Abstract
Maintenance of the integrity of the cell wall in fungi is essential. One mechanism that cells use to maintain cell wall integrity in response to cell wall damage is to up-regulate chitin synthesis. In Candida albicans, the PKC cell wall integrity, Ca2+/calcineurin and high osmolarity glycerol (HOG) signalling pathways co-ordinately regulate chitin synthesis in response to cell wall stress. The transcription factors downstream of these pathways and their DNA binding sites within the promoters of target genes are well characterised in Saccharomyces cerevisiae, but not in C. albicans. The promoters of the C. albicans class I CHS genes (CaCHS2 and CaCHS8) were functionally dissected with the aim of identifying and characterising the transcription factors and promoter elements that mediate the transcriptional up-regulation of CaCHS2 and CaCHS8 in response to cell wall stress. This analysis provided evidence that the PKC cell wall integrity pathway may operate through RLM1-elements in the CaCHS2 and CaCHS8 promoters, but that promoter sequences that respond to the Ca2+/calcineurin and HOG signalling pathways in S. cerevisiae did not directly regulate chitin synthase 2 and 8 gene transcription in C. albicans.
Keywords: Chitin, Transcription, Candida albicans, Cell wall, Signal transduction
Introduction
The cell wall of Candida albicans plays many vital roles. These range from maintaining cell morphology, providing protection from external pressure, aiding in colonisation and pathogenesis of the human host, and in immune-recognition and immune-avoidance (Netea et al. 2008). Damage to the cell wall leads to the activation of a “compensatory” response in yeast (Popolo et al. 2001). Part of this compensatory response includes the up-regulation of chitin synthesis. In C. albicans, chitin is synthesised by four chitin synthase isoenzymes, CaChs1p (class II), CaChs2p and CaChs8p (class I) and CaChs3p (class IV) (Gow et al. 1994; Bulawa et al. 1995; Mio et al. 1996; Munro et al. 2001, 2003). Exposure of C. albicans to cell wall stresses such as CaCl2 or Calcofluor White (CFW) can result in an increase in the chitin synthase activity that can be measured in vitro, and/or in an increase in the amount of chitin in the cell wall (Munro et al. 2007). Treatment with caspofungin, which targets β(1,3)-glucan synthesis, also results in an increase in the level of chitin in the cell wall and an increase in the level of CaChs3p in the cell (Walker et al. 2008).
We have shown indirectly that the PKC cell wall integrity pathway, the Ca2+/calcineurin signalling pathway and the high osmolarity glycerol (HOG) pathway co-ordinately regulate chitin synthesis in response to such stresses. Treatment with CaCl2 stimulates all four CHS promoters in a calcineurin-dependent manner and the deletion of CaCRZ1 results in a loss of the Ca2+ signal to the CaCHS1, CaCHS2, CaCHS8 promoters. Treatment with CFW activates the CaCHS1, CaCHS2 and CaCHS8 promoters, and deletion of CaMKC1, the MAP kinase of the PKC cell wall integrity pathway, results in a loss of this signal to the CaCHS2 and CaCHS8 promoters. Treatment with both CaCl2 and CFW results in the hyper-stimulation of all four CaCHS promoters, which is partially lost when CaHOG1 is deleted (Munro et al. 2007).
The transcription factors downstream of these signalling pathways and the DNA sequences they recognise and bind, in the promoters of target genes, are well characterised in Saccharomyces cerevisiae. The PKC cell wall integrity pathway up-regulates the expression of a number of cell wall related genes through the ScRlm1p transcription factor (Garcia et al. 2004), and can also operate through the ScSwi4p/ScSwi6p transcription factor (Kim et al. 2008). This pathway is activated by mutations in cell wall biosynthetic genes and by treatment with cell wall perturbing agents such as CFW, Congo Red, SDS and caspofungin, and results in the up-regulation of genes involved in cell wall maintenance, biogenesis and remodelling (Lagorce et al. 2003; Reinoso-Martin et al. 2003; Boorsma et al. 2004; Garcia et al. 2004). ScRlm1p binds to RLM1-elements in the promoters of its target genes via the consensus sequence NTAW4TAG (Dodou and Treisman 1997; Jung and Levin 1999; Jung et al. 2002).
The Ca2+/calcineurin pathway is activated by a number of stresses including high extracellular levels of Ca2+ (Yoshimoto et al. 2002; Cyert 2003). The Ca2+/calcineurin pathway operates through the transcription factor ScCrz1p, which regulates the expression of a number of genes including those involved in maintaining cell ion homeostasis (Yoshimoto et al. 2002). The ScCrz1p transcription factor binds to calcineurin-dependent response elements (CDREs) with the consensus sequence AGCCTC (Mendizabal et al. 2001; Yoshimoto et al. 2002; Lagorce et al. 2003). The HOG pathway mediates the up-regulation of genes in response to osmotic stress (de Nadal et al. 2002). ScSko1p is a transcription factor that is phosphorylated by ScHog1p and activates osmotic stress responsive genes (Proft and Struhl 2002; Proft et al. 2005). ScSko1p binds to the ATF/CREB consensus sequence ATKACGTMAT (Proft et al. 2005).
The corresponding transcription factors and their binding sites are less well characterised in C. albicans. Santos and de Larrinoa (2005) showed that CaCrz1p can act through the CDRE in the promoter of the ScENA1 gene when heterologously expressed in S. cerevisiae. Karababa et al. (2006) identified a putative CaCDRE consensus (GYGGT) by analysing the promoters of 60 C. albicans genes that were up-regulated in response to CaCl2 in a CaCrz1p- and calcineurin-dependent manner. This type of analysis has not yet been performed for CaRlm1p and CaSko1p.
We wanted to further characterise the transcriptional response of the CaCHS genes to cell wall stresses by identifying the specific promoter elements and transcription factors that are involved in this response in C. albicans. We identified putative RLM1-elements, CDREs and ATF/CREB-elements in silico in the promoters of the class I CaCHS genes. Their function was assessed by mutating the promoter elements in CHS-promoter-lacZ reporter constructs and monitoring the level of activation in response to cell wall stress with CFW, CaCl2 and sorbitol. Since the mutation of individual promoter elements did not have any effect on the expression of the CHS-promoter-lacZ reporter constructs, a set of nested window deletions of the CaCHS2 and CaCHS8 promoters were constructed and used to identify the regions of the promoters that contained important regulatory sequences. The sequences that were required for the activation of these two promoters by CFW and CaCl2 were localised to narrow regions within the first 347 and 125 base pairs (bp) of the CaCHS2 and CaCHS8 promoters respectively. The PKC cell wall integrity pathway was shown to act on the minimal class I CaCHS promoters. In addition, the Ca2+/calcineurin signalling pathway and the HOG pathway also act on the minimal CaCHS2 promoter. We provide evidence that the PKC cell wall integrity pathway may operate through the CaRlm1p transcription factor binding to RLM1-elements in the CaCHS2 and CaCHS8 promoters, but the Ca2+/calcineurin and HOG signalling pathways may act in a less well established and possibly indirect manner to regulate these genes. This analysis highlights the complex nature of the transcriptional regulation of chitin synthesis in C. albicans.
