon in viral contig 1 and the traG-like gene in contig 2 of the putative conjugative plasmid were selected to detect the appropriate genetic element in host cells. DNA was amplified by PCR using forward and reverse primers 5′-CCCACCTATATCGAATTC-3′ and 5′-GTGTCTCTCATATTTGCAATC-3′, respectively, for the virus and 5′-GCCTTAGCGAGGGCCCAGTTGAACCTGG-3′ and 5′- CTATCCTATCCCTGTCTATCCCTAG-3′, respectively, for the putative conjugative plasmid. DNA from the initial enrichment culture and the S. solfataricus P2 culture were used as positive and negative controls respectively.
DNase I treatment of the virus mixture
The virus mixture obtained from infected S. solfataricus cells was purified in a CsCl density gradient followed by dialysis against 10 mM Tris-HCl, pH 8. One hundred and fifty microlitres of purified virus mixture was digested with DNase I (Fermentas, St. Leon-Rot, Germany) at 37°C. DNA of the digested virus mixture and an undigested 150 µl sample was isolated using DNeasy® Blood&Tissue Kit (Qiagen). DNA concentration was measured using NanoDrop®.
Electron microscopy
Virus particles were adsorbed onto carbon-coated copper grids for 5 min and stained with 2% uranyl acetate. Images were recorded using a Tecnai G2 transmission electron microscope (FEI, Eindhoven, Netherlands), with a CCD camera, at an acceleration voltage of 120 kV.
Growth curves and PCR amplification of leader proximal CRISPR regions
Wild-type P2 strain and the mutant strain lacking CRISPR loci A to D and their associated cas genes, were infected with virus mixture and cultivated, with an uninfected control sample, in 50 ml of TYS medium at 78°C. One millilitre of samples of each culture was taken every 6 h and optical densities were recorded at 600 nm. Two millilitres of samples of each culture was taken every 24 h and cells were harvested by centrifugation (6000 g, 10 min). DNA was isolated using DNeasy® Blood&Tissue Kit (Qiagen). The leader proximal regions of the six CRISPR loci were amplified by PCR using forward primers 5′-AGCTTCTGACCCGCTCCTGA-3′ for locus A, 5′-AGGGGTTTGTGGGATGGGTTGTG-3′, for locus B, 5′-TCGCTTATCTCTCTCATGCGCCATT-3′ for locus C, 5′-AGTTCCACCCCCGAAGCTCCT-3′ for locus D, 5′-ATAGGGAAAGAGTTCCCCCG-3′ for locus E, 5′-CGGCGTTATAATGGGTATCGGAATCGG-3′ for locus F and reverse primers 5′-GCACATCATCAAACAATGGTAAGCC-3′ for locus A, 5′-ACAACTACCACCACTACCACGG-3′ for locus B, 5′-TGTCCCGTTTTTGTAAGTGGGGG-3′ for locus C, 5′-AGCCGGGACAAGTTTCACAAATTGA-3′ for locus D, 5′-TGACTCTAGTGCAATCTTCGA-3′ for locus E, 5′-GCTCACTATCTCACCCCTATCAATACCC-3′ for locus F. Single clones of infected S. solfataricus cells were obtained on Gel-rite plates and cultured in TYS medium (Zillig et al., 1994).
Cloning of PCR products and sequencing
PCR products were separated on 1.5% agarose gels and bands larger than the those of the uninfected control sample were excised from the gel and purified with QIAquick Gel Extraction Kit (Qiagen). Purified PCR products were cloned using InsTAclone™PCR Cloning Kit (Fermentas) following the manufacturers' protocols. Plasmid purification and sequencing were performed by GATC Biotech AG, (Konstanz, Germany).