Materials and methods
Strains, media and growth conditions
Candida albicans strains used in this study are listed in Table 1 and were grown at 30°C in liquid rich medium (YEPD + Uri) containing 10 g/l yeast extract, 20 g/l mycopeptone, 20 g/l D-glucose and 25 ug/ml uridine, or on solid medium containing 20 g/l agar type 3. Ura+ transformants were selected and maintained on minimal medium (SD) containing 20 g/l D-glucose, 6.7 g/l yeast nitrogen base without amino acids and 20 g/l purified agar.
Table 1.
Candida albicans strains used in this study
| Strain | Strain name | Genotype | Source |
|---|---|---|---|
| CAI-4 | CAI-4 | ura3Δ::λimm434/ura3Δ::λimm434 | Fonzi and Irwin (1993) |
| crz1Δ | DSY2842 | crz1Δ::hisG/crz1Δ::hisG, ura3Δ::λimm434/ura3Δ::λimm434 | Karababa et al. (2006) |
| cna1Δ | DSY2101 | cna1Δ::hisG/cna1Δ::hisG, ura3Δ::λimm434/ura3Δ::λimm434 | Sanglard et al. (2003) |
| mkc1Δ | CM1613c | mkc1Δ::hisG/mkc1Δ::hisG, ura3Δ::λimm434/ura3Δ::λimm434 | Navarro-Garcia et al. (1995) |
| hog1Δ | CNC15 | hog1Δ::hisG/hog1Δ::hisG, ura3Δ::λimm434/ura3Δ::λimm434, his1::hisG/his1::hisG | Alonso-Monge et al. (1999) |
E. coli strains DH5α (Invitrogen) and XL-10 Gold (Stratagene) were used in this work. Bacterial strains harbouring plasmids encoding the ampicillin resistance marker were grown selectively in Luria-Bertani (LB) medium containing 5 g/l yeast extract, 10 g/l NaCl and 10 g/l tryptone supplemented with 100 μg/ml ampicillin, or on solid medium containing 15 g/l agar type 3.
In silico analysis of the class I CHS promoters
All in silico analyses were performed using Genomatix Software (http://www.genomatix.de). MatInspector was used to identify any putative promoter elements listed in the MatBase database. CoreSearch was used to identify any novel putative promoter elements. Default search parameters were used in all cases.
Site-directed mutagenesis of specific putative promoter elements
Putative promoter elements in the CaCHS2 and CaCHS8 promoters were mutated using the QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene) as per the manufacturer’s instructions. All mutations were confirmed by DNA sequencing. The mutations that were introduced to the CaCHS2 promoter sequence in the plasmid pCHS2plac (Table 2) and the CaCHS8 promoter sequence in the plasmid pCHS8plac (Table 2) using mutagenic oligonucleotide primers (Table 3) are summarised in Table 4. The mutations that were introduced to the minimal CaCHS2 promoter sequence in the plasmid pCHS2plac-347 (Table 5) and the minimal CaCHS8 promoter sequence in the plasmid pCHS8plac-125 (Table 5) using mutagenic oligonucleotide primers (Table 3) are summarised in Table 4.
Table 2.
Plasmids used in this study
| Plasmid | Features | Source |
|---|---|---|
| placpoly6 | Empty vector; Strep. thermophilus lacZ; CaURA3; CaRPS1; amp r; ColE1 origin | Uhl and Johnson (2001) |
| pCHS2plac | CaCHS2 promoter fused to Strep. thermophilus lacZ; CaURA3; CaRPS1; amp r; ColE1 origin | Munro et al. (2007) |
| pCHS8plac | CaCHS8 promoter fused to Strep. thermophilus lacZ; CaURA3; CaRPS1; amp r; ColE1 origin | Munro et al. (2007) |
Table 3.
Mutagenic oligonucleotide primers used in this study
| Name | Sequence (5′–3′) |
|---|---|
| MDL42 | GCTAATTTAGTTTCAAATCTATTTTTACTTTCTATTGAGCTTTAGCGTCTCATTGTCATTGCCCCCTTTTTTTTTCATTTCAC |
| MDL43 | GTGAAATGAAAAAAAAAGGGGGCAATGACAATGAGACGCTAAAGCTCAATAGAAAGTAAAAATAGATTTGAAACTAAATTAGC |
| MDL44 | GATTTAGCTAATTTAGTTTCAAATCTATTTTTACTTGAGCTTTAGCGTTCTATTCTCATTGTCATTGCCCCCTTTTTTTTTCATTTC |
| MDL45 | GAAATGAAAAAAAAAGGGGGCAATGACAATGAGAATAGAACGCTAAAGCTCAAGTAAAAATAGATTTGAAACTAAATTAGCTAAATC |
| MDL46 | CATTCTTTTAAAAATTGATTTAGCTAATTTAGTTTCAAAGAGCTTTAGCGTTTCTATTTCTATTTCTATTCTCATTGTCATTGCCCCC |
| MDL47 | GGGGGCAATGACAATGAGAATAGAAATAGAAATAGAAACGCTAAAGCTCTTTGAAACTAAATTAGCTAAATCAATTTTTAAAAGAATG |
| MDL48 | CCTAAAAAAAAAAATGTTAAGAAAGGACAAGAAAGAAAGAAATCAATCTGGATTAAATTCTTCTTAGTCGTTGTTCGTTGATTTG |
| MDL49 | CAAATCAACGAACAACGACTAAGAAGAATTTAATCCAGATTGATTTCTTTCTTTCTTGTCCTTTCTTAACATTTTTTTTTTTAGG |
| MDL50 | GAAACAATGATGAATCGACAAATCAAGCAAAAAAGAGAAGATTACTTGGACACGAAAACGAATGAAATAATTCTGATGTCG |
| MDL51 | CGACATCAGAATTATTTCATTCGTTTTCGTGTCCAAGTAATCTTCTCTTTTTTGCTTGATTTGTCGATTCATCATTGTTTC |
| MDL52 | GGCTGAGTGAAACAAAAAAAAACAACTGTTTAGTGAATCTGTGTGCAGACCAATTTGTTTGAAAAAGTCGTGAC |
| MDL53 | GTCACGACTTTTTCAAACAAATTGGTCTGCACACAGATTCACTAAACAGTTGTTTTTTTTTGTTTCACTCAGCC |