Acknowledgments
The research was supported by the Danish Natural Science Research Council and S.E. received grants from Copenhagen University. Daniel Werner helped with sampling and Soley Gudbergsdottir provided the CRISPR-minus mutant. We thank Sven le Moine Bauer for help with the early single virus experiments and for inspiring discussions. We are grateful to Shiraz A. Shah for localizing the CRISPR spacer matches on the viral contigs. Xu Peng is thanked for demonstrating that SIRV2 virus could infect the CRISPR-minus mutant, and discussions with Qunxin She were appreciated.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Aranaz A, Liébana E, Mateos A, Dominguez L, Vidal D, Domingo M, et al. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–2740. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold HP, She Q, Phan H, Stedman K, Prangishvili D, Holz I, et al. The genetic element pSSVx of the extremely thermophilkic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol Microbiol. 1999;34:217–226. doi: 10.1046/j.1365-2958.1999.01573.x. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Basta T, Smyth J, Forterre P, Prangishvili D, Peng X. Novel archaeal plasmid pAH1 and its interactions with the lipothrixvirus AFV1. Mol Microbiol. 2009;71:23–34. doi: 10.1111/j.1365-2958.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- Bize A, Karlsson EA, Ekefjard K, Quax TE, Pina M, Prevost MC, et al. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci USA. 2009;106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Kenchappa CS, Peng X, She Q, Garrett RA. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012;40:2470–2480. doi: 10.1093/nar/gkr1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett RA, Bize A, Shah SA, Stetter K, Prangishvili D, Peng X. Metagenomic analyses of novel viruses, plasmids, and their variants, from an environmental sample of hyperthermophilic neutrophiles cultured in a bioreactor. Environ Microbiol. 2010;12:2918–2930. doi: 10.1111/j.1462-2920.2010.02266.x. [DOI] [PubMed] [Google Scholar]
- Garrett RA, Vestergaard G, Shah SA. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 2011a;19:549–556. doi: 10.1016/j.tim.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Garrett RA, Shah SA, Vestergaard G, Deng L, Gudbergsdottir S, Kenchappa CS, et al. CRISPR-based immune systems of the Sulfolobales – complexity and diversity. Biochem Soc Trans. 2011b;39:51–57. doi: 10.1042/BST0390051. [DOI] [PubMed] [Google Scholar]
- Greve B, Jensen S, Brügger K, Zillig W, Garrett RA. Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea. 2004;1:231–239. doi: 10.1155/2004/151926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen PM, Bunschoten AE, van Soolingen D, van Embden JD. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- Gudbergsdottir S, Deng L, Chen Z, Jensen JVK, Jensen LR, She Q, et al. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häring M, Vestergaard G, Rachel R, Chen L, Garrett RA, Prangishvili D. Independent virus development outside a host. Nature. 2005;436:1101–1102. doi: 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci USA. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- Hermans PW, van Soolingen D, Bik EM, de Haas PE, Dale JW, van Embden JD. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coûté-Monvoisin A-C, Richards M, Deveau H, Moineau S, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, Van Duijn E, Bultema JB, Westra ER, Waghmare SP, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van Agterfeld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestøl RK, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genius Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- Lintner NG, Frankel KA, Tsutakawa SE, Alsbury DL, Copié V, Young MJ, et al. The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J Mol Biol. 2011;405:939–955. doi: 10.1016/j.jmb.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A, Zebec Z, Teichmann D, Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol. 2011;80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Pina M, Bize A, Forterre P, Prangishvili D. The archaeoviruses. FEMS Microbiol Rev. 2011;35:1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- Porter K, Russ BE, Dyall-Smith ML. Virus–host interactions in salt lakes. Curr Opin Microbiol. 2007;10:418–424. doi: 10.1016/j.mib.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Albers SV, Holz I, Arnold HP, Stedman K, Klein T, et al. Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid. 1998;40:190–202. doi: 10.1006/plas.1998.1363. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat Rev Microbiol. 2006a;11:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Vestergaard G, Häring M, Aramayo R, Basta T, Rachel R, et al. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J Mol Biol. 2006b;359:1203–1216. doi: 10.1016/j.jmb.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011;162:27–38. doi: 10.1016/j.resmic.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Shah SA, Hansen NR, Garrett RA. Distributions of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009;37:23–28. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]
- Shah SA, Vestergaard G, Garrett RA. CRISPR/Cas and CRISPR/Cmr immune systems of Archaea. In: Marchfelder A, Hess W, editors. Regulatory RNAs in Prokaryotes. Berlin: Springer Press; 2011. pp. 163–181. [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler N, Marguet E, Verbavatz JM, Forterre P. Virus-like vesicles and extracellular DNA from hyperthermophilic archaea of the order Thermococcales. Res Microbiol. 2008;159:390–399. doi: 10.1016/j.resmic.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MWJ, Brouns SJJ. CRISPR interference directs strand specific spacer acquisition. PLoS ONE. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T-H, Bachellerie J-P, Rozhdestvensky T, Bortolin M-L, Huber H, Drungowski M, et al. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T-H, Polacek N, Zywicki M, Huber H, Brügger K, Garrett R, et al. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Duan Z, Zhu H, Guo X, Wang Z, Zhou J, et al. A novel Sulfolobus non-conjugative extrachromosomal element capable of integration into the host genome and spreading in the presence of a fusellovirus. Virology. 2007;363:124–133. doi: 10.1016/j.virol.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F, Zhu YW, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Chen L, Huang X, Luo Y, She Q, Huang L. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus–host interactions and genomic features. J Virol. 2005;79:8677–8686. doi: 10.1128/JVI.79.14.8677-8686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, et al. Structure and mechanism of the Cmr complex of CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, et al. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol. 1994;16:109–128. [Google Scholar]
- Zillig W, Arnold HP, Holz I, Prangishvili D, Schweier A, Stedman K, et al. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles. 1998;2:131–140. doi: 10.1007/s007920050052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