| MDL54 | GGAGGAGGGGGTGAGACAGTTAGTGAGAAAATCGAGTGAAACAAAAAAAAACAACTGTTTAGTGGG |
| MDL55 | CCCACTAAACAGTTGTTTTTTTTTGTTTCACTCGATTTTCTCACTAACTGTCTCACCCCCTCCTCC |
| MDL58 | CATTCATTCGCTTTCGGTATTTTTTTGGGGAGTGTCGCTTTTGCTCAATCACTATATCCAACCTAGACTAAAGTTCAGTTCCC |
| MDL59 | GGGAACTGAACTTTAGTCTAGGTTGGATATAGTGATTGAGCAAAAGCGACACTCCCCAAAAAAATACCGAAAGCGAATGAATG |
| MDL60 | GAGATTCAGAACAAAAAAAAGAATTTCCTTCACAAGAGCAAAAGCGTAATAATAACAAACAATAAACACTATTGAATTTCCAC |
| MDL61 | GTGGAAATTCAATAGTGTTTATTGTTTGTTATTATTACGCTTTTGCTCTTGTGAAGGAAATTCTTTTTTTTGTTCTGAATCTC |
| MDL62 | GATTATTGTGTCATTTGTGGGAAGAAGTAAAGGAGCATAAGCGGAATGATAATAATAATGTGGTCCAGGCTTATTTGGTAGC |
| MDL63 | GCTACCAAATAAGCCTGGACCACATTATTATTATCATTCCGCTTATGCTCCTTTACTTCTTCCCACAAATGACACAATAATC |
| MDL64 | GCAATTACGAAAGGGAGTAAGCAGGAGAGATTAGGCAAGTTCGGTCCTAGATGCAGGACATG |
| MDL65 | CATGTCCTGCATCTAGGACCGAACTTGCCTAATCTCTCCTGCTTACTCCCTTTCGTAATTGC |
| MDL66 | GATAAATATAGAGAATGATAATAATAATGTGGTCCAAATCTATTTGGTAGCAATTACGAAAGGGAGTAAGCAGGAG |
| MDL67 | CTCCTGCTTACTCCCTTTCGTAATTGCTACCAAATAGATTTGGACCACATTATTATTATCATTCTCTATATTTATC |
| MDL68 | GGACATGTGGTTGGTCTCCGTGTGATCTGCCTAAATAGGAAAGCATGCCACAATAAGAATCAATTCTTTTTTTTTAGAGATTCAG |
| MDL69 | CTGAATCTCTAAAAAAAAAGAATTGATTCTTATTGTGGCATGCTTTCCTATTTAGGCAGATCACACGGAGACCAACCACATGTCC |
| MDL103 | CCTAAAGTTTAACACAAGTGTTGAAACATTCTTTTAAAAATTGATTTAGCTAATTTAGTTTCAAAGAGCTTTAGCGTTGAGCTTGAGCGTTAGCGTCTCATTGTCATTGCCCCCTTTTTTTTTCATTTCACTTCACTTCATTTATTTATTATATTAATTGG |
| MDL104 | CCAATTAATATAATAAATAAATGAAGTGAAGTGAAATGAAAAAAAAAGGGGGCAATGACAATGAGACGCTAACGCTCAAGCTCAACGCTAAAGCTCTTTGAAACTAAATTAGCTAAATCAATTTTTAAAAGAATGTTTCAACACTTGTGTTAAACTTTAGG |
| MDL105 | CATTCGCTTTCGGTATTTTTTTGGGGAGTGTCGCTTTTGCTCAATCACTATATCCAACCTAGACTAAAGTTCAGTTCCCACTC |
| MDL106 | GAGTGGGAACTGAACTTTAGTCTAGGTTGGATATAGTGATTGAGCAAAAGCGACACTCCCCAAAAAAATACCGAAAGCGAATG |
| MDL109 | GATTTAGCTAATTTAGTTTCAAAGAGCTTTAGCGTTGAGCTTTAGCGTTCTATTCTCATTGTCATTGCCCCCTTTTTTTTTCATTTC |
| MDL110 | GAAATGAAAAAAAAAGGGGGCAATGACAATGAGAATAGAACGCTAAAGCTCAACGCTAAAGCTCTTTGAAACTAAATTAGCTAAATC |
Table 4.
Summary of mutations introduced to the pCHS2plac and pCHS8plac reporter plasmids
| Wild type plasmid | Putative element | Positiona | Wild type sequenceb | Mutant sequencec | Mutagenic primers | Resulting plasmid name |
|---|---|---|---|---|---|---|
| pCHS2plac | RLM1−261 | −261 to −251 | TCTATTTCTAT | GAGCTTTAGCG | MDL42; MDL43 | pCHS2placΔRLM1−261 |
| pCHS2plac | RLM1−267 | −267 to −257 | TCTATTTCTAT | GAGCTTTAGCG | MDL44; MDL45 | pCHS2placΔRLM1−267 |
| pCHS2plac | RLM1−280 | −280 to −270 | TCTATTTTTAC | GAGCTTTAGCG | MDL46; MDL47 | pCHS2placΔRLM1−280 |
| pCHS2plac | CDRE−406 | −406 to −403 | GGCT | AATC | MDL48; MDL49 | pCHS2placΔCDRE−406 |
| pCHS2plac | CDRE−543 | −543 to −540 | AGCC | GATT | MDL50; MDL51 | pCHS2placΔCDRE−543 |
| pCHS2plac | CDRE−882 | −882 to −879 | GGCG | AATC | MDL52; MDL53 | pCHS2placΔCDRE−882 |
| pCHS2plac | CDRE−917 | −917 to −914 | AGGCT | AAATC | MDL54; MDL55 | pCHS2placΔCDRE−917 |
| pCHS8plac | RLM1−94 | −94 to −84 | TTATTTTTAGA | CGCTTTTGCTC | MDL58; MDL59 | pCHS8placΔRLM1−94 |
| pCHS8plac | RLM1−713 | −713 to −703 | CCTAAAAATAG | GAGCAAAAGCG | MDL60; MDL61 | pCHS8placΔRLM1−713 |
| pCHS8plac | RLM1−927 | −927 to −917 | ATAAATATAGA | GAGCTTTAGCG | MDL62; MDL63 | pCHS8placΔRLM1−927 |
| pCHS8plac | CDRE−851 | −851 to −848 | AGCC | GATT | MDL64; MDL65 | pCHS8placΔCDRE−851 |
| pCHS8plac | CDRE−892 | −892 to −889 | GGCT | AATC | MDL66; MDL67 | pCHS8placΔCDRE−892 |
| pCHS8plac | ATF/CREB−782 | −782 to −776 | TACGTAA | GCATGCC | MDL68; MDL69 | pCHS8placΔATF/CREB−782 |
| pCHS2plac-347 | RLM1−280 | −280 to −270 | TCTATTTTTAC | GAGCTTTAGCG | MDL46; MDL47 | pCHS2plac −347ΔRLM1−280 |
| pCHS2plac-347 | RLM1−267 | −267 to −257 | TCTATTTCTAT | GAGCTTTAGCG | MDL44; MDL45 | pCHS2plac-347ΔRLM1−267 |
| pCHS2plac-347 | RLM1−261 | −261 to −251 | TCTATTTCTAT | GAGCTTTAGCG | MDL42; MDL43 | pCHS2plac-347ΔRLM1−261 |
| pCHS2plac-347ΔRLM1−280 | RLM1−267 | −267 to −257 | TCTATTTCTAT | GAGCTTTAGCG | MDL109; MDL110 | pCHS2plac-347ΔRLM1−280,−267 |
| pCHS2plac-347ΔRLM1−280,−267 | RLM1−261 | −261 to −251 | TCTATTTCTAT | GAGCTTTAGCG | MDL103; MDL104 | pCHS2plac-347ΔRLM1−280,−267,−261 |
| pCHS8plac-125 | RLM1−94 | −94 to −84 | TTATTTTTAGA | CGCTTTTGCTC | MDL105; MDL106 | pCHS8plac-125ΔRLM1−94 |
aRelative to ATGCHS
bCore sequence indicated in bold
cMutated core sequence is underlined
Table 5.
Window deletion reporter constructs produced in this study
| Plasmid name | Portion of promoter in reporter construct | Nucleotides deleted upstream of KpnI site |
|---|---|---|
| pCHS2plac | −969 to −1 (relative to ATGCHS2) | 0 |
| pCHS2plac-347 | −347 to −1 (relative to ATGCHS2) | 468 |
| pCHS8plac | −969 to −1 (relative to ATGCHS8) | 0 |
| pCHS8plac-513 | −513 to −1 (relative to ATGCHS8) | 387 |
| pCHS8plac-499 | −499 to −1 (relative to ATGCHS8) | 387 |
| pCHS8plac-429 | −429 to −1 (relative to ATGCHS8) | 364 |
| pCHS8plac-125 | −125 to −1 (relative to ATGCHS8) | 432 |
Construction of window deletions of the C. albicans class I CHS promoters
A set of nested window deletions of the CaCHS2 and CaCHS8 promoters was created using the method described in Sambrook et al. (1989). Briefly, 10 μg of plasmid DNA was digested with SalI and then with KpnI. The plasmid DNA was extracted with phenol extraction and precipitated in ethanol after each digestion. The linearised plasmid DNA was treated with exonuclease III (Promega) either at 37 or 30°C. Samples were taken from the digestion reaction at 30 s intervals and treated with SI nuclease (Promega) for 30 min at 30°C. The resulting ends were filled in using the large (Klenow) fragment of DNA Polymerase I (Promega) and ligated together using T4 DNA polymerase (Promega). The ligation mixes were transformed into E. coli strain DH5α and transformants selected on LB plates containing 100 μg/ml ampicillin. Plasmids were extracted and sequenced to identify the region of the promoters that had been deleted.
Assay for β-galactosidiase activity
All reporter constructs were linearised with StuI and transformed into the C. albicans strain CAI-4. Correct integration at CaRPS1 was confirmed by Southern analysis as described previously (Munro et al. 2007). Three independent transformants for each reporter construct were assayed for β-galactosidase activity in triplicate using a method adapted from Rose and Botstein (1983) after growth in YEPD + Uri in the presence and absence of an appropriate stress from an OD600 0.1 to 1.0 at 30°C.
Cultures were harvested in pre-chilled tubes containing 5 ml ice by centrifugation (4 min, 3,350×g, 4°C), washed in 1 ml ice-cold breakage buffer (100 mM Tris–HCl (pH 8.0), 1 mM DTT, 20% (v/v) glycerol) and re-suspended in 250 μl breakage buffer. 5 μl 100 mM PMSF was added and cells were broken in a mini-bead beater in the presence of acid-washed glass beads (2 × 20 s bursts with 2 min incubation on ice between bursts). A further 200 μl of breakage buffer was added, the samples were vortexed briefly and then centrifuged at 14,000×g for 15 min at 4°C. The cleared lysate was used for the protein assays.
The o-nitrophenol (o-NP) assay was scaled down and adapted for use in a microtitre plate. The reaction mixture contained 10 μl cleared lysate and 90 μl Z-buffer (16.1 g/l Na2HPO4·7H2O, 5.5 g/l NaH2PO4·H2O, 0.75 g/l KCl, 0.246 g/l MgSO4·7H2O, 2.7 ml/l 2-mercaptoethanol, pH 7.0). The blank wells contained 10 μl breakage buffer instead of cleared lysate. Plates were equilibrated at 28°C and the reaction started by the addition of 20 μl ONPG (4 mg/ml in Z-buffer) and the start time noted. The reactions were allowed to proceed until a pale-yellow colour developed. The reactions were stopped by the addition of 50 μl 1 M Na2CO3 and the stop time noted. The optical density at 420 nm was measured. Protein concentration was determined using the method described by Bradford (1976) with BSA as the standard. β-galactosidase specific activity was then determined based on measurement of OD420 (the absorbance maximum for o-NP at 420 nm) and was expressed as nmol hydrolysed ONPG per mg protein per min, or nmol/mg/min.
Statistical analyses
Statistically significant differences were determined using SPSS software to perform 1-way ANOVA and post-hoc Bonferroni’s t test, p < 0.05.
Results
Putative transcription factor binding sites of the CaCHS2 and CaCHS8 promoters
In our previous work, the class I CHS genes (CaCHS2 and CaCHS8) showed the highest level of activation in response to cell wall stresses (Munro et al. 2007). For this reason, our analysis concentrated on these two promoters. In silico analysis of the CaCHS2 and CaCHS8 promoters using MatInspector identified a number of putative promoter elements that may be involved in the transcriptional response to cell wall stresses. These included putative CDREs (Crz1p-binding sites), ATF/CREB elements (Sko1p-binding sites), and RLM1-elements (Rlm1p-binding sites). The locations of putative transcription factor binding sites that were identified are shown in Fig. 1. Putative RLM1-elements were identified based on homology with the S. cerevisiae consensus sequence in the MatBase database (Genomatix)(NTAW4TAG; Dodou and Treisman 1997). The putative C. albicans CDRE (NGGCKCA) and ATF/CREB (TACGT) sites had been identified previously (Munro et al. 2007).
Fig. 1.
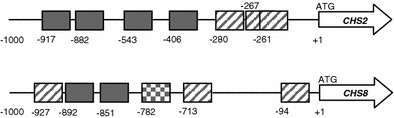
Schematic of the CHS2 and CHS8 promoters. The positions of the putative transcription factor binding sites in the CHS2 (top) and CHS8 (bottom) promoters are indicated relative to ATGCHS (+1). Diagonal lines indicate putative RLM1-elements. Putative CDREs are indicated in solid fill. The putative ATF/CREB-element is checked
Putative promoter elements in the C. albicans class I CHS promoters do not function individually
Reporter constructs pCHS2plac and pCHS8plac were used that had been generated previously to assess the transcriptional response of CaCHS2 and CaCHS8 to various stresses (Munro et al. 2007; Walker et al. 2008). These reporter constructs contain approximately 1 kb of each CaCHS promoter fused to the Streptococcus thermophilus lacZ gene. To assess whether the in silico-identified putative RLM1-elements, CDREs and ATF/CREB-elements were functional, the core sequences of each motif were mutated by site-directed mutagenesis. The mutations to the putative RLM1-elements and ATF/CREB-elements were designed to invert the core sequence by changing the A’s to C’s and T’s to G’s. The core sequences of the putative CDREs were inverted by changing the A’s to G’s and T’s to C’s. If a putative promoter element was functional individually, we predicted that mutation of the element would result in a loss of induction of the lacZ reporter gene in response to an appropriate stress compared to the lacZ reporter gene with the wild type promoter element. We used 100 μg/ml CFW to activate the PKC cell wall integrity pathway and test the function of the RLM1-elements and 100 mM CaCl2 to activate the Ca2+/calcineurin pathway and test the function of the CDREs. Sorbitol at 1 M was used to activate the HOG pathway and test the function of the ATF/CREB-elements.
Candida albicans strains containing the reporter constructs with the wild type and mutated putative promoter elements were assayed for β-galactosidase activity following growth in the presence and absence of an appropriate stress (Fig. 2a, b). Expression mediated by the wild type CaCHS2 promoter was induced 4.6-fold following treatment with CFW (pCHS2plac ± CFW; Fig. 2b) and threefold following treatment with CaCl2 (pCHS2plac ± CaCl2; Fig. 2b). None of the mutations introduced into the CaCHS2 promoter conferred an obvious loss of activation of the lacZ reporters after treatment with CFW or CaCl2 (Fig. 2a). Similarly, expression mediated by the wild type CaCHS8 promoter was induced 2.6-fold by treatment with CFW (pCHS8plac ± CFW; Fig. 2b) and 1.5-fold by treatment with CaCl2 (pCHS8plac ± CaCl2; Fig. 2b). Treatment with sorbitol did not result in an increase in expression conferred by the CaCHS8 promoter (pCHS8plac ± sorbitol; Fig. 2b) and so the role of the ATF/CREB-element was not considered further. Again, mutation of the putative promoter elements in the CaCHS8 promoter did not result in an obvious loss of induction of the lacZ reporters after treatment with CFW, CaCl2 or sorbitol (Fig. 2a). The basal level of expression of the lacZ reporter constructs with the mutant promoter elements in the absence of any cell wall stress (YEPD + Uri) was consistent with the wild type reporters. One exception was observed where the basal level of expression of pCHS8placΔATF/CREB−782 was roughly half that of pCHS8plac (Fig. 2a). These results indicated that no single putative promoter element identified in our in silico analysis mediated the activation of CaCHS2 or CaCHS8 in response to the stresses tested.
Fig. 2.
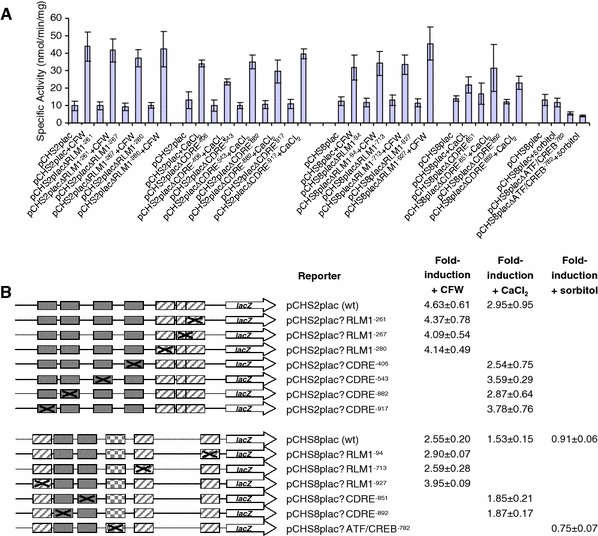
Mutation of putative promoter elements does not effect the activation of CHS2 and CHS8 in response to cell wall stresses. a C. albicans strains containing a single copy of the reporter constructs with the wild type CHS2 and CHS8 promoters and those with mutated RLM1-elements, CDREs, and ATF/CREB-elements were assayed for β-galactosidase activity following growth from an OD600 of 0.1–1.0 in the presence and absence of 100 μg/ml CFW, 100 mM CaCl2 and 1 M sorbitol. Each measurement is the average β-galactosidase activity measured from three independent transformants for each reporter construct assayed in triplicate ± SD (n = 9). b Diagrams representing the reporter constructs containing the individual mutations of the putative promoter elements and summary of the average fold-induction ± the standard deviation for each of the reporter constructs shown above
Identification of regions of the CaCHS2 and CaCHS8 promoters that may contain important regulatory elements
Since no loss of induction in response to CaCl2 or CFW was observed when putative promoter elements were mutated individually, we assessed whether multiple elements had to be deleted to see a loss of the transcriptional response. A set of nested window deletions of the class I CHS promoters were constructed by digesting the full length CaCHS2 and CaCHS8 promoters in pCHS2plac and pCHS8plac (approximately 1 kb) with exonuclease III from their 5′ ends for different lengths of time. Details of the specific deletions are indicated in Table 5. Protruding 3′-termini with more than three unpaired bases are resistant to degradation by exonuclease III except when the terminal base is a C, such as following digestion with KpnI (Sambrook et al. 1989). Since no other suitable restriction sites were available upstream of the CaCHS promoters in the pCHSplac plasmids, the KpnI site was used. As a consequence, a portion of the plasmid backbone sequence upstream of the KpnI site was also deleted. This did not produce any deleterious effects and plasmids with various deletions of the CaCHS promoters were constructed accordingly. The resulting set of window deletion reporter constructs of the CaCHS2 and CaCHS8 promoters is shown in Fig. 3b. All four putative CDREs in the CaCHS2 promoter were deleted in the reporter construct containing only the first 347 bp of the CaCHS2 promoter (pCHS2plac-347; Fig. 3b). Similarly, both putative CDREs and two of three putative RLM1-elements were deleted in all four CaCHS8 window deletion reporter constructs (pCHS8plac-513, -499, -429 and -125; Fig. 3b).
Fig. 3.
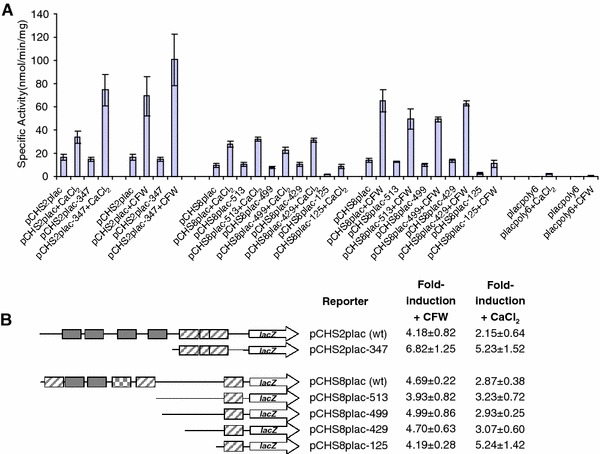
Window deletions of the CHS2 and CHS8 promoter reveal regions containing regulatory sequences. a C. albicans strains containing a single copy of the full length and window deletions of the CHS2 and CHS8 promoters were assayed for β-galactosidase activity following growth from an OD600 of 0.1–1.0 in the presence and absence of 100 mM CaCl2 and 100 μg/ml CFW. Each measurement is the average β-galactosidase activity measured from three independent transformants for each reporter construct assayed in triplicate ± SD (n = 9). The empty vector control (placpoly6) is also included for comparison. b Diagrams representing the set of nested deletions created in this study and a summary of the average fold-induction in response to CFW and CaCl2 for each of the reporter constructs. Errors are the standard deviation
Candida albicans strains containing the wild type, window deletion reporter constructs and empty vector control were assayed for β-galactosidase activity following growth in the presence and absence of 100 mM CaCl2 and 100 μg/ml CFW. The results are summarised in Fig. 3a, b. Two different types of effects were observed, one where the deletion of a region resulted in an increase in the level of expression of the lacZ reporter in response to CaCl2 and CFW with the basal level of expression remaining the same, and one where the deletion of a region reduced the basal level of expression of the reporter. However, a normal level of activation was still observed in response to CaCl2 and CFW.
Expression of the lacZ reporter gene mediated by the full length CaCHS2 promoter was induced 2.2-fold following treatment with CaCl2 (pCHS2plac ± CaCl2; Fig. 3b) and 4.2-fold following treatment with CFW (pCHS2plac ± CFW; Fig. 3b). No loss of induction to either stimulus was observed when only 347 bp of the CaCHS2 promoter was present (pCHS2plac-347) and the basal level of promoter activity in unstressed cells was comparable to the wild type (Fig. 3b). In fact, the level of expression driven by the truncated CaCHS2 promoter in response to CaCl2 doubled relative to that driven by the full length promoter (Fig. 3a) despite deletion of all four putative CDREs. This indicated that the regulatory elements in the CaCHS2 promoter that activate expression of CaCHS2 in response to CaCl2 and CFW are located within the first 347 bp of the promoter, and that there are sequences in the region between −347 and −968 bp relative to ATGCHS2 that attenuate the expression of CaCHS2 in response to CaCl2 and CFW.
The full length CaCHS8 promoter conferred a 2.9-fold and 4.7-fold-induction of the lacZ reporter gene in response to CaCl2 and CFW respectively (Fig. 3b). All truncated versions of the CaCHS8 promoter were able to confer a similar fold-induction of the lacZ reporter gene in response to CaCl2 and CFW, despite the deletion of several putative RLM1-elements and CDREs (Fig. 3b). The most notable difference observed was in the basal level of expression of the lacZ reporter gene mediated by the first 125 bp of the CaCHS8 promoter which was approximately 5.5-times less than that conferred by the full length CaCHS8 promoter (Fig. 3a). Despite the significant reduction in the basal level of expression, this small region of the CaCHS8 promoter was still able to confer a 5.2-fold and 4.2-fold-induction of the lacZ reporter gene in response to CaCl2 and CFW (Fig. 3b). For comparison, the basal level of expression for the empty lacZ reporter construct was typically around 100-fold less than that of the full length CaCHS8 promoter (compare placpoly6 to pCHS8plac; Fig. 3a). These results indicated that some of the promoter elements that are required for the basal level of expression of CaCHS8 are contained in the region of the promoter between −429 and −125 relative to ATGCHS8 and that the regulatory elements that activate expression of CaCHS8 in response to CaCl2 and CFW are located within the first 125 bp of the promoter.
The PKC cell wall integrity, Ca2+/calcineurin and HOG signalling pathways act on the minimal CaCHS promoters
The PKC cell wall integrity, Ca2+/calcineurin and/or the HOG signalling pathways have been shown to act on the full length class I CHS promoters (Munro et al. 2007). In order to determine whether these pathways were still acting on the minimal CaCHS2 and CaCHS8 promoters (CaCHS2-347 and CaCHS8-125), we transformed the crz1Δ, cna1Δ, mkc1Δ and hog1Δ mutants with the minimal CaCHS promoter reporter constructs (pCHS2plac-347 and pCHS8plac-125) and assessed whether the lacZ reporter genes were activated in response to combined treatments of 100 mM CaCl2 and 100 μg/ml CFW. Treatment with CaCl2 and CFW together had previously been shown to hyperstimulate the CaCHS promoters (Munro et al. 2007). The fold-induction of each lacZ reporter in the signalling mutant backgrounds was compared to that in the wild type background (Table 6). A significant reduction in the fold-induction mediated by the CaCHS2-347 promoter was observed in the cna1Δ, mkc1Δ and hog1Δ mutants. No significant reductions in the fold-induction were mediated by the CaCHS8-125 promoter. Instead, a significant increase in the fold-induction was observed in the mkc1Δ mutant. These results indicated that all three signalling pathways contribute towards the activation mediated through the CaCHS2-347 promoter, and that the PKC cell wall integrity pathway may be involved in repression mediated through the CaCHS8-125 promoter.
Table 6.
The PKC cell wall integrity-, Ca2+/calcineurin- and the HOG pathway act on a minimal CaCHS2 promoter
| Reporter | Strain | Fold-induction with CaCl2 and CFWa | p-value |
|---|---|---|---|
| pCHS2plac-347 | CAI-4 | 7.88 ± 0.69 | |
| pCHS2plac-347 | crz1Δ | 7.46 ± 0.47 | |
| pCHS2plac-347 | cna1Δ | 4.33 ± 0.66 | 0.003 |
| pCHS2plac-347 | mkc1Δ | 3.07 ± 0.35 | 1.25 × 10−4 |
| pCHS2plac-347 | hog1Δ | 2.49 ± 0.41 | 3.36 × 10−5 |
| pCHS8plac-125 | CAI-4 | 10.69 ± 0.73 | |
| pCHS8plac-125 | crz1Δ | 7.02 ± 0.65 | |
| pCHS8plac-125 | cna1Δ | 16.27 ± 1.84 | |
| pCHS8plac-125 | mkc1Δ | 26.81 ± 2.88 | 1.06 × 10−4 |
| pCHS8plac-125 | hog1Δ | 16.69 ± 1.63 |
aAverage fold-induction of each lacZ reporter upon treatment with CaCl2 and CFW ± the standard error of the mean from two independent transformants assayed twice (n = 4). Statistically significant changes in the fold-induction in the mutant backgrounds relative to that observed in the wild type background are highlighted in bold
The minimal class I CHS promoters contain functional RLM1-elements
The PKC cell wall integrity pathway was shown to be involved in the regulation of the CaCHS2-347 and CaCHS8-125 promoters. The CaCHS2-347 promoter contained three putative RLM1-elements (RLM1−280, RLM1−267 and RLM1−261) and the CaCHS8-125 promoter contained one putative RLM1-element (RLM1−94). To test whether the putative RLM1-elements were functional in a minimal promoter context, the putative RLM1-elements were mutated by site-directed mutagenesis individually and in combination. C. albicans strains containing the reporter constructs with the minimal wild type and mutated putative RLM1-elements were then assayed for β-galactosidase activity following growth in the presence and absence of CFW (Fig. 4a, b). Mutation of RLM1−280 in the CaCHS2-347 promoter almost completely abolished the normal 8.5-fold-induction of the lacZ reporter in response to CFW (reduced to 1.5-fold). Mutation of RLM1−267 and RLM1−261 individually reduced the fold-induction of the lacZ reporter in response to CFW by half (reduced to 4.6- and 4.3-fold respectively). Mutation of all three RLM1-elements also reduced the level of induction to a quarter of normal (reduced to 2.6-fold). These results indicated that all three RLM1-elements in the CaCHS2-347 promoter were functional and contributed to the activation of CaCHS2. In contrast, mutation of RLM1−94 in the CaCHS8-125 promoter did not reduce the level of induction of the lacZ reporter in response to CFW but instead raised the basal level of expression of the lacZ reporter to an equal level. This indicated that the RLM1-element in the CaCHS8-125 promoter played a role in repression of CaCHS8 in un-stressed conditions.
Fig. 4.
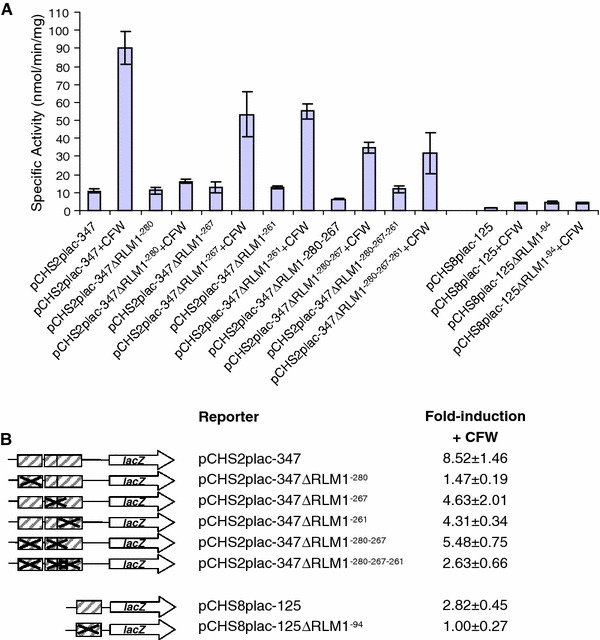
A minimal CaCHS2 promoter contains functional RLM1-elements. a C. albicans strains containing a single copy of the minimal CHS2 and CHS8 promoters with mutated RLM1-elements were assayed for β-galactosidase activity following growth from an OD600 of 0.1–1.0 in the presence and absence of 100 μg/ml CFW. Each measurement is the average β-galactosidase activity measured from two independent transformants for each reporter construct assayed twice ± SD (n = 4). b Diagrams representing the minimal promoter reporters containing the mutations of the RLM1-elements and summary of the average fold-induction in response to CFW for each of the reporter constructs shown above. Errors are the standard deviation
Common promoter elements in the minimal class I CaCHS promoters
Although there were no putative CDREs present in the minimal class I CaCHS promoters, the CaCHS2-347 and CaCHS8-125 promoters were capable of mediating activation of a lacZ reporter gene in response to CaCl2 (Fig. 3). We therefore looked for any novel promoter elements common to the first 347 bp of the CaCHS2 promoter and 125 bp of the CaCHS8 promoter in silico. Analyses of these regions failed to identify any common elements.
Discussion
Fungi respond to cell wall damage by remodelling their cell wall architecture and by up-regulating chitin synthesis (Popolo et al. 1997; Munro et al. 2007; Walker et al. 2008). Transcriptional regulation of chitin synthesis is also thought to be involved in cell wall remodelling during yeast-hypha morphogenesis (Munro et al. 1998; Nino-Vega et al. 2000). In C. albicans, CaCHS2 and CaCHS8 are activated at the level of transcription in response to cell wall stresses. These responses involve the PKC cell wall integrity, Ca2+/calcineurin and HOG signalling pathways (Munro et al. 2007). In this work, we functionally dissected the CaCHS2 and CaCHS8 promoters to further characterise the transcriptional response of the C. albicans class I CHS genes. We took two approaches, the first was based on the assumption the transcription factors that act at the end of the signalling pathways and the promoter elements to which they bind may have been conserved in S. cerevisiae and C. albicans. Therefore, we analysed the CaCHS2 and CaCHS8 promoter sequences for consensus binding sites of the Rlm1p, Crz1p and Sko1p transcription factors. Putative RLM1-elements, CDREs and ATF/CREB-elements were identified in the sequences 1 kb upstream of the CaCHS2 and CaCHS8 start codons (Fig. 1). In the second approach we generated nested deletions of each promoter to identify regions involved in transcriptional regulation. The latter approach was included to take into account evidence that C. albicans signalling pathways can be significantly rewired compared to S. cerevisiae (Kadosh and Johnson 2001; Tsong et al. 2003; Martchenko et al. 2007; Banerjee et al. 2008) and would allow the identification of novel regulatory elements in these promoters.
We assessed the function of the putative promoter elements by measuring the level of expression of a lacZ reporter gene that was conferred by the CaCHS promoters containing mutated versions of the putative promoter elements and comparing them to that conferred by the wild type promoter. We reasoned that if the putative promoter element was functional then we would see a loss of induction of the lacZ reporter gene in response to an appropriate stress, an approach that has been validated for other C. albicans promoter elements (Martchenko et al. 2007). This analysis revealed that the CaCHS2 promoter contained no single RLM1-element or CDRE responsible for induced gene expression in response to CFW or CaCl2 respectively. Similarly no evidence was found that the CaCHS8 promoter contained any RLM1-elements, CDREs or ATF/CREB-elements that functioned alone (Fig. 2).
In order to rule out the possibility that multiple promoter elements acted co-operatively to induce expression in response to particular cell wall stresses, we also constructed a set of nested window deletions of the C. albicans class I CHS promoters. No loss of induction of the lacZ reporter gene in response to CaCl2 was observed when all four putative CDREs were deleted from the CaCHS2 promoter, providing evidence that these putative CDREs were not the regulatory elements that mediate the activation of CaCHS2 in response to CaCl2. This analysis did however allow us to determine that regulatory elements in the CaCHS2 promoter that activate expression of CaCHS2 in response to CaCl2 and CFW were located within the first 347 bp of the promoter (Fig. 3).
Analysis of the expression of a lacZ reporter gene fused to the minimal CaCHS2 promoter (CaCHS2-347) in various signal transduction mutants revealed that the PKC cell wall integrity, Ca2+/calcineurin and HOG signalling pathways all act on the CaCHS2-347 promoter (Table 6). Three putative RLM1-elements lie in this region of the CaCHS2 promoter. We assessed whether these putative RLM1-elements were functional in the minimal CaCHS2 promoter context by measuring the level of expression of a lacZ reporter gene that was conferred by the CaCHS2-347 promoter containing mutated versions of the putative RLM1-elements and comparing them to that conferred by the un-mutated CaCHS2-347 promoter. This analysis revealed that the minimal CaCHS2-347 promoter contained three functionally redundant RLM1-elements that were required for the activation of CaCHS2 in response to CFW. Hence the PCK cell wall integrity pathway may operate through these RLM1-elements, although this was only detectable in a reduced CaCHS2 promoter context.
Since the Ca2+/calcineurin signalling pathway does act on the minimal CaCHS2 promoter, one possibility is that transcription factor and/or the DNA sequence to which it binds may differ from those predicted by the S. cerevisiae paradigm. In support of this, we show that the promoter elements that mediate the transcriptional response to CaCl2 are not the putative CDREs that were indentified in silico, and deletion of CaCRZ1, the gene encoding the transcription factor thought to act at the end of the Ca2+/calcneurin signalling pathway, had little effect on the expression of the lacZ reporter fused to the CaCHS2-347 promoter. It is possible that indirect mechanisms connect the Ca2+/calcineurin signalling pathway to the CaCHS2-347 promoter. In support of this, deletion of CaCNA1, the gene encoding the catalytic subunit of calcineurin upstream of this signalling pathway, resulted in a reduction in the level of activation of the lacZ reporter fused to the CaCHS2-347 promoter.
The HOG signalling pathway also acts on the CaCHS2-347 promoter. Although the CaCHS2 promoter contains no putative ATF/CREB elements which are the binding sites described for the ScSko1p transcription factor, other transcription factors may act the end of the HOG signalling pathway in C. albicans. Indeed, there are four other transcription factors known to act downstream of the HOG pathway in S. cerevisiae, ScMsn2p/Msn4p, ScHot1p and ScSmp1p (reviewed in Hohmann 2002 and Westfall et al. 2004).
The results of the assays performed using the window deletions of the CaCHS8 promoter indicated that the two putative CDREs identified in the CaCHS8 promoter were not the regulatory elements that activate expression of CaCHS8 in response to CaCl2. We were unable to identify any other putative promoter elements in this region of the CaCHS8 promoter to explain the transcriptional response of CaCHS8 to CaCl2. However, these data indicate that the regulatory elements in the CaCHS8 promoter that activate expression of CaCHS8 in response to CaCl2 and CFW are located within the first 125 bp of the promoter (Fig. 3). Analysis of a lacZ reporter gene fused to the minimal CaCHS8 promoter in signalling mutants revealed that the PKC cell wall integrity pathway may be involved in repression of the CaCHS8-125 promoter (Table 6). In support of this, mutation of the one putative RLM1-element in the minimal CaCHS8-125 promoter (RLM1−94) increased the basal level of expression of this reporter in un-stressed conditions (Fig. 4).
In summary, the PKC cell wall integrity pathway may regulate CHS transcription through the traditional CaRlm1p transcription factor binding to RLM1-elements in the CaCHS2 and CaCHS8 promoters. The Ca2+/calcineurin and HOG signalling pathways may act via unknown promoter elements or indirectly to regulate these genes. Post-transcriptional regulatory events may also mediate the increase in chitin we see in response to cell wall stress in C. albicans. One such example is the regulation of ScChs3p by phosphorylation which occurs in response to heat shock in S. cerevisiae (Valdivia and Schekman 2003), and this is mediated by ScPkc1p, a kinase in the cell wall integrity pathway.
Acknowledgments
We acknowledge financial support from the Biotechnology and Biological Sciences Research Council (10161), Medical Research Council (New Investigator Award to C.A.M.), the European Community FUNGALWALL and SIGNALPATH initiatives and the Wellcome Trust.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alonso-Monge R, Navarro-Garcia F, Molero G, Diez-Orejas R, Gustin M, Pla J, Sanchez M, Nombela C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, Kadosh D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell. 2008;19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorsma A, de Nobel H, ter Riet B, Bargmann B, Brul S, Hellingwerf KJ, Klis FM. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast. 2004;21:413–427. doi: 10.1002/yea.1109. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bulawa CE, Miller DW, Henry LK, Becker JM. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc Natl Acad Sci USA. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS. Calcineurin signalling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem Biophys Res Commun. 2003;311:1143–1150. doi: 10.1016/S0006-291X(03)01552-3. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Alepuz PM, Posas F. Dealing with osmostress through MAP kinase activation. EMBO Rep. 2002;3:735–740. doi: 10.1093/embo-reports/kvf158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Treisman R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:1848–1859. doi: 10.1128/mcb.17.4.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, Francois J, Nombela C, Arroyo J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signalling pathway. J Biol Chem. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- Gow NAR, Robbins PW, Lester JW, Brown AJ, Fonzi WA, Chapman T, Kinsman OS. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism, or virulence of Candida albicans. Proc Natl Acad Sci USA. 1994;91:6216–6220. doi: 10.1073/pnas.91.13.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signalling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung US, Levin DE. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- Jung US, Sobering AK, Romeo MJ, Levin DE. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol Microbiol. 2002;46:781–789. doi: 10.1046/j.1365-2958.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol Cell Biol. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karababa M, Valentino E, Pardini G, Coste AT, Bille J, Sanglard D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59:1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- Kim KY, Truman AW, Levin DE. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol Cell Biol. 2008;28:2579–2589. doi: 10.1128/MCB.01795-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagorce A, Hauser NC, Labourdette D, Rodriguez C, Martin-Yken H, Arroyo J, Hoheisel JD, Francois J. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J Biol Chem. 2003;278:20345–20357. doi: 10.1074/jbc.M211604200. [DOI] [PubMed] [Google Scholar]
- Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol. 2007;17:1007–1013. doi: 10.1016/j.cub.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendizabal I, Pascual-Ahuir A, Serrano R, de Larrinoa IF. Promoter sequences regulated by the calcineurin-activated transcription factor Crz1 in the yeast ENA1 gene. Mol Genet Genomics. 2001;265:801–811. doi: 10.1007/s004380100474. [DOI] [PubMed] [Google Scholar]
- Mio T, Yabe T, Sudoh M, Satoh Y, Nakajima T, Arisawa M, Yamada-Okabe H. Role of three chitin synthase genes in the growth of Candida albicans. J Bacteriol. 1996;178:2416–2419. doi: 10.1128/jb.178.8.2416-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro CA, Schofield DA, Gooday GW, Gow NAR. Regulation of chitin synthesis during dimorphic growth of Candida albicans. Microbiology. 1998;144:391–401. doi: 10.1099/00221287-144-2-391. [DOI] [PubMed] [Google Scholar]
- Munro CA, Winter K, Buchan A, Henry K, Becker JM, Brown AJ, Bulawa CE, Gow NAR. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol Microbiol. 2001;39:1414–1426. doi: 10.1046/j.1365-2958.2001.02347.x. [DOI] [PubMed] [Google Scholar]
- Munro CA, Whitton RK, Hughes HB, Rella M, Selvaggini S, Gow NAR. CHS8-a fourth chitin synthase gene of Candida albicans contributes to in vitro chitin synthase activity, but is dispensable for growth. Fungal Genet Biol. 2003;40:146–158. doi: 10.1016/S1087-1845(03)00083-5. [DOI] [PubMed] [Google Scholar]
- Munro CA, Selvaggini S, de Bruijn I, Walker L, Lenardon MD, Gerssen B, Milne S, Brown AJ, Gow NAR. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol Microbiol. 2007;63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Garcia F, Sanchez M, Pla J, Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol Cell Biol. 1995;15:2197–2206. doi: 10.1128/mcb.15.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Brown GD, Kullberg BJ, Gow NAR. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- Nino-Vega GA, Munro CA, San-Blas G, Gooday GW, Gow NAR. Differential expression of chitin synthase genes during temperature-induced dimorphic transitions in Paracoccidioides brasiliensis. Med Mycol. 2000;38:31–39. doi: 10.1080/714030921. [DOI] [PubMed] [Google Scholar]
- Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1delta mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo L, Gualtieri T, Ragni E. The yeast cell-wall salvage pathway. Med Mycol. 2001;39(Suppl 1):111–121. [PubMed] [Google Scholar]
- Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/S1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- Proft M, Gibbons FD, Copeland M, Roth FP, Struhl K. Genomewide identification of Sko1 target promoters reveals a regulatory network that operates in response to osmotic stress in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1343–1352. doi: 10.1128/EC.4.8.1343-1352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinoso-Martin C, Schuller C, Schuetzer-Muehlbauer M, Kuchler K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signalling. Eukaryot Cell. 2003;2:1200–1210. doi: 10.1128/EC.2.6.1200-1210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M, Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. New York: Cold Spring Harbour Press; 1989. [Google Scholar]
- Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol. 2003;48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- Santos M, de Larrinoa IF. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr Genet. 2005;48:88–100. doi: 10.1007/s00294-005-0003-8. [DOI] [PubMed] [Google Scholar]
- Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/S0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- Uhl MA, Johnson AD. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology. 2001;147:1189–1195. doi: 10.1099/00221287-147-5-1189. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Schekman R. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc Natl Acad Sci USA. 2003;100:10287–10292. doi: 10.1073/pnas.1834246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LA, Munro CA, de Bruijn I, Lenardon MD, McKinnon A, Gow NAR. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4:e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Ballon DR, Thorner J. When the stress of your environment makes you go HOG wild. Science. 2004;306:1511–1512. doi: 10.1126/science.1104879. [DOI] [PubMed] [Google Scholar]
- Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D, Brown PO, Cyert MS. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signalling pathway in Saccharomyces cerevisiae. J Biol Chem. 2002;277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]


